Abstract
Genome-wide association studies (GWAS) and candidate gene analyses have led to the discovery of several dozen genetic polymorphisms associated with breast cancer susceptibility, many of which are considered well-established risk factors for the disease. Despite attempts to replicate these same variant-disease associations in African Americans, the evaluable populations are often too small to produce precise or consistent results. We estimated the associations between 83 previously identified single nucleotide polymorphisms (SNPs) and breast cancer among Carolina Breast Cancer Study (1993–2001) participants using maximum likelihood, Bayesian, and hierarchical methods. The selected SNPs were previous GWAS hits (n = 22), near-hits (n = 19), otherwise well-established risk loci (n = 5), or located in the same genes as selected variants (n = 37). We successfully replicated 18 GWAS-identified SNPs in whites (n = 2,352) and 10 in African Americans (n = 1,447). SNPs in the fibroblast growth factor receptor 2 gene (FGFR2) and the TOC high mobility group box family member 3 gene (TOX3) were strongly associated with breast cancer in both races. SNPs in the mitochondrial ribosomal protein S30 gene (MRPS30), mitogen-activated protein kinase kinase kinase 1 gene (MAP3K1), zinc finger, MIZ-type containing 1 gene (ZMIZ1), and H19, imprinted maternally expressed transcript gene (H19) were associated with breast cancer in whites, and SNPs in the estrogen receptor 1 gene (ESR1) and H19 gene were associated with breast cancer in African Americans. We provide precise and well-informed race-stratified odds ratios for key breast cancer–related SNPs. Our results demonstrate the utility of Bayesian methods in genetic epidemiology and provide support for their application in small, etiologically driven investigations.
Keywords: Bayesian analysis, breast cancer, genetics, genome-wide association studies, GWAS replication, race, single nucleotide polymorphisms
As of June 2013, a total of 74 single nucleotide polymorphisms (SNPs) met the criterion for inclusion in the National Human Genome Research Institute's database of genome-wide association study (GWAS) hits, which requires a P value of less than 10−5 in at least 1 of 25 breast cancer GWAS (1). Most of these SNPs were consistently associated with the disease in subsequent investigations among women of European or Asian descent (Web Appendix 1, available at http://aje.oxfordjournals.org/), but attempts to replicate these findings in African-American women have been largely unsuccessful (2–12). In general, the search for African-American–specific risk variants has made slow progress, with few insights to explain the tendency for African-American women to have more aggressive, less treatable disease subtypes (13–16) and markedly higher breast cancer–specific mortality than whites (32.7 vs. 23.7 deaths per 100,000 US women per year in 2000–2009) (17).
Allele frequencies and linkage disequilibrium (LD) structures vary by race, with African Americans exhibiting generally weaker between-SNP correlations and smaller LD blocks (18, 19). Because each SNP represents all correlated variants, SNPs associated with breast cancer in African Americans correspond to a narrower genomic region than SNPs associated with the disease in whites. Therefore, studying African Americans should help us identify causal loci and improve our understanding of disease etiology. Unfortunately, most of the studies of genetic breast cancer risk factors carried out in African Americans have been small and therefore have lacked the necessary statistical power to reliably differentiate between null associations and odds ratios of 1.1–1.3, a typical range for GWAS-identified risk variants in other populations (1, 20).
Given existing knowledge about effect size, as well as information about the genome's correlation structure, Bayesian statistical methods may be useful tools for enhancing the analysis of race-specific genetic risk factors for breast cancer. A variety of Bayesian-based methods have been proposed for use in genetic epidemiology (21–27), but in the current study, we focused on 2 of the most basic applications, hierarchical modeling and Bayesian regression. Our goal was to obtain valid and precise effect estimates by capitalizing on existing information.
To further our understanding of genetic risk factors for the disease, we examined the association between breast cancer and several candidate SNPs by using traditional maximum likelihood methods and both Bayesian approaches. All of the selected SNPs were located on genes with strong prior evidence of an association with breast cancer from GWAS and candidate gene investigations. We assessed the relationship between these SNPs and breast cancer risk using data from the Carolina Breast Cancer Study, a population-based case-control study of white and African-American women.
MATERIALS AND METHODS
Study population
Cases were identified using the North Carolina Central Cancer Registry's rapid case ascertainment program (28). Women were eligible for the study if they were diagnosed with invasive breast cancer in 1993–2001, were 20–74 years of age at diagnosis, and lived in 1 of 24 North Carolina counties. Women with in situ disease were eligible if they had ductal carcinoma in situ with microinvasion of 2 mm or more or lobular carcinoma in situ. To ensure approximately equal representation of African Americans and non–African Americans, as well as both pre- and postmenopausal women, we randomly sampled breast cancer cases at disproportionate rates based on race and age.
Controls aged 20–64 or 65–74 years were selected from records of the North Carolina Department of Motor Vehicles (Raleigh, North Carolina) and the Health Care Financing Administration (Centers for Medicare & Medicaid Services, Baltimore, Maryland), respectively. Controls were probability-matched to cases on race and 5-year age group (29). Women with a history of breast cancer were ineligible. All participants provided informed consent, and study procedures were approved by the institutional review board of the University of North Carolina.
A study nurse administered a questionnaire to each participant during an in-home visit. The survey included questions on basic demographic factors, known or suspected breast cancer risk factors, and medical and family history. Additionally, the nurse drew 30 mL of blood. The overall response rate was 77% for cases and 57% for controls. Of those enrolled, 88% of cases and 90% of controls provided sufficient blood samples for inclusion in genotype analyses, leaving a total sample size of 2,013 cases (1,247 white and 766 African American) and 1,786 controls (1,105 white and 681 African American). We excluded 166 persons who identified themselves as having a race/ethnicity other than African American or non-Hispanic white.
SNP selection
Initially, we selected candidate SNPs with P values less than 10−5 in any of 8 early breast cancer GWAS (30–37) or 2 GWAS follow-up studies (38, 39). We also evaluated several SNPs from these initial studies that had P values less than 10−5 in discovery-phase analyses. Lastly, we retained any SNPs already genotyped in the study population that Zhang et al. (40) determined had “cumulative evidence of an association” with breast cancer or that were located in the same gene as a previously selected variant. In total, we selected 41 GWAS-identified SNPs, including 22 GWAS hits and 19 other strongly associated SNPs, as well as 5 SNPs from the meta-analysis by Zhang et al. (40) and 37 SNPs from included genes. We later excluded 6 SNPs with minor allele frequencies less than 1% in white participants and 10 SNPs with minor allele frequencies less than 1% in African Americans, leaving a total of 77 and 73 SNPs, respectively.
All study participants were genotyped for 144 ancestry informative markers, which were used to estimate each participant's proportion of African ancestry. Inclusion of this variable in regression models should reduce confounding due to population stratification (41, 42).
Genotype analysis
The SNPs included in this analysis were genotyped using either a custom GoldenGate Genotyping Assay (Illumina, Inc., San Diego, California) or a customized TaqMan panel (Applied Biosystems, Inc., Foster City, California). Both processes have been described previously (43, 44). All of the SNPs included in this analysis passed quality control evaluations, including examination of individual call rates and inspection of assay intensity data and genotype clustering images.
Statistical methods
We calculated risk allele frequencies and age and African ancestry distributions for whites and African Americans separately. We tested for departures from Hardy-Weinberg equilibrium in white and African-American controls using Pearson's χ2 test, and we reinspected the genotype clustering images of SNPs with P values less than 0.05 in either population for signs of poor genotype differentiation. SNPs were retained if their genotypes formed discrete clusters with minimal overlap.
The relationship between each risk variant and incident breast cancer was assessed by using logistic regression. We estimated odds ratios and 95% confidence intervals assuming additive genetic models. The risk allele for each SNP was selected a priori based on the meta-analysis by Zhang et al. (40) or previous GWAS (30–39). We assumed that the risk allele was the same for African Americans and whites unless the majority of the statistically significant associations reported indicated otherwise (2–5, 11, 12).
All models were stratified by race and adjusted for proportion of African ancestry and age at diagnosis or selection. We centered age at 50 years and ancestry at race-specific means. An offset term was included to account for unequal sampling by race and age group. We will herein refer to these frequentist estimates as the maximum-likelihood estimation (MLE) odds ratios.
Bayesian analysis
Bayes' theorem states that the posterior probability distribution for the parameter of interest given the observed data, f(β|D), is proportional to the likelihood of the observed data, L(β|D), multiplied by the prior probability distribution f(β) (45, 46). Here, the likelihood function is the same as the MLE likelihood, with the log odds of being a case given exposure Xj and covariates W modeled as follows:
In this application, βj is the estimated log odds ratio of being a breast cancer case for each copy of the risk allele at SNP j, and γ is a vector of estimated log odds ratios for age and ancestry. To incorporate the priors, we added a second stage to the model, 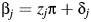 , where zj is a j × 1 vector of 1 second, π is the prior log odds ratio, and δj is assumed to be normally distributed with mean 0 and variance τ2. For each covariate, the prior mean (π), variance (τ2), or both can either be estimated directly from the data or assigned by the investigator. We use the terms “full-Bayes” to indicate that all priors were assigned independently of the data at hand, “empirical Bayes” when all priors were estimated from the data, and “semi-Bayes” when some priors were assigned and others were estimated (47).
, where zj is a j × 1 vector of 1 second, π is the prior log odds ratio, and δj is assumed to be normally distributed with mean 0 and variance τ2. For each covariate, the prior mean (π), variance (τ2), or both can either be estimated directly from the data or assigned by the investigator. We use the terms “full-Bayes” to indicate that all priors were assigned independently of the data at hand, “empirical Bayes” when all priors were estimated from the data, and “semi-Bayes” when some priors were assigned and others were estimated (47).
We conducted both a full-Bayes analysis and a semi-Bayes analysis, but did not use empirical Bayes methods, because the near-0 τ2 generated from this rich data set caused overshrinkage of SNP-specific effect estimates. For full-Bayes analyses, we assigned priors for the mean and τ2 of all covariates. For the semi-Bayes analysis, we assigned a fixed τ2 to the SNP parameters, but used LD block (i.e., haplotype)–level odds ratio estimates to inform the SNPs' prior mean π and did not specify priors for any other covariates.
To obtain Bayesian (i.e., full-Bayes) log odds ratios, we assigned a null-centered, lognormal prior with a mean of 0 and variance τ2 ∼ Γ−1 (3, 0.2) to each SNP, such that 95% of the prior mass for each SNP–breast cancer odds ratio lay between 0.64 and 1.55 when τ2 was equal to its mode (0.05). As discussed previously, this range likely includes the true value for any single SNP–breast cancer association. Each Bayesian model included exactly 1 SNP (j = 1).
We also assigned null-centered, lognormal priors for age and ancestry, giving both parameters prior variances of 0.68. These priors reflect our belief, with moderate uncertainty, that these mean-centered covariates are weakly associated with breast cancer. We placed a β0 ∼ N(0, 1,000) prior on the intercept. In the absence of other information, this vague prior should generate posterior intercept estimates nearly identical to the MLE estimates. We assumed that all priors were independent.
For the semi-Bayes analysis, we used hierarchical modeling to integrate haplotype information (22, 48–50). More specifically, we used the estimated joint effect of all of the SNPs in an LD block to inform the prior mean (π). If there was only 1 genotyped SNP in an LD block, π and βj were identical, and the hierarchical estimate was very similar to the MLE estimate. We assigned fixed prior variances of 0.05 for each SNP.
