Abstract
Cell size, measured as either volume or mass, is a fundamental indicator of cell state. Far more tightly regulated than size is density, the ratio between mass and volume, which can be used to distinguish between cell populations even when volume and mass appear to remain constant. Here we expand upon a previous method for measuring cell density involving a suspended microchannel resonator (SMR). We introduce a new device, the dual SMR, as a high-precision instrument for measuring single-cell mass, volume, and density using two resonators connected by a serpentine fluidic channel. The dual SMR designs considered herein demonstrate the critical role of channel geometry in ensuring proper mixing and damping of pressure fluctuations in microfluidic systems designed for precision measurement. We use the dual SMR to compare the physical properties of two well-known cancer cell lines: human lung cancer cell H1650 and mouse lymphoblastic leukemia cell line L1210.
1. Introduction
At the cellular level, a tradeoff exists between synthesizing biochemical content to perform vital functions and the resulting increase in energy expenditure needed to maintain a larger size. Thus, cell size is a fundamental physical property linked to physiological purpose, overall health, surrounding environment, and metabolic function. Cell size is determined by the aggregate contribution of biochemical content—mainly proteins and lipids—and water, which occur in an approximately 1:3 ratio.1 Size is measured as either mass or volume, and the ratio of these two parameters is density. Whereas cellular mass and volume can vary by as much as 50%, density is far more tightly regulated. Thus, density can often be used to distinguish between cell populations even when volume and mass cannot.2–4
There are few tools available to measure the volume, mass, and density of a single cell. Current methods for determining cell volume include z-stack analysis, flow cytometry, and measurement with a Coulter counter.5–8 Cell mass can be measured with quantitative phase microscopy.9 The gold standard for determining cell density is density gradient centrifugation, which is difficult to precisely calibrate and subjects cells to stresses that may lead to biological artifacts. Despite a multitude of instruments and techniques available for measuring cellular physical properties, few tools are capable of simultaneously measuring multiple physical properties and at the level of a single cell.
A microfluidic approach to measuring mass, volume, and density offers the means to make precise single cell measurements in physiological solutions with minimal perturbation to the cell's native environment. Grover, et al., demonstrated a method for determining single-cell density by measuring the buoyant mass of a single cell in two fluids of different densities.2 In this method, a cell travels through a suspended microchannel resonator (SMR), pauses in a bypass channel containing fluid of a higher density, then travels a second time through the SMR in the reverse direction, to be measured in a higher-density fluid. The throughput of this method is limited by both the requirement that a cell pass through the same resonator twice and the time required to sufficiently mix two fluids by diffusion—up to 15 seconds for larger-sized cells. An instrument with increased throughput could complement current high-throughput cellular analysis methods, such as flow cytometry, thereby providing additional parameters to identify cellular subpopulations important in diagnosis and prognosis decisions. We therefore developed a device for measuring cell density using two resonators arranged in series, each filled with a fluid of a different density and connected by a long serpentine channel. We apply this device—the dual SMR—towards multivariate size analysis of mammalian cell populations.
2. Measurement Concept
The SMR is a microfluidic device that consists of a fluid channel embedded in a vacuum-packaged cantilever.10 The cantilever resonates at a frequency proportional to its total mass, and as an individual cell travels through the embedded microchannel, the total cantilever mass changes. This change in mass is detected as a change in resonance frequency that corresponds directly to the buoyant mass of the cell. In equation form, buoyant mass is:
where mB,1 is the buoyant mass of the cell, Vcell is the cell volume, and ρcell and ρfluid,1 are the density of the cell and the surrounding fluid, respectively. If the same cell is measured a second time in a different density fluid (ρfluid,2), then a second buoyant mass (mB2) is obtained. From these two measurements (Figure 1A) the mass, volume, and density of a single cell are calculated. As measurements are recorded for a population of cells, the distributions of mass, density, and volume are also determined (Figure 1B).
Figure 1.
A Calculating single cell mass, volume, and density. Cell buoyant mass is measured in two fluids of different densities (red dots) to determine the linear relationship between buoyant mass and fluid density. The absolute mass (y-intercept), volume (slope), and density (x-intercept) of the cell can then be calculated. B Buoyant mass measurements of a cell population measured in two different fluids. Cell-to-cell variations in mass, density, and volume are directly observed from the intercepts and slopes created by the pairs of buoyant mass measurements.
