Pre-mRNA alternative splicing is modified in cancer, but the origin and specificity of these changes remain unclear. This study examines the expression pattern of splice variants and associated splicing factors in dissected tumor tissues to identify cancer-specific splicing programs. Together, our results indicate that the architecture of transcripts is systematically and specifically redesigned in the ovarian cancer tumor and its microenvironment.
Keywords: alternative splicing, ovarian cancer, tumor microenvironment, RNA binding proteins, laser capture microdissection
Abstract
Pre-mRNA alternative splicing is modified in cancer, but the origin and specificity of these changes remain unclear. Here, we probed ovarian tumors to identify cancer-associated splicing isoforms and define the mechanism by which splicing is modified in cancer cells. Using high-throughput quantitative PCR, we monitored the expression of splice variants in laser-dissected tissues from ovarian tumors. Surprisingly, changes in alternative splicing were not limited to the tumor tissues but were also found in the tumor microenvironment. Changes in the tumor-associated splicing events were found to be regulated by splicing factors that are differentially expressed in cancer tissues. Overall, ∼20% of the alternative splicing events affected by the down-regulation of the splicing factors QKI and RBFOX2 were altered in the microenvironment of ovarian tumors. Together, our results indicate that the tumor microenvironment undergoes specific changes in alternative splicing orchestrated by a limited number of splicing factors.
INTRODUCTION
Ovarian cancer is the fourth most common cause of cancer-related death in women, largely because it is typically diagnosed at advanced stages (Coleman et al. 2011). Like most solid tumors, the majority of ovarian cancers are epithelial in origin. Ovaries that are normally composed of a thin layer of epithelial cells and a large mass of stromal cells that include fibroblasts, smooth muscle cells, and endothelial cells are dramatically transformed into a large epithelial mass in advanced cancer stages (Auersperg et al. 2001). In addition, the composition of the cells surrounding the tumor (the tumor microenvironment) changes as the tumor grows and gets established (Saad et al. 2010). The tumor microenvironment is composed of extracellular matrix (ECM), genetically stable cancer-associated fibroblasts (CAFs), immune cells, and soluble factors required for cancer progression and metastasis (Agarwal et al. 2010; Mandai et al. 2011). Interplay between tumor and adjoining stromal tissues has been observed to be an important aspect of the tumorigenic process (Yang et al. 2008; Barbolina et al. 2011). Indeed, it was shown that epithelial ovarian cancer cells could directly induce a CAF phenotype (i.e., changes of normal fibroblasts into CAFs) via secretion of transforming growth factor β (Casey et al. 2008; Iwatsuki et al. 2010). At the same time, CAFs may secrete growth factors such as hepatocyte growth factor to promote cancer cell proliferation and invasion (Jing et al. 2011).
Gene expression and, more recently, alternative splicing, a highly regulated and cell-type-specific process, have been found to be globally altered in cancer cells (Sotiriou et al. 2006; Klinck et al. 2008; Venables et al. 2009). Profiling the global expression resulted in several sets of biomarkers capable of detecting cancer subtypes and was particularly successful in differentiating between different breast cancer subtypes (Munirah et al. 2011; Prat et al. 2012). However, most expression profiling techniques focus on changes in the levels of gene expression and simply ignore changes in the transcript architecture due to alternative splicing. Profiling the expression of splicing isoforms identified potential markers for ovarian and breast cancers, and some of these were shown to be required for cell survival in vitro (Klinck et al. 2008; Venables et al. 2008a; Prinos et al. 2011). Hundreds of splicing isoforms associated with the cell cytoskeleton and other tissue-specific genes were associated with ovarian cancer when RNA extracted from whole tumor tissues was compared with that extracted from normal ovaries (Venables et al. 2009). This made the distinction between cancer and tissue-specific markers difficult (Biermann et al. 2007; Venables et al. 2009). Indeed, a recent study suggested that most cancer-associated alternative splicing events (ASEs) identified by comparing whole tumor tissues are not necessarily cancer-specific and mandated the use of different strategies to identify cancer-associated splicing events (Venables et al. 2013).
Recently, enriched cell populations generated by laser capture microdissection (LCM) have been useful in the identification of tumor-specific expression markers (Rogerson et al. 2008; Tone et al. 2008). In this study, we have used this approach to identify a highly specific group of cancer-associated ASEs by isolating tumors and their microenvironment and comparing them with reciprocal normal tissues. Strikingly, ∼20% of the ASEs associated with the tumor microenvironment were regulated by the cancer-associated splicing factors QKI and RBFOX2. Our results indicate that cancer cells reprogram the splicing of a restricted set of functionally related mRNAs in the tumor microenvironment.
RESULTS
Cancer-specific modulation of alternative splicing
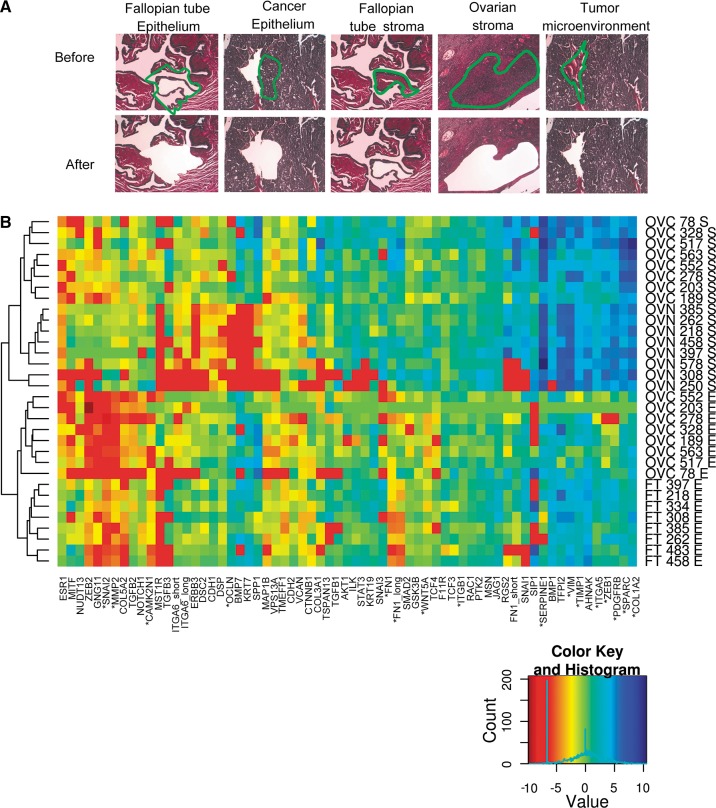
Identifying the molecular characteristics of cancer by comparing tumor and normal tissues is often hampered by tissue heterogeneity. Therefore, we sought to obtain relatively pure cell populations from representative tumor and normal tissues by laser capture microdissection (LCM). Normal ovaries and high-grade serous ovarian tumors were dissected, and visual inspection indicated that homogeneous populations of stromal and epithelial cells were obtained from each tissue (Fig. 1A). Given the low number of epithelial cells in normal ovaries, Fallopian tube, which is considered the origin of serous ovarian cancer (Kurman and Shih Ie 2010; Mehra et al. 2011), was used as a source for normal epithelium. The dissected portions of the tumor microenvironment and normal stroma were mostly composed of fibroblasts (Fig. 1A), and only samples that had a similar number of fibroblasts, immune cells, and endothelial cells were used for RNA extraction (for details, see Materials and Methods). Therefore, despite the overall different cellular composition of normal and tumor-associated stroma, the RNA was extracted from comparable cellular populations. To demonstrate the quality of tissue dissection, the expression levels of established epithelial and stromal markers (Nofech-Mozes et al. 2008) were monitored in the different dissected tissues by quantitative RT-PCR (Fig. 1B; Supplemental Table S1). As expected, all tissues (normal and cancer) clustered according to their stromal and epithelial status, confirming the similar cellular composition of the dissected tissues. Indeed, immunohistochemical analysis of ovarian cancer tissues confirms the predominant expression of stromal markers in the ovarian tumor microenvironment and the expression of the epithelial marker in ovarian cancer cells (http://www.proteinatlas.org; data not shown). Together these tests demonstrate the homogeneity and comparable nature of the normal and cancer cell populations isolated by LCM.
FIGURE 1.