The above grouping approach is valid as long as an exchangeability assumption is met. This assumption states that, before evaluating the relationship between the exposures and the outcome, there was no reason to suspect that any 1 exposure in a group had a true log odds ratio different from the others in that group. Because none of the included SNPs are known causal variants, and all effects are evaluated in terms of risk alleles, we believe this assumption is acceptable in our setting.
For Bayesian methods, we present posterior geometric mean odds ratios (i.e., antilogs of posterior mean log odds ratios) and 95% posterior intervals. For the Bayesian analyses, we ran 30,000 samples for each SNP model, discarding the first 1,000 draws as a burn-in and retaining every fifth draw. We inspected autocorrelation, trace, and density plots to verify that all posterior estimates converged appropriately.
LD blocks were determined by using methods proposed by Gabriel et al. (51) and conducted in Haploview, version 4.2, software (Broad Institute, Cambridge, Massachusetts) (52). Bayesian models were analyzed using PROC MCMC or PROC GLIMMIX in SAS, version 9.3, software (SAS Institute, Inc., Cary, North Carolina). Example code is provided as Web Appendix 2.
GWAS-identified SNPs were considered successfully replicated if their entire 95% posterior intervals fell above the null, as were SNPs identified in the candidate gene meta-analysis. More formal homogeneity tests comparing our findings with the meta-analysis or initial GWAS estimates were inappropriate, because these studies did not consistently report odds ratios from additive genetic models.
RESULTS
As expected, age distributions were similar for cases and controls, regardless of race. White cases and controls were 52 and 53 years of age at selection, on average, and African-American cases and controls were 52 years of age, on average (Web Table 1). Whites had approximately 7% African ancestry and African Americans had 77%. More detailed descriptions of the study population have been published elsewhere (53) (Web Appendix 3).
Table 1 shows race-stratified risk allele frequencies and Hardy-Weinberg equilibrium P values. Seven SNPs were not in Hardy-Weinberg equilibrium by our criterion (P < 0.05). We retained 6 of these, because their clustering images indicated good differentiation, and none failed Hardy-Weinberg equilibrium tests in both races. We excluded the seventh SNP, rs614367 (myeloma overexpressed gene (MYEOV)), after observing disparate clusters for the homozygous rare genotype and finding evidence of allelic dropout.
Table 1.
Frequencies of Breast Cancer Risk Alleles Among Whites and African Americans in the Carolina Breast Cancer Study, by Race and Case Status, North Carolina, 1993–2001
| Gene or Chromosomal Region | SNP | Whites |
African Americans |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Risk Allele |
RAF Casesa (n = 1,247) |
RAF Controlsa (n = 1,105) |
HWE P Value |
Risk Allele |
RAF Casesa (n = 766) |
RAF Controlsa (n = 681) |
HWE P Value |
||
| 1p12 | rs11249433 | G | 0.44 | 0.41 | 0.54 | G | 0.14 | 0.10 | 0.01 |
| CASP8 | rs1045485 | G | 0.88 | 0.87 | 0.63 | C | 0.06 | 0.05 | 0.74 |
| CASP8 | rs17468277 | C | 0.87 | 0.87 | 0.63 | C | 0.95 | 0.95 | 0.95 |
| 2q35 | rs13387042 | A | 0.54 | 0.47 | 0.83 | A | 0.74 | 0.73 | 1.00 |
| 2p | rs4666451 | G | 0.60 | 0.63 | 0.30 | G | 0.78 | 0.77 | 0.12 |
| SLC4A | rs4973768 | T | 0.48 | 0.42 | 0.22 | T | 0.36 | 0.40 | 0.05 |
| 4p | rs12505080 | C | 0.29 | 0.24 | 0.80 | C | 0.17 | 0.17 | 0.64 |
| TLR1 | rs7696175 | T | 0.45 | 0.45 | 0.91 | T | 0.08 | 0.06 | 0.54 |
| MRPS30 | rs4415084 | T | 0.43 | 0.42 | 0.18 | T | 0.64 | 0.58 | 0.70 |
| MRPS30 | rs10941679 | G | 0.29 | 0.30 | 0.76 | G | 0.19 | 0.19 | 0.17 |
| 5p12 | rs981782 | T | 0.53 | 0.59 | 0.26 | T | 0.92 | 0.91 | 0.60 |
| 5q | rs30099 | T | 0.10 | 0.10 | 0.40 | T | 0.16 | 0.12 | 0.75 |
| MAP3K | rs889312 | C | 0.32 | 0.34 | 0.85 | C | 0.33 | 0.36 | 0.08 |
| ESR1 | rs2046210 | A | 0.36 | 0.35 | 0.48 | A | 0.64 | 0.61 | 0.15 |
| ESR1 | rs851974 | G | 0.42 | 0.43 | 0.28 | G | 0.17 | 0.17 | 0.46 |
| ESR1 | rs2077647 | A | 0.51 | 0.49 | 0.64 | A | 0.52 | 0.51 | 0.16 |
| ESR1 | rs2234693 | T | 0.53 | 0.57 | 0.45 | T | 0.47 | 0.48 | 0.63 |
| ESR1 | rs1801132 | C | 0.76 | 0.76 | 0.43 | C | 0.90 | 0.88 | 0.36 |
| ESR1 | rs3020314 | C | 0.36 | 0.34 | 0.15 | C | 0.69 | 0.71 | 0.75 |
| ESR1 | rs3798577 | T | 0.52 | 0.53 | 0.43 | T | 0.57 | 0.54 | 0.27 |
| ECHDC1 | rs2180341 | G | 0.25 | 0.27 | 0.55 | G | 0.31 | 0.33 | 0.83 |
| RELN | rs17157903 | T | 0.13 | 0.12 | 0.06 | T | 0.11 | 0.10 | 0.08 |
| 8q24 | rs13281615 | G | 0.43 | 0.42 | 0.17 | G | 0.44 | 0.43 | 0.58 |
| 8q24 | rs1562430 | T | 0.59 | 0.57 | 0.78 | T | 0.54 | 0.53 | 0.61 |
| CDKN2A/B | rs3731257 | T | 0.23 | 0.23 | 0.24 | T | 0.09 | 0.11 | 0.89 |
| CDKN2A/B | rs3731249 | A | 0.03 | 0.03 | 0.90 | A | 0.01 | 0.00 | 0.95 |
| CDKN2A/B | rs518394 | G | 0.44 | 0.48 | 0.17 | G | 0.08 | 0.08 | 0.06 |
| CDKN2A/B | rs564398 | G | 0.42 | 0.47 | 0.29 | G | 0.08 | 0.08 | 0.02 |
| CDKN2A/B | rs1011970 | T | 0.19 | 0.15 | 0.62 | T | 0.33 | 0.34 | 0.14 |
| CDKN2A/B | rs10757278 | A | 0.54 | 0.55 | 0.18 | A | 0.81 | 0.82 | 0.77 |
| CDKN2A/B | rs10811661 | C | 0.17 | 0.20 | 0.02 | C | 0.07 | 0.07 | 0.24 |
| ANKRD16 | rs2380205 | C | 0.56 | 0.60 | 0.88 | C | 0.42 | 0.46 | 0.72 |
| ZNF365 | rs10995190 | G | 0.86 | 0.82 | 0.76 | G | 0.83 | 0.83 | 0.90 |
| ZMIZ1 | rs704010 | T | 0.43 | 0.42 | 0.93 | T | 0.11 | 0.08 | 0.82 |
| FGFR2 | rs1896395 | A | 0.00 | 0.00 | 0.96 | A | 0.20 | 0.20 | 0.04 |
| FGFR2 | rs3750817 | C | 0.65 | 0.60 | 0.16 | C | 0.91 | 0.88 | 0.83 |
| FGFR2 | rs10736303 | G | 0.54 | 0.49 | 0.19 | G | 0.87 | 0.84 | 0.75 |
| FGFR2 | rs11200014 | A | 0.46 | 0.41 | 0.65 | A | 0.20 | 0.21 | 0.75 |
| FGFR2 | rs2981579 | T | 0.47 | 0.41 | 0.51 | T | 0.62 | 0.61 | 0.10 |
| FGFR2 | rs1078806 | G | 0.45 | 0.41 | 0.53 | G | 0.21 | 0.21 | 0.99 |
| FGFR2 | rs2981578 | C | 0.54 | 0.49 | 0.09 | C | 0.87 | 0.84 | 0.45 |
| FGFR2 | rs1219648 | G | 0.45 | 0.39 | 0.35 | G | 0.44 | 0.41 | 0.57 |
| FGFR2 | rs2912774 | A | 0.45 | 0.40 | 0.26 | A | 0.59 | 0.55 | 0.07 |
| FGFR2 | rs2936870 | T | 0.45 | 0.40 | 0.25 | T | 0.60 | 0.56 | 0.14 |
| FGFR2 | rs2420946 | T | 0.44 | 0.39 | 0.21 | T | 0.54 | 0.52 | 0.03 |
| FGFR2 | rs2162540 | G | 0.44 | 0.39 | 0.28 | G | 0.54 | 0.52 | 0.41 |
| FGFR2 | rs2981582 | T | 0.44 | 0.39 | 0.30 | T | 0.49 | 0.49 | 0.96 |
| FGFR2 | rs3135718 | G | 0.44 | 0.39 | 0.23 | G | 0.58 | 0.54 | 0.65 |
| 10q | rs10510126 | C | 0.89 | 0.89 | 0.38 | C | 0.89 | 0.90 | 0.21 |
| ATM | rs1800054 | G | 0.02 | 0.02 | 0.34 | G | 0.00 | 0.00 | 0.94 |
| ATM | rs4986761 | C | 0.02 | 0.01 | 0.68 | C | 0.00 | 0.00 | 0.98 |
| ATM | rs1800056 | C | 0.02 | 0.01 | 0.67 | C | 0.00 | 0.00 | 0.95 |
| ATM | rs1800057 | G | 0.03 | 0.02 | 0.90 | G | 0.01 | 0.01 | 0.91 |
| ATM | rs1800058 | T | 0.02 | 0.02 | 0.06 | T | 0.01 | 0.01 | 0.91 |
| ATM | rs1801516 | A | 0.15 | 0.14 | 0.17 | A | 0.03 | 0.02 | 0.48 |
| ATM | rs3092992 | C | 0.06 | 0.04 | 0.13 | C | 0.01 | 0.01 | 0.77 |
| ATM | rs664143 | C | 0.58 | 0.57 | 0.70 | C | 0.66 | 0.66 | 0.45 |
| ATM | rs170548 | G | 0.31 | 0.37 | 0.88 | G | 0.09 | 0.12 | 0.07 |
| ATM | rs3092993 | A | 0.15 | 0.14 | 0.19 | A | 0.03 | 0.02 | 0.48 |
| LSP1 | rs3817198 | C | 0.33 | 0.34 | 0.18 | C | 0.17 | 0.17 | 0.16 |
| LSP1 | rs909116 | T | 0.54 | 0.52 | 0.20 | T | 0.71 | 0.72 | 0.96 |
| MYEOV | rs614367 | T | 0.18 | 0.11 | 0.05 | T | 0.13 | 0.15 | 0.33 |
| H19 | rs2107425 | C | 0.71 | 0.68 | 0.74 | C | 0.48 | 0.53 | 0.42 |
| TOX3 | rs8049149 | T | 0.00 | 0.00 | 0.98 | T | 0.02 | 0.02 | 0.32 |
| TOX3 | rs16951186 | T | 0.01 | 0.01 | 0.75 | T | 0.17 | 0.19 | 0.95 |
| TOX3 | rs8051542 | T | 0.46 | 0.44 | 0.43 | T | 0.35 | 0.30 | 0.12 |
| TOX3 | rs12443621 | G | 0.51 | 0.41 | 0.39 | G | 0.47 | 0.51 | 1.00 |
| TOX3 | rs3803662 | T | 0.32 | 0.24 | 0.73 | C | 0.48 | 0.46 | 0.65 |
| TOX3 | rs4784227 | T | 0.29 | 0.22 | 0.62 | T | 0.08 | 0.07 | 0.59 |
| TOX3 | rs3104746 | A | 0.03 | 0.02 | 0.48 | A | 0.26 | 0.18 | 0.87 |
| TOX3 | rs3112562 | G | 0.22 | 0.20 | 0.45 | G | 0.52 | 0.46 | 0.88 |
| TOX3 | rs9940048 | A | 0.26 | 0.24 | 0.50 | A | 0.31 | 0.30 | 0.64 |
| TP53 | rs9894946 | G | 0.82 | 0.84 | 0.48 | G | 0.95 | 0.95 | 0.25 |
| TP53 | rs1614984 | T | 0.41 | 0.39 | 0.22 | T | 0.40 | 0.40 | 0.03 |
| TP53 | rs4968187 | T | 0.00 | 0.00 | 0.93 | T | 0.01 | 0.00 | 0.92 |
| TP53 | rs12951053 | C | 0.07 | 0.06 | 0.47 | C | 0.11 | 0.11 | 0.09 |
| TP53 | rs17880604 | C | 0.02 | 0.01 | 0.21 | C | 0.00 | 0.00 | 0.95 |
| TP53 | rs1800372 | G | 0.02 | 0.02 | 0.54 | G | 0.00 | 0.00 | 0.98 |
| TP53 | rs2909430 | G | 0.15 | 0.13 | 0.66 | G | 0.27 | 0.24 | 0.64 |
| TP53 | rs1042522 | C | 0.75 | 0.77 | 0.64 | C | 0.39 | 0.43 | 0.77 |
| TP53 | rs8079544 | C | 0.95 | 0.95 | 1.00 | C | 0.89 | 0.89 | 0.83 |
| COX11 | rs7222197 | G | 0.71 | 0.75 | 0.60 | G | 0.66 | 0.65 | 0.70 |
| COX11 | rs6504950 | G | 0.71 | 0.75 | 0.59 | G | 0.67 | 0.65 | 0.66 |
Abbreviations: HWE, Hardy-Weinberg equilibrium; RAF, risk allele frequency; SNP, single nucleotide polymorphism.