3. Device Design
To measure the buoyant mass of single cells in two different density fluids in a continuous flow format, we fabricated and tested devices with two fluidically connected and simultaneously operated SMRs (Figure 2A). During operation of the dual SMR, a dilute cell population suspended in cell media, Fluid 1, is delivered to the sample bypass via pressure-driven flow (Figure 2A, Supplementary Figure 1), and single cells flow into the first SMR (SMR1) for the first buoyant mass measurement. The cells then travel through a microchannel to a cross-junction, where a second fluid of different density is introduced. After the cross-junction, cells continue through a long serpentine channel, which facilitates mixing of the two fluids. The cells next enter a second cantilever (SMR2) for a buoyant mass measurement in the mixed fluid, Fluid 2. As cells flow through each cantilever, a change in resonance frequency is recorded (Figure 2B), which is determined by each cell's buoyant mass in each cantilever's corresponding fluid.
Figure 2.
Dual SMR schematic and measurement. A A single cell flows from the sample bypass channel into the first SMR (SMR1) for a buoyant mass measurement in the cell's culture media (fluid 1, blue). The cell then continues to a cross-junction where a high density fluid (light red) is introduced and mixes with fluid 1 via diffusion in the serpentine channel. The second buoyant mass measurement is recorded as the particle flows through the second SMR (SMR2) in this mixed fluid (fluid 2, dark red). B SMR buoyant mass measurements are determined by the change in resonance frequency (Δfr) from the baseline as a cell traverses the cantilever channel. The direction of this frequency change depends on the density of the cell relative to the surrounding fluid. A slope in the baseline of SMR2 is observed due to a ~0.01% change in density of the fluid along the length of the cantilever microchannel.
Although the dual SMR design is amenable to increased throughput, several non-obvious challenges to precision measurements in a low Reynold's number (Re<1) environment were evident during testing of preliminary designs. Three critical design features address these challenges and facilitate the measurement: (1) differently-sized cantilevers to prevent signal cross-talk; (2) a microfluidic cross-junction to steadily introduce a second fluid; and (3) a narrow serpentine channel to facilitate mixing the two fluids.
The first design feature, differently-sized cantilevers, minimizes crosstalk of the signals measured from SMR1 and SMR2. Crosstalk results from mechanical coupling between the vibrations of similarly sized cantilevers with their out-of-phase neighbors. If the two cantilevers in the dual SMR have similar dimensions, their resonance frequencies are similar; thus, the mechanical vibrations of one will apply an auxiliary driving force on its neighbor. Significantly altering the geometry of one cantilever (300 and 360μm length for SMR1 and SMR2, respectively) ensures that the two resonance frequencies are different, thereby eliminating crosstalk.
The dual SMR's second critical design feature is a microfluidic cross-junction that consistently introduces a second fluid of higher density. The addition of this high density fluid may occur by either a cross-junction (Figure 2A) or a T-junction (Supplementary Figure 2). The time required for two fluids to mix across a channel is approximately four times lower in a cross-junction design relative to a T-junction because mixing occurs at two interfaces rather than just one. What is not readily apparent is how differently the two configurations (Supplementary Table 1) perform in the presence of cells. Variations in pressure occur as large-sized cells pass the microfluidic junctions and enter the high resistance serpentine channel. These pressure changes alter the relative amount of high density fluid introduced at the junction and create changes to fluid density along the serpentine channel, which adversely affect the SMR2 baseline stability at the time of the large cell's measurement. However, baseline stability for cells already in the vicinity of SMR2 is not adversely affected. The cross-junction design better dampens these effects due to its larger interface between the two fluid streams, as compared to the T-junction design (Supplementary Figure 2, measurement in SMR2). We selected the cross-junction design for all cell measurements. In this design, SMR2 baseline changes in the vicinity of a cell measurement are typically ~1 × 10−5 g/mL, a value which corresponds to a <0.01% change in the ratio between the two fluids.