Laser capture microdissection (LCM) of normal and cancer ovarian tissues. (A) Hematoxylin and eosin stain samples from Fallopian tubes, ovaries, and high-grade serous ovarian tumors visualized by light microscopy before and after dissection at 10× magnification. (Green) The limits of dissected tissues; (top) the type of tissues examined. Dissected tissues with similar cell population in both normal and cancer samples were chosen for RNA extraction. Typically, dissected Fallopian tube epithelium was composed of 90% epithelial and 10% fibroblast cells; dissected cancer epithelium was composed of >95% epithelial and <5% fibroblast and other nonepithelial cells (e.g., endothelial and inflammatory cells); dissected ovarian stroma and dissected tumor microenvironment were composed of 60%–80% fibroblast, 10%–20% endothelial, and 10%–20% inflammatory cells (for details, see Materials and Methods). (B) Quality of the dissected tissues. The expression of 65 stromal and epithelial markers was monitored in eight high-grade serous ovarian cancer, eight Fallopian tube epithelia, and eight normal ovarian stroma dissected samples. Relative values were normalized to housekeeping genes as previously described (Prinos et al. 2011). The results are displayed in the form of a heatmap representing the log-transformed gene expression value of epithelial and stromal markers (x-axis) in the different tissues (y-axis). Tissue samples were classified using unsupervised clustering of log-transformed gene expression values using Manhattan distance. (Left) The clustering dendrograms; (right) the type of tissues; (bottom right) the color code representing the gene expression. (Red) Low expression; (blue) high expression. (*) The most predictive markers (P < 0.0001, t-test). Detailed description (Supplemental Table S13) and individual expression values (Supplemental Table S1) of the dissected tissues are provided as Supplemental Material.
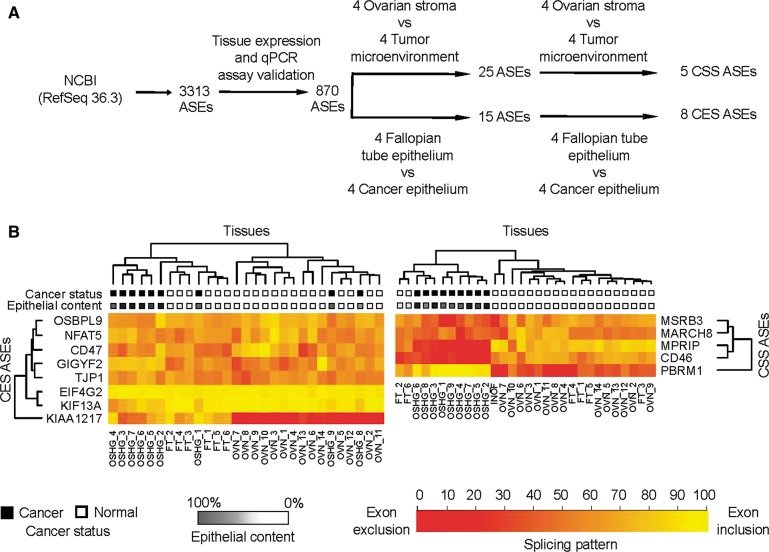
To investigate cancer-specific alternative splicing, we examined the splicing profiles of 870 simple ASEs (cassette exons, alternative 5′ or 3′ splice sites, and intron retention events) found in the RefSeq database (Pruitt et al. 2007) in dissected normal and cancer tissues by quantitative RT-PCR (Fig. 2A). Comparison of the splicing patterns obtained from the Fallopian tube and cancer epithelia identified eight cancer-specific ASEs, referred to hereafter as the cancer epithelial signature (CES) (Supplemental Table S2). On the other hand, comparison of the tumor microenvironment to normal stroma splicing patterns revealed five cancer-specific ASEs, referred to hereafter as the cancer stromal signature (CSS) (Supplemental Table S3). The splicing profiles obtained from dissected tissues displayed distinct patterns that cannot be explained by the average of the patterns obtained from the undissected samples (Supplemental Fig. S1). Consistently, the expression pattern of the CSS splicing events in the normal fibroblast cell line (INOF) was similar to that obtained in normal stroma, confirming that the dissected tissues obtained from normal stroma represent normal fibroblast cells (Fig. 2B, right panel). We conclude that the changes in alternative splicing not only distinguish tumor from normal epithelial tissues, but also permit discrimination between the tumor microenvironment and normal stroma.
FIGURE 2.
Identification of alternative splicing events associated with ovarian tumor and its microenvironment. (A) Strategy for the identification of cancer-specific alternative splicing patterns. Expression of the 3313 simple ASEs (i.e., cassette exons, alternative 5′ and 3′ splice sites, and intron retention) annotated in the RefSeq database 36.3 (Pruitt et al. 2007) was examined using endpoint PCR amplification of five RNA samples extracted from normal Fallopian tube tissue and ovarian cancer tissues (for details, see Materials and Methods; see raw data in http://palace.lgfus.ca/pcrreactiongroup/list/226). Validated quantitative RT-PCR assays were developed for a total of 870 expressed ASEs, and the resulting PCR values were used to calculate the mean quantitative splicing shift (Prinos et al. 2011) (ΔQψ = QψTUMOR − QψNORMAL) in the different normal and cancer tissues described in Figure 1 and Supplemental Table S13. ASEs were ranked based on a t-test of the mean quantitative splicing shift, and ASEs displaying statistically significant differences between normal and cancer tissues (P < 0.05, t-test) and a ΔQψ > 15 were reexamined in an independent set of dissected tissues (for details, see Materials and Methods). The final number of validated ASEs capable of discriminating between normal and cancer epithelium (CES) or stroma (CSS) is indicated. (B) Splicing markers detected in undissected cancer tissues and immortalized normal ovarian fibroblast (INOF) cell line. The quantitative percent splicing pattern (Qψ) (Prinos et al. 2011) of the CES and CSS ASEs was determined by quantitative RT-PCR in an independent set of undissected ovarian tissues containing nine high-grade serous ovarian cancers, six Fallopian tubes, and 14 normal ovarian tissues (Supplemental Table S14). In addition, we also monitored splicing in INOF as a pure source of normal ovarian fibroblast. The results are displayed in the form of two heatmaps representing the splicing patterns of the CES (left panel) and the CSS (right panel) in the different tissues. Gene names and the gene clusters are shown on the y-axis. The type of tissue—(OSHG) ovarian serous high grade, (OVN) normal ovary, (FT) Fallopian tube—is indicated at the bottom, and the tissue clusters are shown on top. (Black boxes) Cancer and (white boxes) normal tissues are indicated. (Grayscale) The epithelial content of tissues; (dark gray) high epithelial content; (light gray) low epithelial content. The color code representing the different splicing patterns is indicated at the bottom; (dark orange) complete exon exclusion; (bright yellow) complete exon inclusion.
The potential clinical usefulness of the newly identified cancer-specific signatures was further evaluated by monitoring their capacity to distinguish between undissected normal and cancer tissues. RNA was extracted from 14 normal ovarian tissues, six Fallopian tubes, and nine high-grade ovarian cancer tumors (for details, see Supplemental Table S14). As shown in Figure 2B (left panel), unsupervised clustering of the tissues based on the splicing pattern of the CES ASEs separated epithelial and stromal tissues into two distinct groups. Indeed, the tissues extracted from normal Fallopian tubes were clustered closer to cancer than normal tissues, consistent with the low epithelial contents in normal ovary. This indicates that while CES may identify cancer tissues, it is sensitive to changes in tissue composition (epithelial/mesenchymal). In general, the CES events discriminated between Fallopian tubes and cancer tissues with P-values >0.005 (t-test) with the exception of KIF13A, which distinguished between cancer and Fallopian tube tissues with a P-value of 3 × 10−4 (t-test) (Supplemental Table S2). We conclude that at least one of the epithelial markers identified using dissected tissues could also efficiently discriminate between undissected normal and cancer tissues.
Interestingly, the stromal signature (CSS) events clustered most Fallopian tubes with the mostly stromal normal ovarian tissues in one group and cancer tissues in another (Fig. 2B, right panel), suggesting that CSS events are not biased by the epithelial and stromal contents, which are very different in tissues extracted from Fallopian tubes and normal ovary. Consistently, three out of the five CSS splicing events (CD46, MPRIP, and PBRM1) distinguished between normal and cancer tissues with P-values <1.0 × 10−7 (t-test) (Supplemental Table S3). This group of splicing events was named the ovarian cancer signature (OCS) and was further tested for its capacity to identify ductal breast cancer (Supplemental Table S4). Two of these ASEs (CD46 and PBRM1) were able to discriminate between 18 normal samples and 20 ductal breast cancers with P-values <1.0 × 10−7 (t-test), further confirming the value of the OCS (Supplemental Table S4). These results suggest that splicing signatures identified in the tumor microenvironment could serve as cancer biomarkers.
