Durairaj et al. demonstrate that an F-box protein Mdm30 promotes proteasomal degradation of the Sub2 subunit of the Transcription/Export complex (TREX). Reduced abundance of Sub2 in the presence of Mdm30 enhances the recruitment of the mRNA export adaptor, Yra1, to active genes, and promotes mRNA export. Collectively, these results indicate that Mdm30 facilitates mRNA export by reducing Sub2 levels and consequently promoting Yra1 recruitment.
Keywords: Mdm30, Sub2, Yra1, TREX, mRNA export, transcription
Abstract
Mdm30, an F-box protein in yeast, has been recently shown to promote mRNA export. However, it remains unknown how Mdm30 facilitates mRNA export. Here, we show that Mdm30 targets the Sub2 component of the TREX (Transcription/Export) complex for ubiquitylation and subsequent proteasomal degradation. Such a targeted degradation of Sub2 enhances the recruitment of the mRNA export adaptor, Yra1, to the active genes to promote mRNA export. Together, these results elucidate that Mdm30 promotes mRNA export by lowering Sub2's stability and consequently enhancing Yra1 recruitment, thus illuminating new regulatory mechanisms of mRNA export by Mdm30.
INTRODUCTION
In eukaryotes, mRNA is exported from the nucleus to the cytoplasm for translation into protein. Nuclear export of mRNA is a highly regulated process, and its misregulation is linked to an altered pattern of gene expression. The transcription/export (TREX) complex plays a key role in nuclear export of mRNA (Strässer et al. 2002; Carmody and Wente 2009). It is evolutionarily conserved among eukaryotes and consists of two mRNA export factors and the THO complex. In yeast, these export factors are Sub2 (human homolog, UAP56) and Yra1 (human homolog, ALY). On the other hand, the THO complex that is involved in transcriptional elongation contains four nonessential subunits, namely, Hpr1, Thp2, Tho2, and Mft1 (Chavez et al. 2000; Jimeno et al. 2002; Rondón et al. 2003; Rehwinkel et al. 2004). In addition, Tex1, a nonessential protein of unknown function, has been shown to be associated with THO (Strässer et al. 2002). Thus, THO, Tex1, Sub2, and Yra1 constitute the TREX complex. However, Hurt et al. (2004) have also implicated two poly(A)+ RNA-binding proteins, namely, Hrb1 and Gbp2, as TREX subunits.
The THO complex is recruited to the genes during transcriptional elongation and plays an important role to load Sub2 and Yra1 (Hammell et al. 2002; Strässer et al. 2002; Zenklusen et al. 2002; Rondón et al. 2003; Kim et al. 2004). Sub2 interacts directly with the Hpr1 component of THO, and hence, Hpr1 helps to recruit Sub2 onto the active genes (Zenklusen et al. 2002). Sub2, in turn, recruits Yra1 to the actively transcribing genes. Yra1 functions as an export adaptor for transferring mRNA to the export receptor, Mex67 (Strässer and Hurt 2000; Stutz et al. 2000; Kashyap et al. 2005). Once Yra1 is recruited, Sub2 dissociates from it, thereby allowing the interaction of Yra1 with Mex67 (Strässer et al. 2002). This interaction is mutually exclusive as Sub2 and Mex67 compete for binding to the two domains of Yra1 (Strässer and Hurt 2001). However, one Yra1 molecule could also theoretically interact with Sub2 and Mex67 at the same time, since each of the two domains of Yra1 can independently interact with Sub2 and Mex67 (Strässer and Hurt 2001). Following interaction with Yra1, Mex67 exports mRNA through nuclear pore complex. Thus, transcription and nuclear export of mRNA are tightly coupled, and Sub2 and Yra1 play key regulatory roles in this transcription-coupled process of mRNA export. An additional link between transcription and mRNA export has also been established with the identification of another transcription-coupled export complex, TREX-2, in yeast (Fischer et al. 2002; Dieppois and Stutz 2010). TREX-2 is composed of Sac3, Thp1, Cdc31, and Sus1. However, Sus1 has also been identified as a bona fide component of the transcriptional coactivator SAGA (Spt3-Ada-Gcn5-acetyltransferase), and has been demonstrated to serve as a link between transcriptional activation and mRNA export. Therefore, mRNA export is coupled to both transcriptional activation and elongation.
Since ubiquitylation has been implicated in transcription, transcription-coupled mRNA export is likely to be regulated by ubiquitylation directly and/or indirectly. Indeed, previous studies (Duncan et al. 2000; Neumann et al. 2003; Rodriguez et al. 2003; Gwizdek et al. 2005; Iglesias et al. 2010) have demonstrated the direct roles of two HECT domain-containing E3 ubiquitin ligases, namely Tom1 and Rsp5, in mRNA export in yeast. Recently, we have demonstrated that Mdm30, an F-box protein, is also involved in stimulation of nuclear export of mRNA in yeast (Durairaj et al. 2009; Shukla et al. 2009). An F-box is a conserved protein motif of ∼50 amino acids (Bai et al. 1994, 1996; Jonkers and Rep 2009) and functions as a component of SCF (Skp1-Cul1-F-box-protein) ubiquitin ligase complex to facilitate the interaction between substrate and ubiquitin conjugating enzyme for covalent transfer of ubiquitin onto substrate (Bai et al. 1996; Skowyra et al. 1997; Reed 2003; Yen and Elledge 2008; Jonkers and Rep 2009). Like other F-box proteins, Mdm30 copurifies with SCF components (Uetz et al. 2000; Kus et al. 2004; Cohen et al. 2008; Lackner and Nunnari 2009) and is involved in ubiquitylation and proteasomal degradation of Fzo1 and Mdm34 for the maintenance of fusion competent mitochondria (Fritz et al. 2003; Meeusen and Nunnari 2005; Cohen et al. 2008; Ota et al. 2008; Livnat-Levanon and Glickman 2011). Additionally, Mdm30 is involved in regulation of transcription via ubiquitylation and consequent proteasomal degradation of transcriptional activator (Gal4) or its negative regulator (Muratani et al. 2005; Ang et al. 2012). However, it is yet unknown how Mdm30 facilitates mRNA export. Here, we show that Mdm30 promotes mRNA export by enhancing the recruitment of Yra1 to the active genes via ubiquitylation and proteasomal degradation of Sub2, thus deciphering new regulatory mechanisms of mRNA export.
