Abstract
Background
Nuclear factor IA (NFIA), a transcription factor and essential regulator in embryonic glial development, is highly expressed in human glioblastoma (GBM) compared with normal brain, but its contribution to GBM and cancer pathogenesis is unknown. Here we demonstrate a novel role for NFIA in promoting growth and migration of GBM and establish the molecular mechanisms mediating these functions.
Methods
To determine the role of NFIA in glioma, we examined the effects of NFIA in growth, proliferation, apoptosis, and migration. We used gain-of-function (overexpression) and loss-of-function (shRNA knockdown) of NFIA in primary patient-derived GBM cells and established glioma cell lines in culture and in intracranial xenografts in mouse brains.
Results
Knockdown of native NFIA blocked tumor growth and induced cell death and apoptosis. Complementing this, NFIA overexpression accelerated growth, proliferation, and migration of GBM in cell culture and in mouse brains. These NFIA tumor-promoting effects were mediated via transcriptional repression of p53, p21, and plasminogen activator inhibitor 1 (PAI1) through specific NFIA-recognition sequences in their promoters. Importantly, the effects of NFIA on proliferation and apoptosis were independent of TP53 mutation status, a finding especially relevant for GBM, in which TP53 is frequently mutated.
Conclusion
NFIA is a modulator of GBM growth and migration, and functions by distinct regulation of critical oncogenic pathways that govern the malignant behavior of GBM.
Keywords: glioblastoma (GBM), glioma, nuclear factor IA (NFIA), p53, p21, PAI1
Glioblastomas (GBM) represent the most common primary and deadly form of human brain tumors, with median survival of 15 months despite aggressive therapy.1 Many molecular aberrations contributing to gliomagenesis are being uncovered.2,3 Growing evidence also indicates possible links between glial fate determinants and glioma formation. For example, Olig2, an oligodendrocyte fate determinant, is a critical factor in the generation of gliomas in a genetically defined mouse neurosphere model.4,5 Also, studies involving astrocyte fate determinants have indicated that inhibition of STAT3 reduces tumorigenicity of gliomas in vitro and in vivo.6–8 Here, we asked whether nuclear factor IA (NFIA), a glialfate determinant, has a functional role in glioma pathogenesis.
The nuclear factor I (NFI) family of transcription factors are site-specific DNA-binding proteins that bind to a specific motif (TGGC(N5)GCCA) and function as activators or repressors of gene expression.9 Among their functions, NFI genes also participate in control of cell proliferation in vertebrate systems.9,10 NFIA, a member of the NFI family, plays an essential role in glial development in the central nervous system: it specifies glial identity, maintains glial progenitors, and regulates astrocyte differentiation in part through transcriptional regulation of glial fibrillary acidic protein (GFAP).11–14 More recently, our group and others discovered an association between NFIA and human gliomas15,16 and showed that NFIA is abundantly expressed in astroglial tumors compared with non-neoplastic brains.16 It was unknown, however, whether NFIA is involved in regulation of glioma growth.
Here, we report novel tumor-promoting functions of NFIA in GBM, demonstrating that NFIA increases cell proliferation and survival by repressing p53 and p21. Furthermore, NFIA promoted GBM cell migration by repression of PAI1, an important regulator of cell migration. Importantly, these effects were mediated via specific NFIA-recognition sequences in the promoters of p53, p21, and plasminogen activator inhibitor 1 (PAI1). Collectively, these data suggest that NFIA is a critical component of the oncogenic network that contributes to GBM aggressiveness.
Materials and Methods
Materials
ZVAD-FMK was purchased from Calbiochem, etoposide from Sigma, and rhPAI1 from PeproTech. Anti-NFIA rabbit polyclonal antibody (1:1000) was from V. Dawson, Johns Hopkins University, and Active Motif and specifically detects NFIA but not other NFI family members.17,18 Other primary antibodies for immunoblotting/immunohistochemistry were anti-GFAP, mouse monoclonal (Millipore, 1:500); anti-polyadenosine diphosphate-ribose polymerase (PARP, 1:1000); anti-caspase-8 (1:500), anti-cleaved caspase-9 (1:500), anti-CKD2 (1:1000), anti-phospho-CDK2 (Thr160, 1:1000), anti-Rb (1:1000), anti-phospho-Rb (Ser780, 1:1000), rabbit polyclonal (Cell Signaling Technology); anti-GAPDH (1:10 000), mouse monoclonal (Meridian Life Science); anti-p53 (1:500), anti-MMP2 (1:1000), and anti-PAI1 (1:500), rabbit polyclonal (Santa Cruz); and anti-p21 (1:500), mouse monoclonal (BD Pharmingen).