Cancer-specific splicing factors
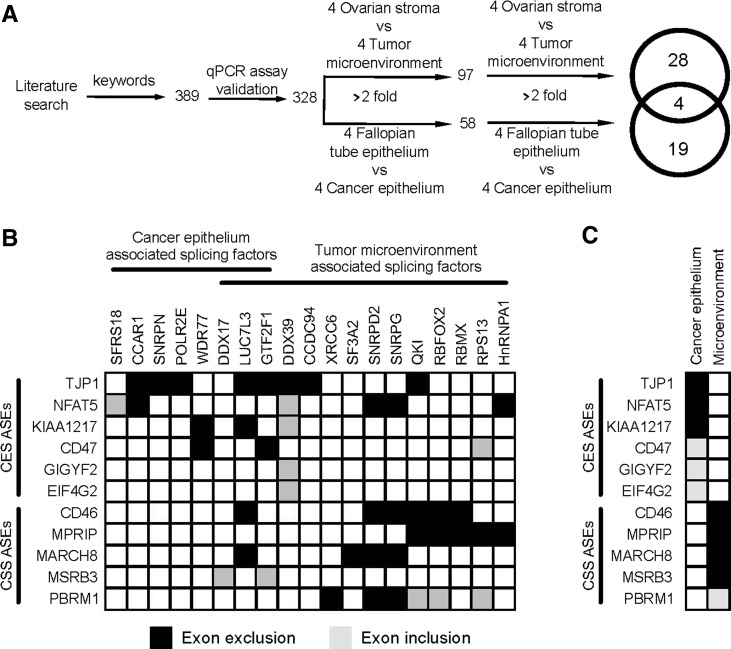
To understand how the CES and CSS are produced, we examined the expression of 328 putative splicing factors in epithelial and stromal cells isolated from normal and cancer tissues using quantitative RT-PCR (raw data are accessible through http://palace.lgfus.ca/pcrreactiongroup/list/227). Comparison of the RNA levels in normal and cancer tissues identified 23 and 32 splicing factors whose expression was changed in the cancer epithelium and the tumor microenvironment, respectively (Fig. 3A; Supplemental Tables S5, S6). The overlap between the tumor epithelium and its microenvironment was small (8%, four out of 51) (Fig. 3A), as would be expected from tissues with distinct profiles of alternative splicing (Fig. 2A). The relatively small numbers of splicing factors associated with the tumor (7%, 23 out of 328) and its microenvironment (10%, 32 out of 328) indicates that the changes in the expression of splicing factors is not due to global perturbation of gene expression in cancer tissues or due to differences in the cell composition of the cancer tissues (Fig. 1A). Together these observations suggest that cancer alters the expression of a subset of splicing factors to reprogram the splicing of a specific group of alternative exons associated with the tumor microenvironment.
FIGURE 3.
The expression of cancer-associated splicing isoforms is regulated by a small group of splicing factors. (A) Strategy for the identification of cancer-associated splicing factors. The expression of all splicing factors identified in both the RefSeq database and the NCBI PubMed database (Lu 2011) was evaluated by quantitative RT-PCR in the different normal and cancer tissues described in Figure 1. The Venn diagram illustrates the tissue distribution of the splicing factors capable of discriminating between cancer and normal tissues by at least twofold (listed in Supplemental Table S6). (B) Depletion of the tumor-associated splicing factors alters the splicing pattern of the cancer stromal signature (CSS). The cancer-associated splicing factors were depleted using siRNA in the ovarian cancer cell line SKOV3ip1. The impact on the CES and CSS ASEs identified in Figure 2 was evaluated using quantitative RT-PCR. Exons inclusion (gray boxes) or exclusion (black boxes) generating a quantitative splicing shift (Prinos et al. 2011) (ΔQψ = QψKNOCKDOWN − QψCONTROL) of at least 10 was considered significant and presented in the form of a table. For simplicity, only the ASEs regulated by at least one splicing factor and the splicing factors regulating at least one ASE are shown. The expression of all splicing factors except DDX39 was down-regulated in cancer tissues, and therefore the illustrated in vitro knockdown of these factors are expected to induce a splicing pattern similar to that detected in cancer tissues. (C) Illustration of the CES and CSS exon exclusion and inclusion events in the cancer epithelium and tumor microenvironment as detected by quantitative RT-PCR (see Supplemental Tables S2, S3). The cancer-associated genes were listed in the same order used in B, and their expression in cancer tissues is indicated as exon exclusion (black boxes) and inclusion (gray boxes).
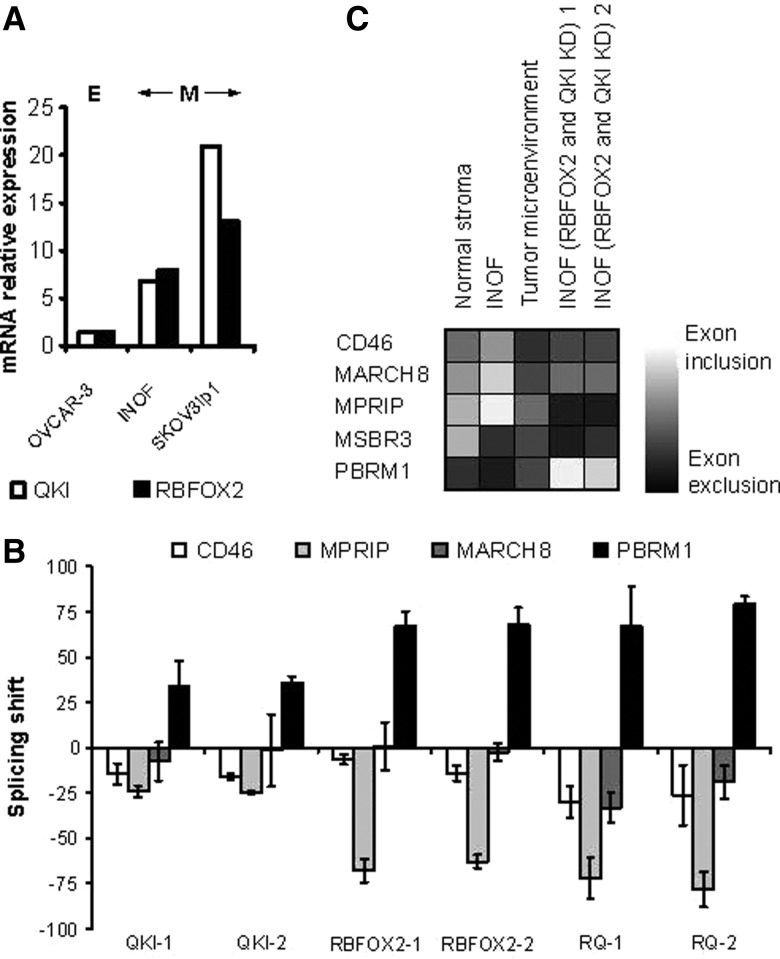
Most of the exons associated with CES (five out of eight) or CSS (four out of five) are excluded in cancer samples (Supplemental Tables S2, S3), and all the cancer-associated splicing factors presented in Figure 3B, with the exception of DDX39, are down-regulated in cancer. Therefore, silencing splicing factors in cancer cell lines is expected to create a splicing pattern similar to that observed in cancer tissues. Accordingly, we depleted 41 out of 51 cancer-associated splicing factors by at least twofold using two independent siRNAs in the model ovarian cancer cell line SKOV3ip1 (Supplemental Table S7; raw data are accessible through http://palace.lgfus.ca/pcrreactiongroup/list/232) and tested the impact on the splicing pattern of both CES and CSS ASEs. The SKOV3ip1 cell line was chosen as a model because it is an epithelial cell line that exhibits mesenchymal character in culture and expresses many of the CES and CSS splicing events (Chen et al. 2001). Indeed, the expression level of the tumor microenvironment–associated splicing factors QKI and RBFOX2 was highest in SKOV3ip1 followed by the ovarian fibroblast cell line INOF and was lowest in the epithelial cancer cell line OVCAR-3 (Fig. 4A). The knockdown of the splicing factors linked to cancer epithelium affected mostly CES events (Fig. 3B), while those repressed in the tumor microenvironment affected CSS events in both SKOV3ip1 and INOF (Figs. 3B, 4B). As expected, in 78% (18 out of 23) of the cases, the depletion of splicing factors in SKOV3ip1 altered splicing in the same direction as that observed in cancer tissues (cf. Fig. 3, B and C). These results demonstrate that the change in cancer-associated splicing is due at least in part to changes in the expression of splicing factors.
FIGURE 4.
RBFOX2 and QKI regulate the splicing of the CSS ASEs in a normal ovarian fibroblast cell line. (A) RBFOX2 and QKI expression levels in epithelial and fibroblast cell line. Global expression levels of QKI and RBFOX2 were monitored using quantitative RT-PCR in the immortalized normal ovarian fibroblast cell line (INOF) and compared with that obtained from the epithelial tumor cell lines OVCAR-3 and SKOV3ip1. SKOV3ip1 is an ovarian cancer cell line from epithelial origin that displays mesenchymal characters in cell culture. (B) Impact of RBFOX2 and QKI on the splicing of the CSS ASEs in the INOF cell line. The RNA was extracted from INOF cells transfected by two different siRNAs against QKI (QKI-1 and QKI-2), RBFOX2 (RBFOX2-1 and RBFOX2-2), or both QKI and RBFOX2 (RQ-1 and RQ-2). Shown is a bar graph representing the splicing shift of the different CSS events 72 h after transfection relative to mock-transfected cells by quantitative RT-PCR. The results are the average of three independent experiments. (C) Illustration of the CSS exon exclusion and inclusion events in the normal stroma and tumor microenvironment (see Supplemental Table S3) and INOF cell line (see B) as detected by quantitative RT-PCR. Grayscale with (black boxes) total exon exclusion and (white boxes) total exon inclusion is presented in the form of a table.