RESULTS AND DISCUSSION
The stability/abundance of the Sub2 component of the TREX complex is enhanced in the absence of Mdm30
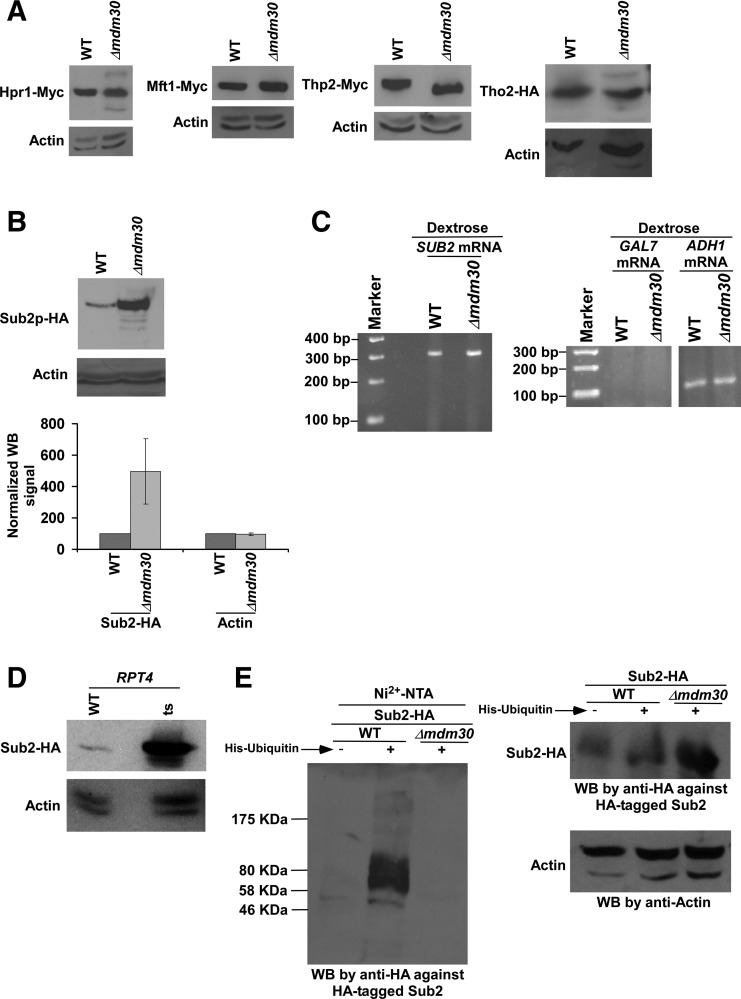
Mdm30 is involved in mitochondrial fusion via ubiquitylation and proteasomal degradation of Fzo1 and Mdm34 (Fritz et al. 2003; Meeusen and Nunnari 2005; Cohen et al. 2008; Ota et al. 2008; Livnat-Levanon and Glickman 2011). Likewise, Mdm30 might be targeting some component of mRNA export machinery for ubiquitylation and subsequent proteasomal degradation to promote mRNA export. If Mdm30 is involved in degradation of a specific component of mRNA export machinery, the stability of that particular component would be enhanced in the absence of Mdm30. To test this possibility, we analyzed the global levels of the TREX components in the presence and absence of Mdm30. In this direction, we tagged the TREX components by Myc or HA epitope at the C-terminals in their chromosomal loci in the Δmdm30 strain and its isogenic wild-type equivalent, and then determined their global levels by Western blot assay. We found that the global levels of Hpr1, Mft1, Tho2, and Thp2 were not altered in the Δmdm30 strain in comparison to the wild-type equivalent (Fig. 1A). The level of actin was monitored as a loading control, and its level was not changed in the Δmdm30 strain (Fig. 1A). Thus, Mdm30 does not regulate the abundances of Hpr1, Mft1, Tho2, and Thp2. Similarly, the global level of Yra1 was not altered in the Δmdm30 strain (Shukla et al. 2009). Intriguingly, we found that the global level of Sub2 was significantly elevated in the absence of Mdm30 (Fig. 1B). However, the level of actin was not increased in the Δmdm30 strain in comparison to the wild-type equivalent (Fig. 1B). These results support that the stability/abundance of the Sub2 component of the TREX complex is enhanced in the absence of Mdm30. However, such an increased stability of Sub2 in the absence of Mdm30 could be due to an enhanced transcription of SUB2. To test this possibility, we next analyzed the SUB2 mRNA levels in the Δmdm30 and wild-type strains by reverse transcriptase–PCR (RT-PCR) assay. We found that the SUB2 mRNA level was not altered in the Δmdm30 strain in comparison to the wild-type equivalent (Fig. 1C). As a loading control, we analyzed the ADH1 mRNA levels in the Δmdm30 and wild-type strains (Fig. 1C) since transcription of ADH1 is not changed in the absence of Mdm30 (Shukla et al. 2009). We found that the ADH1 mRNA level was not altered in the Δmdm30 strain (Fig. 1C). Further, we analyzed GAL7 as a control for possible DNA contamination as done previously (Malik et al. 2013), since GAL7 gene is not expressed in dextrose-containing growth medium (Malik et al. 2013). We did not observe a PCR signal, when the primer pair targeted to the GAL7 coding sequence was used in amplifying cDNA in the RT-PCR analysis (Fig. 1C). These results indicate the absence of DNA contamination in our mRNA analysis. Collectively, our results support that Mdm30 regulates the abundance of Sub2 at the protein level, but not at the level of mRNA or transcription.
FIGURE 1.
The abundance of Sub2 is enhanced in the absence of Mdm30. (A,B) Analysis of the global levels of Hpr1, Mft1, Thp2, Tho2, Sub2, and actin in the Δmdm30 and wild-type strains. Yeast strains expressing HA/Myc-tagged proteins were grown in YPD at 30°C up to an OD600 of 1.0 prior to harvesting for Western blot analysis. An anti-Myc, anti-HA, or anti-actin antibody was used in the Western blot analysis. Western blot signal of Sub2 in wild-type strain was set to 100. Western blot signal of Sub2 in the Δmdm30 strain was normalized with respect to 100. Likewise, actin levels were normalized. Normalized Western blot signals were plotted in the form of a histogram. Error bar denotes standard deviation (STDEV; Microsoft Excel 2003) from three biologically independent experiments. (WB) Western blot. (C) RT-PCR analysis of SUB2 mRNA levels in the Δmdm30 and wild-type strains using oligo-dT primer in the cDNA synthesis. Yeast strains were grown as in A. GAL7 and ADH1 mRNA levels were analyzed as controls. (D) Analysis of Sub2's abundance in the rpt4-ts and wild-type strains. Both the wild-type and ts mutant strains expressing HA-tagged Sub2 were grown in YPD at 23°C up to an OD600 of 0.85, and then switched to 39°C for 1 h before harvesting for Western blot analysis. (E) Ubiquitylation analysis of Sub2. Yeast strains expressing HA-tagged Sub2 and/or hexahistidine-tagged ubiquitin were grown in synthetic complete medium at 30°C up to an OD600 of 0.7, and then treated with CuSO4 at a final concentration of 0.1 M for 6 h. Precipitation was carried out by Ni2+-NTA agarose beads, and Western blot analysis was performed using an anti-HA antibody against HA-tagged Sub2.
Sub2 is ubiquitylated by Mdm30 for proteasomal degradation
Mdm30 has been shown to be copurified with other components of SCF ubiquitin ligase to ubiquitylate its targets such as Fzo1 and Mdm34 for proteasomal degradation (Fritz et al. 2003; Meeusen and Nunnari 2005; Cohen et al. 2008; Ota et al. 2008; Livnat-Levanon and Glickman 2011). Likewise, Mdm30 might be regulating Sub2's stability via ubiquitylation and proteasomal degradation. To test this, we first analyzed whether Sub2 is degraded by the 26S proteasome. For this purpose, we tagged Sub2 by HA epitope at its C-terminal in the chromosomal locus in the wild-type and temperature-sensitive (ts) mutant strains of Rpt4, an essential component of the 26S proteasome for its proteolytic activity (Rubin et al. 1998; Russell et al. 2001; Bhaumik and Malik 2008). Using these strains, we determined the global levels of Sub2 in the rpt4-ts (sug2-ts) mutant and wild-type strains at the nonpermissive temperature. We found that the global level of Sub2 was dramatically enhanced in the rpt4-ts mutant strain (Fig. 1D). However, the level of actin was not changed in the rpt4-ts mutant strain in comparison to the wild-type equivalent (Fig. 1D). Thus, Sub2 has much higher turnover than actin, and such turnover is impaired in the rpt4-ts mutant strain. Therefore, our results indicate that the proteasome is involved in the degradation of Sub2, and hence, the stability/abundance of Sub2 is increased in the rpt4-ts mutant strain.
