Abstract
Background
Genetic variation around interleukin-28B (IL28B), encoding IFN-λ3, predict non-responders to pegylated interferon-α/ribavirin (Peg-IFN/RBV) therapy in chronic hepatitis C (CHC). However, it remains unclear the expression and the role of IL28B itself. The aim of this study is to develop easy and useful methods for the prediction of treatment outcomes.
Methods
The mRNA and protein levels of IFN-λ3 induced by ex vivo stimulation of peripheral blood mononuclear cells (PBMC) or magnetically selected dendritic cells (DCs) with toll-like receptor agonists (TLR3; poly I:C, TLR7; R-837) were measured by the quantitative real-time polymerase chain reaction and our newly developed chemiluminescence enzyme immunoassays, respectively, and compared with the clinical data.
Results
We found that BDCA-4+ plasmacytoid and BDCA-3+ myeloid DCs were the main producers of IFN-λs when stimulated with R-837 and poly I:C, respectively. Detectable levels of IFN-λs were inducible even in a small amount of PBMC, and IFN-λ3 was more robustly up-regulated by R-837 in PBMC of CHC patients with favorable genotype for the response to Peg-IFN/RBV (TT in rs8099917) than those with TG/GG. Importantly, the protein levels of IFN-λ3 induced by R-837 clearly differentiated the response to Peg-IFN/RBV treatment (p = 1.0 × 10−10), including cases that IL28B genotyping failed to predict the treatment response. The measurement of IFN-λ3 protein more accurately predicted treatment efficacies (95.7 %) than that of IL28B genotyping (65.2 %).
Conclusions
Genetic variations around IL28B basically affect IFN-λ3 production, but different amounts of IFN-λ3 protein determines the outcomes of Peg-IFN/RBV treatment. This study, for the first time, presents compelling evidence that IL28B confer a functional phenotype.
Electronic supplementary material
The online version of this article (doi:10.1007/s00535-013-0814-1) contains supplementary material, which is available to authorized users.
Keywords: Chronic hepatitis C, IL28B, IFN-λ3, Peg-IFN/RBV
Introduction
Recently, we and others independently identified single nucleotide polymorphisms (SNPs) on chromosome 19 associated with the interleukin-28B gene (IL28B), encoding IFN-λ3, that were strongly associated with the response to pegylated interferon-α/ribavirin (Peg-IFN/RBV) in chronic hepatitis C (CHC) patients, through a genome-wide association study (GWAS) [1–3]. According to our results, about 80 % of CHC patients with the TT genotype (rs8099917) showed viral virologic response (VR), including SVR (sustained virologic response) or TVR (transient virologic response), whereas only about 20 % of HCV patients with the TG/GG genotype showed VR [1]. Thus, by genotyping of IL28B, we can predict the efficacy of Peg-IFN/RBV before beginning treatment, avoiding unnecessary side effects and the high cost of Peg-IFN/RBV treatment. However, it is still unknown whether genetic variation of IL28B is a functional phenotype for Peg-IFN/RBV treatment. In addition, genotyping of IL28B alone failed to predict about 20 % of the response [1], which would be reasonable because final products of the genes are affected by DNA methylation or chromatin modifications as well as genetic variations [4].
Type III IFNs, consisting of IFN-λ1, λ2, and λ3 (also known as IL29, IL28A and IL28B, respectively), have recently been characterized [5, 6]. IFN-λs up-regulate IFN-stimulated genes (ISGs) via Janus kinase/signal transducer and activator of transcription (Jak/STAT) intracellular signaling, inhibiting hepatitis B virus (HBV) or hepatitis C virus (HCV) replication [7]. Antiviral responses evoked by toll-like receptor (TLR)3 or TLR9 agonists are attenuated in IL28RA−/− mice [8], indicating the central role of IFN-λs in antiviral protection. Clinically, early virologic response by Peg-IFN/RBV is associated with a high probability of SVR in HCV patients [9]. Genetic variations of IL28B influence spontaneous clearance of HCV [10], or on-treatment viral kinetics [11]. These results suggest a mechanistic link between innate immunity and genetic variations of IL28B.
To recognize viruses and trigger innate antiviral responses, mammals have 2 independent receptors, retinoic acid-induced gene-I (RIG-I)-like receptors (RLRs) and TLRs, distinct families of pattern recognition receptors that sense nucleic acids derived from viruses [12]. RIG-I is a double-stranded RNA-binding DExD/H box RNA helicase that is essential for initiating the intracellular response to RNA viral infection [13]. However, NS3/4A, the major serine protease expressed by HCV, disrupts the RIG-I pathway through proteolysis of essential signaling components of IFN regulatory factor 3 (IRF-3) activation [13, 14], reducing immune response. Alternatively, the TLR-families play an important role in innate immune responses in mammals [15]. Among them, TLR3 recognizes viral double-stranded RNA, whereas TLR7 recognize single-stranded RNA. Because some TLRs ligands induce IFN-λ in human macrophages [16], contributing to antiviral defense and HCV is a single-stranded RNA [8], we hypothesized that IFN-λ3 induced via the TLR pathway might contribute to early antiviral response against HCV, which could lead to accurate prediction of treatment efficacy. Therefore, we investigated IFN-λs production in peripheral blood mononuclear cells (PBMC) in healthy volunteers or CHC patients by ex vivo stimulation with TLR agonists, and analyzed whether this method could predict the responses to Peg-IFN/RBV treatment in clinical practice.
Patients, materials, and methods
Study population
Blood samples were obtained from 12 healthy volunteers and 100 consecutive Japanese outpatients with CHC (genotype 1b and high viral load) who visited our hospital between April 2011 and March 2012. The study protocol was conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the ethical committee of our institutes (NCGM-G-001023-01). Written informed consent was obtained from all volunteers and patients. All subjects were negative for HBV and human immunodeficiency virus, and did not have hepatocellular carcinoma. IFN treatment was not being given to any patient at the time blood samples were taken. The subjects were all evaluated for SNP near IL28B (rs8099917, rs12979860) using the InvaderPlus assay (Invader Chemistry, Madison, WI, USA) as previously reported [17].
Definition of treatment responses
Non-virologic response (NVR) was defined as less than a 2-log-unit decline in the serum level of HCV RNA from the pre-treatment baseline value within the first 12 weeks, and detectable viremia 24 weeks after initiation of treatment. VR was defined as achieving SVR or TVR. SVR was defined as undetectable HCV RNA in the serum 6 months after the end of treatment, whereas TVR was defined as reappearance of HCV RNA in the serum during or after completion of treatment.