Only three CES splicing events (TJP1, NFAT5, KIAA1217) were affected by splicing factors that were associated with the splicing epithelium and not the tumor microenvironment. Furthermore, no more than one CES event was affected by multiple splicing factors, suggesting that this set of CES events is not regulated by common factors (Fig. 3B). Closer examination of the effects of knocking down cancer-associated splicing factors on CSS ASEs revealed two groups of splicing factors. The first group includes core spliceosomal factors such as SNRPD2 (Schumperli and Pillai 2004) and SNRPG (Ma et al. 2005), while the second includes alternative splicing regulators RBFOX2 (Sun et al. 2012) and QKI (Novikov et al. 2011). Both groups are underexpressed in the tumor microenvironment and modified the splicing of three out of five CSS ASEs in the SKOV3ip1 cell line (Fig. 3B), but splicing factors from the first group also induced apoptosis (data not shown), as would be expected from the knockdown of essential splicing factors. Induction of apoptosis and general perturbation of splicing patterns make the distinction between direct and indirect effects very difficult, and as such this group of splicing factors was not pursued further. In contrast, reproducing the tumor microenvironment–associated down-regulation of RBFOX2 or QKI in SKOV3ip1 and the immortalized normal ovarian fibroblast (INOF) cells (Lawrenson et al. 2012), replicated the cancer-associated splicing profile of three out of five CSS ASEs without dramatically affecting cell viability (cf. Figs. 3B,C and 4C; data not shown). This indicates that a part of the splicing profiles observed in the tumor microenvironment is reproduced by the inhibition of RBFOX2 and QKI expression.
The tumor microenvironment–associated splicing factors QKI and RBFOX2 regulate common splicing targets
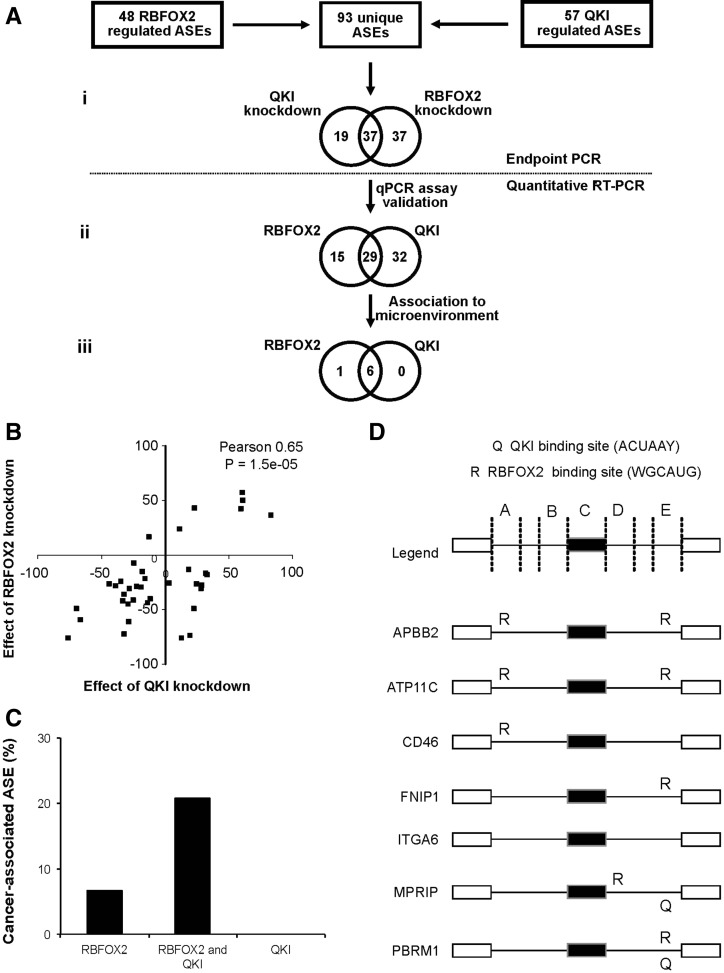
To determine the extent of overlap between RBFOX2 and QKI splicing targets and uncover potentially new cancer-associated splicing events, we compared the effects of depleting the expression of these two proteins on a larger set of ASEs. We monitored the splicing profiles of a total of 93 unique ASEs (Fig. 5Ai) containing 48 pre-established RBFOX2-regulated ASEs (Venables et al. 2009) and 57 newly identified QKI-regulated ASEs (for details, see Materials and Methods) after QKI or RBFOX2 knockdown in SKOV3ip1 (Supplemental Table S8, columns 3–5). As indicated in Figure 5A, 66% (37 out of 56) of ASEs modified by RBFOX2 knockdown were also affected by the depletion of QKI, and half (37 out of 74) of the QKI-regulated ASEs were perturbed by RBFOX2 knockdown. The effect of RBFOX2 and QKI knockdown on the common group of ASEs (Fig. 5A) displayed a Pearson correlation of 0.65 (P-value = 1.5 × 10−5), confirming their status as shared splicing targets (Fig. 5B). This indicates that there is a broad overlap between QKI and RBFOX2 splicing targets. In the tumor microenvironment, the expression of QKI and RBFOX2 is down-regulated (Supplemental Table S5), and their expression levels are correlated with the changes in alternative splicing. Therefore, while RBFOX2 and QKI may individually affect different splicing targets, collectively they converge to regulate the splicing of a subset of the tumor microenvironment–associated ASEs.
FIGURE 5.
RBFOX2 and QKI regulate the expression of common splicing isoforms in the tumor microenvironment. (A) Identification of common RBFOX2 and QKI splicing targets. RBFOX2 and QKI were individually knocked down using two independent siRNAs in SKOV3ip1. The effect of the different knockdowns was evaluated by endpoint PCR using a set of 48 pre-established RBFOX2 targets and a set of 57 newly identified QKI targets (for details, see Materials and Methods). The 93 unique ASEs responding to the knockdown of at least one splicing factor are presented in the form of Venn diagram (i) (Supplemental Table S8, column 5). Quantitative RT-PCR primers were designed and validated for 76 out of 93 ASEs (ii) (Supplemental Table S8, column 6), and their splicing pattern in the tumor microenvironment or the epithelial and stromal normal tissues was tested (iii). (B) Comparison of the impact of RBFOX2 and QKI knockdown on the splicing of 37 common ASEs. The impact of RBFOX2 and QKI knockdown on the common set of 37 splicing targets discovered in A was plotted as a quantitative splicing shift (Prinos et al. 2011) (ΔQψ = QψKNOCKDOWN − QψCONTROL) to generate a scatter graph. The Pearson correlation between the effect of RBFOX2 and QKI knockdown on splicing pattern and its P-value is indicated on the top right of the graph. (C) Common RBFOX2 and QKI targets are more likely to be associated with the tumor microenvironment than those affected by only one splicing factor. The bar graphs represent the percentage of the RBFOX2, QKI, or common RBFOX2 and QKI splicing targets (identified in A) associated with the tumor microenvironment as described in Figure 2A. (D) Schematic representation of the protein binding sites near RBFOX2- and QKI-responsive exons. The position of RBFOX2 and QKI binding sites (WGCAUG and ACUAAY) in seven common RBFOX2 and QKI splicing targets associated with the tumor microenvironment (identified in Fig. 3B and in panel Aiii) are indicated as “R” and “Q,” respectively. The existence of binding sites was verified in five regions: 250 nucleotides (nt) from the 5′ splice site of the upstream intron (a); 250 nt from the 3′ splice site of the upstream intron (b); within the exon (c); 250 nt from the 5′ splice site of the downstream intron (d); or 250 nt from the 3′ splice site of the downstream intron (e).
If RBFOX2 and QKI regulate splicing in the tumor microenvironment, we expect at least some of the alternative splicing events affected by the knockdown of these proteins in vitro to be associated with cancer in vivo. Accordingly, we monitored the splicing profiles of RBFOX2- and QKI-dependent ASEs in the tumor microenvironment. Out of 29 ASEs tested, six ASEs including two that overlap with Figure 2A (PBRM1 and MPRIP) were associated with the tumor microenvironment (Supplemental Table S9). Indeed, these four additional ASEs (APBB2, ATP11C, ITGA6, FNIP1) could be considered potential markers for ovarian cancer. However, for simplicity, they were not incorporated in the CSS, which refer only to ASEs identified in the original association screen using dissected tissues (Fig. 2A). Overall, 21% (six out of 29) of the RBFOX2- and QKI-dependent ASEs were associated with the tumor microenvironment, whereas <7% (one out of 15) of the ASEs affected by only one of the two splicing factors were linked to cancer (Fig. 5C). This indicates that while a substantial fraction of ASEs under the control of RBFOX2 and QKI are associated with the tumor microenvironment, a large number of RBFOX2 and QKI in vitro splicing targets are not. Together, these results suggest that while RBFOX2 and QKI are important regulators of cancer-associated transcript variation, other factors influence their capacity to modulate the splicing of their target genes in the tumor microenvironment. Examining the sequence surrounding the RBFOX2- and QKI-dependent ASEs revealed consensus binding motifs (Galarneau and Richard 2005; Auweter et al. 2006) for at least one of the two splicing factors, in all cases except ITGA6 (Fig. 5D). This suggests that RBFOX2 and QKI directly contribute to the splicing decisions of a subset of tumor microenvironment–associated events.