Since the proteasome is involved in targeted degradation of the ubiquitylated-proteins (Bhaumik and Malik 2008), Sub2 is thus likely to be ubiquitylated for its proteasomal degradation in the presence of Mdm30. To test this possibility, we analyzed the ubiquitylation status of Sub2 in the Mdm30 wild-type strain. In this direction, we introduced a plasmid expressing hexahistidine-tagged ubiquitin under the CUP1 promoter (pUB221) in the wild-type strain expressing HA-tagged Sub2. Using this strain, we performed the ubiquitylation assay as described previously (Muratani et al. 2005). Ubiquitin and ubiquitylated proteins were precipitated from the whole-cell extract using Ni2+-NTA agarose beads that bind to hexahistine tag attached to ubiquitin. The precipitate was analyzed by Western blot assay for the presence of Sub2 using an anti-HA antibody against HA-tagged Sub2. We observed Sub2 in the precipitate (Fig. 1E). As control, we performed similar experiments using the yeast strain that did not have the plasmid expressing hexahistidine-tagged ubiquitin (but expressed HA-tagged Sub2). We did not find Sub2 in the precipitate (Fig. 1E). These results support that Sub2 is ubiquitylated in the presence of Mdm30. Likewise, its mammalian homolog, UAP56, has recently been shown to be ubiquitylated (Xiong et al. 2012). To determine whether Mdm30 is involved in ubiquitylation of Sub2, we next analyzed the ubiquitylation status of Sub2 in the absence of Mdm30. We found that ubiquitylation of Sub2 was impaired in the Δmdm30 strain (Fig. 1E). The equal amounts of whole-cell extracts were used for the Δmdm30 and wild-type strains, as evident by equal levels of actin (Fig. 1E). However, the Δmdm30 strain contained a significantly high level of Sub2 in comparison to the wild-type equivalent (Fig. 1E), as the stability of Sub2 is enhanced in the absence of Mdm30 (Fig. 1B). Even in the presence of a high level/abundance of Sub2 in the Δmdm30 strain, Sub2 was not found in the precipitate (Fig. 1E). Together, these results support that Mdm30 is involved in ubiquitylation and proteasomal degradation of Sub2.
Enhanced stability of Sub2 lowers mRNA export
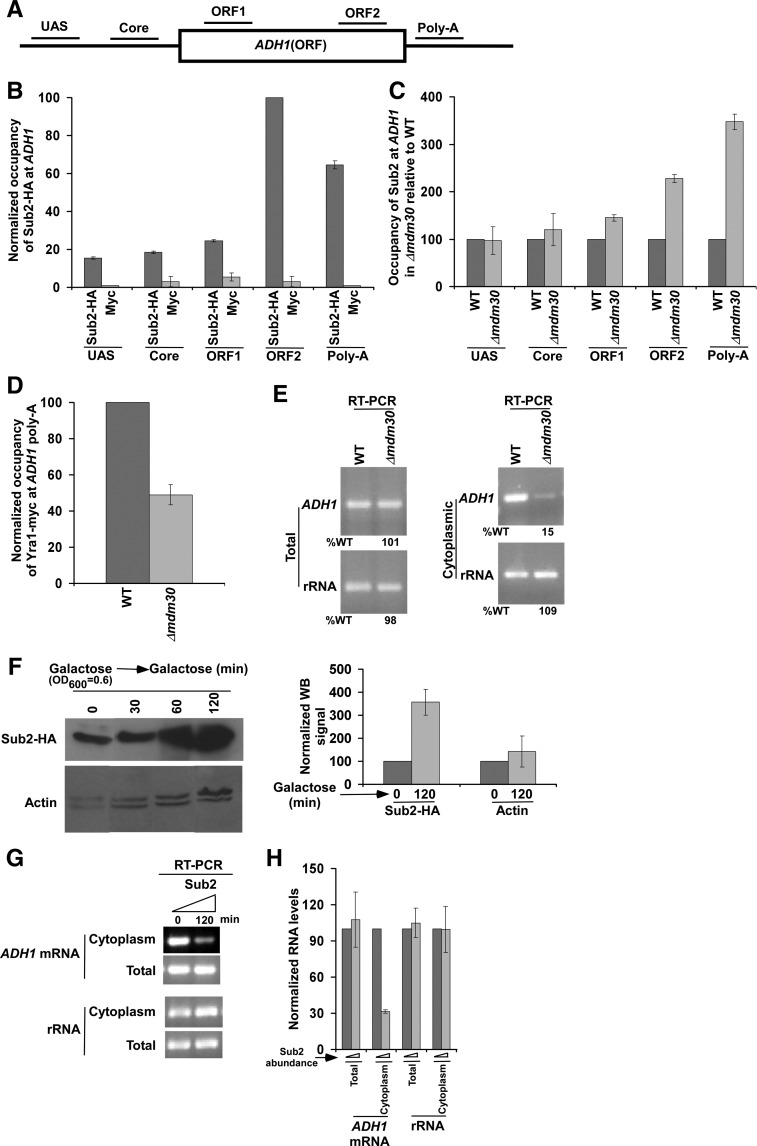
We find here that the stability/abundance of Sub2 is enhanced in the absence of Mdm30. Further, we have recently demonstrated the stimulatory role of Mdm30 in mRNA export of ADH1, PHO84, and RPS5 (Shukla et al. 2009). These results indicate that enhanced stability of Sub2 in the absence of Mdm30 is correlated with reduced mRNA export. Consistently, Luo et al. (2001) have demonstrated in Xenopus oocytes that a significantly high level of Sub2 homolog, UAP56, reduces the recruitment of Yra1 homolog, ALY, and hence, mRNA export. To test whether the recruitment of Yra1 is also similarly decreased in yeast when the abundance of Sub2 is significantly increased in the absence of Mdm30, we analyzed the association of Sub2 and Yra1 to ADH1 in the Δmdm30 and wild-type strains. We found that the recruitment of Sub2 toward the 3′-end of the ADH1 gene was significantly increased in the Δmdm30 strain in comparison to the wild-type equivalent (Fig. 2A–C). Thus, enhanced abundance of Sub2 in the Δmdm30 strain increases the targeted association of Sub2 with the 3′-end of ADH1. Intriguingly, such enhanced recruitment of Sub2 in the Δmdm30 strain decreased the association of Yra1 with the 3′-end of ADH1 (Fig. 2D; Supplemental Fig. S1A) (Yra1 is predominantly associated with the 3′-end of the active genes) (Johnson et al. 2009, 2011; MacKellar and Greenleaf 2011). Consistently, the export of ADH1 mRNA from nucleus to cytoplasm was significantly decreased in the Δmdm30 strain (Fig. 2E; Supplemental Fig. S1B). As a loading control, we analyzed the level of rRNA (18S rRNA) and found that rRNA level was not altered in the absence of Mdm30 (Fig. 2E). Further, as quality control, we analyzed the presence/absence of nuclear GAL1 gene in the cytoplasmic fraction. If the cytoplasmic fractionation is of good quality, nuclear GAL1 gene would not be found in the cytoplasmic fraction. We observed nuclear GAL1 gene in the cytoplasmic fraction following vortexing of the spheroplasts with glass beads for 45 and 60 sec in both the wild-type and Δmdm30 strains (Supplemental Fig. S1C,D). However, GAL1 was not observed in the cytoplasmic fraction following 15 or 30 sec of vortexing, but rather was present in the nuclear fraction (Supplemental Fig. S1C,D). Therefore, spheroplasts were vortexed for 15 sec in our mRNA analysis in the cytoplasmic fraction to study mRNA export.