Preparation of PBMC and selection of plasmacytoid or myeloid dendritic cells (DCs)
Whole blood anti-coagulated with EDTA was obtained from healthy volunteers and CHC patients. PBMC were isolated by Ficoll-Hypaque (Mediatech, Herndon, VA, USA) density gradient centrifugation. BDCA-1, 3, 4+DCs were negatively or positively selected by BDCA-1+DC isolation kit, BDCA-3 MicroBead kit and BDCA-4/Neuropilin-1 MicroBead kit, respectively (Miltenyi Biotec, Auburn, CA, USA) according to the manufacturer’s instructions.
Ex vivo induction of IFN-λ1, IFN-λ2, and IFN-λ3
After pre-treatment with or without 100 U/ml of IFN-α (Hayashibara Co. Ltd., Okayama, Japan) in 200 μl of Roswell Park Memorial Institutes (RPMI) medium supplemented with 10 % fetal bovine serum for 16 h, 100,000 of mononuclear cells were stimulated with 30 μg/ml of poly I:C (TLR3 agonist; Imgenex, San Diego, CA, USA), or 5 μg/ml of imiquimod (R-837; TLR7 agonist, Imgenex) as previously reported [16]. For chemiluminescence enzyme immunoassays (CLEIA), 200,000 cells were subjected to the same stimulation protocol.
RNA isolation and cDNA synthesis
After stimulation with TRL-agonists for 4 h, the PBMC were lysed with ISOGEN-II (Nippon Gene, Tokyo, Japan). In some experiments, PBMC were harvested at each indicated time point. The lysate was supplemented with chloroform, incubated for 15 min on ice, and centrifuged at 22,000 g for 15 min. The aqueous layer was removed and precipitated with isopropanol. The RNA was pelleted by centrifugation, washed with ethanol, and dissolved in 20 μl of water. Reverse transcription was performed using the SuperScript III first-strand synthesis system (Invitrogen, Carlsbad, CA, USA).
Real-Time quantitative polymerase chain reaction (PCR)
Quantitative real-time PCR was performed to estimate IFN-λ1, IFN-λ2, and IFN-λ3 mRNA expression based on SYBR green fluorescence (Roche Diagnostics Japan), using TaqMan Universal PCR master mix (Roche Diagnostics Japan), according to the manufacturer’s protocol. Relative gene expression was calculated as a fold induction. Data were analyzed using the 2-∆∆C(t) method with Sequence Detector version 1.7 software (Applied Biosystems, Carlsbad, CA, USA) and were normalized using human hypoxanthine phosphoribosyltransferase (HPRT). A standard curve was prepared by serial 10-fold dilutions of human cDNA. The curve was linear over 7 log units with a 0.998 correlation coefficient. Quantitative mRNA expression was determined by triplicate real-time PCR.
Chemiluminescence enzyme immunoassays
We recently developed a CLEIA system for IFN-λ3 that showed a wide detection range of 0.1–10,000 pg/ml with little or no cross-reactivity to IFN-λ1 or IFN-λ2 [18]. In addition, this CLEIA system can correctly detect IFN-λ3 from different IL28B genotypes.
Acoustic radiation force impulse (ARFI) elastography
For non-invasive evaluation of liver fibrosis, ARFI elastography was performed using a Siemens Acuson S2000™ ultrasound system (Mochida Siemens Medical System Co, Ltd, Tokyo, Japan) as previously reported [19]. We performed 5 measurements for each patient, and a median value was calculated. Liver stiffness was expressed as the shear wave velocity (m/s) and has been reported to be well correlated with histological liver fibrosis [19].
Statistical analyses
Continuous variables between groups were compared using the Mann–Whitney U test, and categorical data were compared using the Chi square test or Fisher’s exact test. Correlations between continuous variables were searched using the Pearson correlation test. Values of p < 0.05 were considered significant.
Results
Genetic variation in IL28B
In CHC patients (n = 100), only 1 patient showed discrepancy between rs8099917 and rs12979860 with the same prediction for the treatment response by genotyping (TG in rs8099917, and TT in rs12979860). In addition, we recently reported that rs8099917 has the greatest accuracy in determining the outcome of Peg-IFN/RBV treatment in Japanese patients [17]. Therefore, rs8099917 is used in the following analyses. The major homologous (TT) in rs8099917 is considered a predictive factor for a favorable response to Peg-IFN/RBV treatment, while having minor alleles (TG or GG) is considered predictive for non-responders. Seven of 12 healthy volunteers had the TT genotype of IL28B and 5 had TG genotype. In CHC patients, 59 patients had the TT genotype, 36 had TG, and 5 had GG in rs8099917.
BDCA-4+ plasmacytoid DCs are the major producers of IFN-λs in response to R-837
Since Lauterbach et al. [20] found that human DCs expressing BDCA3 (CD141) in myeloid DC subsets were the primary producers of IFN-λs, we sought which cell types are the main producers of IFN-λs when stimulated with R-837. Because DCs from the different IL28B genotype are supposed to produce different amounts of IFN-λs, we used DCs from healthy volunteers with TT genotype. After negative or positive magnetic selection of BDCA-1, 3, 4+DCs using 100 ml of peripheral blood, each collection was stimulated with IFN-α, following poly I:C or R-837 as previously reported [16], and evaluated the mRNA of IFN-λs or the protein levels of IFN-λ3. We confirmed that BDCA-3+DCs were the main producers of IFN-λs when stimulated with poly I:C as previously reported (Fig. 1a) [20]. Interestingly, when stimulated with R-837, positive selection of BDCA-4+DCs (plasmacytoid DCs), not BDCA-3+DCs, produced IFN-λs whereas depletion of BDCA-4+DCs showed marked reduction of IFN-λs (Fig. 1b). Therefore, BDCA-4+DCs were the main producers of IFN-λs when stimulated with R-837. Thus, different stimulation targeted different DC subsets to induce IFN-λs.
Fig. 1.