The ovarian tumor microenvironment–associated splice variants are differentially expressed in breast cancer
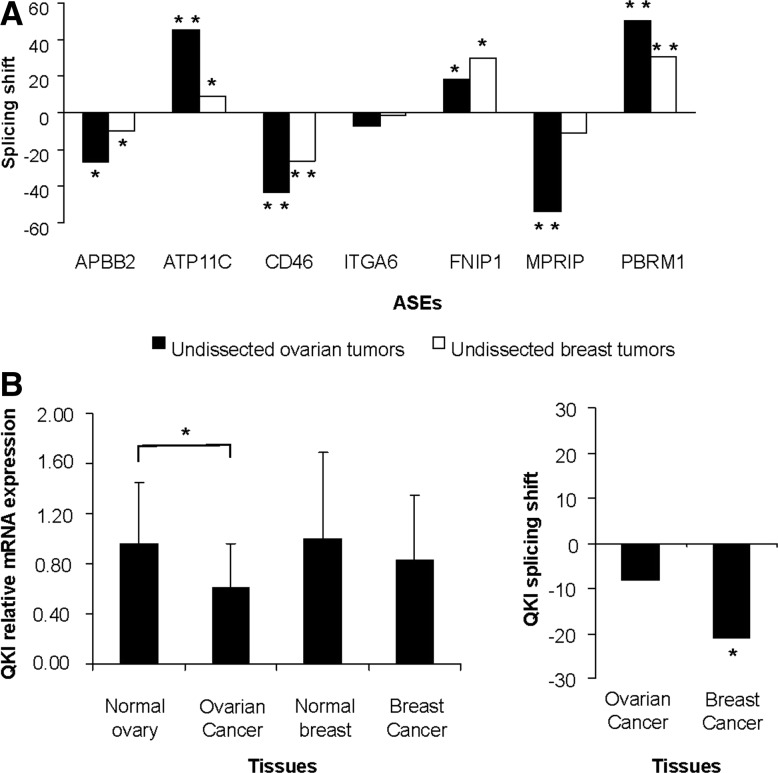
It was previously suggested that RBFOX2 might be inactivated in breast cancer through the skipping of a 40-nt C-terminal cassette exon (exon 11) (Underwood et al. 2005; Venables et al. 2009). Therefore, at least some of the RBFOX2 splicing targets, including those associated with the ovarian tumor microenvironment, are likely to be similarly modified in breast cancer. To examine this possibility, we compared the splicing pattern of the set of RBFOX2- and QKI-dependent ASEs associated with the tumor microenvironment in undissected ovarian and breast tumors using quantitative RT-PCR. As expected, the splicing patterns in both ovarian and breast cancers were tightly correlated, indicating that RBFOX2 and QKI splicing targets are also repressed in high-grade breast cancer (Fig. 6A; Supplemental Table S4), and suggesting that breast and ovarian cancer use similar mechanisms for modifying the splicing patterns of tumor-associated ASEs. Consistently, an independent study using a large number of breast cancer tissues concluded that QKI expression is reduced in breast tumors (P-value <3.0 × 10−3, t-test) (Novikov et al. 2011). In our hands, however, the most striking difference between tumor and normal breast tissues was not the change in global expression but rather in the pattern of QKI splicing (Fig. 6B; Supplemental Tables S10, S11). In the tumor, the QKI-6 isoform was preferentially expressed over the QKI-7 isoform. This change was previously shown to affect QKI activity (Pilotte et al. 2001). Therefore, both changes in the expression and splicing of QKI are altered depending on the type of cancer or cancer subtype examined. Together the data suggest that RBFOX2 and QKI play an important role in regulating the splicing of a subset of tumor microenvironment–associated events in both ovarian and breast cancer.
FIGURE 6.
The common RBFOX2 and QKI splicing targets are deregulated in both ovarian and breast cancer. (A) The behavior of seven common RBFOX2 and QKI splicing targets associated with the tumor microenvironment was monitored using quantitative RT-PCR in 14 normal and 13 serous high-grade ovarian cancer tissues and their quantitative splicing pattern (Qψ) (Prinos et al. 2011) compared with that detected in 18 normal and 20 ductal breast cancer tissues (Supplemental Table S4). Shown are histograms representing the mean quantitative splicing shift (Prinos et al. 2011) (ΔQψ = QψTUMOR − QψNORMAL). (B) QKI expression is altered by different mechanisms in ovarian and breast cancer. The expression (left panel) (Supplemental Table S11) and splicing pattern (right panel) (Supplemental Table S10) of QKI was monitored by quantitative RT-PCR in breast and ovarian samples as described in A. The global expression pattern was calculated relative to housekeeping genes as previously described (Prinos et al. 2011), and the relative value is presented in the form of a bar graph. The splicing pattern of QKI isoforms 6 (short) and 7 (long) was calculated as the mean quantitative splicing shift (Prinos et al. 2011) (ΔQψ = QψTUMOR − QψNORMAL) as previously described in A and is plotted in the form of bar graphs. When significant, the P-value (t-test) of difference in expression or splicing shift is displayed on top of the histogram by asterisks ([*] P < 0.05; [**] P < 5.0 × 10−7).
DISCUSSION
Recent studies suggested that the majority of splicing events identified by comparing whole normal with tumor tissues are mostly tissue-specific events that are not necessarily linked to cancer (Venables et al. 2013). This finding raised the questions of whether or not a true cancer-specific splicing program exists and mandated a more carefully designed procedure to identify cancer-specific splice markers. In this study, we compared the splicing program of a histologically homogeneous population of normal and cancer cells with the goal of identifying cancer-specific splicing isoforms that are independent of tissue composition. The results indicate that differences between normal and cancer tissues are much smaller than previously thought. Analyzing the expression of a subset of the RefSeq alternative splicing events in RNA extracted from whole ovarian tumors identified 336 cancer-associated ASEs (Klinck et al. 2008; Venables et al. 2009), while comparison of normal and cancer epithelial or stromal tissues identified a total of 17 cancer-specific ASEs (Supplemental Tables S2, S3, S9). Most of the 336 whole tumor markers affected cell-type-specific genes including those linked to cell plasticity (MAP3K7) and cell movement (CEACAM1) (Venables et al. 2009). On the other hand, the splicing events identified in genes with known function using dissected cancer epithelium were associated with tumorigenesis (Supplemental Table S12). Splicing events associated with the tumor microenvironment did not display common cancer-related function (Supplemental Table S12). Therefore, while whole tumor screens identified cell-type-specific genes, comparison of samples with similar cell composition produced gene signatures that reflect the contribution of each tissue to tumor function (e.g., cell growth and migration in the epithelium and cell metabolism and immune response in the tumor microenvironment). Consistently, the overlap between the cancer markers identified by the screens conducted using whole and dissected tissue was limited to five ASEs. Surprisingly, the overlap between the ASEs identified using whole tissues and dissected stromal tissue was greater than that between whole tissues and dissected cancer epithelium. The relatively low overlap between epithelium and whole tissue–based screens suggests that most of the ASEs identified in whole tumor screens are generated by the marked difference in the level of epithelial content extracted from tumor and normal ovarian tissues.
Combinatorial regulation of alternative splicing in cancer
Reducing the expression of splicing factors in any given cell line leads to changes in a large number of ASEs, few of which can be associated with a given condition in vivo. In our hands, repression of QKI or RBFOX2 in vitro changed the splicing pattern of a total of 39 common splicing targets in the SKOV3ip1 ovarian adenocarcinoma cell line (Figs. 3B, 5A; Supplemental Table S8, columns 3–5). However, only a small subset of these was associated with the tumor microenvironment (Figs. 2A, 5C). This may reflect differences between cell lines grown in vitro and from patient tissues. The splicing events affected by QKI and RBFOX2 knockdowns in vitro might be affected by other cancer-associated splicing factors not present in the cell lines tested. Indeed, the outcome of any splicing target is determined by many splicing factors that may vary depending on the cell line and conditions tested. For examples, RBFOX2 and RBFOX1 splicing activities are affected by hnRNP H1 and TFG in brain (Sun et al. 2012). On the other hand, RBFOX2 and the neuron-specific splicing factor NOVA regulate common splicing targets in neuronal tissues (Zhang et al. 2010). It was also shown that the splicing factor QKI controls the expression of other splicing factors like hnRNP A1, suggesting that one splicing factor may lead to a cascade of changes in splicing that goes beyond its direct splicing targets (Zearfoss et al. 2011). In the case of ovarian cancer, the effect of QKI on splicing appears not to be mediated by hnRNP A1 because, despite the fact that the expression level of hnRNP A1 is altered in the tumor microenvironment (Supplemental Table S5, column 3), its depletion affects the splicing of only one common target (MPRIP) of RBFOX2 and QKI in SKOV3ip1 (Fig. 3B). In addition, knocking down QKI in SKOV3ip1 did not significantly increase the level of RBFOX2 cancer-associated isoform and vice versa (Supplemental Fig. S5). Therefore, while our data point to a group of splicing targets that are specifically regulated by QKI and RBFOX2 in the tumor microenvironment, it is likely that the generation of the cancer splicing program involves a larger number of factors that together define the splicing outcome in the tumor microenvironment.