FIGURE 2.
Enhanced stability of Sub2 in the Δmdm30 strain reduces the recruitment of mRNA export adaptor to ADH1, and hence mRNA export. (A) A schematic diagram showing the locations of the primer pairs at ADH1 for the ChIP analysis. (B) Analysis of association of Sub2 with ADH1. Yeast strain expressing HA-tagged Sub2 was grown in YPD up to an OD600 of 1.0 at 30°C prior to formaldehyde-based in vivo crosslinking. Immunoprecipitation was performed as described previously (Bhaumik and Green 2002, 2003; Bhaumik et al. 2004; Shukla et al. 2006), using an anti-HA antibody (F-7; Santa Cruz Biotechnology, Inc.) against HA-tagged Sub2 or anti-myc (9E10; Santa Cruz Biotechnology, Inc.) as a control. Immunoprecipitated DNA was analyzed by PCR using the primer pairs located at different locations of ADH1 as schematically shown in A. The ratio of the immunoprecipitate over the input in the autoradiogram (termed as a ChIP signal) was measured. The maximum ChIP signal was set to 100, and other ChIP signals were normalized with respect to 100. The normalized ChIP signals (represented as normalized or relative occupancy) were plotted in the form of a histogram. Error bars denote standard deviations. (C) Analysis of Sub2 association with ADH1 in the presence and absence of Mdm30. Yeast cells were grown, crosslinked, and immunoprecipitated as in B. The ChIP signal of the wild-type strain was set to 100, and the signal from the mutant strain was normalized with respect to 100 at different locations of ADH1. (D) Analysis of Yra1 association with the poly(A) site of ADH1 in the presence and absence of Mdm30. Both the Δmdm30 and wild-type strains expressing myc-tagged Yra1 were grown, crosslinked, and immunoprecipitated as in B. (E) Analysis of total and cytoplasmic ADH1 mRNAs in the Δmdm30 and wild-type strains by RT-PCR assay. Yeast strains were grown as in B. The level of rRNA (18S rRNA) was analyzed as a loading control. (F) Sub2 expression analysis under the control of GAL1 promoter in galactose-containing growth medium (or YPG) by Western blot assay. Yeast cells were initially grown at 30°C up to an OD600 of 0.6, and then were collected at different time points (0, 30, 60, and 120 min). Western blot signal of Sub2 at 0 min was set to 100. Western blot signal of Sub2 at 120 min in galactose was normalized with respect to 100. Likewise, actin levels were normalized. Normalized Western blot signals were plotted in the form of a histogram. Error bar denotes standard deviation from three biologically independent experiments. (G) Analysis of total and cytoplasmic ADH1 mRNAs in the presence of a high level/abundance of Sub2 by RT-PCR assay. (H) The results of G were plotted in the form of a histogram. The PCR signal at 0 min was set to 100, and the signal at 120 min was normalized with respect to 100.
The above mRNA export experiments revealed a significantly decreased level of ADH1 mRNA in the cytoplasm in the Δmdm30 strain in comparison to the wild-type equivalent (Fig. 2E). However, the level of total ADH1 mRNA was not altered in the Δmdm30 strain (Fig. 2E). Thus, Mdm30 facilitates nuclear export of ADH1 mRNA, but not transcription. Therefore, enhanced stability/abundance of Sub2 in the absence of Mdm30 increases Sub2 recruitment and decreases the association of mRNA export adaptor Yra1, hence leading to reduced mRNA export. To further complement that a high level/abundance of Sub2 in the Δmdm30 strain decreases mRNA export, we next analyzed nuclear export of ADH1 mRNA in the presence of a high abundance of Sub2. In this direction, we replaced the endogenous promoter of SUB2 by GAL1, and expressed Sub2 in galactose-containing growth medium (Fig. 2F). Subsequently, we analyzed the levels of total and cytoplasmic ADH1 mRNAs by RT-PCR assay. We found that the level of ADH1 mRNA in the cytoplasm was significantly decreased in the presence of a high abundance of Sub2, while total ADH1 mRNA levels remained unaltered (Fig. 2F–H). The rRNA level (i.e., loading control) was not altered in the presence of a high abundance of Sub2 (Fig. 2G,H). Taken together, our results support that enhanced abundance of Sub2 in the Δmdm30 strain decreases ADH1 mRNA export.
Although previous studies in Xenopus oocyte (Luo et al. 2001) have demonstrated that an excess of Sub2's homolog, UAP56, inhibits mRNA export, it remains unknown how UAP56's abundance is regulated to control mRNA export. Our data support that Sub2 is ubiquitylated by Mdm30, and subsequently degraded by the proteasome to promote mRNA export, thus providing a new regulatory pathway of mRNA export. Similarly, UAP56 is likely to be regulated in Xenopus oocytes and mammals. In support of this possibility, a recent study (Xiong et al. 2012) demonstrated ubiquitylation of UAP56 in mammalian cells. However, it remains to be determined whether the abundance of UAP56 is regulated by F-box and proteasome to control mRNA export, similar to the results in yeast.