a Weighted by inverse sampling probability.
MLE odds ratios for whites and African Americans are displayed in Tables 2 and 3, respectively. Confidence limit ratios and posterior limit ratios for each model are displayed to facilitate comparisons of model precision.
Table 2.
Comparison of Odds Ratios and Confidence Limit Ratios or Posterior Limit Ratios for Maximum Likelihood Estimation, Bayesian, and Hierarchical Regression Models Among White Women (1,247 Cases and 1,105 Controls) in the Carolina Breast Cancer Study, North Carolina, 1993–2001
| Locus and LD Block | SNP | Reference No. | Reference |
Model Type |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MLE |
Bayesian |
Hierarchical |
|||||||||||
| ORa | 95% CI | ORb | 95% CI | CLR | ORb | 95% PI | PLR | ORc | 95% PI | PLR | |||
| 1p12 | rs11249433d | 28 | 1.14 | 1.10, 1.19 | 1.09 | 0.96, 1.24 | 1.28 | 1.09 | 0.96, 1.22 | 1.28 | 1.09 | 0.96, 1.24 | 1.28 |
| CASP8 block1 | rs1045485e | 33 | 1.12 | 1.08, 1.18 | 1.13 | 0.94, 1.35 | 1.43 | 1.11 | 0.93, 1.29 | 1.39 | 1.08 | 0.78, 1.49 | 1.90 |
| CASP8 block1 | rs17468277 | 1.12 | 0.93, 1.34 | 1.43 | 1.11 | 0.93, 1.29 | 1.38 | 1.04 | 0.76, 1.44 | 1.90 | |||
| 2q35 | rs13387042d | 27 | 1.20 | 1.14, 1.26 | 1.08 | 0.96, 1.22 | 1.28 | 1.08 | 0.96, 1.21 | 1.27 | 1.08 | 0.96, 1.22 | 1.28 |
| 2p | rs4666451f | 23 | 1.03 | 1.00, 1.06 | 1.02 | 0.90, 1.16 | 1.28 | 1.02 | 0.90, 1.14 | 1.27 | 1.02 | 0.90, 1.16 | 1.28 |
| SLC4A7 | rs4973768d | 29 | 1.16 | 1.10, 1.24 | 1.04 | 0.92, 1.17 | 1.28 | 1.04 | 0.92, 1.17 | 1.28 | 1.04 | 0.92, 1.17 | 1.28 |
| 4p | rs12505080f | 25 | 1.15g | 1.03, 1.28 | 1.06 | 0.92, 1.23 | 1.33 | 1.06 | 0.92, 1.21 | 1.31 | 1.06 | 0.92, 1.23 | 1.33 |
| TLR1 | rs7696175f | 25 | 1.12g | 1.00, 1.26 | 1.09 | 0.96, 1.23 | 1.28 | 1.09 | 0.96, 1.22 | 1.27 | 1.09 | 0.96, 1.23 | 1.28 |
| MRPS30 | rs4415084d | 32 | 1.16 | 1.10, 1.21 | 1.23 | 1.08, 1.40 | 1.30 | 1.22 | 1.07, 1.37 | 1.28 | 1.23 | 1.08, 1.40 | 1.30 |
| MRPS30 | rs10941679f | 32 | 1.19 | 1.13, 1.26 | 1.18 | 1.03, 1.36 | 1.32 | 1.17 | 1.01, 1.33 | 1.31 | 1.18 | 1.03, 1.36 | 1.32 |
| 5p12 | rs981782d | 23 | 1.04 | 1.01, 1.08 | 0.98 | 0.86, 1.11 | 1.28 | 0.98 | 0.87, 1.10 | 1.27 | 0.98 | 0.86, 1.11 | 1.28 |
| 5q | rs30099f | 23 | 1.05 | 1.01, 1.10 | 1.04 | 0.85, 1.28 | 1.52 | 1.04 | 0.84, 1.24 | 1.48 | 1.04 | 0.85, 1.28 | 1.52 |
| MAP3K1 | rs889312d | 23 | 1.13 | 1.10, 1.16 | 1.19 | 1.04, 1.35 | 1.30 | 1.18 | 1.03, 1.32 | 1.29 | 1.19 | 1.04, 1.35 | 1.30 |
| ESR1 | rs2046210d | 30 | 1.29 | 1.21, 1.37 | 1.09 | 0.96, 1.24 | 1.30 | 1.08 | 0.95, 1.22 | 1.28 | 1.09 | 0.96, 1.24 | 1.30 |
| ESR1 | rs851974 | 0.91 | 0.80, 1.03 | 1.29 | 0.91 | 0.81, 1.03 | 1.27 | 0.91 | 0.80, 1.03 | 1.29 | |||
| ESR1 | rs2077647 | 0.97 | 0.86, 1.10 | 1.28 | 0.97 | 0.86, 1.10 | 1.27 | 0.97 | 0.86, 1.10 | 1.28 | |||
| ESR1 | rs2234693 | 0.95 | 0.84, 1.07 | 1.28 | 0.95 | 0.84, 1.06 | 1.27 | 0.95 | 0.84, 1.07 | 1.28 | |||
| ESR1 | rs1801132e | 33 | 1.05 | 1.00, 1.11 | 0.92 | 0.80, 1.06 | 1.34 | 0.93 | 0.80, 1.05 | 1.31 | 0.92 | 0.80, 1.06 | 1.34 |
| ESR1 | rs3020314e | 33 | 1.12 | 1.06, 1.18 | 1.05 | 0.92, 1.19 | 1.29 | 1.05 | 0.93, 1.18 | 1.27 | 1.05 | 0.92, 1.19 | 1.29 |
| ESR1 | rs3798577 | 1.03 | 0.91, 1.17 | 1.28 | 1.03 | 0.91, 1.16 | 1.27 | 1.03 | 0.91, 1.17 | 1.28 | |||
| ECHDC1 | rs2180341d | 24 | 1.41 | 1.25, 1.59 | 1.04 | 0.90, 1.20 | 1.34 | 1.04 | 0.89, 1.19 | 1.33 | 1.04 | 0.90, 1.20 | 1.34 |
| RELN | rs17157903f | 25 | 1.11 | 1.00, 1.23 | 0.87 | 0.73, 1.04 | 1.42 | 0.89 | 0.75, 1.05 | 1.40 | 0.87 | 0.73, 1.04 | 1.42 |
| 8q24 | rs13281615d | 23 | 1.08 | 1.05, 1.11 | 1.11 | 0.98, 1.26 | 1.28 | 1.11 | 0.98, 1.24 | 1.26 | 1.11 | 0.98, 1.26 | 1.28 |
| 8q24 | rs1562430d | 29 | 1.17 | 1.10, 1.25 | 1.13 | 0.99, 1.28 | 1.29 | 1.12 | 0.99, 1.26 | 1.27 | 1.13 | 0.99, 1.28 | 1.29 |
| CDKN2A/B | rs3731257 | 0.93 | 0.81, 1.07 | 1.32 | 0.94 | 0.81, 1.07 | 1.31 | 0.93 | 0.81, 1.07 | 1.32 | |||
| CDKN2A/B | rs3731249 | 0.90 | 0.63, 1.28 | 2.04 | 0.94 | 0.68, 1.20 | 1.78 | 0.90 | 0.63, 1.29 | 2.04 | |||
| CDKN2A/B block 1 | rs518394 | 1.03 | 0.91, 1.16 | 1.28 | 1.03 | 0.91, 1.15 | 1.26 | 0.98 | 0.76, 1.27 | 1.28 | |||
| CDKN2A/B block 1 | rs564398 | 1.04 | 0.92, 1.17 | 1.28 | 1.04 | 0.91, 1.17 | 1.28 | 1.05 | 0.82, 1.36 | 1.67 | |||
| CDKN2A/B | rs1011970d | 29 | 1.20 | 1.11, 1.30 | 1.13 | 0.96, 1.33 | 1.38 | 1.12 | 0.95, 1.30 | 1.36 | 1.13 | 0.96, 1.33 | 1.38 |
| CDKN2A/B | rs10757278 | 1.17 | 1.04, 1.33 | 1.28 | 1.16 | 1.01, 1.30 | 1.28 | 1.17 | 1.04, 1.33 | 1.28 | |||
| CDKN2A/B | rs10811661 | 1.00 | 0.85, 1.18 | 1.38 | 1.01 | 0.85, 1.16 | 1.36 | 1.00 | 0.85, 1.18 | 1.38 | |||
| ANKRD16 | rs2380205d | 29 | 1.06 | 1.02, 1.10 | 1.01 | 0.89, 1.14 | 1.28 | 1.01 | 0.89, 1.14 | 1.27 | 1.01 | 0.89, 1.14 | 1.28 |
| ZNF365 | rs10995190d | 29 | 1.16 | 1.10, 1.22 | 1.00 | 0.84, 1.20 | 1.43 | 1.00 | 0.85, 1.17 | 1.38 | 1.00 | 0.84, 1.20 | 1.43 |
| ZMIZ1 | rs704010d | 29 | 1.07 | 1.03, 1.11 | 1.24 | 1.09, 1.41 | 1.29 | 1.23 | 1.08, 1.39 | 1.28 | 1.24 | 1.09, 1.41 | 1.29 |
| FGFR2 block 1 | rs3750817 | 1.24 | 1.09, 1.40 | 1.29 | 1.22 | 1.08, 1.37 | 1.28 | 0.97 | 0.81, 1.16 | 1.43 | |||
| FGFR2 block 1 | rs10736303f | 23 | 1.25 | 1.18, 1.32 | 1.33 | 1.17, 1.50 | 1.28 | 1.31 | 1.15, 1.47 | 1.28 | 1.08 | 0.79, 1.48 | 1.88 |
| FGFR2 block 1 | rs11200014 | 1.30 | 1.15, 1.48 | 1.28 | 1.29 | 1.13, 1.44 | 1.27 | 0.94 | 0.66, 1.35 | 2.05 | |||
| FGFR2 block 1 | rs2981579d | 28 | 1.17g | 1.07, 1.27 | 1.33 | 1.18, 1.51 | 1.28 | 1.31 | 1.16, 1.48 | 1.27 | 1.20 | 0.85, 1.72 | 2.03 |
| FGFR2 block 1 | rs1078806f | 24 | 1.26 | 1.13, 1.40 | 1.29 | 1.14, 1.46 | 1.28 | 1.28 | 1.14, 1.44 | 1.26 | 0.95 | 0.67, 1.34 | 1.99 |