To ensure that each cell is immersed in a near-homogeneous solution when measured in SMR2, the dual SMR has a 5000 μm long serpentine channel, and flow rates are set such that the lag time for cells traveling from SMR1 to SMR2 is greater than ten seconds. In a 25 μm wide serpentine channel, the time required for the fluid mixture to reach 95% homogeneity is approximately six seconds, and in principle, the dual SMR enables cell mass, volume, and density measurements at the same rate as a single SMR, approximately two cells per second. Increased flow rate, higher data acquisition rate, a longer serpentine channel, and lower viscosity fluids would improve throughput without sacrifice to measurement resolution. Cell rupture and other negative effects on cell viability are not expected to occur at increased flow rate.11 In the same way that junction design affects baseline stability, serpentine channel geometry is also important; a wider serpentine channel introduces even greater baseline instability than a narrow channel. In the wide T-junction design (Supplementary Figure 2B), the baseline frequency instabilities are more than 10 times those observed in other designs. Thus, pressure damping features (Supplementary Table 1) at the point of fluid introduction and high downstream channel resistances are critical to achieving a stable system when particles are sized close to that of the channel. These features are included in the cross-junction design.
4. Device Operation
4.1 Dual SMR Calibration
The dual SMR system must be calibrated for (1) fluid density, measured as the baseline resonance frequency, and (2) particle buoyant mass, measured as peak height, or the change in resonance frequency as a cell traverses the cantilever.
For a fluid density calibration, each of three sodium chloride solutions of known densities is loaded into the dual SMR. The baseline resonance frequency of SMR1 and SMR2 filled with each solution is measured. A linear relationship can be approximated between the change in resonance frequency and the density of each salt solution (Fig. 3A). This relationship converts the experimentally-recorded baseline frequency to fluid density.
Figure 3.
A Fluid density calibration for SMR1 and SMR2. The baseline frequency for different density fluids is measured to determine each SMR's fluid density calibration factor (kHz/g mL−1). B Distribution of peak heights of a population of nominal 10 μm beads measured in SMR1 and SMR2. The mean peak height is used in determining each SMR's point mass calibration factor. The dark black curve is a simulated bead population based on CV reported by manufacturer (1.2%; Duke Scientific).
To calibrate peak height in each SMR, a monodisperse population of polystyrene beads of known diameter (10.61±0.05 μm) and density (1.05 g/mL) (Duke Scientific) is measured (Figure 3B). The buoyant mass calibration factor is determined by the ratio of the mean population peak height to calculated buoyant mass of the beads.
4.2 Fluidic Set-up and Operation
At the start of a cell density measurement, the system is first flushed with filtered Percoll media, which serves as the high density fluid. Next, the sample bypass is filled with a dilute cell sample, and the vial heights at the sample inlet and outlet are adjusted to direct fluid flow into SMR1. Pressure at the high density fluid inlet is used to set the density of Fluid 2, and pressure at the waste outlet controls the overall flow speed in the device. The arrangement of fluidic components external to the SMR is illustrated in Supplementary Figure 1. To minimize the likelihood of size biasing due to heavier cells settling at the bottom of the sample vial or tubing, a fresh sample is introduced at regular intervals by flushing the sample bypass channel. Data is acquired via LabVIEW and processed with MATLAB.
5. Data Analysis
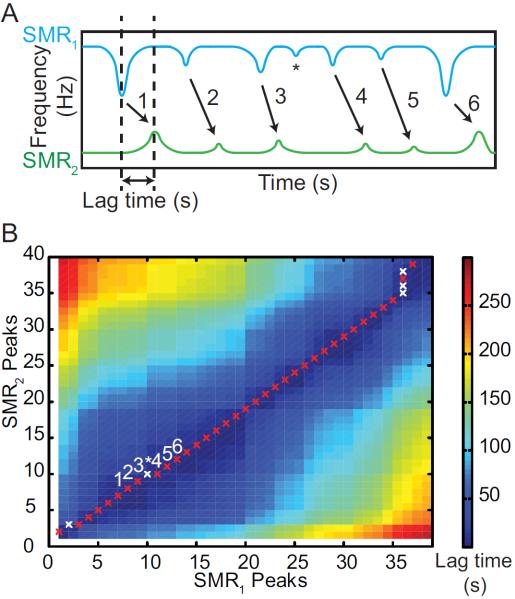
An automated method for pairing a specific cell's SMR1 and SMR2 buoyant mass measurements is required for determining cell density. Simultaneously collected resonance frequency datasets from the two cantilevers each can have hundreds of single cell measurements, and pairing these measurements is complicated by subtle changes in flow rates and other anomalies (Figure 4A). In addition to a gradually shifting time delay, or lag time, between SMR1 and SMR2 measurements, datasets typically have different numbers of measurements, due to a variety of events. Particles can stick to walls within the serpentine channel and be lost, and thus only be measured in SMR1; contaminants in Fluid 2 can appear as extra peaks in SMR2; and cells can enter SMR1 as a doublet that generates only a single peak, be separated into two discrete particles when traveling through the serpentine channel, and appear as two peaks in SMR2. These complications render a simple time offset between the two SMR datasets insufficient to successfully assign peak pairs.