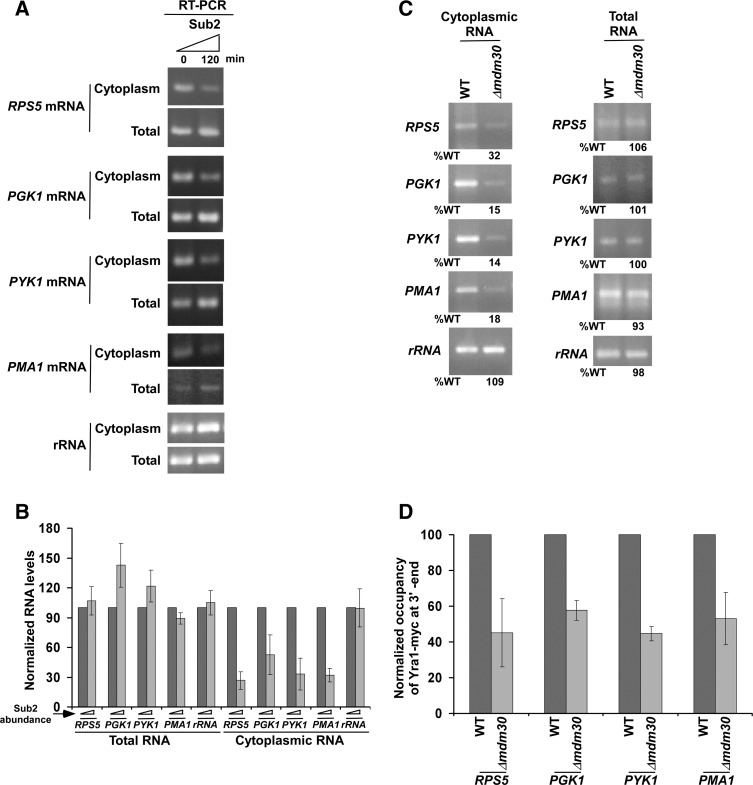
To determine whether Mdm30 also regulates mRNA export of other genes, we analyzed nuclear export of mRNAs of several genes such as RPS5, PGK1, PYK1, and PMA1. We found that nuclear export of RPS5, PGK1, PYK1, and PMA1 mRNAs was significantly decreased in the presence of a high abundance of Sub2 (Fig. 3A,B). However, total levels of RPS5, PGK1, PYK1, and PMA1 mRNAs were not altered (Fig. 3A,B). Thus, a significantly high abundance of Sub2 does not appear to alter the synthesis/transcription of RPS5, PGK1, PYK1, and PMA1, but rather lowers nuclear export of their mRNAs. Therefore, a high abundance of Sub2 in the absence of Mdm30 (Fig. 1B) decreases mRNA export of RPS5, PGK1, PYK1, and PMA1 (Fig. 3C). Such a decrease in mRNA export is mediated via reduced recruitment of export adaptor Yra1 in the Δmdm30 strain (Fig. 3D). Thus, like the results at ADH1, we find that enhanced abundance of Sub2 in the Δmdm30 strain (Fig. 1B) decreases the recruitment of Yra1 (Fig. 3D), which subsequently reduces nuclear export of RPS5, PGK1, PYK1, and PMA1 mRNAs (Fig. 3A–C). An abundant Sub2 might be squelching Yra1, hence impairing the recruitment of Yra1. Additionally, an excess helicase activity of Sub2 might lower the recruitment of Yra1. Further, Yra1 has been recently shown to be ubiquitylated by Tom1 (Iglesias et al. 2010). Thus, Mdm30 and Tom1 are likely to crosstalk to control the recruitment of Yra1, and hence mRNA export. These possibilities remain to be further elucidated in order to shed much light into this new regulatory process of mRNA export.
FIGURE 3.
Enhanced abundance of Sub2 in the absence of Mdm30 reduces the recruitment of Yra1 to the 3′-ends of RPS5, PGK1, PYK1, and PMA1 coding sequences, and hence mRNA export. (A) Analysis of total and cytoplasmic RPS5, PGK1, PYK1, and PMA1 mRNAs, and 18S rRNA in the presence of a high abundance of Sub2 by RT-PCR assay. (B) The results of A were plotted together in the form of a histogram. (C) Analysis of total and cytoplasmic RPS5, PGK1, PYK1, and PMA1 mRNAs, and 18S rRNA in the wild-type and Δmdm30 strains. (D) Analysis of Yra1 association with the 3′-ends of RPS5, PGK1, PYK1, and PMA1 coding sequences in the presence and absence of Mdm30. Both the Δmdm30 and wild-type strains expressing myc-tagged Yra1 were grown, crosslinked, and immunoprecipitated as in Figure 2B.
Enhanced stability of Sub2 does not alter transcription
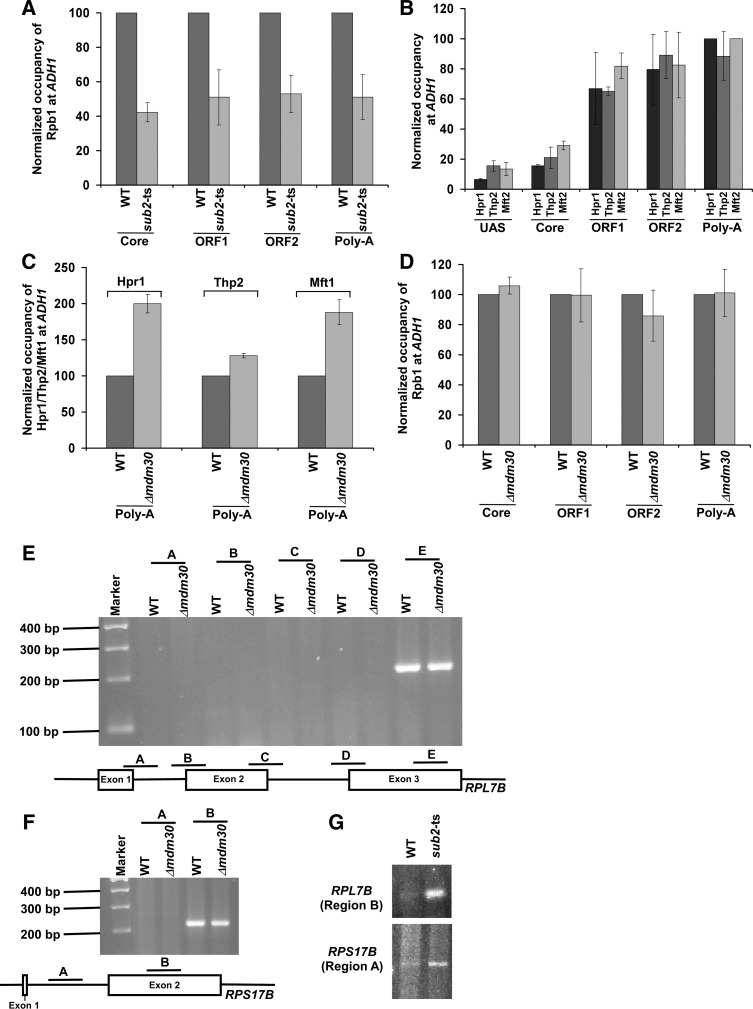
In addition to its stimulatory role in mRNA export, Sub2 also promotes transcriptional elongation (Chavez et al. 2000; Hammell et al. 2002; Jimeno et al. 2002; Strässer et al. 2002; Zenklusen et al. 2002; Rondón et al. 2003; Kim et al. 2004; Rehwinkel et al. 2004; Carmody and Wente 2009). Consistently, we found that the association of RNA polymerase II with ADH1 was significantly impaired in the ts strain of Sub2 (which carries mutations in the ATP binding/hydrolysis motif) (Kistler and Guthrie 2001) in comparison to the wild-type equivalent (Fig. 4A). However, it remains unknown whether a high abundance of Sub2 alters transcription. To address this, we analyzed the recruitment of Sub2, THO, and RNA polymerase II to ADH1 in the wild-type strain and Δmdm30 mutant (which has a high abundance of Sub2 in comparison to the wild-type equivalent) (Fig. 1B). As presented above, the recruitment of Sub2 to the 3′-end of ADH1 was enhanced by around threefold in the Δmdm30 strain (Fig. 2A–C), while the association of the Hpr1 and Mft2 components of THO with ADH1 was increased by around twofold (Fig. 4B,C). However, the recruitment of the Thp2 component of THO to ADH1 was not similarly altered in the absence of Mdm30 (Fig. 4B,C). Such differential recruitment of the THO components and Sub2 to ADH1 in the Δmdm30 strain did not alter the association of RNA polymerase II with ADH1 (Fig. 4D), and hence total ADH1 mRNA level was not changed in the absence of Mdm30 (Fig. 2E). Therefore, an enhanced abundance of Sub2 in the Δmdm30 strain (Fig. 1B) does not appear to alter RNA polymerase II association with ADH1, and thus transcription (Figs. 2E, 4D). Likewise, transcription of RPS5, PGK1, PYK1, and PMA1 was not altered in the presence of a high abundance of Sub2 (Fig. 3A–C).