| FGFR2 block 1 | rs2981578f | 23 | 1.26 | 1.19, 1.34 | 1.32 | 1.17, 1.50 | 1.28 | 1.30 | 1.15, 1.45 | 1.26 | 1.11 | 0.81, 1.51 | 1.86 |
| FGFR2 block 1 | rs1219648d | 25 | 1.27 | 1.18, 1.36 | 1.31 | 1.16, 1.48 | 1.28 | 1.29 | 1.14, 1.45 | 1.27 | 1.04 | 0.72, 1.51 | 2.10 |
| FGFR2 block 1 | rs2912774f | 23 | 1.26 | 1.19, 1.34 | 1.30 | 1.15, 1.47 | 1.28 | 1.28 | 1.13, 1.44 | 1.27 | 0.96 | 0.65, 1.40 | 2.13 |
| FGFR2 block 1 | rs2936870f | 23 | 1.26 | 1.19, 1.34 | 1.30 | 1.15, 1.47 | 1.28 | 1.29 | 1.13, 1.44 | 1.28 | 0.98 | 0.67, 1.44 | 2.14 |
| FGFR2 block 1 | rs2420946f | 25 | 1.25 | 1.18, 1.36 | 1.30 | 1.15, 1.48 | 1.28 | 1.28 | 1.14, 1.45 | 1.27 | 0.99 | 0.67, 1.46 | 2.16 |
| FGFR2 block 1 | rs2162540 | 1.31 | 1.15, 1.48 | 1.28 | 1.29 | 1.13, 1.44 | 1.27 | 1.04 | 0.71, 1.52 | 2.10 | |||
| FGFR2 block 1 | rs2981582d | 23 | 1.26 | 1.23, 1.30 | 1.30 | 1.15, 1.48 | 1.28 | 1.29 | 1.13, 1.44 | 1.28 | 1.01 | 0.69, 1.48 | 2.09 |
| FGFR2 block 1 | rs3135718f | 23 | 1.15 | 1.07, 1.23 | 1.31 | 1.16, 1.48 | 1.28 | 1.29 | 1.14, 1.45 | 1.27 | 1.05 | 0.72, 1.51 | 2.04 |
| 10q | rs10510126f | 25 | 1.20 | 1.08, 1.35 | 1.11 | 0.91, 1.35 | 1.47 | 1.10 | 0.91, 1.31 | 1.43 | 1.11 | 0.91, 1.35 | 1.47 |
| ATM | rs1800054 | 1.01 | 0.65, 1.58 | 2.45 | 1.03 | 0.70, 1.42 | 2.03 | 1.01 | 0.65, 1.58 | 2.45 | |||
| ATM | rs1800057e | 33 | 1.20h | 1.01, 1.44 | 1.09 | 0.76, 1.56 | 2.06 | 1.08 | 0.78, 1.39 | 1.79 | 1.09 | 0.76, 1.56 | 2.06 |
| ATM | rs1800058 | 0.82 | 0.54, 1.25 | 2.33 | 0.90 | 0.62, 1.18 | 1.90 | 0.82 | 0.54, 1.25 | 2.34 | |||
| ATM block 1 | rs1801516 | 0.98 | 0.82, 1.17 | 1.43 | 0.99 | 0.84, 1.16 | 1.39 | 0.95 | 0.69, 1.32 | 1.94 | |||
| ATM block 1 | rs3092992 | 1.19 | 0.89, 1.60 | 1.80 | 1.16 | 0.87, 1.46 | 1.67 | 1.15 | 0.89, 1.49 | 1.68 | |||
| ATM block 1 | rs664143 | 1.02 | 0.90, 1.15 | 1.28 | 1.02 | 0.90, 1.14 | 1.27 | 1.10 | 0.92, 1.31 | 1.42 | |||
| ATM block 1 | rs170548 | 0.98 | 0.86, 1.12 | 1.31 | 0.98 | 0.86, 1.11 | 1.29 | 0.92 | 0.76, 1.11 | 1.50 | |||
| ATM block 1 | rs3092993 | 0.98 | 0.82, 1.18 | 1.43 | 0.99 | 0.83, 1.15 | 1.39 | 0.97 | 0.70, 1.35 | 1.94 | |||
| LSP1 | rs3817198d | 23 | 1.07 | 1.04, 1.11 | 1.08 | 0.95, 1.24 | 1.30 | 1.08 | 0.95, 1.22 | 1.28 | 1.08 | 0.95, 1.24 | 1.30 |
| LSP1 | rs909116d | 29 | 1.17 | 1.10, 1.24 | 1.14 | 1.01, 1.30 | 1.28 | 1.13 | 0.99, 1.27 | 1.28 | 1.14 | 1.01, 1.30 | 1.28 |
| H19 | rs2107425f | 23 | 1.04 | 1.01, 1.08 | 1.54 | 1.30, 1.82 | 1.31 | 1.49 | 1.25, 1.74 | 1.29 | 1.54 | 1.30, 1.82 | 1.31 |
| TOX3 | rs16951186 | 1.15 | 1.00, 1.31 | 3.49 | 1.14 | 1.01, 1.30 | 2.26 | 1.15 | 1.00, 1.31 | 3.49 | |||
| TOX3 | rs8051542f | 23 | 1.09 | 1.06, 1.13 | 1.12 | 0.99, 1.26 | 1.28 | 1.11 | 0.97, 1.23 | 1.27 | 1.12 | 0.99, 1.26 | 1.28 |
| TOX3 | rs1244362f | 23 | 1.11 | 1.08, 1.14 | 1.17 | 1.04, 1.33 | 1.28 | 1.16 | 1.03, 1.31 | 1.27 | 1.17 | 1.04, 1.33 | 1.28 |
| TOX3 block 1 | rs3803662d | 23 | 1.20 | 1.16, 1.24 | 1.27 | 1.11, 1.46 | 1.31 | 1.25 | 1.10, 1.42 | 1.29 | 1.14 | 0.91, 1.43 | 1.58 |
| TOX3 block 1 | rs4784227d | 26 | 1.25 | 1.20, 1.31 | 1.26 | 1.09, 1.44 | 1.32 | 1.23 | 1.07, 1.41 | 1.31 | 1.11 | 0.88, 1.41 | 1.60 |
| TOX3 | rs3104746 | 1.66 | 1.10, 2.51 | 2.29 | 1.42 | 0.97, 1.94 | 2.01 | 1.66 | 1.10, 2.51 | 2.29 | |||
| TOX3 | rs3112562 | 0.99 | 0.86, 1.15 | 1.34 | 0.99 | 0.86, 1.13 | 1.32 | 0.99 | 0.86, 1.15 | 1.34 | |||
| TOX3 | rs9940048 | 1.03 | 0.89, 1.19 | 1.33 | 1.03 | 0.89, 1.17 | 1.32 | 1.03 | 0.89, 1.19 | 1.33 | |||
| TP53 | rs9894946 | 0.84 | 0.72, 0.99 | 1.38 | 0.86 | 0.73, 1.00 | 1.36 | 0.84 | 0.72, 0.99 | 1.38 | |||
| TP53 | rs1614984 | 1.03 | 0.91, 1.17 | 1.28 | 1.03 | 0.92, 1.15 | 1.26 | 1.03 | 0.91, 1.17 | 1.28 | |||
| TP53 | rs12951053e | 33 | 1.15 | 1.04, 1.26 | 1.09 | 0.85, 1.39 | 1.63 | 1.08 | 0.83, 1.32 | 1.58 | 1.09 | 0.85, 1.39 | 1.63 |
| TP53 | rs17880604 | 0.82 | 0.51, 1.33 | 2.62 | 0.92 | 0.61, 1.26 | 2.06 | 0.82 | 0.51, 1.33 | 2.62 | |||
| TP53 block 1 | rs1800372 | 0.88 | 0.55, 1.40 | 2.57 | 0.95 | 0.64, 1.30 | 2.03 | 0.95 | 0.68, 1.32 | 1.94 | |||
| TP53 block 1 | rs2909430 | 1.11 | 0.93, 1.33 | 1.43 | 1.10 | 0.92, 1.29 | 1.41 | 1.16 | 0.91, 1.49 | 1.51 | |||
| TP53 block 1 | rs1042522 | 0.98 | 0.85, 1.13 | 1.33 | 0.99 | 0.85, 1.12 | 1.31 | 1.06 | 0.89, 1.26 | 1.40 | |||
| TP53 | rs8079544 | 1.24 | 0.95, 1.63 | 1.72 | 1.19 | 0.92, 1.50 | 1.63 | 1.24 | 0.95, 1.63 | 1.72 | |||
| COX11 block 1 | rs7222197f | 29 | 1.12 | 1.04, 1.20 | 0.98 | 0.85, 1.12 | 1.32 | 0.98 | 0.86, 1.12 | 1.29 | 0.99 | 0.72, 1.36 | 1.89 |
| COX11 block 1 | rs6504950f | 31 | 1.05 | 1.03, 1.09 | 0.98 | 0.85, 1.12 | 1.32 | 0.98 | 0.84, 1.11 | 1.31 | 0.99 | 0.72, 1.36 | 1.89 |
Abbreviations: CI, confidence interval; CLR, confidence limit ratio; GWAS, genome-wide association study; LD, linkage disequilibrium; MLE, maximum likelihood estimated; OR, odds ratio; PI, posterior interval; PLR, posterior limit ratio; SNP, single nucleotide polymorphism.
a Odds ratios from initial GWAS or candidate gene meta-analyses (if met criteria for cumulative evidence of association); all odds ratios for log-additive genetic models, unless otherwise specified. Those without values are not GWAS hits and were not included in the candidate gene meta-analysis.
b Adjusted for age at diagnosis (cases) or selection (controls) and proportion of African ancestry.
c Adjusted for age at diagnosis (case) or selection (controls), proportion of African ancestry, and other SNPs in LD block.
d Previous GWAS hit.
e Cumulative evidence of an association in the meta-analysis by Zhang et al. (40).
f Other GWAS-identified gene.
g Odds ratio estimated using general genetic model.
h Odds ratio estimated using dominant genetic model.