Figure 4.
A Sample frequency reading for a population of cells. Peak heights in each dataset are identified and the time at which each peak occurs is recorded. Lag time reflects the amount of time required for a single cell to travel from the first cantilever, through the serpentine channel, to the second cantilever. The asterisk indicates an SMR1 measurement that does not have a match in SMR2, as would occur if debris stuck within the serpentine channel. B A heat map representing the implementation of the Needleman-Wunsch algorithm for pairing peaks. The X and Y axes indicate the ordinal peak number recorded from SMR1 and SMR2, respectively, and the colors reflect absolute lag time calculated between a given peak in SMR1 with one in SMR2. Red Xs represent peaks paired by their optimal lag time. White Xs correspond to a pairing that has been rejected based on the lag time. Labeled Xs refer to the example peaks shown in Fig. 4A.
To address these issues, an approach based on dynamic programming was developed. Dynamic programming recursively scores solutions to subproblems in order to find an optimal solution to a larger problem.12 The Needleman-Wunsch algorithm is a dynamic programming method for DNA sequence alignment that optimizes alignment by maximizing the number of perfect base matches and minimizing the number of gaps in an aligned sequence. It is often used to locate a DNA sequence within an organism genome; here, we present an adaptation in which it is used to align peaks from the dual SMR's datasets.
The algorithm determines optimal alignment of the datasets by first calculating a matrix in which every possible pairing between peaks from SMR1 and SMR2 is scored. For visualization purposes, this matrix is represented as a heat map in Figure 4B. Pairs are made by starting at the lower left hand corner of the matrix and moving to the upper, upper right, or right neighboring value that is most similar to a predicted lag time. A diagonal motion within the scoring matrix indicates a match and a vertical or horizontal motion causes the algorithm to discard either the current or previous pair based on proximity to the predicted lag time. This procedure ensures that each peak is used in no more than one pair. The predicted lag time is adjusted through the pairing procedure to account for changes to flow rates during an experiment. This lag time-based approach is particularly effective when considering that peak pairs are formed without knowledge of peak height—the most naïve approach. The pairing result is relatively insensitive to the initial value set for the lag time, but very high particle concentrations can result in fewer successful pairings.
6. Materials and Methods
6.1 Cell culture
Human lung carcinoma cells (H1650) were grown at 37° C in RPMI (Invitrogen) supplemented with 10% (vol/vol) fetal bovine serum (FBS) (Invitrogen), 100 IU penicillin, and 100 μg/mL streptomycin (Invitrogen). Cells were seeded at approximately 5×104 in a 25 cm2 flask and passaged at approximately 70% confluency (106 cells on a 25 cm2 flask). Cell measurements were performed on cultures grown to approximately 50% confluency.
Mouse lymphocytic leukemia cells (L1210) were grown in suspension at 37° C in Leibovitz's L15 media (Invitrogen) supplemented with 10% (vol/vol) FBS, 0.4% (w/vol) glucose (Sigma), 100 IU penicillin, and 100 μg/mL streptomycin. Cells were seeded at approximately 5×104 cells/mL in a 25 cm2 flask, and diluted to fresh media after having reached a concentration of 1×106 cells/mL. Cell concentration was monitored using a Coulter counter. Cell measurements were performed on cultures grown to 5×105–1×106 cells/mL.
6.2 Percoll media formulation
High density fluid introduced for measurement in SMR2 was formulated as a solution of 50% (v/v) Percoll (Sigma), 1.38% (w/v) powdered L15 media (Sigma), 0.4% (w/v) glucose, 100 IU penicillin, and 100 μg/mL streptomycin. Media pH was adjusted to 7.2. This Percoll media was stored at 4°C and filtered immediately prior to use in the dual SMR.