Impact of cancer types on the expression of the ovarian tumor microenvironment–associated splicing events
The cancer stromal signature (CSS) discovered in ovarian cancer was also detected in breast cancer, indicating a similarity in the splicing patterns between these types of cancer. The CSS was tested in a variety of breast cancer types including high-grade estrogen-receptor-positive and -negative ductal breast cancer samples (Fig. 6). Overall, the splicing profile of these ASEs was similar to that detected in the microenvironment of ovarian tumors regardless of the estrogen receptor status or cancer subtype, underlining the generality of the newly discovered signature. Earlier study of luminal (estrogen-receptor-positive) and basal (estrogen-receptor-negative) breast cancer cell lines identified cancer-subtype-specific splicing signatures (Lapuk et al. 2010; Shapiro et al. 2011). In contrast, we did not find any statistically significant differences in the splicing patterns of RBFOX2- and QKI-dependent ASEs in estrogen-receptor-positive and -negative breast cancer subtypes (Supplemental Table S4). This contradiction might be due to differences between the minimalistic, in vitro growth conditions of monolayer cell cultures and the intricate, three-dimensional complex growth of cells in patient tissues in vivo or the number and nature of splicing events examined. The expression pattern of splicing factors was also perturbed in both ovarian and breast cancer but in different ways: In ovarian cancer, the expression of RBFOX2 and QKI was down-regulated, while their splicing patterns were altered in breast cancer (Fig. 6; Venables et al. 2009). Other studies of QKI expression in breast cancer suggested that QKI might also be down-regulated in breast cancer (Novikov et al. 2011), suggesting that the expression of QKI varies between breast cancer tumors. Therefore, while there is similarity between the breast and ovarian cancer splicing patterns, the way these patterns are produced in each type of cancer may be different.
Regulation of alternative splicing in the tumor microenvironment
Oncogenesis and tumor progression rely on a reciprocal autocrine communication program with the tumor microenvironment (Bhowmick et al. 2004). This communication results in a tumor microenvironment replete with inflammatory mediators, growth factors, matrix remodeling enzymes, and angiogenic factors (Tlsty and Coussens 2006). Growth factors produced by many kinds of cancer cells including breast and ovarian carcinomas attract fibroblasts and stimulate their proliferation (Orimo and Weinberg 2007). Differentiation of fibroblasts in the vicinity of cancer cells to myofibroblasts (i.e., activated fibroblasts) is important for tumor growth and has been observed in several in vitro studies using ovarian cells (Schauer et al. 2011). This transformation is required for tumor development since fibroblasts derived from the omentum, the richly vascularized fatty subperitoneal layer surrounding the ovaries, increased ovarian cancer cell adhesion and invasive behavior, whereas omentum-derived mesothelial cells reduced growth of ovarian cancer cells (Kenny et al. 2007). In this study, we uncovered a new layer of tumor-dependent modification of stromal cells defined by changes in alternative splicing. These changes occur preferentially in the tumor microenvironment and include differences in the splicing of genes related to diverse functions linked to cancer development and were associated with changes in the expression of splicing factors. Interestingly, the cancer-specific splicing markers that are least sensitive to tissue composition are found in the tumor microenvironment and not the tumor itself, suggesting that cancer can be detected more readily by changes in the microenvironment that may even take place early during tumorigenesis. However, it is currently unclear whether these changes in stromal cells precede tumor development or occur after the tumor is established. In vitro studies using cell and animal models will be required to clarify this issue. In any case, the results presented here pave the way for in-depth studies of a new level of tumor-dependent modification of gene expression in stromal cells. Altogether, it suggests that cancer cells modulate their environment in a specific and programmed fashion and that the monitoring of splicing provides sensitive and specific markers that may prove to have clinical value.
MATERIALS AND METHODS
Tissue selection
Clinically homogeneous (high-grade, high-stage, and chemonaïve) tumors were selected for both dissected and undissected analyses. Tissue sourcing was performed as previously described (Klinck et al. 2008; Venables et al. 2008a) and separated into six sample sets (see Supplemental Tables S13–S15):
the active ASEs set containing one Fallopian tube, one low malignant potential, one serous ovarian cancer grade 1, one serous ovarian cancer grade 3, and one ovarian clear cell carcinoma;
the LCM discovery set containing four normal ovarian stroma, four Fallopian tube epithelia, four cancer epithelia, and four tumor microenvironment samples;
the LCM validation set containing four normal ovarian stroma, four Fallopian tube epithelia, four cancer epithelia, and four tumor microenvironment samples;
the fresh frozen (FF) and formalin-fixed, paraffin-embedded (FFPE) matched sample sets containing mirror images of four undissected normal ovaries and four undissected ovarian serous carcinomas;
the undissected serous ovarian sample set composed of 14 undissected normal ovaries, six Fallopian tube epithelia (FT), and 13 undissected ovarian serous carcinomas; and
the undissected ductal breast sample set composed of 18 normal breast tissues and 20 ductal breast cancer samples.
Dissected regions were selected based on the epithelium (hematoxylin) and stromal cell (eosin) specific staining patterns. Cancer epithelial cells were selected based on their morphology (cylindrical or cubic cells with round to ovoid nuclei) and the absence of necrosis. Cells in the tumor microenvironment were selected based on their morphology (spindle cells with spindle nuclei) from the region adjacent to tumors visualized at 10× magnification.
Cell culture, transfection, RNA extraction, and quantitative RT-PCR
Cell culture, transfection, RNA extraction, and quantitative RT-PCR were performed as previously described (Brosseau et al. 2010; Prinos et al. 2011) except for FFPE and LCM samples. LCM samples isolated using several successive 8-µm cuts from FFPE blocs of each LCM sample were deposited on membrane slides (Molecular Machines & Industries). In general, one to three cuts were used for ovarian stroma and cancer epithelia, while three to five and eight to 15 cuts were used for tumor microenvironment and Fallopian tube, respectively. Dissected tissues with similar cell populations in both normal and cancer samples were chosen for RNA extraction. Typically, dissected Fallopian tube epithelium was composed of 90% epithelial and 10% fibroblast cells; dissected cancer epithelium was composed of >95% epithelial and <5% fibroblast and other nonepithelial cells (e.g., endothelial and inflammatory cells); dissected Fallopian tube stroma, dissected ovarian stroma, and dissected tumor microenvironment were composed of 60%–80% fibroblast, 10%–20% endothelial, and 10%–20% inflammatory cells. LCM slides were stained with hematoxylin and eosin (Fischer) and were dissected using MMICellCut (Molecular & Machines Industries). Samples with the same cell type and tissue source were pooled. RNA extraction was performed using the RNeasy FFPE kit (Qiagen) with the following modifications: Dissected materials were incubated in Aqua DePar 1× (Davis Diagnostics) for 15 min at 95°C, then lysed by incubation in tris-guanidinium thiocyanate 4 M (Sigma-Aldrich) and 10 µL of proteinase K (Qiagen) for 15 min at 55°C followed by 15 min of incubation at 80°C. Finally, demodification was achieved by the addition of NH4Cl (Sigma-Aldrich) to a final concentration of 2.5 M and incubation for 20 min at 95°C. DNase treatment was performed using an RNase-free DNase set (Qiagen). The resulting RNA (250 ng/reaction) was subjected to a single round of 25 PCR cycles of amplification using the Sigma-Aldrich whole transcriptome amplification kit (Sigma-Aldrich). The resulting DNA libraries were purified using the QIAquick PCR purification kit (Qiagen) and diluted to a final volume of 12.5 mL. Global (Prinos et al. 2011) and isoform-specific (Brosseau et al. 2010) quantitative RT-PCR primers were designed and validated as previously described (Brosseau et al. 2010). The reliability of the gene expression assay using RNA samples from archived material was evaluated by comparing global gene expression (Supplemental Fig. S2A), technical duplicates of amplification (Supplemental Fig. S2B), and splicing pattern (Supplemental Fig. S3) obtained from the fresh-frozen (FF) and fresh-frozen formalin-fixed, paraffin-embedded (FFPE) matched sample set and different methodology (e.g., quantitative RT-PCR vs. endpoint PCR) (see Supplemental Figs. S2C, S3). The quality of the tissue dissection was verified visually (Fig. 1A) and through the expression of known cell-type markers (Fig. 1B). The expression levels of the different markers did not vary greatly between tissues of the same type, confirming the absence of random sample-to-sample variations in cell type (Fig. 1B; Supplemental Table S1). Furthermore, the changes in gene expression observed in tumor samples were gene-specific and not marker type-specific (e.g., epithelial or stromal) (see Fig. 1; Supplemental Table S1), confirming that any changes observed in cancer tissues are not due to variation in the level of cell type contaminations. All raw PCR data are available through http://palace.lgfus.ca/data/related/844.