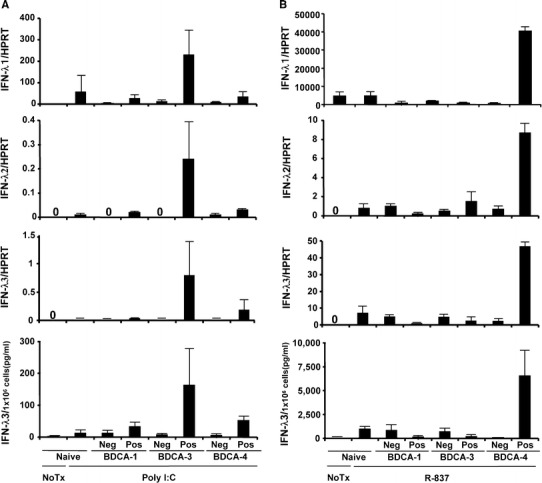
IFN-λs were produced from different subsets of dendritic cells (DCs) when stimulated with different TLR agonists. BDCA-3+ or BDCA-4+DCs was negatively or positively selected using peripheral blood mononuclear cells (PBMC) from healthy volunteers (n = 5). PBMC or DCs were stimulated with IFN-α, following poly I:C (a) or R-837 (b). The mRNA and the protein levels of IFN-λs were determined by real-time PCR and CLEIA, respectively. Neg negative selection, Pos positive selection of each DCs
Induction of IFN-λs (IFN-λ1, IFN-λ2, and IFN-λ3) in PBMC from healthy volunteers
We confirmed that the main producers of IFN-λs were DCs. However, analyses of IFN-λs using DC subsets need a lot of blood, which cannot apply to patients, because BDCA-3+ or 4+DCs are very minor subsets in peripheral blood (0.03, 0.5 %, respectively) [21]. Therefore, we examined if a small amount of PBMC, using 2–3 ml of whole blood from healthy volunteers with negative anti-HCV (Supplementary Table 1), still induced detectable levels of IFN-λs. We confirmed that, even with a small amount of PBMC, detectable levels of IFN-λs were induced by R-837 and the levels of IFN-λ3 were different between IL28B genotypes (Fig. 2a, b). Therefore, whole PBMC from healthy volunteers with the TT genotype were used for the initial experiments (Fig. 2a–d). IFN-α or TLR-agonists alone failed to induce significant amounts of IFN-λs. All IFN-λs mRNA were strongly induced by R-837 along with IFN-α (Fig. 2a). Poly I:C also induced detectable levels of IFN-λs, but those were not prominent in our setting. Protein levels of IFN-λ3 were detectable only by R-837 (Fig. 2b). Therefore, we focused on R-837 in the following experiments. Next, we sought to confirm the kinetics of IFN-λs after stimulation with R-837. When PBMC from volunteers with the TT genotype (n = 7) were stimulated with R-837, an IFN-λ mRNA peak was observed 4 h after incubation, rapidly decreasing to undetectable levels at 12 h (Fig. 2c). In the CLEIA results, IFN-λ3 was detected in the supernatant beginning 3 h after stimulation with R-837, peaking at 9–12 h, then plateauing (Fig. 2d). From these observations, mRNA levels of IFN-λs and protein levels of IFN-λ3 induced by R-837 were measured at 4 and 24 h after R-837, respectively.
Fig. 2.
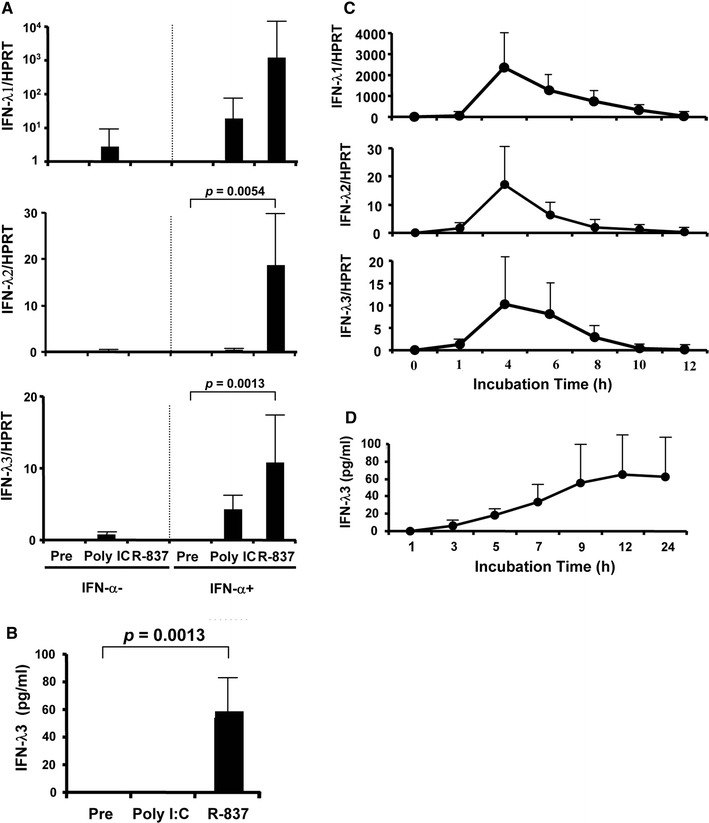
Ex vivo induction of IFN-λs in PBMC from healthy volunteers. a mRNA expression levels of IFN-λs by real-time quantitative PCR. After pre-treatment with or without 100 U/ml of IFN-α for 16 h, 100,000 of mononuclear cells were stimulated with 30 μg/ml of poly I:C (a TLR3 agonist) or 5 μg/ml of R-837 (a TLR7 agonist). After stimulation with TRL-agonists for 4 h, the PBMC were harvested. b Protein levels of IFN-λs. After pre-treatment with 100 U/ml of IFN-α, 200,000 of mononuclear cells were stimulated with 30 μg/ml of poly I:C (a TLR3 agonist) or 5 μg/ml of R-837 (a TLR7 agonist), and the supernatant was harvested 24 h after stimulation with TLR agonists. c Kinetics of IFN-λs mRNA levels. After pre-incubation with IFN-α for 16 h, PBMC was stimulated with 5 μg/ml of R-837. Real-time quantitative PCR was conducted at each time point. d CLEIA results for IFN-λ3 protein in the supernatant at each time point
Patients’ characteristics
Consecutive 100 HCV-RNA positive CHC patients with genotype 1b (35 male, 65 female) were enrolled in this study (Supplementary Table 2). Our patients with the TG/GG genotype were more prevalent (41.0 %, 41/100) than in the normal population (about 20 %), which was expected given that patients who had previously failed to benefit from HCV treatment tended to visit our center. Each patient who had histories of Peg-IFN/RBV (n = 38) was treated with Peg-IFN-α2b (1.5 μg per kg body weight (μg/kg) subcutaneously once a week) or PEG-IFN- α2a (180 μg once a week) plus RBV (600–1,000 mg daily depending on body weight) (Fig. 3). Since a reduction in the dose of PEG-IFN-α and RBV can contribute to less SVR [22], 2 patients with an adherence of <80 % dose for either drugs during the first 12 weeks who had shown NVR were excluded from “the known treatment-response” group. Patients who had been treated with IFN monotherapy (n = 5) were also excluded because IL28B was identified in patients having had Peg-IFN/RBV treatment, but not IFN monotherapy, through the GWAS. Among them (n = 36), 19 had shown TVR whereas 17 had shown NVR. After enrollment to this study, 10 treatment-naïve patients started Peg-IFN/RBV therapy. One patient showed undetectable HCV-RNA during therapy, and the therapy is ongoing (this patient was categorized in VR and/or TVR). Seven patients achieved SVR (categorized in VR and/or SVR), whereas 2 patients showed NVR (categorized in NVR). Therefore, 47 CHC patients were treatment-naïve and 46 CHC patients had the known response to Peg-IFN/RBV treatment. There were no differences in the characteristic backgrounds between patients with the TT and TG/GG genotypes in treatment-naïve CHC patients (n = 47) except for their serum γ-GTP level (32 ± 19 and 59 ± 53, respectively, p = 0.017) (Table 1), which were consistent with our recent report [23].