7. Results and Discussion
7.1 Measurement Error Analysis
Density variation measured in a cell population is a result of natural biological heterogeneity and error in the measurement technique. One source of error in the measurement technique arises from the value of the density of Fluid 2 relative to the density of the measured cells. Though Fluid 1 is almost always cell media, the composition of Fluid 2 can be adjusted by changing the concentration of Percoll in the high density fluid, or by adjusting the pressure ratio between the channels meeting at the cross-junction. The effect of the Fluid 2 density value on measurement error was estimated by applying multiplicative and additive errors to average L1210 cell buoyant masses in Fluid 1 and a range of Fluid 2 values. Multiplicative error results from an uncertainty in determining the cell's exact lateral position in the tip of the cantilever channel13. This error, estimated from the buoyant mass distribution of polystyrene beads (Figure 3), is inversely dependent on particle radius and directly proportional to buoyant mass. Thus, minimizing this error involves measuring either larger particles or adjusting the fluid density to reduce buoyant mass. In the theoretical case of pure multiplicative error, uncertainty in determining the density of the cell will be at a minimum when the density of Fluid 2 matches that of the cell (Supplementary Figure 3A). Here the cell buoyant mass is zero, as is the associated error, and measuring Fluid 2 density is sufficient to determine the density of the cell. As the density of Fluid 2 deviates from that of the cell, the magnitude of the cell's buoyant mass in Fluid 2 will increase, as will the associated density measurement error (Supplementary Figure 3A). Interestingly, multiplicative error in the volume measurement continually decreases for higher Fluid 2 densities (Supplementary Figure 3A). This decrease is graphically indicated as a decreasing standard error in the slope (Figure 1A) when the x-axis (fluid density) distance increases between the two buoyant mass measurements.
A second form of error is additive error, which results from a constant baseline noise and leads to uncertainty in determining peak height and thus cell buoyant mass.14 In the theoretical case of pure additive error, the minimum uncertainty in determining cell density occurs when the density of Fluid 2 is greater than the density of the cell (Supplementary Figure 3B). Under the conditions of our simulation, the minimum value occurs when the fluid density is approximately 1.15 g/mL. Beyond this minimum, the uncertainty increases at a relatively slow rate. Similarly to the case of multiplicative error, uncertainty in the volume measurement due to additive error decreases as the difference between Fluid 1 and Fluid 2 increases (Supplementary Figure 3B).
When multiplicative and additive errors are both present, as is the case with the dual SMR, each dominate different measurement regimes. Multiplicative error dominates when buoyant mass is relatively large and additive error dominates when buoyant mass is relatively small. When both forms of error are present, the error in the cell density measurement is minimized where the Fluid 2 density is slightly greater than cell density (Figure 5). Here multiplicative error is small and additive error dominates, meaning density measurement error is mainly determined by noise in the instrument baseline. When Fluid 2 density deviates from this minimum, multiplicative error dominates, and density measurement error increases. Volume measurement error decreases asymptotically as the difference between Fluid 1 and Fluid 2 increases. Thus, to optimize the measurement error for both density and volume, the Fluid 2 density should be somewhat greater than that of the cell.
Figure 5.
Measurement uncertainty of cell density (blue) and cell volume (green) as a function of fluid 2 density, assuming no uncertainty in measuring fluid density. The simulation is calculated using fixed values for buoyant mass in Fluid 1, average L1210 cell density, and Fluid 1 density, along with a range of experimentally relevant values for Fluid 2 density, which correspond to a range of buoyant masses in Fluid 2. Multiplicative error was applied to the simulated measurements using the variation in buoyant masses of polystyrene beads, and additive error was applied using the magnitude of the baseline noise in each cantilever. The fluid 2 density of a typical experiment was adjusted to approximately 1.07 g/mL. The pink asterisk indicates the cell density, and the black circle corresponds to the point of minimum uncertainty in the density measurement.
As an approximation of the contribution of measurement error to the distribution of cell density in our measurement, the variation in buoyant mass measured for polystyrene beads was multiplicatively applied, and the relative baseline noise was additively applied to the mean buoyant masses and fluid densities of experimental cell data (Supplementary Figure 4). The width of this simulated density distribution is narrower than that obtained from the cells, which indicates that the variation in the cell measurement results primarily from natural biological variation.