Cell Culture, Growth, and Soft Agar Assay
U251MG, LN18 GBM, and U87MG cells (ATCC) were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum and L-glutamine (2 mM) and maintained in 5% CO2 at 37°C. GBM1 human primary GBM cells were isolated from a freshly resected tumor obtained from NYU Human Brain Tumor Bank after IRB (7658) approval. Tumor was dissociated using Papain Dissociation System (Worthington Biochemical) according to the manufacturer's instructions. Dissociated cells were resuspended and maintained as neurospheres in serum-free Neurobasal media supplemented with N2, B27, and L-glutamine (Invitrogen), and growth factors EGF and bFGF (20 ng/mL each; Sigma-Aldrich). A161T mutation in TP53 was identified in GBM1 cells by PCR and sequencing. U87 cells have wild-type TP53 and U251, and LN18 contains mutated TP53.19 Cells were used according to the needs of the experimental design (ie, transfection efficacy, growth as monolayer, p53 mutation status, etc). For cell growth analysis, glioma cells were enumerated by direct microscopic cell and counted using trypan blue exclusion at day 3 post transduction. Soft agar assay was performed using glioma cells freshly transduced with shRNAs or stably expressing NFIA or vector. Briefly, 5 × 103 cells were seeded in soft agar (20% serum), incubated for a period of 4 weeks, and photographed at 28 days. Colonies larger than 0.1 mm were quantified using Photoshop.
Senescence-associated β-Galactosidase Assay
Three days after cells were infected with lentivirus containing shCont or shNFIA, senescence-associated (SA)β-galactosidase was assayed at pH 6.0 according to the manufacturer's instructions (InvivoGen). Three pictures using phase contrast light microscopy were taken of each well, and the number of b-galactosidase stained cells was counted.
Apoptosis Assays, Cell Cycle Analysis, and BrdU Incorporation
Apoptosis was evaluated by labeling DNA breaks with fluorescein isothiocyanate–deoxyuridine triphosphate (FITC-dUTP) followed by flow cytometry using Apo-Direct kit (BD Biosciences Pharmingen) according to the manufacturer's instructions. For cell cycle analysis, cells were stained with propidium iodide (PI)/RNase buffer for 30 min, and fluorescence was measured using a FACS-Calibur flow cytometer. DNA histograms were generated using CellQuest software (Becton Dickinson). Cell proliferation was measured using BrdU (5-bromo-2′-deoxyuridine) incorporation according to manufacturer's instructions (Calbiochem). Absorbance at 450 nm was determined after 12 hours of labeling, and backgrounds were corrected by subtracting absorbance at 590 nm.
Caspase-3 Activity
Caspase activity was measured using ApoTarget Caspase-3 Colorimetric Protease Assay (BioSource) in 200 μg of lysate proteins according to the manufacturer's instructions. Absorbance at 400/405 nm was determined after 16 hours of incubation (37°C) with the substrate.
Plasmids
To express NFIA, we used pools of glioma cells transduced by lentiviral vector containing HA-NFIA cDNA13 compared with vector control. These cells continued to stably express HA-NFIA. To achieve efficient knockdown of endogenous NFIA, we used Promega siRNA target designer. The selected sequences were integrated into oligos containing a stem-loop sequence and sites to facilitate cloning. Oligonucloetides were annealed and cloned into a Bluescript-hU6 plasmid (EcoRI/HindIII). Clones were sequence-verified, and the hU6-shRNA cassette was cloned into FG12 vector (XbaI/XhoI). Orientation was confirmed via restriction digestion and sequence analysis. For a nonsilencing control (shCont), we created 6 point mutations to the shRNA sequence to disrupt annealing to endogenous NFIA transcripts. NFIA expression in the transduced pools of cells was examined by qPCR and immunoblotting and was stable for the duration of the cells' viability. Among the 4 sequences tested, *shNFIA3 (shNFIA) resulted in ∼80% knockdown of NFIA (Supplementary material, Fig S2) and was used for subsequent experiments. The sequences of shNFIA and shCont are: 5′-AGGCACATGGAGAACTAAAT-3′ and 5′-ATGCACGTGAAGACGTATAT-3′, respectively. The plasmid containing p21 promoter (from −2341 to +9; GenBank #U24170.1) was purchased from Addgene (Cat# 16462). p53 (from −532 to −1; GenBank #J04238) and PAI1 (from −816 to +104; GenBank #NG_013213) promoters were cloned from human genomic DNA by PCR using the primers F: 5′-GGGAGAAAACGTTAGGGTG-3′, R: 5′-CCAATCCAGGGAAGCGTG-3′ (p53) and F: 5′-CCTGTTTCCTTACCAAGCTTTTAC-3′, R: 5′-AGGGGGGCGTGTGGGTCTTC-3′ (PAI1), respectively. Inserts were ligated into pGL3-basic vector (Promega) using KpnI/XhoI sites. Mutagenesis was performed using QuikchangeII site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer's instructions. pJP1520-CDKN1A (clone ID, HsCD00074777) and vector (clone ID, EvNO00023114) were purchased from Arizona State University. See Supplemental Methods for details.
Transient Transfection and Promoter Reporter Assays
Transient transfection was performed using calcium phosphate. To knockdown p53, siRNA for p53 or control (Cell Signaling; cat# 6231 and 6568, respectively; 100 nM) were transfected. After 1 day of lentiviral infection with NFIA, shNFIA, or controls, cells were cotransfected with Renilla luciferase reporter plasmid (20 ng) and pGL3 Firefly luciferase plasmid (50 ng). After 2 days of transfection, luciferase activities were measured using Dual Luciferase Reporter assay system (Promega) according to the manufacturer's instructions.