FIGURE 4.
Transcription and splicing are not altered in the absence of Mdm30. (A) Analysis of RNA polymerase II (Rpb1) association with ADH1 in the sub2-ts and wild-type strains. Both the wild-type and ts mutant strains were grown at 23°C up to an OD600 of 0.85, and then switched to 37°C for 1 h prior to crosslinking. (B) Analysis of association of Hpr1, Thp2, and Mft2 with ADH1. Yeast strains expressing myc-tagged Hpr1, Thp2, or Mft2 were grown, crosslinked, and immunoprecipitated as in Figure 2B. (C) Analysis of association of Hpr1, Thp2, and Mft2 with ADH1 in the presence and absence of Mdm30. (D) Analysis of RNA polymerase II association with ADH1 in the Δmdm30 and wild-type strains. (E) Mdm30 does not impair mRNA splicing of RPL7B. Both the Δmdm30 and wild-type strains were grown as in Figure 2B. The cDNA was synthesized using oligo-dT primer and amplified by different primer pairs as schematically shown in the bottom panel. (F) Mdm30 does not impair mRNA splicing of RPS17B. (G) Sub2 promotes splicing of RPL7B and RPS17B mRNAs.
As presented above, our ChIP analysis revealed that the recruitment of the Sub2, Hpr1, and Mft2 components of TREX to ADH1 was differentially increased in the absence of Mdm30 (Figs. 2A–C, 4B,C), while Thp2 recruitment was not altered (Fig. 4B,C). Further, the recruitment of the Yra1 component of TREX to ADH1 was decreased in the Δmdm30 strain (Fig. 2D; Supplemental Fig. S1A). Thus, the TREX components are differentially associated with ADH1 in the Δmdm30 strain in comparison to the wild-type equivalent. A similar pattern of differential association of the TREX components with another gene, PYK1, was also observed in the Δmdm30 strain in comparison to the wild-type equivalent (Fig. 3D; Supplemental Fig. S2). These results suggest that the TREX complex may have been differently assembled or adopted an altered conformation in the absence of Mdm30. Otherwise, a differential association pattern of the TREX components with the active gene would not have been observed in the Δmdm30 strain.
We show above that transcription of ADH1, RPS5, PGK1, PYK1, and PMA1 is not altered in the presence of a high abundance of Sub2 in the Δmdm30 strain, while mRNA export of these genes is significantly decreased, but not completely impaired. Such a decrease in mRNA export might have an indirect effect on transcription. However, we find that transcription of ADH1, RPS5, PGK1, PYK1, and PMA1 is not altered in the Δmdm30 strain. It may be possible that the steady-state levels of mRNAs of these constitutively active genes might not be altered in the Δmdm30 mutant, as observed in other mutants (e.g., Rad26 and Cbp80 null mutants) in our previous studies (Lahudkar et al. 2011; Malik et al. 2012). However, transcription of the inducible genes at an early induction period might be altered in the Δmdm30 strain, similar to other mutants (e.g., Rad26 and Cbp80 null mutants) in our previous studies (Lahudkar et al. 2011; Malik et al. 2012). To test this, we analyzed transcription of the GAL1 inducible gene in the Δmdm30 and wild-type strains under transcriptionally inducible conditions. We found that transcription of GAL1 was reduced in the Δmdm30 strain following induction (Supplemental Fig. S3). Further, we observed that GAL1 mRNA export was dramatically decreased in the absence of Mdm30 (Supplemental Fig. S4A). Consistently, recruitment of export adaptor Yra1 to GAL1 was reduced in the Δmdm30 strain, while Sub2 recruitment is increased (Supplemental Fig. S4B,C). Therefore, Mdm30 plays a stimulatory role in transcription of inducible gene at an early induction stage in addition to its function in promoting mRNA export.
Enhanced stability of Sub2 does not alter splicing
In addition to its role in transcription and mRNA export, Sub2 has also been implicated in mRNA splicing (Kistler and Guthrie 2001; Libri et al. 2001; Zhang and Green 2001). Thus, enhanced abundance of Sub2 in the Δmdm30 strain (Fig. 1B) may alter mRNA splicing. To test this possibility, we analyzed splicing of RPL7B (that contains two introns and three exons) (Fig. 4E) in the Δmdm30 and wild-type strains. In this direction, we isolated total RNA from the wild-type and Δmdm30 strains, and prepared cDNAs using oligo-dT-based reverse transcription assay. The cDNAs were amplified using different sets of primer pairs targeted to the exon and intron–exon junction regions of RPL7B as schematically shown in Figure 4E. The PCR signal from the intron–exon junction would indicate impaired splicing. We did not observe the PCR signal from the intron–exon junctions (Regions A, B, C, and D) in the Δmdm30 and wild-type strains (Fig. 4E). However, the PCR signal was found at the exon region (Region E) (Fig. 4E). These results demonstrate that RPL7B mRNA is spliced in the Δmdm30 and wild-type strains. Further, we found that the level of the PCR signal at the exon region E in the Δmdm30 strain remained the same as that of the wild-type equivalent (Fig. 4E). These results indicate that Mdm30 does not regulate transcription of RPL7B, consistent with the results at ADH1 and other genes (Figs. 2E, 3C, 4D; Shukla et al. 2009). Likewise, Mdm30 does not alter transcription and splicing of another gene, RPS17B (Fig. 4F). Together, our results support that enhanced abundance of Sub2 in the Δmdm30 strain (Fig. 1B) does not impair the splicing of RPL7B and RPS17B mRNAs. However, the splicing of RPL7B and RPS17B mRNAs was impaired in the sub2-ts mutant strain (Fig. 4G), consistent with previous studies (Libri et al. 2001; Zhang and Green 2001).
In summary, we demonstrate here that the stability/abundance of Sub2 is enhanced in the absence of Mdm30. Such increased abundance of Sub2 does not impair constitutive transcription and mRNA splicing. However, the recruitment of Yra1 onto the active genes is decreased when the abundance of Sub2 is enhanced in the absence of Mdm30. Reduced recruitment of Yra1 to the active genes decreases mRNA export. Collectively, our results support that enhanced abundance of Sub2 in the Δmdm30 strain reduces the recruitment of export adaptor Yra1 which subsequently decreases mRNA export, thus providing a new regulatory pathway of mRNA export.
MATERIALS AND METHODS
Plasmids and strains
The plasmids and yeast strains used in this study are described in the Supplemental Information section.