7.2 Stability and Throughput
There are several challenges associated with operating the dual SMR, most of which relate to its sensitivity to changes in pressure and high channel resistances. The pressures at the start of an experiment must be carefully balanced to ensure proper direction and speed of fluid flow at all inlets and to maintain the desired composition of Fluid 2. During the course of the experiment, the fluid height in each of the vials gradually changes, and so the pressures must be monitored and adjusted periodically. Pressure adjustments are implemented by either changing the setting on an electronically controlled pressure regulator (resolution = 0.006 PSI) or by manually adjusting the vertical height of the fluid vials. These methods allow changes to fluid flow rates by ~0.02%. Large-sized cells introduce baseline instabilities (discussed in Device Design), and bubbles and small pieces of debris also upset the pressure balance. Filtering all fluids and a lengthy flushing procedure (five to seven minutes) prior to the start of an experiment helps mitigate this problem, but debris still occasionally disrupts the system. Although the current system is sufficient for proof of concept, implementing electronic flow sensors to monitor fluid flow rates as feedback to electronic pressure regulators would better automate system stabilization.
One practical consideration when operating the dual SMR relates to selecting a cell concentration that allows for a reasonably steady baseline. When a cell passes through the cross-junction into the serpentine channel, it causes a local fluctuation in the composition of Fluid 2. Thus, when many cells are measured in quick succession, the baseline becomes less steady, which increases the uncertainty in determining the fluid density. One approach to solving this problem is to increase the fraction of high density fluid delivered to the serpentine channel. This requires the high density fluid to be delivered with higher pressure, which makes pressure fluctuations from cells less significant. So as not to sacrifice measurement accuracy, the increased pressure also requires adjustments to slow the passage of cells, which slows fluid flow in the system overall and results in an overall steadier baseline. These adjustments, however, reduce the rate at which cells can be measured. Although in principle the dual SMR should be able to measure approximately two cells per second, the most reliable operation is achieved when cells enter SMR1 at approximately one cell every ten seconds, which is comparable in throughput to the fluid-switching method for measuring density presented by Grover, et al2. Thus, practical considerations associated with the existing design currently limit its overall performance.
7.3 Mass, Volume, and Density Measurements for Mammalian Cell Populations
To demonstrate single-cell mass, volume, and density measurements of a biological sample, we measured H1650 and L1210 cells (Figure 6A–B). H1650 cells are an adherent cell line originating from human lung tissues, and are commonly used as a model for studying lung cancer.15 We compared these cells to L1210 cells, a mouse lymphocytic leukemia cell line. As expected, the variations in H1650 cell mass (41%) and volume (41%) are much greater than that of cell density (0.3%). We observed a similar trend with variation in L1210 cell mass (55%), volume (56%), and density (1.5%). When compared to a commercial Coulter counter, the dual SMR cell volume measurements are nearly identical (Figure 6B). Since the Coulter counter measurements were made prior to the start of the SMR measurement, this outcome suggests that dual SMR measurement conditions do not alter cell volume.
Figure 6.
A Density versus mass of H1650 (n = 148) and L1210 (n = 136) cells. Although these homogeneous cell populations exhibit large variation in mass (~50%), density is a much more tightly regulated parameter. B Dual SMR volume distribution compared to results from a Coulter counter. A small aliquot of cells was measured on the Coulter counter prior to the SMR measurement.
Interestingly, though both the mass and the volume of the L1210 cells are lower than that of the H1650 cells, the density is higher. This result suggests that the concentration of high-density biochemical components— proteins and nucleic acids—is higher in L1210 cells than in H1650 cells. In particular, this outcome agrees with the high nuclear to cytoplasmic ratio characteristic to hematopoietic cells relative to epithelial cells.16–18 Alternatively, a higher density can reflect a higher basal protein concentration, which may alter rates of transcription, protein-protein interactions, and enzymatic processes.19 Future aims include exploring how these physical properties change during specific cellular processes, such as cell growth, division, and death.