Table 3.
Comparison of Odds Ratios and Confidence Limit Ratios or Posterior Limit Ratios for Frequentist, Basic Hierarchical, and Bayesian Regression Models Among African American Women (766 Cases and 681 Controls) in the Carolina Breast Cancer Study, North Carolina, 1993–2001
| Locus and LD Block | SNP | Reference No. | Reference |
Model Type |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MLE |
Bayesian |
Hierarchical |
|||||||||||
| ORa | 95% CI | ORb | 95% CI | CLR | ORb | 95% PI | PLR | ORc | 95% PI | PLR | |||
| 1p12 | rs11249433d | 28 | 1.14 | 1.10, 1.19 | 1.26 | 0.99, 1.60 | 1.61 | 1.22 | 0.96, 1.48 | 1.53 | 1.26 | 0.99, 1.60 | 1.61 |
| CASP8 block 1 | rs1045485e | 33 | 1.12 | 1.08, 1.18 | 0.93 | 0.67, 1.29 | 1.93 | 0.96 | 0.69, 1.22 | 1.77 | 1.12 | 0.39, 3.17 | 8.05 |
| CASP8 block 1 | rs17468277 | 1.09 | 0.78, 1.54 | 1.99 | 1.07 | 0.78, 1.38 | 1.78 | 1.20 | 0.41, 3.53 | 8.68 | |||
| 2q35 | rs13387042d | 27 | 1.20 | 1.14, 1.26 | 1.02 | 0.86, 1.22 | 1.43 | 1.02 | 0.86, 1.20 | 1.39 | 1.02 | 0.86, 1.22 | 1.43 |
| 2p | rs4666451f | 23 | 1.03 | 1.00, 1.06 | 1.15 | 0.96, 1.39 | 1.45 | 1.13 | 0.95, 1.34 | 1.41 | 1.15 | 0.96, 1.39 | 1.45 |
| SLC4A7 | rs4973768d | 29 | 1.16 | 1.10, 1.24 | 0.90 | 0.77, 1.06 | 1.38 | 0.91 | 0.77, 1.04 | 1.35 | 0.90 | 0.77, 1.06 | 1.38 |
| 4p | rs12505080f | 25 | 1.15g | 1.03, 1.28 | 1.09 | 0.88, 1.34 | 1.52 | 1.08 | 0.88, 1.30 | 1.48 | 1.09 | 0.88, 1.34 | 1.52 |
| TLR1 | rs7696175f | 25 | 1.12g | 1.00, 1.26 | 1.39 | 1.04, 1.86 | 1.79 | 1.29 | 0.99, 1.66 | 1.68 | 1.39 | 1.04, 1.86 | 1.80 |
| MRPS30 | rs4415084d | 32 | 1.16 | 1.10, 1.21 | 1.13 | 0.97, 1.33 | 1.38 | 1.13 | 0.96, 1.30 | 1.35 | 1.13 | 0.97, 1.33 | 1.38 |
| MRPS30 | rs10941679f | 32 | 1.19 | 1.13, 1.26 | 1.00 | 0.82, 1.22 | 1.49 | 1.01 | 0.83, 1.19 | 1.43 | 1.00 | 0.82, 1.22 | 1.49 |
| 5p12 | rs981782d | 23 | 1.04 | 1.01, 1.08 | 1.11 | 0.84, 1.46 | 1.74 | 1.09 | 0.84, 1.36 | 1.61 | 1.11 | 0.84, 1.46 | 1.74 |
| 5q | rs30099f | 23 | 1.05 | 1.01, 1.10 | 1.22 | 0.98, 1.52 | 1.55 | 1.19 | 0.96, 1.44 | 1.50 | 1.22 | 0.98, 1.52 | 1.55 |
| MAP3K1 | rs889312d | 23 | 1.13 | 1.10, 1.16 | 0.95 | 0.80, 1.13 | 1.41 | 0.96 | 0.81, 1.11 | 1.37 | 0.95 | 0.80, 1.13 | 1.41 |
| ESR1 | rs2046210d | 30 | 1.29 | 1.21, 1.37 | 1.22 | 1.04, 1.43 | 1.38 | 1.20 | 1.03, 1.39 | 1.35 | 1.22 | 1.04, 1.43 | 1.38 |
| ESR1 | rs851974 | 0.93 | 0.76, 1.14 | 1.50 | 0.94 | 0.78, 1.13 | 1.45 | 0.93 | 0.75, 1.14 | 1.50 | |||
| ESR1 | rs2077647 | 1.07 | 0.92, 1.25 | 1.37 | 1.07 | 0.92, 1.23 | 1.34 | 1.07 | 0.92, 1.25 | 1.37 | |||
| ESR1 | rs2234693 | 0.96 | 0.82, 1.13 | 1.37 | 0.97 | 0.83, 1.12 | 1.35 | 0.96 | 0.82, 1.13 | 1.37 | |||
| ESR1 | rs1801132e | 33 | 1.05 | 1.00, 1.11 | 1.19 | 0.93, 1.52 | 1.64 | 1.16 | 0.91, 1.42 | 1.55 | 1.19 | 0.93, 1.52 | 1.64 |
| ESR1 | rs3020314e | 33 | 1.12 | 1.06,1.18 | 1.00 | 0.84, 1.19 | 1.41 | 1.01 | 0.85, 1.17 | 1.37 | 1.00 | 0.84, 1.19 | 1.41 |
| ESR1 | rs3798577 | 1.02 | 0.88, 1.19 | 1.36 | 1.02 | 0.89, 1.18 | 1.33 | 1.02 | 0.87, 1.19 | 1.36 | |||
| ECHDC1 | rs2180341d | 24 | 1.41 | 1.25, 1.59 | 0.98 | 0.83, 1.15 | 1.39 | 0.98 | 0.84, 1.14 | 1.36 | 0.98 | 0.83, 1.15 | 1.39 |
| RELN | rs17157903f | 25 | 1.11 | 1.00, 1.23 | 1.07 | 0.83, 1.37 | 1.64 | 1.07 | 0.85, 1.31 | 1.54 | 1.07 | 0.83, 1.37 | 1.64 |
| 8q24 | rs13281615d | 23 | 1.08 | 1.05, 1.11 | 1.00 | 0.86, 1.18 | 1.38 | 1.01 | 0.85, 1.17 | 1.37 | 1.00 | 0.86, 1.18 | 1.38 |
| 8q24 | rs1562430d | 29 | 1.17 | 1.10, 1.25 | 1.00 | 0.86, 1.17 | 1.36 | 1.00 | 0.87, 1.16 | 1.34 | 1.00 | 0.86, 1.17 | 1.36 |
| CDKN2A/B | rs3731257 | 0.88 | 0.67, 1.15 | 1.71 | 0.91 | 0.71, 1.14 | 1.60 | 0.88 | 0.67, 1.15 | 1.71 | |||
| CDKN2A/B block 1 | rs518394 | 1.01 | 0.76, 1.35 | 1.76 | 1.02 | 0.78, 1.28 | 1.64 | 1.01 | 0.73, 1.40 | 1.76 | |||
| CDKN2A/B block 1 | rs564398 | 1.01 | 0.75, 1.35 | 1.79 | 1.01 | 0.77, 1.27 | 1.66 | 1.00 | 0.72, 1.40 | 1.96 | |||
| CDKN2A/B | rs1011970d | 29 | 1.20 | 1.11, 1.30 | 0.95 | 0.81, 1.11 | 1.38 | 0.96 | 0.82, 1.10 | 1.34 | 0.95 | 0.81, 1.11 | 1.38 |
| CDKN2A/B | rs10757278 | 0.91 | 0.75, 1.12 | 1.50 | 0.93 | 0.76, 1.09 | 1.44 | 0.91 | 0.75, 1.12 | 1.50 | |||
| CDKN2A/B | rs10811661 | 1.00 | 0.74, 1.35 | 1.83 | 1.01 | 0.76, 1.28 | 1.67 | 1.00 | 0.74, 1.35 | 1.83 | |||
| ANKRD16 | rs2380205d | 29 | 1.06 | 1.02, 1.10 | 0.97 | 0.83, 1.13 | 1.37 | 0.98 | 0.84, 1.13 | 1.34 | 0.97 | 0.83, 1.13 | 1.37 |
| ZNF365 | rs10995190d | 29 | 1.16 | 1.10, 1.22 | 1.06 | 0.87, 1.29 | 1.49 | 1.05 | 0.86, 1.23 | 1.44 | 1.06 | 0.87, 1.29 | 1.49 |
| ZMIZ1 | rs704010d | 29 | 1.07 | 1.03, 1.11 | 1.05 | 0.80, 1.36 | 1.69 | 1.04 | 0.80, 1.28 | 1.61 | 1.05 | 0.80, 1.36 | 1.69 |
| FGFR2 | rs1896395 | 1.01 | 0.83, 1.23 | 1.48 | 1.02 | 0.84, 1.20 | 1.44 | 1.01 | 0.83, 1.23 | 1.48 | |||
| FGFR2 block 1 | rs3750817 | 1.74 | 1.34, 2.26 | 1.69 | 1.61 | 1.22, 2.02 | 1.66 | 1.38 | 1.05, 1.83 | 1.74 | |||
| FGFR2 block 1 | rs10736303f | 23 | 1.25 | 1.18, 1.32 | 1.39 | 1.12, 1.74 | 1.56 | 1.33 | 1.07, 1.61 | 1.51 | 1.08 | 0.83, 1.39 | 1.67 |
| FGFR2 block 1 | rs11200014 | 1.04 | 0.86, 1.26 | 1.47 | 1.04 | 0.85, 1.23 | 1.45 | 0.97 | 0.70, 1.34 | 1.92 | |||
| FGFR2 block 1 | rs2981579d | 28 | 1.17g | 1.07, 1.27 | 1.22 | 1.04, 1.42 | 1.36 | 1.19 | 1.03, 1.37 | 1.33 | 1.10 | 0.93, 1.31 | 1.41 |
| FGFR2 block 1 | rs1078806f | 24 | 1.26 | 1.13, 1.40 | 1.06 | 0.88, 1.29 | 1.47 | 1.06 | 0.87, 1.25 | 1.43 | 1.00 | 0.72, 1.38 | 1.92 |
| FGFR2 block 2 | rs2981578f | 23 | 1.26 | 1.19, 1.34 | 1.42 | 1.14, 1.77 | 1.56 | 1.36 | 1.10, 1.65 | 1.51 | 1.23 | 0.99, 1.53 | 1.54 |
| FGFR2 block 2 | rs1219648d | 25 | 1.27 | 1.18, 1.36 | 1.19 | 1.02, 1.39 | 1.37 | 1.18 | 1.01, 1.35 | 1.33 | 1.01 | 0.82, 1.24 | 1.51 |
| FGFR2 block 2 | rs2912774f | 23 | 1.26 | 1.19, 1.34 | 1.27 | 1.09, 1.49 | 1.37 | 1.25 | 1.06, 1.43 | 1.35 | 1.09 | 0.80, 1.49 | 1.85 |
| FGFR2 block 2 | rs2936870f | 23 | 1.26 | 1.19, 1.34 | 1.27 | 1.09, 1.48 | 1.37 | 1.25 | 1.07, 1.44 | 1.34 | 1.09 | 0.80, 1.48 | 1.84 |
| FGFR2 block 3 | rs2420946f | 25 | 1.25 | 1.18, 1.36 | 1.17 | 1.00, 1.37 | 1.37 | 1.16 | 1.00, 1.35 | 1.35 | 0.95 | 0.72, 1.25 | 1.73 |
| FGFR2 block 3 | rs2162540 | 1.23 | 1.05, 1.44 | 1.36 | 1.21 | 1.04, 1.40 | 1.34 | 1.27 | 0.96, 1.66 | 1.37 | |||
| FGFR2 | rs2981582d | 23 | 1.26 | 1.23, 1.30 | 1.19 | 1.02, 1.39 | 1.37 | 1.18 | 1.00, 1.35 | 1.35 | 1.19 | 1.02, 1.39 | 1.37 |
| FGFR2 | rs3135718f | 23 | 1.15 | 1.07, 1.23 | 1.26 | 1.08, 1.46 | 1.35 | 1.24 | 1.07, 1.43 | 1.33 | 1.26 | 1.08, 1.46 | 1.35 |