Growth media
The yeast strains were grown in YPD (yeast extract peptone plus 2% dextrose) up to an OD600 of 1.0 at 30°C for the studies at the ADH1, RPS5, PGK1, PYK1, and PMA1 genes. For ubiquitylation analysis, yeast cells were grown in synthetic complete medium (yeast nitrogen base and complete amino acid mixture plus 2% dextrose) at 30°C up to an OD600 of 0.7, and then treated with 0.1 mM of CuSO4 for 6 h. The wild-type and ts mutant strains were grown at 23°C up to an OD600 of 0.9, and then switched to 37°C (for sub2-ts mutant) or 39°C (for rpt4-ts mutant) for 1 h prior to harvesting. For studies at GAL1, yeast cells were grown in YPR (yeast extract, peptone plus 2% raffinose) up to an OD600 of 0.9 at 30°C, and then switched to YPG (yeast extract, peptone plus 2% raffinose) for 60 min.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed as described previously (Bhaumik and Green 2002, 2003; Bhaumik et al. 2004; Shukla et al. 2006). For analysis of Sub2, Hpr1, Thp2, Mft2, and Yra1 recruitment to the active genes, we modified the ChIP protocol as described previously (Malik et al. 2010). The primer pairs used for PCR analysis of immunoprecipitated DNA samples are described in the Supplemental Information section.
Whole-cell extract preparation and Western blot analysis
To analyze the global levels of the TREX components, yeast strains expressing myc/HA epitope-tagged TREX components were grown in 25 mL YPD up to an OD600 of 1.0. Yeast cells were then harvested, lysed, and sonicated to prepare the whole-cell extract with solubilized chromatin following the protocol as described previously for the ChIP assay (Bhaumik and Green 2002, 2003; Bhaumik et al. 2004; Shukla et al. 2006). The whole-cell extract was run on SDS–polyacrylamide gel, and then analyzed by Western blot assay. An anti-myc (9E10; Santa Cruz Biotechnology, Inc.), anti-HA (F-7; Santa Cruz Biotechnology, Inc.), or anti-actin (A2066; Sigma) antibody was used for Western blot analysis.
Isolation of total and cytoplasmic RNAs
The total RNA was prepared from yeast cell culture as described previously (Peterson et al. 1991). Cytoplasmic mRNA was prepared as described previously (Shukla et al. 2009). Briefly, harvested yeast cells from 10 mL culture were suspended in 400 μL sorbitol solution (0.9 M sorbitol, 0.1 M EDTA, and 14 mM β-mercaptoethanol), and then incubated with 20 μL zymolase (10 mg/mL) for 25 min at 37°C, followed by centrifugation for 5–10 sec. The supernatant was carefully removed, and spheroplasts were gently suspended in 100 μL RNA preparation buffer along with 20 μL volume equivalent of glass beads. The spheroplast suspension was mildly vortexed (∼15 sec on the VWR mini-vortexer with a low speed of 5). Following centrifugation, the supernatant was carefully collected and used for cytoplasmic mRNA preparation following phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation.
RT-PCR analysis
RT-PCR analysis was performed as described previously (Ausubel et al. 2001; Shukla et al. 2009; Malik et al. 2013). The primer pairs used for PCR analysis of cDNAs are described in the Supplemental Information section.
Ubiquitylation assay
The ubiquitylation assay was performed as described previously (Muratani et al. 2005). Briefly, the expression of hexahistidine-tagged ubiquitin from plasmid pUB221 was induced for 6 h by addition of CuSO4 to a final concentration of 0.1 mM. Cells were harvested, suspended in buffer A2 (6 M guanidine-HCl, 100 mM Na2HPO4/NaH2PO4 at pH 8.0, 10 mM imidazole, 250 mM NaCl, 0.5% NP-40), and lysed by glass beads. Cell lysate was clarified by centrifugation, and supernatant was incubated with Ni2+-NTA agarose resin (Qiagen) for 2 h at room temperature. Following incubation, Ni2+-NTA resin was washed twice by buffer A2, twice by buffer A2/T2 (1 volume of buffer A2 and 3 volumes of buffer T2), and twice by buffer T2 (50 mM Na2HPO4/NaH2PO4 at pH 8.0, 250 mM NaCl, 20 mM imidazole, 0.5% NP-40). Subsequently, the resin was washed by buffer T2 containing 50 mM histidine to reduce the level of nonspecific binding to the resin. Finally, hexahistidine-tagged ubiquitin/ubiquitylated-proteins were eluted with 2X SDS loading buffer containing 250 mM imidazole and analyzed by Western blot assay.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Stephen Johnston and Thomas Kodadek (University of Texas–Southwestern Medical Center) and Christine Guthrie (University of California–San Francisco) for yeast strains; Daniel Finley (Harvard Medical School) for plasmid; and Sarah Frankland-Searby for editorial assistance. The work in the Bhaumik laboratory was supported by a National Institutes of Health grant (1R15GM088798-01), a grant-in-aid (10GRNT4300059) from the American Heart Association (Greater Midwest Affiliate), a Mallinckrodt Foundation grant, and Excellence in Academic Medicine (EAM) grants from the School of Medicine of Southern Illinois University.