RNA Extraction and RT-PCR
RNA was extracted using TRIzol, and first-strand cDNA was synthesized using Super Script™ II Reverse Transcriptase (Invitrogen). qPCR analysis was done with the StepOne real time PCR system (Applied Biosystems) using SYBR Green PCR Master Mix (Invitrogen) and StepOne Software v2.1, according to the manufacturer's instructions. See Supplementary material, Table S1 for the primers used.
Transwell Assay
Cell migration across Transwells was analyzed using Boyden polycarbonate Transwell membrane chambers with 8 μm pores (Corning). DMEM with 10% FBS was added to lower chambers, and cells in serum-free media were seeded onto the filter of upper chambers. After 12 hours, migrated cells were visualized using MTT staining (5 mg/mL, 3 h incubation) and photographed. The number of migrated cells was counted, and the extent of migration was expressed as cell number fold changes compared with controls. Where noted, conditioned media or rhPAI1 were added to cells.
Wound-healing Assay
Twelve hours following seeding, confluent cell monolayers were treated with mitomycin-C (10 μg/mL, 1 h) in serum-free DMEM to inhibit cell proliferation and then washed with phosphate-buffered saline. Fresh DMEM containing 10% FBS and rhPAI1 (1 ng/mL) was added to cells, and then wound was created with a 10 μL pipette-tip. Pictures of the central region of wound were taken immediately and at various time points after wound creation using a phase-contrast light microscopy. Quantification was performed on the images using Photoshop and calculated as percent of wound closed at each time point.
In Vivo Tumor Formation
Intracranial orthotopic transplantation of glioma cells into mice and monitoring by MRI and bioluminescence were performed as previously described20,21 and in accordance with animal-research protocol at Children's Hospital Los Angeles. Glioma cells (2 × 105 viable cells verified by trypan blue exclusion), freshly transduced with shRNAs or stably expressing NFIA or vector, were implanted into the forebrains of 6–8 week old athymic mice. MRI was performed 21 and 28 days after transplantation,21,22 and tumor volume was quantified using Metamorph.
Immunohistochemistry
Cells or tissue sections were fixed with 4% paraformaldehyde and incubated at 4°C overnight with primary antibodies, followed by incubation at room temperature with AlexaFluor-conjugated secondary antibodies (Invitrogen). After nuclei labeling (Hoechst), cells or sections were mounted with Vectashield mounting media and analyzed using EclipseE800 microscope (Nikon Instruments) and Nikon FDX-35 camera (Axiovision software, Carl Zeiss), and Olympus IX71 microscope and Olympus U-CMAD3 camera (cellSens software, Olympus).
Immunoblotting
Whole cell lysates were fractionated on 10 or 12% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Milipore). Blots were incubated overnight (4°C) with primary antibodies, probed with horseradish peroxidase-conjugated secondary antibodies, and visualized by enhanced chemiluminescence (Amersham). Immunodensitometry was measured using ImageJ (National Institutes of Health).
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 5.0c for MacIntosh (GraphPad Software). All experiments were performed in triplicate and repeated at least 3 times unless indicated otherwise. Data are expressed as mean ± standard deviation (SD). P-values were calculated by unpaired Student' t test. Significance level was set at P < .05.
Results
Expression of NFIA Modulates Glioma Cell Growth
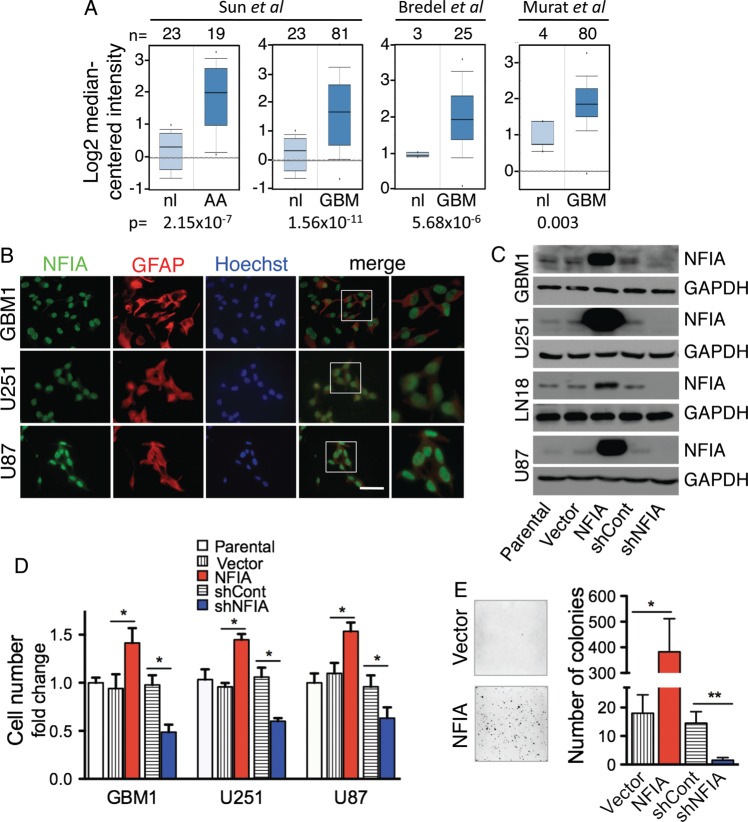
We previously showed by immunohistochemistry that NFIA is highly expressed in human astrocytomas of all grades compared with non-neoplastic brains.16 Microarray data from Oncomine23–25 confirms that NFIA expression is indeed elevated in malignant gliomas (Fig. 1A). Further, in silico analysis of GBMs in The Cancer Genome Atlas (TCGA)26 demonstrates that NFIA is high in proneural GBMs, a subgroup in which TP53 mutations are more common, as well as in classical GBMs, which lack TP53 mutations (Supplementary material, Fig. S1).