| 10q | rs10510126f | 25 | 1.20 | 1.08, 1.35 | 1.04 | 0.82, 1.32 | 1.61 | 1.04 | 0.82, 1.25 | 1.53 | 1.04 | 0.82, 1.32 | 1.61 |
| ATM | rs1801516 | 1.22 | 0.77, 1.95 | 2.54 | 1.14 | 0.76, 1.58 | 2.08 | 1.22 | 0.77, 1.95 | 2.54 | |||
| ATM | rs664143 | 0.95 | 0.81, 1.11 | 1.38 | 0.96 | 0.82, 1.10 | 1.35 | 0.95 | 0.81, 1.11 | 1.38 | |||
| ATM | rs170548 | 0.86 | 0.67, 1.11 | 1.66 | 0.90 | 0.70, 1.10 | 1.57 | 0.86 | 0.67, 1.11 | 1.66 | |||
| ATM | rs3092993 | 1.22 | 0.77, 1.95 | 2.54 | 1.14 | 0.76, 1.58 | 2.08 | 1.22 | 0.77, 1.95 | 2.54 | |||
| LSP1 | rs3817198d | 23 | 1.07 | 1.04, 1.11 | 1.01 | 0.82, 1.24 | 1.52 | 1.01 | 0.84, 1.21 | 1.47 | 1.01 | 0.82, 1.24 | 1.52 |
| LSP1 | rs909116d | 29 | 1.17 | 1.10, 1.24 | 1.00 | 0.84, 1.19 | 1.42 | 1.01 | 0.84, 1.17 | 1.39 | 1.00 | 0.84, 1.19 | 1.42 |
| H19 | rs2107425f | 23 | 1.04 | 1.01, 1.08 | 0.84 | 0.71, 0.98 | 1.38 | 0.86 | 0.73, 0.98 | 1.35 | 0.84 | 0.71, 0.98 | 1.38 |
| TOX3 | rs8049149 | 0.92 | 0.54, 1.56 | 2.87 | 0.98 | 0.64, 1.35 | 2.10 | 0.92 | 0.54, 1.56 | 2.87 | |||
| TOX3 | rs16951186 | 0.90 | 0.74, 1.09 | 1.49 | 0.92 | 0.76, 1.10 | 1.45 | 0.90 | 0.74, 1.09 | 1.49 | |||
| TOX3 | rs8051542f | 23 | 1.09 | 1.06, 1.13 | 1.14 | 0.97, 1.35 | 1.39 | 1.13 | 0.97, 1.30 | 1.35 | 1.14 | 0.97, 1.35 | 1.39 |
| TOX3 | rs12443621f | 23 | 1.11 | 1.08, 1.14 | 0.86 | 0.74, 1.01 | 1.36 | 0.88 | 0.75, 1.00 | 1.34 | 0.86 | 0.74, 1.01 | 1.36 |
| TOX3 block 1 | rs3803662d | 23 | 1.20 | 1.16, 1.24 | 1.06 | 0.90, 1.23 | 1.37 | 1.05 | 0.91, 1.21 | 1.34 | 1.11 | 0.94, 1.31 | 1.39 |
| TOX3 block 1 | rs4784227d | 26 | 1.25 | 1.20, 1.31 | 1.25 | 0.93, 1.67 | 1.80 | 1.19 | 0.90, 1.51 | 1.67 | 1.28 | 0.96, 1.71 | 1.77 |
| TOX3 block 2 | rs3104746 | 1.54 | 1.27, 1.86 | 1.46 | 1.49 | 1.22, 1.75 | 1.43 | 1.39 | 1.14, 1.71 | 1.50 | |||
| TOX3 block 2 | rs3112562 | 1.28 | 1.09, 1.50 | 1.37 | 1.26 | 1.09, 1.46 | 1.35 | 1.12 | 0.94, 1.33 | 1.41 | |||
| TOX3 | rs9940048 | 1.10 | 0.93, 1.31 | 1.40 | 1.10 | 0.92, 1.29 | 1.39 | 1.10 | 0.93, 1.31 | 1.41 | |||
| TP53 | rs9894946 | 0.96 | 0.68, 1.36 | 2.01 | 0.98 | 0.70, 1.28 | 1.82 | 0.96 | 0.68, 1.37 | 2.01 | |||
| TP53 | rs1614984 | 1.07 | 0.92, 1.25 | 1.36 | 1.07 | 0.91, 1.22 | 1.34 | 1.07 | 0.92, 1.25 | 1.36 | |||
| TP53 block 1 | rs12951053e | 33 | 1.15 | 1.04, 1.26 | 1.03 | 0.80, 1.32 | 1.64 | 1.03 | 0.81, 1.25 | 1.55 | 1.01 | 0.77, 1.32 | 1.71 |
| TP53 block 1 | rs2909430 | 1.06 | 0.89, 1.25 | 1.40 | 1.06 | 0.89, 1.22 | 1.37 | 1.04 | 0.84, 1.28 | 1.52 | |||
| TP53 block 1 | rs1042522 | 0.98 | 0.83, 1.15 | 1.38 | 0.98 | 0.83, 1.13 | 1.36 | 1.00 | 0.82, 1.22 | 1.50 | |||
| TP53 | rs8079544 | 0.89 | 0.69, 1.15 | 1.66 | 0.92 | 0.72, 1.12 | 1.57 | 0.89 | 0.69, 1.15 | 1.66 | |||
| COX11 block 1 | rs7222197f | 29 | 1.12 | 1.04, 1.20 | 1.14 | 0.97, 1.33 | 1.38 | 1.12 | 0.95, 1.29 | 1.36 | 1.07 | 0.77, 1.47 | 1.90 |
| COX11 block 1 | rs6504950f | 31 | 1.05 | 1.03, 1.09 | 1.13 | 0.96, 1.32 | 1.37 | 1.12 | 0.95, 1.28 | 1.35 | 1.07 | 0.77, 1.47 | 1.90 |
Abbreviations: CI, confidence interval; CLR, confidence limit ratio; GWAS, genome-wide association study; LD, linkage disequilibrium; MLE, maximum likelihood estimated; OR, odds ratio; PI, posterior interval; PLR, posterior limit ratio; SNP, single nucleotide polymorphism.
a Odds ratios from initial GWAS or candidate gene meta-analyses (if met criteria for cumulative evidence of association); all odds ratios for log-additive genetic models, unless otherwise specified. Those without values are not GWAS hits and were not included in the candidate gene meta-analysis.
b Adjusted for age at diagnosis (cases) or selection (controls) and proportion of African ancestry.
c Adjusted for age at diagnosis (cases) or selection (controls), proportion of African ancestry, and other SNPs in the LD block.
d Previous GWAS hit.
e Cumulative evidence of an association in the meta-analysis by Zhang et al. (40).
f Other GWAS-identified gene.
g Odds ratio estimated using general genetic model.
Among whites in our study, 18 of the GWAS-identified SNPs successfully replicated. All of the fibroblast growth factor receptor 2 gene (FGFR2) SNPs had relatively strong, positive associations with breast cancer (odds ratios (ORs) > 1.15), as did both of the mitochondrial ribosomal protein S30 gene (MRPS30) SNPs, 2 of the TOX high mobility group box family member 3 gene (TOX3) SNPs (rs3803662 and rs4784227), rs889312 in mitogen-activated protein kinase kinase kinase 1 gene (MAP3K1), rs704010 in zinc finger, MIZ-type containing 1 gene (ZMIZ1), and rs2107425 in H19, imprinted maternally expressed transcript gene (H19). The 95% confidence interval for rs909116 in lymphocyte-specific protein 1 gene (LSP1) excluded the null, but the Bayesian odds ratio was attenuated and did not meet our replication criteria. Three other FGFR2 SNPs (rs3750817, rs11200014, and rs2162540) were strongly associated with breast cancer (OR > 1.2) in whites.
None of the SNPs selected from the candidate gene meta-analysis replicated, though several SNPs in ataxia telangiectasia mutated gene (ATM) and tumor protein p53 gene (TP53) were strongly associated with disease (|ln OR| > 0.15). The original GWAS and meta-analysis odds ratios are provided in Table 2 for further reference.
The most extreme example of the difference between MLE and Bayesian estimates in whites in our study was for rs3104746 in TOX3, a rare SNP (risk allele frequency = 2%) with the highest MLE odds ratio (OR = 1.66). Here, the Bayesian estimate was closer to the null (OR = 1.42) and was more precise (MLE confidence limit ratio = 2.29 vs. Bayesian posterior limit ratio = 2.01; Web Appendix 4, Web Figure 1).
Ten of the GWAS-identified SNPs successfully replicated in African Americans (Table 3 and Web Figure 2). This included 9 SNPs in FGFR2 (ORs > 1.15) and rs2046210 in estrogen receptor 1 gene (ESR1). Two other GWAS-identified SNPs, rs2107425 (in H19) and rs12443621 (in TOX3), had 95% posterior intervals that excluded the null, but both were inversely associated with breast cancer and, thus, inconsistent with original reports. Two additional TOX3 SNPs (rs3104746 and rs3112562) had odds ratios greater than 1.25 via either analysis method. Some of the ATM and ESR1 MLE odds ratios were relatively strong, but none of the SNPs from the candidate gene meta-analysis successfully replicated in African Americans.