Fig. 3.
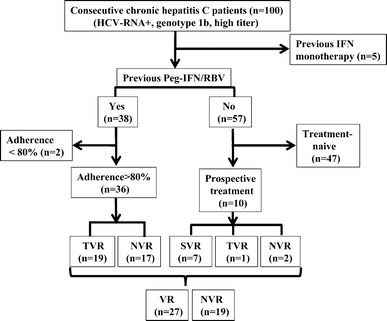
Enrolled chronic hepatitis C patients with or without histories of treatment against HCV
Table 1.
Patients’ characteristics of HCV treatment-naïve patients in TT or TG/GG genotype (n = 47)
| TT (n = 28) | TG/GG (n = 19) | p value | |
|---|---|---|---|
| Age | 66 ± 9 | 62 ± 14 | ns |
| M:F | 12:16 | 6:13 | ns |
| WBC | 4,493 ± 1,240 | 4,337 ± 1,096 | ns |
| Hb | 13.7 ± 1.7 | 13.7 ± 1.3 | ns |
| Plt | 15.5 ± 5.5 | 16.8 ± 6.0 | ns |
| TP | 7.7 ± 0.5 | 7.8 ± 0.5 | ns |
| Alb | 4.2 ± 0.5 | 4.4 ± 0.4 | ns |
| AST | 52 ± 30 | 44 ± 22 | ns |
| ALT | 56 ± 35 | 49 ± 29 | ns |
| γ-GTP | 32 ± 19 | 59 ± 53 | 0.017 |
| ChE | 280 ± 100 | 315 ± 87 | ns |
| T-cho | 170 ± 26 | 177 ± 37 | ns |
| LDL | 89 ± 23 | 97 ± 31 | ns |
| HCV RNA | 6.5 ± 0.6 | 6.3 ± 0.8 | ns |
| ARFIa | 1.50 ± 0.51 | 1.33 ± 0.37 | ns |
aARFI (acoustic radiation force impulse) represents shear wave velocity (m/s)
Association of IFN-λ3 induction with genetic variations around IL28B in healthy volunteers or treatment-naïve CHC patients
R-837 induced higher levels of IFN-λ2 or IFN-λ3 mRNA in healthy volunteers with the TT genotype than in those with the TG/GG genotype whereas no differences were observed in IFN-λ1 (Fig. 4a). However, no statistical differences were observed in the protein levels of IFN-λ3 between TT and TG genotype. The protein levels of IFN-λ3 were robustly induced by R-837 (Fig. 4b). These findings were similarly observed when using PBMC from treatment-naïve CHC patients (Fig. 4c, d). The protein levels of IFN-λ3 in PBMC from treatment-naïve CHC patients with TT genotype were significantly higher than those with TG/GG genotype. The IFN-λ3 mRNA levels were well correlated with those protein levels (Fig. 4e). Similar findings were obtained in all CHC patients (n = 100) (Supplementary Fig. 1).
Fig. 4.
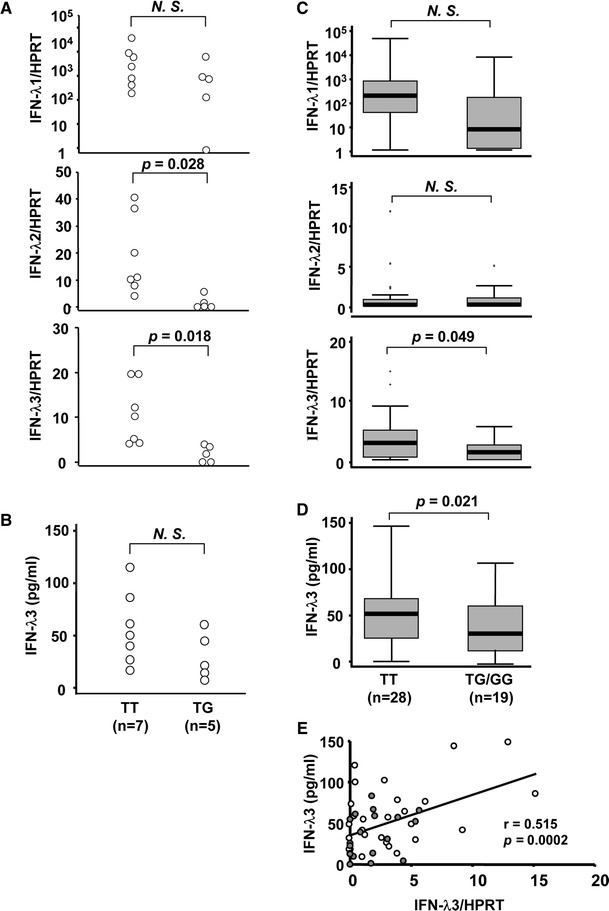
Ex vivo induction of IFN-λs in PBMC from healthy volunteers and treatment-naïve CHC patients. a Differences of IFN-λs mRNA levels between each IL28B genotype in healthy volunteers (n = 12). After pre-treatment with 100 U/ml of IFN-α for 16 h, 100,000 of mononuclear cells were stimulated with 5 μg/ml of R-837. After stimulation with R-837 for 4 h, the PBMC were harvested. b Differences of IFN-λ3 protein levels between each IL28B genotype in healthy volunteers (n = 12) with or without R-837. c Differences of IFN-λs mRNA levels between each IL28B genotype in treatment-naïve CHC patients (n = 47). d Differences of IFN-λ3 protein levels between each IL28B genotypes in treatment-naïve CHC patients (n = 47) with or without R-837. e Correlation between mRNA and protein levels of IFN-λ3 in treatment-naïve CHC patients (n = 47). Each open circle represents TT genotype whereas each closed circle represents TG/GG genotype
Predictiveness of IFN-λ3 induction for response to Peg-IFN/RBV treatment