Fig. 1.
NFIA promotes growth of glioma cells in culture. (A) NFIA expression is elevated in malignant gliomas. Whisker plots of NFIA mRNA levels in malignant gliomas (AA, n=19; GBM, n=186) compared with normal brains were analyzed using 4 different datasets available from Oncomine (http://www.oncomine.org). The numbers above the plots indicate the number of samples. Also see Figure S1. (B) NFIA is expressed in glioma cells. Immunocytochemical staining for NFIA (green), GFAP (red), and Hoechst (blue). Scale bar: 50 µm. (C) Overexpression and knockdown of NFIA by shRNA (shNFIA) was verified by immunoblotting of whole cell lysates on day 3 after infection with NFIA, empty vector, shNFIA, or shRNA control (shCont). (D) Overexpression of NFIA increases, and shNFIA decreases the number of cultured glioma cells 3 days after infection with lentivirus NFIA (red), shNFIA (blue), and controls (vector only and shCont). *P<.05: P-values between parental cells and controls were not significant. (E) 3D colony formation in soft agar is increased in glioma cells overexpressing NFIA. U87 cells expressing NFIA, shNFIA, or controls 3 days after lentiviral transduction were plated in soft agar, and colonies were evaluated after 28 days. Shown are representative fields (left) and quantification of colonies larger than 0.1 mm (right) on day 28; n = 3, means ± SD; *P < .05, **P = .01.
We found endogenous NFIA expression in cultured cells in both patient-derived primary human GBM cells (GBM1) and in established GBM cell lines (U251, LN18, U87) (Fig. 1B and C). Manipulation of NFIA expression level affected cell growth such that ectopic expression of NFIA increased the number of GBM cells in culture, whereas shRNA knockdown of native NFIA (shNFIA) decreased cell number compared with control shRNA (shCont) (Figs 1C, D and S2). Despite the relatively lower overexpression level of NFIA in the GBM1 neurospheres due to their lower infection efficiency, the increase in cell numbers in response to NFIA was similar in the 3 cell lines. This may be due to GBM1 being inherently more responsive to the increase in NFIA level. Another possible explanation for increased response to NFIA expression in GBM1 cells could be yet-unidentified paracrine factors secreted in response to NFIA expression, which would be anticipated to have more marked effect in neurospheres and thus contribute to the increased proliferation. Furthermore, colony formation in soft agar was increased by NFIA overexpression by ∼20 times and was decreased by NFIA knockdown (Figs 1E and S3). This suggests that NFIA may have a novel tumor-promoting role in glioma cells.
NFIA Regulates GBM Proliferation and Cell Death
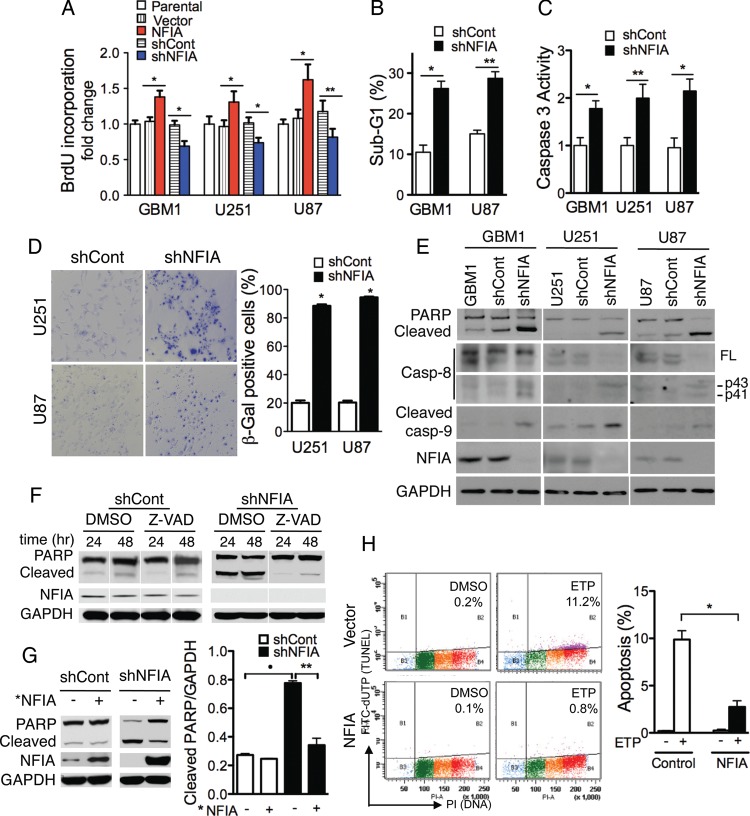
To evaluate the nature of NFIA's effect on GBM cell number, we assessed the effect of changing NFIA level on BrdU uptake in both primary GBM cells and GBM cell lines. NFIA overexpression increased BrdU incorporation by ∼30%–50% while shNFIA decreased it (Fig. 2A), indicating that NFIA promoted GBM proliferation. In addition, knockdown of NFIA increased glioma cell death, as reflected by increased number of cells in sub-G1 phase (Fig. 2B), and increased caspase-3 activity (Fig. 2C). Furthermore, knockdown of NFIA increased SA β-galactosidase staining of glioma cells (Fig. 