The 77 SNPs evaluable in whites separated into 55 LD blocks. The thirteen SNPs in FGFR2 formed the largest block, followed by ATM (5 SNPs) and TP53 (3 SNPs). LD blocks consisting of 2 highly correlated SNPs were also genotyped in caspase 8, apoptosis-related cysteine peptidase gene (CASP8), cyclin-dependent kinase inhibitor 2A/B genes (CDKN2A/CDKN2B), TOX3, and cytochrome c oxidase assembly homolog 11 gene (COX11). In African Americans, the FGFR2 SNPs formed 3 separate blocks of 5, 4, and 2 SNPs, respectively, whereas the other 3 SNPs were not strongly linked. One of the unlinked SNPs, rs1896395, was not evaluable in whites (risk allele frequency = 0%). TOX3 contained two 2-SNP LD blocks, and TP53 contained a single 3-SNP block. The 2 SNPs within CASP8, CDKN2A, and COX11 were again in high LD. None of the ATM SNPs was strongly correlated in African Americans. In total, the 73 SNPs evaluable in African Americans formed 58 LD blocks.
Semi-Bayes odds ratios and 95% posterior intervals for the hierarchical models are presented in Tables 2 and 3 and Web Figures 3 and 4. In general, the hierarchical-based estimates had comparable or slightly lower precision than the MLE odds ratios and consistently lower precision than the Bayesian estimates. According to hierarchically derived estimates, many of the SNPs in the larger LD blocks were not associated with breast cancer. For example, MLE and Bayesian odds ratios indicated that all 13 of the highly correlated FGFR2 SNPs were strongly associated with breast cancer among whites, whereas the hierarchical model generated near-null estimates for these SNPs. Of the 13, rs2981579 had the strongest association (OR = 1.20, 95% posterior interval: 0.85, 1.72). Similarly, MLE and Bayesian models indicated that 10 of the 14 FGFR2 SNPs were associated with breast cancer in African Americans, whereas hierarchical modeling produced elevated associations for 1 SNP in each LD block (rs3750817, rs2981578, and rs2420946) and for 2 of 3 unlinked SNPs.
DISCUSSION
Because several of the SNPs analyzed here were previously reported for this study population (54), we will limit our discussion to novel findings. Among whites, statistically significant associations for rs10757278 (in CDKN2A/CDKN2B) and rs3104746 (in TOX3) have never before been reported. We also corroborated previously observed associations between breast cancer and several well-validated GWAS-identified SNPs, including 2 MRPS30 SNPs (rs4415084 and rs10941679) (4, 35, 39, 55–61), rs1562430 in 8q24 (35, 36, 57), and rs4784227 in TOX3 (33, 62). Additionally, we replicated several less-established GWAS-identified SNPs, including rs704010 in ZMIZ1 (36) and rs3750817, rs10736303, rs1078806, and rs2981578 in FGFR2 (30, 31). The only CASP8, ATM, or TP53 SNP to demonstrate a statistically significant association (rs9894986 in TP53) was not associated with disease in the study by Zhang et al. (40).
We are the first to report a statistically significant association for rs3750817 in FGFR2 in African Americans. Previous investigations of rs2046210 (in ESR1) in African Americans produced mostly near-null odds ratios (2–6, 11, 63), but several of the FGFR2 and TOX3 SNPs were associated with breast cancer in 1 or more prior investigations. This includes rs10736303 and rs2981578 (in FGFR2) (5, 8) and rs3104746 and rs3112562 (in TOX3) (9). Both rs2981578 and rs3104746 were positively associated with disease in a pooled analysis by Chen et al. (2), but approximately 20% of these participants were drawn from our study population. In general, the SNPs identified in the candidate gene meta-analysis were rarer and had weaker associations than the GWAS-identified SNPs, making it difficult to detect meaningful associations.
As expected given our choice of null-centered priors, the Bayesian estimates were closer to the null than the MLE estimates. Although the 2 odds ratio estimates were very similar for many of the SNPs assessed here, the degree of attenuation was strongest when the SNP's minor allele frequency was low. These results demonstrate how Bayesian methods are less vulnerable to extreme observations, and why Bayesian methods are particularly advantageous when data are sparse or there is a high probability of spurious associations. In this way, Bayesian methods may be a less conservative alternative to standard multiple comparisons adjustment methods.
Because the hierarchical models included more parameters than the MLE or Bayesian models, they did not improve precision. However, these methods can help to differentiate individual effects of highly correlated SNPs. For example, it is unlikely that all 13 evaluable FGFR2 SNPs are strongly associated with breast cancer in whites. Rather, 1 or 2 causal variants within the LD block probably drive all of the observed associations. In such scenarios, hierarchical models can effectively accommodate correlated exposures and provide stable SNP- and haplotype-level odds ratios, whereas models that evaluate all the SNPs simultaneously in a single-level model will often produce unstable or nonconvergent estimates (22, 48, 64).
Unfortunately, although we believe these methods would be beneficial in larger studies, we did not have sufficient power to reliably differentiate between the strongest FGFR2 SNP odds ratios and the null when so many SNPs were assessed simultaneously. Analyses of SNPs in the other multi-SNP LD blocks were relatively more precise, but also largely inconclusive. The hierarchical models performed better in African Americans, with rs3750817, rs2981578, rs2420946, and rs3104746 demonstrating notably stronger associations than the other SNP(s) in their respective LD blocks. This performance improvement is likely attributable to the anticipated racial differences in LD block size.
We believe our specifications of prior means and variances are reasonable. First, aside from mutations in BRCA1/BRCA2, it is unlikely that a single SNP has a large effect on breast cancer risk (20). Second, as long as the covariate priors are appropriately specified, Bayesian analysis with null-centered priors will bias effect estimates toward the null (65). Lastly, we believe that correlated SNPs within an LD block meet the criteria for exchangeability.
After we accounted for our sampling mechanisms, the only observed discrepancy between study cases and other North Carolina cases was that African Americans with later stage disease were underrepresented in our study (66). Therefore, odds ratios could be biased if the evaluated SNP is related to disease aggressiveness or medical care utilization. With regard to genotyping, whites were more likely to provide blood samples than African Americans, but blood donation status did not differ by case status, disease stage, or other known risk factors.
The inclusion of in situ cases could bias estimates of SNPs associated with disease aggressiveness or progression, but given strong evidence that invasive and in situ tumors have similar risk profiles (16, 67), we chose to retain these individuals. Analyses limited to invasive cases yielded similar results (Web Table 2).
This was a racially diverse, population-based sample with well-validated data. The inclusion of a relatively large sample of African-American women allowed us to investigate racial differences in genetic risk factors and, accordingly, provide information that may help pinpoint causal variants. Although the results for SNPs that violated Hardy-Weinberg equilibrium should be interpreted with caution, the quality control measures used during the genotyping process should have reduced the number and impact of genotype misclassification.
In this analysis, we replicated several previously identified breast cancer susceptibility loci in whites and African Americans by using both MLE and Bayesian methods. Our findings offer additional evidence that these SNPs or chromosomal regions play an important role in breast cancer etiology. The SNPs that replicated in African Americans are especially instructive, because they refine the genomic region containing the causal variant. Our use of Bayesian methods to incorporate external information further augments the utility of these results. We believe that fine-mapping studies and smaller, etiologically driven investigations may derive even greater benefit from these better-informed, more stable approaches.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: Department of Epidemiology, University of North Carolina Gillings School of Global Public Health, Chapel Hill, North Carolina (Katie M. O'Brien, Stephen R. Cole, Charles Poole, Jeannette T. Bensen, Lawrence S. Engel, Robert C. Millikan); Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina (Katie M. O'Brien, Jeannette T. Bensen, Robert C. Millikan); and Department of Biostatistics and Carolina Population Center, University of North Carolina Gillings School of Global Public Health, Chapel Hill, North Carolina (Amy H. Herring).
This work was supported by the University Cancer Research Fund of North Carolina, the National Cancer Institute Specialized Program of Research Excellence in Breast Cancer (grant NIH/NCI P50-CA58223 to K.M.O., J.T.B., and R.C.M.), the Lineberger Comprehensive Cancer Center Core Grant (NIH/NCI P30-CA16086 to K.M.O., J.T.B., and R.C.M.), and a Cancer Education and Career Development Program institutional training grant from the National Cancer Institute at the National Institutes of Health (NIH/NCI 5R25CA057726-20 to K.M.O.).
We would like to acknowledge the University of North Carolina BioSpecimen Processing Facility for our DNA extractions, blood processing, storage, and sample disbursement, the University of North Carolina Mammalian Genotyping Core for sample genotyping, and Jessica Tse for her technical assistance. We also thank the staff of the Carolina Breast Cancer Study.
Conflict of interest: none declared.