Among 46 HCV-RNA positive patients who had the known response to Peg-IFN/RBV, 27 patients showed VR and 19 showed NVR. Lower platelet counts and higher shear wave velocity were significantly observed in the NVR group (Table 2). The mRNA levels of IFN-λ3 were significantly higher in patients with VR than in those with NVR (Fig. 5c, p = 0.0002). The expression levels of IFN-λ1 (Fig. 5a) and IFN-λ2 (Fig. 5b) were also significantly higher in patients with VR although the statistical differences were markedly bigger than IFN-λ3. The protein levels of IFN-λ3 confirmed these results and more clearly differentiated between VR and NVR in patients with previous therapy (n = 36), in treatment-naïve patients with prospective therapy (n = 10), or combined (n = 46) (Fig. 5d, p = 6.9 × 10−9, p = 0.014, p = 1.0 × 10−10, respectively). Interestingly, 7 patients with the TG/GG genotype who showed VR demonstrated high IFN-λ3, whereas 8 patients with the TT genotype who showed NVR demonstrated low IFN-λ3 induction. Taken together, the response to Peg-IFN/RBV was mainly dependent on the capacity of IFN-λ3 production in the PBMC rather than on genetic variations in IL28B. Our method more accurately predicted treatment efficacies (44/46, 95.7 %) compared to IL28B genotyping (30/46, 65.2 %) when cut-off of IFN-λ3 protein levels was set at the median value (47.6 pg/ml) (Table 3).
Table 2.
Patients’ characteristics categorized in the response to treatment (n = 46)
| VR (n = 27) | NVR (n = 19) | p value | |
|---|---|---|---|
| Age | 63 ± 9 | 63 ± 9 | ns |
| M:F | 9:18 | 5:14 | ns |
| TT:TG:GG | 20:7:0 | 9:9:1 | ns |
| WBC | 4,181 ± 1,299 | 3,947 ± 1,127 | ns |
| Hb | 12.5 ± 2.0 | 13.1 ± 1.7 | ns |
| Plt | 18.1 ± 5.9 | 13.0 ± 3.6 | 0.002 |
| TP | 7.7 ± 0.6 | 7.8 ± 0.4 | ns |
| Alb | 4.4 ± 0.4 | 4.2 ± 0.3 | ns |
| AST | 46 ± 42 | 61 ± 35 | ns |
| ALT | 44 ± 44 | 73 ± 58 | ns |
| γ-GTP | 28 ± 22 | 51 ± 32 | ns |
| ChE | 293 ± 76 | 271 ± 75 | ns |
| T-cho | 178 ± 37 | 159 ± 23 | ns |
| LDL | 95 ± 22 | 88 ± 19 | ns |
| HCV RNA | 6.3 ± 0.6 | 6.3 ± 1.0 | ns |
| Core 70 (W:M) | 18:6 | 8:9 | ns |
| ISDR (0:>0) | 12:12 | 8:9 | ns |
| ARFIa | 1.20 ± 0.26 | 1.72 ± 0.34 | <0.001 |
aARFI (acoustic radiation force impulse) represents shear wave velocity (m/s)
Fig. 5.
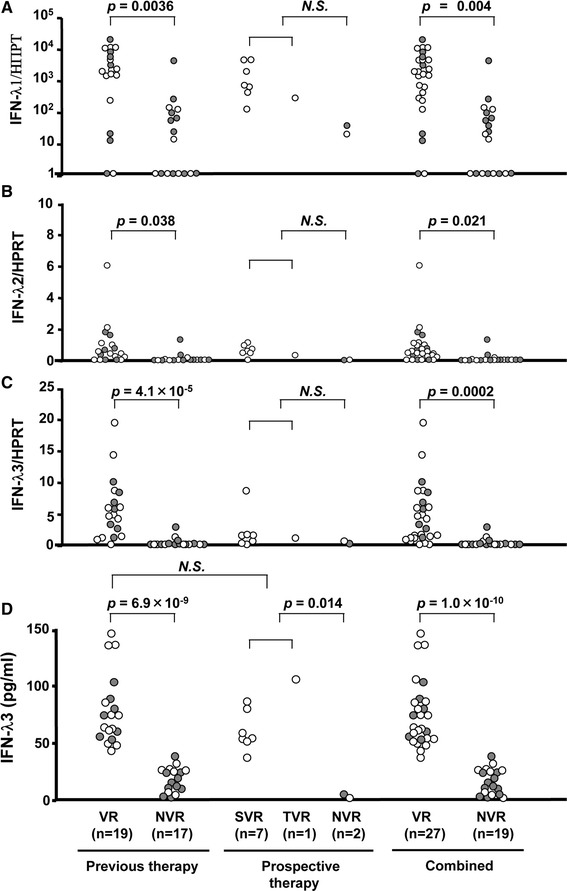
The mRNA and protein levels of IFN-λ3 ex vivo induced by R-837 and treatment responses in CHC patients with “the known response to Peg-IFN/RBV”. The mRNA levels of IFN-λ1 (a), IFN-λ2 (b), and IFN-λ3 (c) of virologic responders (VR) (n = 27) and non-virologic responders (NVR) (n = 19) were shown. (d) Protein levels of IFN-λ3 in CHC patients who had previously failed Peg-IFN/RBV therapy (previous therapy, n = 36) and who prospectively treated with Peg-IFN/RBV (prospective therapy, n = 10). Combined figures were also shown (n = 46). Each open circle represents TT genotype whereas each closed circle represents TG/GG genotype
Table 3.