2D), consistent with increased senescence. NFIA knockdown also increased cleavage of PARP, caspase-8, and caspase-9 (Fig. 2E), which is indicative of apoptosis.27 The broad-spectrum caspase inhibitor, Z-VAD-fmk, effectively prevented PARP cleavage in glioma cells expressing shNFIA, further supporting the caspase requirement in NFIA-dependent apoptosis (Fig. 2F). A “rescue” NFIA (*NFIA), comprising the coding sequence of NFIA but lacking the 3′UTR and thus resistant to the 3′UTR-targeted shNFIA, reversed the shNFIA-induced PARP cleavage, demonstrating the specificity of the targeted NFIA knockdown (Fig. 2G). Lastly, etoposide-induced apoptosis28,29 was reduced by greater than 4 times in NFIA-overexpressing cells (Fig. 2H), suggesting that high NFIA expression may allow glioma cells to become resistant to chemotherapy-induced apoptosis. Taken together, these data demonstrate that NFIA promotes GBM cell proliferation and survival and that loss of NFIA induces cell death and apoptosis.
Fig. 2.
NFIA controls proliferation and cell death. (A) NFIA promotes and shNFIA inhibits proliferation (BrdU uptake) of glioma cells freshly transduced with lentivirus expressing NFIA (red), shNFIA (blue), and controls. Means ± SD of 3 different experiments performed in 3 to 8 replicates; *P < .0001, **P < .05. (B) Knockdown of NFIA increases sub-G1 fraction of glioma cells. GBM1 or U87 glioma cells transduced with shNFIA or control analyzed by propidium iodide (PI) on day 3 after lentiviral infection; n = 3; *P < .0005, **P < .0001. (C) NFIA silencing increases caspase-3 activity. Caspase-3 activity was determined on day 3 after infection with shNFIA or shCont; n = 3; *P < .005, **P < .01. (D) Glioma cells infected with shNFIA or control shRNA were stained with SA-β-gal (x400) on day 3 of transduction. Quantification of SA-β-gal positive cells is on the right panel; n = 3, *P < .0001. Knockdown of NFIA is shown in panel E. (E) Loss of NFIA causes apoptosis evidenced by cleavage of PARP, caspase-8, and caspase-9. Whole cell lysates of glioma cells analyzed on day 3 after infection with shNFIA, vector control, or uninfected parental cells (immunoblotting). Shown is a representative experiment of 3 experiments. (F) Caspase inhibitor, Z-VAD, blocks shNFIA-induced PARP cleavage. Whole-cell lysates of shNFIA and control-infected U251 glioma cells 3 days after lentiviral infection and cultured with Z-VAD-fmk (20 μM) or vehicle (DMSO) for additional 24 and 48 hours were analyzed by immunoblotting. (G) *NFIA (protein coding domain only, resistant to the shNFIA) reverses shNFIA-induced PARP cleavage in U251 glioma cells. After 24 hour infection with shNFIA or control in U251, cells were reinfected with *NFIA for 48 hours and analyzed for PARP cleavage (left). Right panel: densitometric quantification; *P<.001, **P<.01. (H) NFIA protects glioma cells from etoposide-induced apoptosis. U251 glioma cells stably expressing NFIA or vector were treated with vehicle (DMSO) or etoposide (ETP, 1 μg/mL) in serum-free medium for 24 hours. Apoptosis was assessed using the Apo-Direct kit. Left panel: representative experiment of 3 experiments. The percentage of apoptotic cells (FITC-dUTP+ cells in top quadrants) is indicated for each condition. Right panel: means ± SD from 3 experiments; *P < .0001.