REFERENCES
- 1.Hindorff LA, MacArthur J, Morales J, et al. Bethesda, MD: National Human Genome Research Institute, National Institutes of Health; 2013. A catalog of published genome-wide association studies (www.genome.gov/gwastudies. ). (Accessed January 13, 2013) [Google Scholar]
- 2.Chen F, Chen GK, Millikan RC, et al. Fine-mapping of breast cancer susceptibility loci characterizes genetic risk in African Americans. Hum Mol Genet. 2011;20(22):4491–4503. doi: 10.1093/hmg/ddr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hutter CM, Young AM, Ochs-Balcom HM, et al. Replication of breast cancer GWAS susceptibility loci in the Women's Health Initiative African American SHARe Study. Cancer Epidemiol Biomarkers Prev. 2011;20(9):1950–1959. doi: 10.1158/1055-9965.EPI-11-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campa D, Kaaks R, Le Marchand L, et al. Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium. J Natl Cancer Inst. 2011;103(16):1252–1263. doi: 10.1093/jnci/djr265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng W, Cai Q, Signorello LB, et al. Evaluation of 11 breast cancer susceptibility loci in African-American women. Cancer Epidemiol Biomarkers Prev. 2009;18(10):2761–2764. doi: 10.1158/1055-9965.EPI-09-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stacey SN, Sulem P, Zanon C, et al. Ancestry-shift refinement mapping of the C6orf97-ESR1 breast cancer susceptibility locus. PLoS Genet. 2010;6(7):e1001029. doi: 10.1371/journal.pgen.1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rebbeck TR, DeMichele A, Tran TV, et al. Hormone-dependent effects of FGFR2 and MAP3K1 in breast cancer susceptibility in a population-based sample of postmenopausal African-American and European-American women. Carcinogenesis. 2009;30(2):269–274. doi: 10.1093/carcin/bgn247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Udler MS, Meyer KB, Pooley KA, et al. FGFR2 variants and breast cancer risk: fine-scale mapping using African American studies and analysis of chromatin conformation. Hum Mol Genet. 2009;18(9):1692–1703. doi: 10.1093/hmg/ddp078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruiz-Narvaez EA, Rosenberg L, Rotimi CN, et al. Genetic variants on chromosome 5p12 are associated with risk of breast cancer in African American women: the Black Women's Health Study. Breast Cancer Res Treat. 2010;123(2):525–530. doi: 10.1007/s10549-010-0775-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruiz-Narváez EA, Rosenberg L, Cozier YC, et al. Polymorphisms in the TOX3/LOC643714 locus and risk of breast cancer in African-American women. Cancer Epidemiol Biomarkers Prev. 2010;19(5):1320–1327. doi: 10.1158/1055-9965.EPI-09-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer JR, Ruiz-Narvaez EA, Rotimi CN, et al. Genetic susceptibility loci for subtypes of breast cancer in an African American population. Cancer Epidemiol Biomarkers Prev. 2013;22(1):127–134. doi: 10.1158/1055-9965.EPI-12-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huo D, Zheng Y, Ogundiran TO, et al. Evaluation of 19 susceptibility loci of breast cancer in women of African ancestry. Carcinogenesis. 2012;33(4):835–840. doi: 10.1093/carcin/bgs093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 14.Huo D, Ikpatt F, Khramtsov A, et al. Population differences in breast cancer: survey in indigenous African women reveals over-representation of triple-negative breast cancer. J Clin Oncol. 2009;27(27):4515–4521. doi: 10.1200/JCO.2008.19.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lund MJ, Trivers KF, Porter PL, et al. Race and triple negative threats to breast cancer survival: a population-based study in Atlanta, GA. Breast Cancer Res Treat. 2009;113(2):357–370. doi: 10.1007/s10549-008-9926-3. [DOI] [PubMed] [Google Scholar]
- 16.Millikan RC, Newman B, Tse C, et al. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat. 2008;109(1):123–139. doi: 10.1007/s10549-007-9632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Surveillance Epidemiology and End Results. Bethesda, MD: National Cancer Institute, National Institutes of Health; 2013. US cancer morality statistics http://seer.cancer.gov/canques/mortality.html. (Accessed January 9, 2013) [Google Scholar]
- 18.Haiman CA, Stram DO. Exploring genetic susceptibility to cancer in diverse populations. Curr Opin Genet Dev. 2010;20(3):330–335. doi: 10.1016/j.gde.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinch AG, Tandon A, Patterson N, et al. The landscape of recombination in African Americans. Nature. 2011;476(7359):170–175. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter DJ. Lessons from genome-wide association studies for epidemiology. Epidemiology. 2012;23(3):363–367. doi: 10.1097/EDE.0b013e31824da7cc. [DOI] [PubMed] [Google Scholar]
- 21.Hung RJ, Brennan P, Malaveille C, et al. Using hierarchical modeling in genetic association studies with multiple markers: application to a case-control study of bladder cancer. Cancer Epidemiol Biomarkers Prev. 2004;13(6):1013–1021. [PubMed] [Google Scholar]
- 22.Conti DV, Witte JS. Hierarchical modeling of linkage disequilibrium: genetic structure and spatial relations. Am J Hum Genet. 2003;72(2):351–363. doi: 10.1086/346117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephens M, Balding DJ. Bayesian statistical methods for genetic association studies. Nat Rev Genet. 2009;10(10):681–690. doi: 10.1038/nrg2615. [DOI] [PubMed] [Google Scholar]
- 24.Newcombe PJ, Reck BH, Sun J, et al. A comparison of Bayesian and frequentist approaches to incorporating external information for the prediction of prostate cancer risk. Genet Epidemiol. 2012;36(1):71–83. doi: 10.1002/gepi.21600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quintana MA, Berstein JL, Thomas DC, et al. Incorporating model uncertainty in detecting rare variants: the Bayesian risk index. Genet Epidemiol. 2011;35(7):638–649. doi: 10.1002/gepi.20613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakefield J. Bayes factors for genome-wide association studies: comparison with P-values. Genet Epidemiol. 2009;33(1):79–86. doi: 10.1002/gepi.20359. [DOI] [PubMed] [Google Scholar]
- 27.Fridley BL, Serie D, Jenkins G, et al. Bayesian mixture models for the incorporation of prior knowledge to inform genetic association studies. Genet Epidemiol. 2010;34(5):418–426. doi: 10.1002/gepi.20494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aldrich TE, Vann D, Moorman PG, et al. Rapid reporting of cancer incidence in a population-based study of breast cancer: one constructive use of a central cancer registry. Breast Cancer Res Treat. 1995;35(1):61–64. doi: 10.1007/BF00694746. [DOI] [PubMed] [Google Scholar]
- 29.Weinberg CR, Sandler DP. Randomized recruitment in case-control studies. Am J Epidemiol. 1991;134(4):421–432. doi: 10.1093/oxfordjournals.aje.a116104. [DOI] [PubMed] [Google Scholar]
- 30.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gold B, Kirchhoff T, Stefanov S, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci U S A. 2008;105(11):4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39(7):870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long J, Cai Q, Shu X, et al. Identification of a functional genetic variant at 16q12.1 for breast cancer risk: results from the Asia Breast Cancer Consortium. PLoS Genet. 2010;6(6):e1001002. doi: 10.1371/journal.pgen.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor–positive breast cancer. Nat Genet. 2007;39(7):865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 35.Thomas G, Jacobs KB, Kraft P, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41(5):579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turnbull C, Ahmed S, Morrison J, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42(6):504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng W, Long J, Gao Y, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41(3):324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmed S, Thomas G, Ghoussaini M, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41(5):585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor–positive breast cancer. Nat Genet. 2008;40(6):703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 40.Zhang B, Beeghly-Fadiel A, Long J, et al. Genetic variants associated with breast-cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Lancet Oncol. 2011;12(5):477–488. doi: 10.1016/S1470-2045(11)70076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas DC, Witte JS. Point: Population stratification: A problem for case-control studies of candidate-gene associations? Cancer Epidemiol Biomarkers Prev. 2002;11(6):505–512. [PubMed] [Google Scholar]
- 42.Barnholtz-Sloan JS, McEvoy B, Shriver MD, et al. Ancestry estimation and correction for population stratification in molecular epidemiologic association studies. Cancer Epidemiol Biomarkers Prev. 2008;17(3):471–477. doi: 10.1158/1055-9965.EPI-07-0491. [DOI] [PubMed] [Google Scholar]
- 43.Nyante SJ, Gammon MD, Kaufman JS, et al. Common genetic variation in adiponectin, leptin, and leptin receptor and association with breast cancer subtypes. Breast Cancer Res Treat. 2011;129(2):593–606. doi: 10.1007/s10549-011-1517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bortsov AV, Millikan RC, Belfer I, et al. μ-Opioid receptor gene A118G polymorphism predicts survival in patients with breast cancer. Anesthesiology. 2012;116(4):896–902. doi: 10.1097/ALN.0b013e31824b96a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gill J. Bayesian Methods: A Social and Behavioral Sciences Approach. 2nd ed. New York, NY: CRC Press; 2002. [Google Scholar]
- 46.Greenland S. Bayesian perspectives for epidemiological research. II. Regression analysis. Int J Epidemiol. 2007;36(1):195–202. doi: 10.1093/ije/dyl289. [DOI] [PubMed] [Google Scholar]
- 47.Greenland S. Bayesian perspectives for epidemiological research: I. Foundations and basic methods. Int J Epidemiol. 2006;35(3):765–775. doi: 10.1093/ije/dyi312. [DOI] [PubMed] [Google Scholar]
- 48.Chen GK, Witte JS. Enriching the analysis of genomewide association studies with hierarchical modeling. Am J Hum Genet. 2007;81(2):397–404. doi: 10.1086/519794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenland S. Principles of multilevel modelling. Int J Epidemiol. 2000;29(1):158–167. doi: 10.1093/ije/29.1.158. [DOI] [PubMed] [Google Scholar]
- 50.Hung RJ, Baragatti M, Thomas D, et al. Inherited predisposition of lung cancer: a hierarchical modeling approach to DNA repair and cell cycle control pathways. Cancer Epidemiol Biomarkers Prev. 2007;16(12):2736–2744. doi: 10.1158/1055-9965.EPI-07-0494. [DOI] [PubMed] [Google Scholar]
- 51.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 52.Barrett JC. Haploview: visualization and analysis of SNP genotype data. Cold Spring Harb Protoc. 2009;2009(10):71. doi: 10.1101/pdb.ip71. [DOI] [PubMed] [Google Scholar]
- 53.Hall IJ, Moorman PG, Millikan RC, et al. Comparative analysis of breast cancer risk factors among African-American women and white women. Am J Epidemiol. 2005;161(1):40–51. doi: 10.1093/aje/kwh331. [DOI] [PubMed] [Google Scholar]
- 54.Barnholtz-Sloan JS, Shetty PB, Guan X, et al. FGFR2 and other loci identified in genome-wide association studies are associated with breast cancer in African-American and younger women. Carcinogenesis. 2010;31(8):1417–1423. doi: 10.1093/carcin/bgq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milne RL, Goode EL, García-Closas M, et al. Confirmation of 5p12 as a susceptibility locus for progesterone-receptor–positive, lower grade breast cancer. Cancer Epidemiol Biomarkers Prev. 2011;20(10):2222–2231. doi: 10.1158/1055-9965.EPI-11-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J, Humphreys K, Heikkinen T, et al. A combined analysis of genome-wide association studies in breast cancer. Breast Cancer Res Treat. 2011;126(3):717–727. doi: 10.1007/s10549-010-1172-9. [DOI] [PubMed] [Google Scholar]
- 57.Fletcher O, Johnson N, Orr N, et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J Natl Cancer Inst. 2011;103(5):425–435. doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 58.Huang Y, Ballinger DG, Dai JY, et al. Genetic variants in the MRPS30 region and postmenopausal breast cancer risk. Genome Med. 2011;3(6):42. doi: 10.1186/gm258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Milne RL, Gaudet MM, Spurdle AB, et al. Assessing interactions between the associations of common genetic susceptibility variants, reproductive history and body mass index with breast cancer risk in the breast cancer association consortium: a combined case-control study. Breast Cancer Res. 2010;12(6):R110. doi: 10.1186/bcr2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bhatti P, Doody MM, Rajaraman P, et al. Novel breast cancer risk alleles and interaction with ionizing radiation among US radiologic technologists. Radiat Res. 2010;173(2):214–224. doi: 10.1667/RR1985.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Higginbotham KS, Breyer JP, McReynolds KM, et al. A multistage genetic association study identifies breast cancer risk loci at 10q25 and 16q24. Cancer Epidemiol Biomarkers Prev. 2012;21(9):1565–1573. doi: 10.1158/1055-9965.EPI-12-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Udler MS, Ahmed S, Healey CS, et al. Fine scale mapping of the breast cancer 16q12 locus. Hum Mol Genet. 2010;19(12):2507–2515. doi: 10.1093/hmg/ddq122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cai Q, Wen W, Qu S, et al. Replication and functional genomic analyses of the breast cancer susceptibility locus at 6q25.1 generalize its importance in women of Chinese, Japanese, and European ancestry. Cancer Res. 2011;71(4):1344–1355. doi: 10.1158/0008-5472.CAN-10-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.MacLehose RF, Dunson DB, Herring AH, et al. Bayesian methods for highly correlated exposure data. Epidemiology. 2007;18(2):199–207. doi: 10.1097/01.ede.0000256320.30737.c0. [DOI] [PubMed] [Google Scholar]
- 65.Hamra GB, Maclehose RF, Cole SR. Sensitivity analyses for sparse-data problems—using weakly informative bayesian priors. Epidemiology. 2013;24(2):233–239. doi: 10.1097/EDE.0b013e318280db1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Furberg H, Millikan R, Dressler L, et al. Tumor characteristics in African American and white women. Breast Cancer Res Treat. 2001;68(1):33–43. doi: 10.1023/a:1017994726207. [DOI] [PubMed] [Google Scholar]
- 67.Kerlikowske K. Epidemiology of ductal carcinoma in situ. J Natl Cancer Inst Monogr. 2010;2010(41):139–141. doi: 10.1093/jncimonographs/lgq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