Correct prediction rate by genotyping of IL28B or IFN-λ3 value (cut-off 47.6 pg/ml)
| IL28B genotype | IFN-λ3 | ||||||
|---|---|---|---|---|---|---|---|
| TT | TG/GG | High | Low | ||||
| VR | 20 | 7 | 27 | VR | 25 | 2 | 27 |
| NVR | 9 | 10 | 19 | NVR | 0 | 19 | 19 |
| 29 | 17 | 46 | 25 | 21 | 46 | ||
Bold values indicated correct prediction in each category
Discussion
The present study demonstrated that the amount of endogenous IFN-λ3 in PBMC induced by the TLR7 agonist determines the outcome of Peg-IFN/RBV therapy, though its induction was basically dependent on the IL28B genotype. Since this method can evaluate the final gene products, the genetic factor or epigenetic factor is not necessary for consideration, which could provide more accurate prediction of the response to Peg-IFN/RBV therapy than IL28B genotyping. In addition, an annoying informed consent about gene handling is not necessary, and the cost is lower than genomic analyses. IFN-based treatment with novel drugs such as protease inhibitors or polymerase inhibitors could be still predictive by this method because genetic variations in IL28B are strongly associated with response to telaprevir/Peg-IFN/RBV treatment [24].
IFN-λs were shown to inhibit the replication of a number of viruses in vitro, including HBV or HCV [5–7], by up-regulation of ISGs. Several GWAS studies suggested that IFN-λ3 played a key role in the response to HCV [1–3, 10, 11]. Meanwhile, type III IFNs, produced by hepatocytes, in response to HCV infection, predominantly lead functional ISGs induction in comparison to type I IFNs [25]. The combination of IL28B genotype between recipients and donors determines the outcome of Peg-IFN/RBV therapy for recurrent hepatitis C after liver transplantation [26]. Therefore, a favorable response to anti-HCV treatment might be dependent on the amount of endogenous IFN-λ3 produced by both lymphocytes and hepatocytes.
Unsuccessful induction of IFN-λs by IFN-α or TLR-agonists alone (Fig. 1a) may explain controversial results of IFN-λ3 expression in either PBMC or intrahepatic lymphocytes from pre-treatment patients [1, 3, 23, 27]. IFN-α up-regulates the expression of TLR, TRIF, and MyD88, common adaptor molecules associated with TLR signaling [16]. UV-inactivated viruses, as well as infectious viruses, can induce IFN-λ, suggesting viruses can be sensed through a non-infectious route such as endocytosis of infectious or non-infectious viral particles [28]. TLR7 appears to play an important role in the induction of antiviral responses against single-stranded RNA viruses [29–31], and imidazoquinoline activates immune cells via the TLR7-MyD88-dependent signaling pathway [32]. Synthetic TLR7 agonists have recently been characterized with respect to their ability to induce cytokines [33]. Furthermore, in our study, IFN-λ3 induced by R-837 clearly differentiated the response to Peg-IFN/RBV treatment. Therefore, sequential stimulation with IFN-α following a TLR7 agonist (R-837) may mimic IFN therapy in CHC patients in terms of IFN-λs induction, and our sensitive CLEIA system may contribute to clear differentiation among the responses to Peg-IFN/RBV treatment in this study. It has recently been reported that expression levels and function of TLR7 were impaired in HCV-infected human hepatoma cells [34]. However, in the present study, a TLR7 agonist was able to induce IFN-λs in PBMC from HCV patients, although the response was slightly impaired in HCV patients compared with that in healthy volunteers. Therefore, impairment of expression or function of TLR7 in HCV patients may not be a critical factor for whole innate immunity.
IFN-λs display high sequence homology. In particular, IFN-λ2 and IFN-λ3 are virtually identical with 96 % amino acid homology [6]. However, the GWAS revealed that only SNPs near the IL28B, not IL28A or IL29, showed strong associations with response to Peg-IFN/RBV treatment [1–3]. In the current study, we found that the expression level of IFN-λ3 in the PBMCs was better correlated with genetic variations in IL28B or response to Peg-IFN/RBV treatment. Furthermore, the antiviral effect of recombinant IFN-λ3 was more potent than those of IFN-λ1 or IFN-λ2 [35]. Collectively, these findings may support our GWAS data. Alternatively, there are significant differences in IFN-λ2 as well as IFN-λ3 between genotypes or treatment response. The specific primers and probe sets for the IFN-lambda family that we previously developed achieved approximately 107-fold specificity in each IFN-λ [18]. However, Osterlund et al. [36] reported that both IFN-λ2 and IFN-λ3 were regulated by a similar pathway of IRF7 resembling those of IFN-α gene expression. Therefore, it is possible that the upregulation of IFN-λ3 could affect that of IFN-λ2 because the homology of regulatory sequence between IFN-λ2 and IFN-λ3 is pretty high. On the other hand, IFN-λ1 and λ2, λ3 show rather low homology (81 %), which could explain the different responses among them.
About 20 % of the HCV patients with the TT genotype failed to respond to the treatment [1]. Indeed, in the current study, 8 of 28 (28.6 %) patients with favorable IL28B genotype (TT) showed NVR in Peg-IFN/RBV therapy. Interestingly, all these cases who showed NVR despite their favorable IL28B genotype demonstrated low IFN-λ3 production (Fig. 5a–d). Generally, poor response to HCV treatment is related to a number of factors that are unlikely to be solely due to the IL28B genotype, such as greater age, male gender, viral factors and liver fibrosis [37, 38]. Therefore, we have tried to calculate multivariate logistic regression to find the most predictive factors affecting the treatment response including clinical backgrounds and the current data. However, the calculations were impossible because of “complete separation” between the treatment-response groups on ELISA data. Liver fibrosis may be attributed to hyporesponsiveness to Peg-IFN/RBV because surrogate markers of liver fibrosis (low platelet counts and high shear wave velocity) were significantly observed in our NVR cases (Table 2). However, both of surrogate markers showed substantial overlap between VR and NVR (Supplementary Fig. 2). Other factors including genetic deficiency of molecules in the TLR7 signaling pathway [39] may affect IFN-λs production. Meanwhile, 7 of 18 (38.9 %) patients with the unfavorable IL28B genotype (TG/GG) showed VR in Peg-IFN/RBV therapy (Table 3) and all of these VR patients showed high IFN-λ3 production. Because recombinant IFN-λ from any IL28B genetic variant similarly represses HCV RNA [40], the amount of IFN-λ, whatever the genetic variations of IL28B are, would be important for antiviral effects. Enhanced transcriptional regulations of IL28B could partially be attributed to high IFN-λ3 production in patients with TG/GG [41]. Alternatively, the number of BDCA-4+DCs was well correlated with IFN-λ3 production when PBMC from healthy volunteers was stimulated with R-837 (Supplementary Fig. 3), which suggest that the number of BDCA-4+DC strongly affect IFN-λ3 production in our setting. However, the precise mechanisms of regulating IFN-λ3 production or the number of BDCA-4+DCs in CHC patients should be addressed in the future. Importantly, our methods clearly predict the effectiveness of Peg-IFN/RBV including these exceptional cases.