Apoptosis Induced by Loss of NFIA Is Mediated Through p53
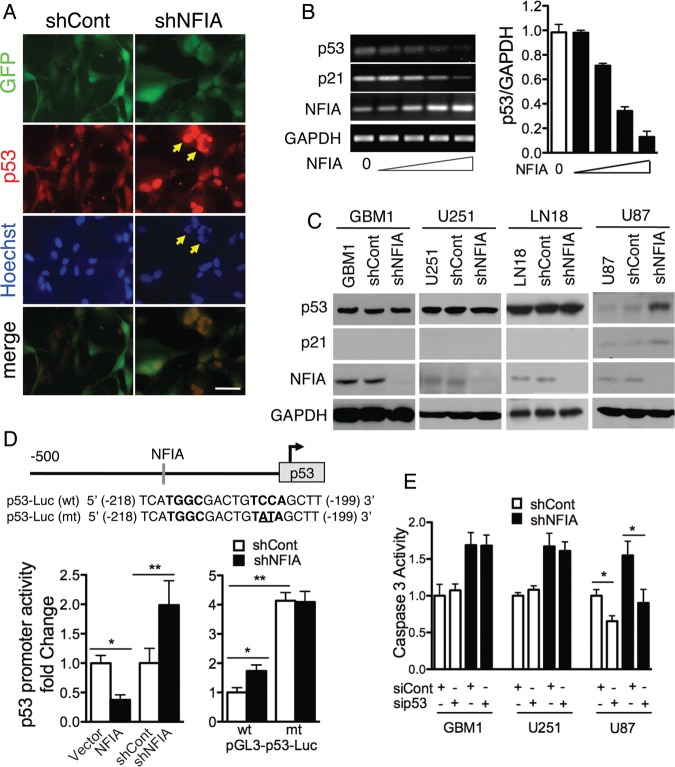
Loss of the TP53 tumor suppressor gene or loss of its function is a common oncogenic event in GBM and increases resistance to apoptosis.30–33 The increased apoptosis in shNFIA-treated cells (Fig. 2) prompted us to ask whether p53 was involved. Indeed, p53-wild-type cells (U87) treated with shNFIA showed increase in nuclear p53 and nuclear fragmentation, characteristics of apoptotic cells (Fig. 3A, arrowheads). Furthermore, transfection of NFIA repressed p53 mRNA level in a dose-dependent manner (Fig. 3B). Complementing this, shNFIA increased p53 protein expression in these p53 wild-type cells (Fig. 3C). However, shNFIA did not affect p53 protein expression in the p53-mutant cells, GBM1, U251, and LN18 (Fig. 3C),19 which natively express stabilized (and increased) mutant p53 protein compared with p53-wild-type cells.34 To determine if the effect of NFIA on wild-type p53 expression was through transcriptional regulation, we measured p53 promoter activity using p53 reporter constructs. These constructs contained either a wild-type p53 basal promoter (p53-Luc-wt) or a mutant counterpart (p53-Luc-mt), which we mutated in the NFIA-binding site.35,36 This NFIA binding site in the promoter conforms to the predicted binding site of other NFI-family members (Fig. 3D, top panel).35,36 Wild-type p53 promoter reporter activity was repressed in U87 cells overexpressing NFIA and increased in cells treated with shNFIA (Fig. 3D, left panel and S4A). Furthermore, use of p53-Luc-mt showed maximal increase in luciferase activity, irrespective of knockdown by shNFIA treatment (Fig. 3D, right panel), indicating that this mutation freed the p53 promoter from NFIA-induced repression. To determine whether shNFIA-induced apoptosis was indeed mediated by p53, we tested if knockdown of p53 (sip53) would prevent shNFIA-induced increase in caspase-3 activity (siCont; Fig S5 shows sip53 efficacy). Knockdown of p53 attenuated shNFIA-induced caspase-3 activity as well as baseline caspase-3 activity in shCont U87 cells (Fig. 3E), consistent with the wild-type functional p53 in U87. Conversely, p53 knockdown did not suppress baseline or shNFIA-induced caspase-3 activity in GBM1 and U251 (Fig. 3E), consistent with their mutant p53 status. Notably, NFIA knockdown itself increased caspase-3 activity even in the GBM1 and U251 cells, suggesting that caspase-3 activation induced by NFIA knockdown is mediated via both p53-dependent and p53-independent mechanisms. Taken together, these findings demonstrate that NFIA regulation of apoptosis is mediated in part by transcriptional repression of p53 and thus suggest the presence of a p53-independent pathway.
Fig. 3.
Apoptosis induced by loss of NFIA is mediated by p53. (A) Immunofluorescence of U87 cells expressing shNFIA and control (shCont) 4 days after transduction; p53 - red, Hoechst - blue. Arrowheads: examples of fragmented nuclei with high nuclear p53. Scale bar: 50 µm. (B) NFIA represses p53 mRNA. p53, p21, and NFIA mRNA (RT-PCR) from U87 cells transduced for 2 days with 0, 1/27, 1/9, 1/3, 1 µg lentiviral plasmid containing NFIA cDNA and adjusted to total 1 µg transfected DNA using empty-vector plasmid. Right panel: p53 mRNA normalized to GAPDH; n = 3; P < .0001 by 1-way ANOVA. (C) Loss of NFIA induces p53 in p53-wild-type U87 cells but not in the p53-mutant GBM1, U251 and LN18 cells. Glioma cells 3 days post lentiviral transduction with shNFIA or control shRNA were assessed for p53 and p21 expression by immunoblotting. (D) NFIA negatively regulates the p53 promoter. Top: NFIA consensus-binding sequence (−199 bp) in the p53 promoter. Left: relative luciferase activity 48 hours after transfection of p53 luciferase reporter into U87 cells stably expressing NFIA or vector or 3 days post lentiviral transduction with shNFIA or controls; *P < .005, **P < .05, see also Figure S4A. Right: relative luciferase assay using a wild-type p53 reporter or a mutated counterpart with destroyed NFIA binding site (see top panel) in U87 cells 3 days post lentiviral transduction with shNFIA or shCont; n = 3; *P < .01, **P < .001. (E) Knockdown of p53 attenuates shNFIA-induced caspase-3 activity in U87 cells but not in p53-mutant cells (GBM1 and U251). Glioma cells 3 days post lentiviral transduction with shNFIA or shCont were treated with p53 siRNA or control siRNA (100 nM) for 48 hours, and caspase-3 activity was measured; n = 3, *P < .05; Representative protein expression and densitometric analysis are shown in Figure S5.