Conclusion
Though high resistance channels and a sensitive measurement introduced complexities that ultimately limited actual throughput, the presented dual SMR serves to demonstrate the potential for a high-throughput measurement system that combines physical property measurements for a powerful multi-parameter index that more accurately describes the state of the cell.
Supplementary Material
Acknowledgements
This work was supported by the Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute, Physical Sciences Oncology Center U54CA143874, Cell Decision Process Center (P50GM68762) and contract R01GM085457 from the US National Institutes of Health. The authors would like to thank Scott M. Knudsen and Nathan Cermak for valuable discussion. V.C.H. acknowledges support through an NSF graduate fellowship.
References
- 1.Alberts B, Bray D, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Essential Cell Biology: An Introduction to the Molecular Biology of the Cell. Vol. 27. Garland Publishing, Inc; New York: 1998. [Google Scholar]
- 2.Grover WH, Bryan AK, Diez-Silva M, Suresh S, Higgins JM, Manalis SR. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10992–6. doi: 10.1073/pnas.1104651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park J-M, Lee J-Y, Lee J-G, Jeong H, Oh J-M, Kim YJ, Park D, Kim MS, Lee HJ, Oh JH, Lee SS, Lee W-Y, Huh N. Analytical chemistry. 2012;84:7400–7. doi: 10.1021/ac3011704. [DOI] [PubMed] [Google Scholar]
- 4.Friedman SL, Roll FJ. Analytical biochemistry. 1987;161:207–18. doi: 10.1016/0003-2697(87)90673-7. [DOI] [PubMed] [Google Scholar]
- 5.Mir M, Wang Z, Shen Z, Bednarz M, Bashir R, Golding I, Prasanth SG, Popescu G. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13124–9. doi: 10.1073/pnas.1100506108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzur A, Moore JK, Jorgensen P, Shapiro HM, Kirschner MW. PloS one. 2011;6:e16053. doi: 10.1371/journal.pone.0016053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirsch J, Gallian E. Journal of lipid research. 1968;9:110–9. [PubMed] [Google Scholar]
- 8.Kim YR, Ornstein L. Cytometry. 1983;3:419–27. doi: 10.1002/cyto.990030606. [DOI] [PubMed] [Google Scholar]
- 9.Barer R, Ross K, Tkaczyk S. Nature. 1953;171:720–724. doi: 10.1038/171720a0. [DOI] [PubMed] [Google Scholar]
- 10.Burg TP, Godin M, Knudsen SM, Shen W, Carlson G, Foster JS, Babcock K, Manalis SR. Nature. 2007;446:1066–1069. doi: 10.1038/nature05741. [DOI] [PubMed] [Google Scholar]
- 11.Byun S, Son S, Amodei D, Cermak N, Shaw J, Kang JH, Hecht VC, Winslow MM, Jacks T, Mallick P, Manalis SR. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7580–5. doi: 10.1073/pnas.1218806110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cormen TH, Leiserson CE, Rivest RL, Stein C. Introduction to Algorithms. 2nd edn. MIT Press; 2001. [Google Scholar]
- 13.Lee J, Bryan AK, Manalis SR. Review of Scientific Instruments. 2011 doi: 10.1063/1.3534825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knudsen SM, von Muhlen MG, Manalis SR. Analytical chemistry. 2012;84:1240–2. doi: 10.1021/ac300034r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao Z, Fenoglio S, Gao DC, Camiolo M, Stiles B, Lindsted T, Schlederer M, Johns C, Altorki N, Mittal V, Kenner L, Sordella R. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15535–40. doi: 10.1073/pnas.1009472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gualberto A, Dolled-Filhart M, Gustavson M, Christiansen J, Wang Y-F, Hixon ML, Reynolds J, McDonald S, Ang A, Rimm DL, Langer CJ, Blakely J, Garland L, Paz-Ares LG, Karp DD, Lee AV. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:4654–65. doi: 10.1158/1078-0432.CCR-10-0089. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Santos IM, Franzon CMR, Koga AH. Revista brasileira de hematologia e hemoterapia. 2012;34:242–4. doi: 10.5581/1516-8484.20120058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao Q, Chu S, Ghanta S, Padbury JF, De Paepe ME. Respiratory research. 2013;14:37. doi: 10.1186/1465-9921-14-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou H-X, Rivas G, Minton AP. Annual review of biophysics. 2008;37:375–97. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.