In the present study, neither the mRNA nor the protein levels of IFN-λ3 were much different among genetic variations in IL28B (Fig. 4c, d). It is possible that the exceptional cases of IFN-λ3 levels (e.g., low IFN-λ3 levels in the TT genotype and high levels in the TG/GG genotype) affected the statistical differences. Substantial overlap in the mRNA levels of IFN-λ3 was observed between the different treatment response groups whereas the protein levels of IFN-λ3 clearly differentiated (Fig. 5a–d). One possibility would be that our newly developed CLEIA system [18] is more sensitive than real-time PCR. Moreover, IFN-λs mRNA levels were rapidly induced by R-837, and these effects were limited to several hours after stimulation (Fig. 2c) whereas protein levels of IFN-λ3 were stable from 9 to 24 h after R-837 stimulation (Fig. 2d). In the prospective study, there were no statistical differences in the mRNA levels of IFN-λs. However, the increase of the number of these patients would clarify the differences because the trend of these levels in the prospective study was similar to those in the previously treated group.
In conclusion, our findings suggest that genetic variations in IL28B basically affect IFN-λ3 production; however, the amount of endogenous IFN-λ3 determines the outcome of Peg-IFN/RBV therapy. This study, for the first time, presents compelling evidence that genetic variations in IL28B confer a functional phenotype and potentially explains our GWAS data. In addition, these results may explain discrepant cases related to IL28B genotyping in the response to Peg-IFN/RBV treatment. Thus, ex vivo induction of IFN-λ3 in PBMC by a TLR7-agonist may be a more accurate predictive method for determining the outcome of Peg-IFN/RBV therapy.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Ex vivo induction of IFN-λs in PBMC from all CHC patients (n = 100). (a) Differences of IFN-λs mRNA levels between each IL28B genotype. After pre-treatment with 100 U/ml of IFN-α for 16 h, 100,000 of mononuclear cells were stimulated with 5 μg/ml of R-837. After stimulation with R-837 for 4 h, the PBMC were harvested. (b) Differences of IFN-λ3 protein levels between each IL28B genotype with or without R-837. (c) Correlation between mRNA and protein levels of IFN-λ3 (n = 100). Each open circle represents the TT genotype whereas each closed circle represents the TG/GG genotype. (TIFF 273 kb)
Platelet counts and the shear wave velocity, and treatment responses in CHC patients with “the known response to Peg-IFN/RBV”. (TIFF 171 kb)
The association between IFN-λ3 protein production and the number of BDCA-4+DCs when stimulated with poly I:C or R-837, respectively. BDCA-4+DCs in healthy volunteers with TT genotype (n = 6) were analyzed by fluorescence activated cell sorting (FACS) before any stimulations. (TIFF 214 kb)
Acknowledgments
This study was supported by grants (23-105) from the National Center for Global Health and Medicine in Japan.
Conflict of interest
Tatsuji Kimura is an employee of the Institute of Immunology Co., Ltd. All other authors have nothing to declare.
Abbreviations
- ARFI
Acoustic radiation force impulses
- CHC
Chronic hepatitis C
- GWAS
Genome-wide association study
- IL28B
Interleukin-28B
- Peg-IFN/RBV
Pegylated interferon-α/ribavirin
- PBMC
Peripheral blood mononuclear cells
- SNP
Single nucleotide polymorphisms
- SVR
Sustained viral response
- TLR
Toll-like receptor
- TVR
Transient viral response
- VR
Viral response
References
- 1.Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-α and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 2.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 3.Suppiah V, Moldovan M, Ahlenstiel G, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 4.Ng HH, Bird A. DNA methylation and chromatin modification. Curr Opin Genet Dev. 1999;9:158–163. doi: 10.1016/S0959-437X(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 5.Kotenko SV, Gallagher G, Baurin VV, et al. IFN-λs mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 6.Sheppard P, Kindsvogel W, Xu W, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 7.Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C replication. J Virol. 2005;79:3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ank N, Iversen MB, Bartholdy C, et al. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J Immunol. 2008;180:2474–2485. doi: 10.4049/jimmunol.180.4.2474. [DOI] [PubMed] [Google Scholar]
- 9.Davis GL, Wong JB, McHutchison JG, et al. Early virologic response to treatment with peginterferon alfa-2b plus ribavirin in patients with chronic hepatitis C. Hepatology. 2003;38:645–652. doi: 10.1053/jhep.2003.50364. [DOI] [PubMed] [Google Scholar]
- 10.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C. Nature. 2009;46:798–802. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson AJ, Muir AJ, Sulkowski MS, et al. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;139:120–129. doi: 10.1053/j.gastro.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 12.Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann NY Acad Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 13.Loo YM, Owen DM, Li K, et al. Viral and therapeutic control of IFN-β promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci USA. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meylan E, Curran J, Hofmann K, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 15.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 16.Siren J, Pirhonen J, Julkunen I, et al. IFN-α regulates TLR-dependent gene expression of IFN-α, IFN-β, IL-28, and IL-29. J Immunol. 2005;174:1932–1937. doi: 10.4049/jimmunol.174.4.1932. [DOI] [PubMed] [Google Scholar]
- 17.Ito K, Higami K, Masaki N, et al. The rs8099917 polymorphism, determined by a suitable genotyping method, is a better predictor for response to pegylated interferon-α/ribavirin therapy in Japanese patients than other SNPs associated with IL28B. J Clin Microbiol. 2011;49:1853–1860. doi: 10.1128/JCM.02139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugiyama M, Kimura T, Naito S, et al. Development of interferon lambda 3 specific quantification assay for its mRNA and serum/plasma specimens. Hepatol Res. 2012;42:1089–1099. doi: 10.1111/j.1872-034X.2012.01032.x. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi H, Ono N, Eguchi Y, et al. Evaluation of acoustic radiation force impulse elastography for fibrosis staging of chronic liver disease: a pilot study. Liver Int. 2010;30:538–545. doi: 10.1111/j.1478-3231.2009.02130.x. [DOI] [PubMed] [Google Scholar]