Growth Promotion Induced by NFIA Is Mediated Through Repression of p21
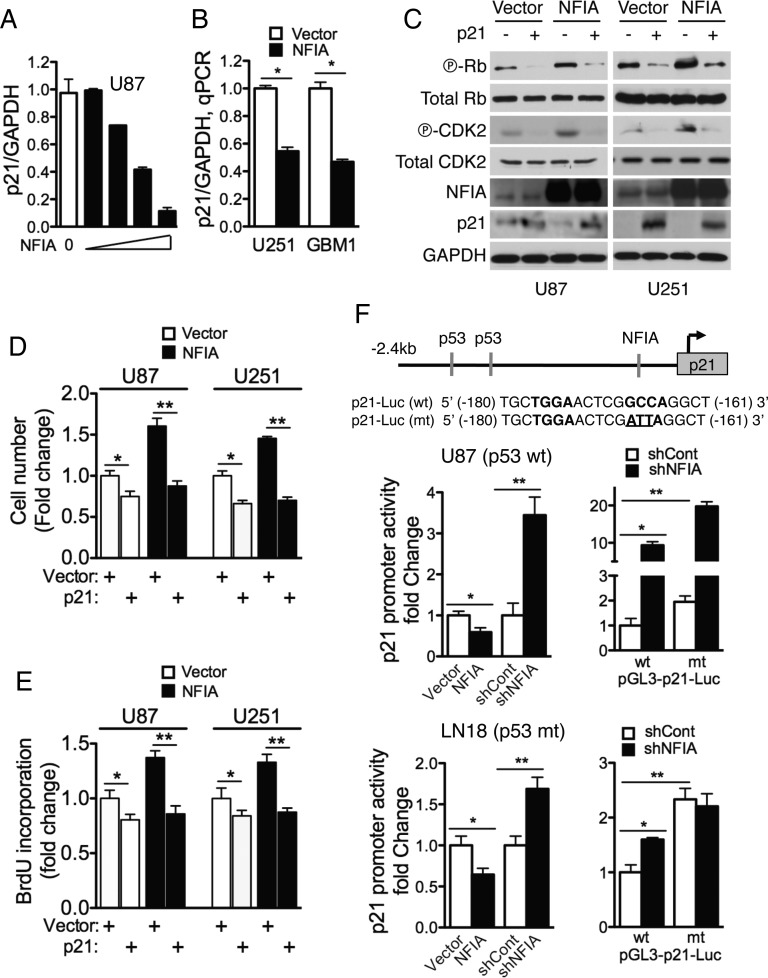
The proliferative effect of NFIA in glioma cells and transcriptional repression of wild-type p53 by NFIA prompted us to examine whether p21 (CDKN1A, WAF1/CIP1), an important cell cycle regulator and downstream transcriptional target of p53, was also regulated by NFIA. Overexpression of NFIA effectively repressed p21 mRNA levels in a dose-dependent manner (Figs 3B and 4A–B). Furthermore, phosphorylation of CDK2 and Rb, key cell cycle proteins regulated by p21, was also increased by NFIA overexpression (Fig. 4C). To test the role of p21 in mediating NFIA-induced GBM proliferation and cell growth, we transiently expressed exogenous p21 in NFIA-overexpressing cells or their controls. As expected, ectopic p21 expression repressed baseline proliferation and cell growth of control glioma cells (Fig. 4D and E). Importantly, ectopic p21 also completely abolished the NFIA-induced increase in cell growth and proliferation (Fig. 4D and E) as well as phosphorylation of CDK2 and Rb (Figs 4C and S6A–B). Using a p21 luciferase reporter assay, we found that p21 promoter activity was repressed by overexpression of NFIA and increased by knockdown of native NFIA using shNFIA (Fig. 4F, left panels, and S4B). Mutation of the NFIA binding site in the p21 promoter (p21-Luc-mt)37 itself increased p21 promoter activity in shCont cells (Fig. 4F, right panels), suggesting that this NFIA binding site is functional. Interestingly, in p21-Luc-mt cells, knockdown of native NFIA (shNFIA) further increased reporter activity in U87 cells (>10 times), but not in the LN18 p53-mutant cells (Fig. 4F, right panels). These results suggest that the NFIA effect on p21 transcription is mediated through both p53-dependent (NFIA→p53→p21) and p53-independent (NFIA→p21) pathways in p53 wild-type cells, while NFIA repression of p21 is p53-independent in p53-mutated cells (NFIA→p21; schema in Fig. S6C). Taken together, these findings suggest that NFIA is a critical regulator of p21 in glioma cell proliferation via p53-dependent as well as p53-independent pathways.