Primer design and alternative splicing screens
The previously described LISA platform (LISA) (Klinck et al. 2008) was updated with RefSeq database build 36.3 (Pruitt et al. 2007) and used for automatic identification of ASEs and the design of isoform-specific PCR primers as described before (Venables et al. 2009). ASEs with strong evidence of the expression of both splicing isoforms (active ASEs) were identified by endpoint PCR using four individual ovarian tissues (one clear cell, one low malignant potential, one serous grade 1, and one serous grade 3) and one Fallopian tube tissue as previously described (Venables et al. 2009). Isoform-specific quantitative RT-PCR primers were designed for all active ASEs and validated using a universal human reference RNA as described earlier (Brosseau et al. 2010). Primer pairs showing no PCR amplifications were subsequently tested using RNAs extracted from the FFPE pool that include two normal ovaries and two serous ovarian cancer tissues, totalizing 1740 validated primer pairs (covering 870 ASEs). The screen for cancer-specific ASE markers was performed in two steps: (1) the discovery screen, which consists of surveying 870 ASEs in the LCM discovery sample set and (2) the validation screen, which assesses the expression of all ASEs obtained from the discovery screen in an independent LCM sample set. Splicing pattern (Qψ) values were calculated as described elsewhere (Prinos et al. 2011). In the discovery screen, ASEs were selected for further studies if they were expressed with a splicing shift (ΔQψ = QψTUMOR MICROENVIRONMENT − QψOVARIAN STROMA or QψCANCER EPITHELIUM − QψFALLOPIAN TUBE EPITHELIUM) of at least 15 in at least three out of four tissues. A t-test was used to rank ASEs, and an arbitrary cut-off was applied at P < 0.05. In the validation screen, ASEs were considered cancer-associated if they were expressed in at least three out of four tissues with a splicing shift (ΔQψ = QψTUMOR MICROENVIRONMENT − QψOVARIAN STROMA or QψCANCER EPITHELIUM − QψFALLOPIAN TUBE EPITHELIUM) of at least 15. The validity of the markers identified in this screen and their capacity to detect cancer were further evaluated using a third independent set of undissected ovarian serous tumors. ASEs association with cancer was considered specific if they displayed a splicing shift (ΔQψ = QψOVARIAN CANCER − QψNORMAL OVARY or QψOVARIAN CANCER − QψNORMAL FALLOPIAN TUBE) of at least 15 and a P < 0.005 (t-test). Data clustering were performed using R statistical software (http://www.r-project.org).
Splicing factors screen
Search of the NCBI literature database PubMed (Lu 2011) retrieved 389 spliceosome-associated genes. Validated quantitative RT-PCR assays were successfully developed for 328 out of the 389 splicing factors tested. The splicing factors discovery screen consisted of surveying the global expression of all 328 splicing factors in the LCM discovery sample set by quantitative RT-PCR. Gene expression levels relative to housekeeping genes (Prinos et al. 2011) were determined, and those that vary between the tumor microenvironment and ovarian stroma, between cancer epithelium and Fallopian tube epithelium by at least twofold in at most one normal and at least three cancer samples or at most one cancer and at least three normal tissues were selected and validated using LCM validation sample set (Supplemental Table S5). Genes with expression that varies by at least twofold in tumor microenvironment/ovarian stroma or cancer epithelium/Fallopian tube epithelium in at most one normal and at least three cancers or at most one cancer and at least three normal tissues were considered associated.
Identification of splicing factor–dependent ASEs in cell lines
The expression levels of the 51 cancer-associated splicing factors were determined using quantitative RT-PCR in the SKOV3ip1 cell line (Chen et al. 2001). A raw Ct of 30 or less for a 1-ng cDNA input was considered as “expressed.” Forty-one splicing factors were expressed and successfully knocked down with two nonoverlapping siRNAs in SKOV3ip1 or immortalized normal ovarian fibroblast (INOF) (Lawrenson et al. 2012) (see Supplemental Table S7 for sequences). RNA was harvested 72 h after transfection, and the quality of knockdown was evaluated by quantitative RT-PCR (Supplemental Table S7) or Western blot (Supplemental Fig. S4). RNA extracted from cells where splicing factors were knocked down by at least twofold or more was used to monitor the impact on 13 cancer-associated ASEs by quantitative RT-PCR. ASEs displaying quantitative splicing shift (ΔQψ) of at least 10 between the knockdowns and mock transfection were considered significant. All cancer-associated ASEs regulated by at least one splicing factor and splicing factors regulating at least one ASE are shown in Figure 3B (the raw data are accessible through http://palace.lgfus.ca/pcrreactiongroup/list/229). The splicing patterns of ASEs affected by QKI and RBFOX2 knockdown in SKOV3ip1 were retested in the INOF cell line using the same method.
Identification of RBFOX2- and QKI-dependent ASEs
To determine QKI-responsive ASEs, QKI was knocked down using two independent siRNAs in the SKOV3ip1 cell line. Effective knockdown of at least twofold was confirmed by quantitative RT-PCR, and the splicing patterns (ψ values) of 382 alternative cassette exons (ACE) were monitored by high-throughput endpoint PCR. A splicing shift (Δψ) of at least 10 between the mock transfection and each siRNA was considered significant (Supplemental Table S8). The effect of QKI on RBFOX2 splicing targets was evaluated using an established set of 48 RBFOX2 splicing targets (Venables et al. 2009) as described above (Supplemental Table S8). The tendency of RBFOX2 and QKI co-regulated exons to be associated with the tumor microenvironment was evaluated as described in Figure 2A.
Western blot analysis
Proteins were extracted from SKOV3ip1 cells 72 h post-transfection and separated on SDS-PAGE and visualized via immunoblotting as described previously (Venables et al. 2008b). QKI detection was performed using polyclonal rabbit anti-QKI 1:1000 (Novus Biologicals). RBFOX2 detection was performed using Flag-Tag M2 (CloneM2) (1 µg/mL) (Sigma-Aldrich). Each blot was decorated with an anti-GAPDH antibody (Novus Biologicals) to correct for total protein content in different lanes.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank the “Réseau de Recherche sur le Cancer FRSQ” tissue bank for ovarian and breast normal and tumor tissues; David Huntsman and the British Columbia tumor bank for the Fallopian tube samples; the Alberta tumor bank for the low malignant potential sample; and Vincent Normandeau-Babin for clinical data management. We also thank Kate Lawrenson for the INOF cell line and David Huntsman for discussions and advice in the early stages of this work. We thank Fernand-Pierre Gendron, the “Service de phénotypage,” and members of the Department of Pathology from the Université de Sherbrooke for help with LCM and tissue preparation. J.-P.P. holds the Chaire de Recherche de l'Université de Sherbrooke on RNA Structure and Genomics. S.A.-E. is the Canada Research Chair in RNA Biology and Cancer Genomics. B.C. is the Canada Research Chair in Functional Genomics. This research project was supported by grants from the Canadian Cancer Society and the Canadian Institute of Health Research (CIHR).
Footnotes
Freely available online through the RNA Open Access option.