- 20.Lauterbach H, Bathke B, Gilles S, et al. Mouse CD8α+ DCs and human BDCA3+ DCs are major producers of IFN-λ in response to poly IC. J Exp Med. 2009;207:2703–2717. doi: 10.1084/jem.20092720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dzionek A, Fuchs A, Schmidt P, et al. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J Immunol. 2000;165:6037–6046. doi: 10.4049/jimmunol.165.11.6037. [DOI] [PubMed] [Google Scholar]
- 22.McHutchison JG, Manns M, Patel K, et al. Adherence to combination therapy enhances sustained response in genotype-1-infected patients with chronic hepatitis C. Gastroenterology. 2002;123:1061–1069. doi: 10.1053/gast.2002.35950. [DOI] [PubMed] [Google Scholar]
- 23.Saito H, Ito K, Sugiyama M, et al. Factors responsible for the discrepancy between IL28B polymorphism prediction and the viral clearance to peginterferon plus ribavirin therapy in Japanese chronic hepatitis C patients. Hepatol Res. 2012;42:958–965. doi: 10.1111/j.1872-034X.2012.01013.x. [DOI] [PubMed] [Google Scholar]
- 24.Akuta N, Suzuki F, Hirakawa M, et al. Amino acid substitution in hepatitis C virus core region and genetic variation near the interleukin 28B gene predict viral response to telaprevir with peginterferon and ribavirin. Hepatology. 2010;52:421–429. doi: 10.1002/hep.23690. [DOI] [PubMed] [Google Scholar]
- 25.Thomas E, Gonzalez VD, Li Q, et al. HCV infection induces a unique hepatic innate responses associated with robust production of type III interferons. Gastroenterology. 2012;142:978–988. doi: 10.1053/j.gastro.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukuhara T, Taketomi A, Motomura T, et al. Variants in IL28B in liver recipients and donors correlate with response to Peg-interferon and ribavirin therapy for recurrent hepatitis C. Gastroenterology. 2010;139:1577–1585. doi: 10.1053/j.gastro.2010.07.058. [DOI] [PubMed] [Google Scholar]
- 27.Honda M, Sakai A, Yamashita T, et al. Hepatic interferon-stimulated genes expression is associated with genetic variation in interleukin 28B and the outcome of interferon therapy for chronic hepatitis C. Gastroenterology. 2010;139:499–509. doi: 10.1053/j.gastro.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 28.Yin Z, Dai J, Deng J, et al. Type III IFNs are produced by and stimulate human plasmacytoid dendritic cells. J Immunol. 2012;189:2735–2745. doi: 10.4049/jimmunol.1102038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heil F, Hemmi H, Hochrein H, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 30.Diebold SS, Kaisho T, Hemmi H, et al. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 31.Lund JM, Alexopoulou L, Sato A, et al. Recognition of single-stranded RNA viruses by toll-like receptor 7. Proc Natl Acad Sci USA. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hemmi H, Kaisho T, Takeuchi O, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 33.Gorden KB, Gorski KS, Gibson SJ, et al. Synthetic TLR agonists reveal functional differences between human TLR7 and TLR8. J Immunol. 2005;174:1259–1268. doi: 10.4049/jimmunol.174.3.1259. [DOI] [PubMed] [Google Scholar]
- 34.Chang S, Kodys K, Szabo G. Impaired expression and function of toll-like receptor 7 in hepatitis C virus infection in human hepatoma cells. Hepatology. 2010;51:35–42. doi: 10.1002/hep.23256. [DOI] [PubMed] [Google Scholar]
- 35.Dellgren C, Gad HH, Hamming OJ, et al. Human interferon-λ3 is a potent member of the type III interferon family. Genes Immun. 2009;10:125–131. doi: 10.1038/gene.2008.87. [DOI] [PubMed] [Google Scholar]
- 36.Osterlund PI, Pietilä TE, Veckman V, et al. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-λ) genes. J Immunol. 2007;179:3434–3442. doi: 10.4049/jimmunol.179.6.3434. [DOI] [PubMed] [Google Scholar]
- 37.O’Brien TR. Interferon-alfa, interferon-lambda and hepatitis C. Nat Genet. 2009;41:1048–1050. doi: 10.1038/ng.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manns MP, McHutchison JG, Gordon SC, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomized trial. Lancet. 2001;358:958–965. doi: 10.1016/S0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 39.Picard C, Puel A, Bonnet M, et al. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science. 2003;299:2076–2079. doi: 10.1126/science.1081902. [DOI] [PubMed] [Google Scholar]
- 40.Urban TJ, Thompson AJ, Bradrick SS, et al. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010;52:1888–1896. doi: 10.1002/hep.23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugiyama M, Tanaka Y, Wakita T, et al. Genetic variation of the IL28B promoter affecting gene expression. PLoS ONE. 2011;6:e26620. doi: 10.1371/journal.pone.0026620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ex vivo induction of IFN-λs in PBMC from all CHC patients (n = 100). (a) Differences of IFN-λs mRNA levels between each IL28B genotype. After pre-treatment with 100 U/ml of IFN-α for 16 h, 100,000 of mononuclear cells were stimulated with 5 μg/ml of R-837. After stimulation with R-837 for 4 h, the PBMC were harvested. (b) Differences of IFN-λ3 protein levels between each IL28B genotype with or without R-837. (c) Correlation between mRNA and protein levels of IFN-λ3 (n = 100). Each open circle represents the TT genotype whereas each closed circle represents the TG/GG genotype. (TIFF 273 kb)
Platelet counts and the shear wave velocity, and treatment responses in CHC patients with “the known response to Peg-IFN/RBV”. (TIFF 171 kb)
The association between IFN-λ3 protein production and the number of BDCA-4+DCs when stimulated with poly I:C or R-837, respectively. BDCA-4+DCs in healthy volunteers with TT genotype (n = 6) were analyzed by fluorescence activated cell sorting (FACS) before any stimulations. (TIFF 214 kb)