REFERENCES
- Ang K, Ee G, Ang E, Koh E, Siew WL, Chan YM, Nur S, Tan YS, Lehming N 2012. Mediator acts upstream of the transcriptional activator Gal4. PLoS Biol 10: e1001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Struhl K 2001. Current protocols in molecular biology. Wiley, New York [Google Scholar]
- Bai C, Richman R, Elledge SJ 1994. Human cyclin F. EMBO J 13: 6087–6098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274 [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Green MR 2002. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol Cell Biol 22: 7365–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik SR, Green MR 2003. Interaction of Gal4p with components of transcription machinery in vivo. Methods Enzymol 370: 445–454 [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Malik S 2008. Diverse regulatory mechanisms of eukaryotic transcriptional activation by the proteasome complex. Crit Rev Biochem Mol Biol 43: 419–433 [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Raha T, Aiello DP, Green MR 2004. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev 18: 333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody SR, Wente SR 2009. mRNA nuclear export at a glance. J Cell Sci 122: 1933–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A 2000. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J 19: 5824–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MM, Leboucher GP, Livnat-Levanon N, Glickman MH, Weissman AM 2008. Ubiquitin–proteasome-dependent degradation of a mitofusin, a critical regulator of mitochondrial fusion. Mol Biol Cell 19: 2457–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieppois G, Stutz F 2010. Connecting the transcription site to the nuclear pore: A multi-tether process that regulates gene expression. J Cell Sci 15: 1989–1999 [DOI] [PubMed] [Google Scholar]
- Duncan K, Umen JG, Guthrie C 2000. A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr Biol 10: 687–696 [DOI] [PubMed] [Google Scholar]
- Durairaj G, Garg P, Bhaumik SR 2009. Nuclear export of mRNA and its regulation by ubiquitylation. RNA Biol 6: 531–535 [DOI] [PubMed] [Google Scholar]
- Fischer T, Strasser K, Racz A, Rodriguez-Navarro S, Oppizzi M, Ihrig P, Lechner J, Hurt E 2002. The mRNA export machinery requires the novel Sac3p–Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J 21: 5843–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz S, Weinbach N, Westermann B 2003. Mdm30 is an F-box protein required for maintenance of fusion competent mitochondria in yeast. Mol Biol Cell 14: 2303–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwizdek C, Hobeika M, Kus B, Ossareh-Nazari B, Dargemont C, Rodriguez MS 2005. The mRNA nuclear export factor Hpr1 is regulated by Rsp5-mediated ubiquitylation. J Biol Chem 280: 13401–13405 [DOI] [PubMed] [Google Scholar]
- Hammell CM, Gross S, Zenklusen D, Heath CV, Stutz F, Moore C, Cole CN 2002. Coupling of termination, 3′ processing, and mRNA export. Mol Cell Biol 22: 6441–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E, Luo MJ, Röther S, Reed R, Strässer K 2004. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc Natl Acad Sci 101: 1858–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N, Tutucci E, Gwizdek C, Vinciguerra P, Von Dach E, Corbett AH, Dargemont C, Stutz F 2010. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev 24: 1927–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno S, Rondón AG, Luna R, Aguilera A 2002. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 21: 3526–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Cubberley G, Bentley DL 2009. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol Cell 33: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Kim H, Erickson B, Bentley DL 2011. The export factor Yra1 modulates mRNA 3′ end processing. Nat Struct Mol Biol 18: 1164–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers W, Rep M 2009. Lessons from fungal F-box proteins. Eukaryot Cell 8: 677–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap AK, Schieltz D, Yates J III, Kellogg DR 2005. Biochemical and genetic characterization of Yra1p in budding yeast. Yeast 22: 43–56 [DOI] [PubMed] [Google Scholar]
- Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J 23: 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler AL, Guthrie C 2001. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev 15: 42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kus BM, Caldon CE, Andorn-Broza R, Edwards AM 2004. Functional interaction of 13 yeast SCF complexes with a set of yeast E2 enzymes in vitro. Proteins 54: 455–467 [DOI] [PubMed] [Google Scholar]
- Lackner LL, Nunnari JM 2009. The molecular mechanism and cellular functions of mitochondrial division. Biochim Biophys Acta 1792: 1138–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahudkar S, Shukla A, Bajwa P, Durairaj G, Stanojevic N, Bhaumik SR 2011. The mRNA cap-binding complex stimulates the formation of pre-initiation complex at the promoter via its interaction with Mot1p in vivo. Nucleic Acids Res 39: 2188–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D, Graziani N, Saguez C, Boulay J 2001. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev 15: 36–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livnat-Levanon N, Glickman MH 2011. Ubiquitin-proteasome system and mitochondria - reciprocity. Biochim Biophys Acta 1809: 80–87 [DOI] [PubMed] [Google Scholar]
- Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413: 644–647 [DOI] [PubMed] [Google Scholar]
- MacKellar AL, Greenleaf AL 2011. Cotranscriptional association of mRNA export factor Yra1 with C-terminal domain of RNA polymerase II. J Biol Chem 286: 36385–36395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Chaurasia P, Lahudkar S, Durairaj G, Shukla A, Bhaumik SR 2010. Rad26p, a transcription-coupled repair factor, is recruited to the site of DNA lesion in an elongating RNA polymerase II-dependent manner in vivo. Nucleic Acids Res 38: 1461–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Chaurasia P, Lahudkar S, Uprety B, Bhaumik SR 2012. Rad26p regulates the occupancy of histone H2A–H2B dimer at the active genes in vivo. Nucleic Acids Res 40: 3348–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Durairaj G, Bhaumik SR 2013. Mechanisms of antisense transcription initiation from the 3′ end of the GAL10 coding sequence in vivo. Mol Cell Biol 33: 3549–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen SL, Nunnari J 2005. How mitochondria fuse. Curr Opin Cell Biol 17: 389–394 [DOI] [PubMed] [Google Scholar]
- Muratani M, Kung C, Shokat KM, Tansey WP 2005. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell 120: 887–899 [DOI] [PubMed] [Google Scholar]
- Neumann S, Petfalski E, Brugger B, Grosshans H, Wieland F, Tollervey D, Hurt E 2003. Formation and nuclear export of tRNA, rRNA and mRNA is regulated by the ubiquitin ligase Rsp5p. EMBO Rep 4: 1156–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota K, Kito K, Okada S, Ito T 2008. A proteomic screen reveals the mitochondrial outer membrane protein Mdm34p as an essential target of the F-box protein Mdm30p. Genes Cells 13: 1075–1085 [DOI] [PubMed] [Google Scholar]
- Peterson CL, Kruger W, Herskowitz I 1991. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell 64: 1135–1143 [DOI] [PubMed] [Google Scholar]
- Reed SI 2003. Ratchets and clocks: The cell cycle, ubiquitylation and protein turnover. Nat Rev Mol Cell Biol 4: 855–864 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Herold A, Gari K, Kocher T, Rode M, Ciccarelli FL, Wilm M, Izaurralde E 2004. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol 11: 558–566 [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Gwizdek C, Haguenauer-Tsapis R, Dargemont C 2003. The HECT ubiquitin ligase Rsp5p is required for proper nuclear export of mRNA in Saccharomyces cerevisiae. Traffic 4: 566–575 [DOI] [PubMed] [Google Scholar]
- Rondón AG, Jimeno S, García-Rubio M, Aguilera A 2003. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J Biol Chem 278: 39037–39043 [DOI] [PubMed] [Google Scholar]
- Rubin DM, Glickman MH, Larsen CN, Dhruvakumar S, Finley D 1998. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J 17: 4909–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SJ, Gonzalez F, Joshua-Tor L, Johnston SA 2001. Selective chemical inactivation of AAA proteins reveals distinct functions of proteasomal ATPases. Chem Biol 8: 941–950 [DOI] [PubMed] [Google Scholar]
- Shukla A, Stanojevic N, Duan Z, Sen P, Bhaumik SR 2006. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol Cell Biol 26: 3339–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Durairaj G, Schneider J, Duan Z, Shadle T, Bhaumik SR 2009. Stimulation of mRNA export by an F-box protein, Mdm30p, in vivo. J Mol Biol 389: 238–247 [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91: 209–219 [DOI] [PubMed] [Google Scholar]
- Strässer K, Hurt E 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J 19: 410–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strässer K, Hurt E 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413: 648–652 [DOI] [PubMed] [Google Scholar]
- Strässer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondón AG, Aguilera A, Struhl K, Reed R, et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308 [DOI] [PubMed] [Google Scholar]
- Stutz F, Bachi A, Doerks T, Braun IC, Séraphin B, Wilm M, Bork P, Izaurralde E 2000. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6: 638–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. 2000. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- Xiong F, Lin Y, Han Z, Shi G, Tian L, Wu X, Zeng Q, Zhou Y, Deng J, Chen H 2012. Plk1-mediated phosphorylation of UAP56 regulates the stability of UAP56. Mol Biol Rep 39: 1935–1942 [DOI] [PubMed] [Google Scholar]
- Yen HC, Elledge SJ 2008. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science 322: 923–929 [DOI] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Wyss JC, Stutz F 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol Cell Biol 22: 8241–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Green MR 2001. Identification and characterization of yUAP/Sub2p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56. Genes Dev 15: 30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]