REFERENCES
- Agarwal A, Tressel SL, Kaimal R, Balla M, Lam FH, Covic L, Kuliopulos A 2010. Identification of a metalloprotease–chemokine signaling system in the ovarian cancer microenvironment: Implications for antiangiogenic therapy. Cancer Res 70: 5880–5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auersperg N, Wong AS, Choi KC, Kang SK, Leung PC 2001. Ovarian surface epithelium: Biology, endocrinology, and pathology. Endocr Rev 22: 255–288 [DOI] [PubMed] [Google Scholar]
- Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S, Allain FH 2006. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J 25: 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbolina MV, Burkhalter RJ, Stack MS 2011. Diverse mechanisms for activation of Wnt signalling in the ovarian tumour microenvironment. Biochem J 437: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick NA, Neilson EG, Moses HL 2004. Stromal fibroblasts in cancer initiation and progression. Nature 432: 332–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biermann K, Heukamp LC, Steger K, Zhou H, Franke FE, Sonnack V, Brehm R, Berg J, Bastian PJ, Muller SC, et al. 2007. Genome-wide expression profiling reveals new insights into pathogenesis and progression of testicular germ cell tumors. Cancer Genomics Proteomics 4: 359–367 [PubMed] [Google Scholar]
- Brosseau JP, Lucier JF, Lapointe E, Durand M, Gendron D, Gervais-Bird J, Tremblay K, Perreault JP, Elela SA 2010. High-throughput quantification of splicing isoforms. RNA 16: 442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey TM, Eneman J, Crocker A, White J, Tessitore J, Stanley M, Harlow S, Bunn JY, Weaver D, Muss H, et al. 2008. Cancer associated fibroblasts stimulated by transforming growth factor β1 (TGF-β1) increase invasion rate of tumor cells: A population study. Breast Cancer Res Treat 110: 39–49 [DOI] [PubMed] [Google Scholar]
- Chen J, Feng Y, Zhou X 2001. [Experimental study on resistance to cisplatin and taxol in human ovarian cancer SKOV3ip1 multicellular aggregates]. Zhonghua Fu Chan Ke Za Zhi 36: 489–492 [PubMed] [Google Scholar]
- Coleman MP, Forman D, Bryant H, Butler J, Rachet B, Maringe C, Nur U, Tracey E, Coory M, Hatcher J, et al. 2011. Cancer survival in Australia, Canada, Denmark, Norway, Sweden, and the UK, 1995–2007 (The International Cancer Benchmarking Partnership): An analysis of population-based cancer registry data. Lancet 377: 127–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau A, Richard S 2005. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat Struct Mol Biol 12: 691–698 [DOI] [PubMed] [Google Scholar]
- Iwatsuki M, Mimori K, Yokobori T, Ishi H, Beppu T, Nakamori S, Baba H, Mori M 2010. Epithelial–mesenchymal transition in cancer development and its clinical significance. Cancer Sci 101: 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y, Han Z, Zhang S, Liu Y, Wei L 2011. Epithelial–mesenchymal transition in tumor microenvironment. Cell Biosci 1: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny HA, Krausz T, Yamada SD, Lengyel E 2007. Use of a novel 3D culture model to elucidate the role of mesothelial cells, fibroblasts and extra-cellular matrices on adhesion and invasion of ovarian cancer cells to the omentum. Int J Cancer 121: 1463–1472 [DOI] [PubMed] [Google Scholar]
- Klinck R, Bramard A, Inkel L, Dufresne-Martin G, Gervais-Bird J, Madden R, Paquet ER, Koh C, Venables JP, Prinos P, et al. 2008. Multiple alternative splicing markers for ovarian cancer. Cancer Res 68: 657–663 [DOI] [PubMed] [Google Scholar]
- Kurman RJ, Shih Ie M 2010. The origin and pathogenesis of epithelial ovarian cancer: A proposed unifying theory. Am J Surg Pathol 34: 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapuk A, Marr H, Jakkula L, Pedro H, Bhattacharya S, Purdom E, Hu Z, Simpson K, Pachter L, Durinck S, et al. 2010. Exon-level microarray analyses identify alternative splicing programs in breast cancer. Mol Cancer Res 8: 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrenson K, Grun B, Gayther SA 2012. Heterotypic three-dimensional in vitro modeling of stromal–epithelial interactions during ovarian cancer initiation and progression. J Vis Exp 2012: e4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z 2011. PubMed and beyond: A survey of web tools for searching biomedical literature. Database (Oxford) 2011: baq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Dostie J, Dreyfuss G, Van Duyne GD 2005. The Gemin6–Gemin7 heterodimer from the survival of motor neurons complex has an Sm protein-like structure. Structure 13: 883–892 [DOI] [PubMed] [Google Scholar]
- Mandai M, Matsumura N, Baba T, Yamaguchi K, Hamanishi J, Konishi I 2011. Ovarian clear cell carcinoma as a stress-responsive cancer: Influence of the microenvironment on the carcinogenesis and cancer phenotype. Cancer Lett 310: 129–133 [DOI] [PubMed] [Google Scholar]
- Mehra K, Mehrad M, Ning G, Drapkin R, McKeon FD, Xian W, Crum CP 2011. STICS, SCOUTs and p53 signatures; a new language for pelvic serous carcinogenesis. Front Biosci 3: 625–634 [DOI] [PubMed] [Google Scholar]
- Munirah MA, Siti-Aishah MA, Reena MZ, Sharifah NA, Rohaizak M, Norlia A, Rafie MK, Asmiati A, Hisham A, Fuad I, et al. 2011. Identification of different subtypes of breast cancer using tissue microarray. Rom J Morphol Embryol 52: 669–677 [PubMed] [Google Scholar]
- Nofech-Mozes S, Khalifa MA, Ismiil N, Saad RS, Hanna WM, Covens A, Ghorab Z 2008. Immunophenotyping of serous carcinoma of the female genital tract. Mod Pathol 21: 1147–1155 [DOI] [PubMed] [Google Scholar]
- Novikov L, Park JW, Chen H, Klerman H, Jalloh AS, Gamble MJ 2011. QKI-mediated alternative splicing of the histone variant MacroH2A1 regulates cancer cell proliferation. Mol Cell Biol 31: 4244–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orimo A, Weinberg RA 2007. Heterogeneity of stromal fibroblasts in tumors. Cancer Biol Ther 6: 618–619 [DOI] [PubMed] [Google Scholar]
- Pilotte J, Larocque D, Richard S 2001. Nuclear translocation controlled by alternatively spliced isoforms inactivates the QUAKING apoptotic inducer. Genes Dev 15: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A, Ellis MJ, Perou CM 2012. Practical implications of gene-expression-based assays for breast oncologists. Nat Rev Clin Oncol 9: 48–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinos P, Garneau D, Lucier JF, Gendron D, Couture S, Boivin M, Brosseau JP, Lapointe E, Thibault P, Durand M, et al. 2011. Alternative splicing of SYK regulates mitosis and cell survival. Nat Struct Mol Biol 18: 673–679 [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR 2007. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35: D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogerson L, Darby S, Jabbar T, Mathers ME, Leung HY, Robson CN, Sahadevan K, O'Toole K, Gnanapragasam VJ 2008. Application of transcript profiling in formalin-fixed paraffin-embedded diagnostic prostate cancer needle biopsies. BJU Int 102: 364–370 [DOI] [PubMed] [Google Scholar]
- Saad AF, Hu W, Sood AK 2010. Microenvironment and pathogenesis of epithelial ovarian cancer. Horm Cancer 1: 277–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer IG, Sood AK, Mok S, Liu J 2011. Cancer-associated fibroblasts and their putative role in potentiating the initiation and development of epithelial ovarian cancer. Neoplasia 13: 393–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumperli D, Pillai RS 2004. The special Sm core structure of the U7 snRNP: Far-reaching significance of a small nuclear ribonucleoprotein. Cell Mol Life Sci 61: 2560–2570 [DOI] [PubMed] [Google Scholar]
- Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, Burge CB, Gertler FB 2011. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet 7: e1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, et al. 2006. Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst 98: 262–272 [DOI] [PubMed] [Google Scholar]
- Sun S, Zhang Z, Fregoso O, Krainer AR 2012. Mechanisms of activation and repression by the alternative splicing factors RBFOX1/2. RNA 18: 274–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlsty TD, Coussens LM 2006. Tumor stroma and regulation of cancer development. Annu Rev Pathol 1: 119–150 [DOI] [PubMed] [Google Scholar]
- Tone AA, Begley H, Sharma M, Murphy J, Rosen B, Brown TJ, Shaw PA 2008. Gene expression profiles of luteal phase Fallopian tube epithelium from BRCA mutation carriers resemble high-grade serous carcinoma. Clin Cancer Res 14: 4067–4078 [DOI] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL 2005. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol 25: 10005–10016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Klinck R, Bramard A, Inkel L, Dufresne-Martin G, Koh C, Gervais-Bird J, Lapointe E, Froehlich U, Durand M, et al. 2008a. Identification of alternative splicing markers for breast cancer. Cancer Res 68: 9525–9531 [DOI] [PubMed] [Google Scholar]
- Venables JP, Koh CS, Froehlich U, Lapointe E, Couture S, Inkel L, Bramard A, Paquet ER, Watier V, Durand M, et al. 2008b. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol Cell Biol 28: 6033–6045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, Durand M, Couture S, Froehlich U, Lapointe E, et al. 2009. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol 16: 670–676 [DOI] [PubMed] [Google Scholar]
- Venables JP, Brosseau JP, Gadea G, Klinck R, Prinos P, Beaulieu JF, Lapointe E, Durand M, Thibault P, Tremblay K, et al. 2013. RBFOX2 is an important regulator of mesenchymal tissue-specific splicing in both normal and cancer tissues. Mol Cell Biol 33: 396–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Huang J, Greshock J, Liang S, Barchetti A, Hasegawa K, Kim S, Giannakakis A, Li C, O'Brien-Jenkins A, et al. 2008. Transcriptional regulation of PIK3CA oncogene by NF-κB in ovarian cancer microenvironment. PLoS ONE 3: e1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zearfoss NR, Clingman CC, Farley BM, McCoig LM, Ryder SP 2011. Quaking regulates Hnrnpa1 expression through its 3′ UTR in oligodendrocyte precursor cells. PLoS Genet 7: e1001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB 2010. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science 329: 439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]