Fig. 4.
Growth properties induced by NFIA is mediated by repression of p21. (A) NFIA represses p21 transcription. Normalized p21 mRNA relative to GAPDH in U87 cells transduced with increasing NFIA plasmid as described in Figure 3B (RT-PCR). P < .0001 by 1-way ANOVA; n = 3. (B) GAPDH-normalized p21 mRNA in U251 and GBM1 cells stably expressing NFIA or vector control; qPCR; *P < .0001. (C) Overexpression of NFIA regulates phosphorylation of cell-cycle dependent proteins, CDK2 (T160) and Rb (S780). Western blot of whole cell lysates of 18 hour serum-starved (ie, synchronized) glioma cells stably expressing NFIA or vector control. (D and E) Overexpression of p21 inhibits NFIA-induced cell growth and proliferation. Cell growth (D) or BrdU incorporation (E) were measured in U87 (p53-wild-type) or U251 (p53-mutant) cells stably expressing NFIA or vector that was transfected with p21 or empty-vector plasmid for 48 h. (assessed as in Figs 1D and 2A). Representative protein expression and densitometric analysis are shown in Figure S6A and B. *P < .01, **P < .0005 (D); *P = .001, **P < .0001 (E). (F) NFIA represses p21 promoter activity. Top: wild-type and mutant NFIA binding sites (161 bp) in the p21 promoter luciferase reporter. Left: luciferase activity was measured 48 hours after transfection of p21 luciferase reporter into U87 (p53-wild-type; top panels) or LN18 (p53-mutant; bottom panels) cells stably expressing NFIA or vector or 3 days post lentiviral transduction with shNFIA or controls; *P < .01, **P = .001; n = 3. Right: Relative luciferase activity of pGL3 wild-type (wt) or mutant (mt) p21 promoter transfected into U87 or LN18 cells expressing shNFIA or shCont similar to the left panel; see also Figure S6C for a schema; *P = .0001, **P < .05 (U87); *P < .005, **P < .001 (LN18).
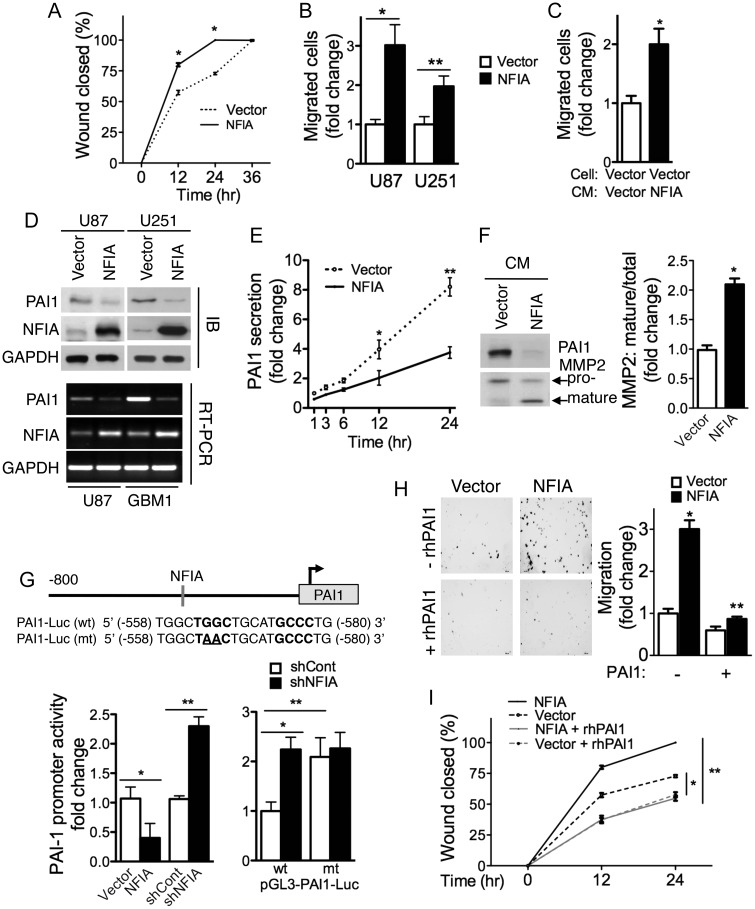
NFIA Enhances Migration of GBM Cells by Repression of PAI1
Local invasive growth and potent angiogenesis are 2 major challenges of GBMs that hamper effective cure. Our earlier study showed that NFIA expression is prominent in the perivascular infiltrating tumor cells in gliomas,16 suggesting that NFIA may play a role in tumor cell migration and/or invasion. Indeed, in a tissue culture wound-healing assay, which is dependent on cell migration, U251 cells overexpressing NFIA closed the wound more rapidly than vector-expressing cells (Fig. 5A). Increased migration of NFIA overexpressing U87 and U251 cells was also observed in a Transwell assay (Figs 5B and S7A). These observations demonstrate a novel promigratory role of NFIA in glioma cells.
Fig. 5.
NFIA enhances migration in glioma cells through PAI1. (A) Wound healing in U251 cells stably expressing NFIA or vector was assessed at 0, 12, 24, and 36 hours after wound was created. Migration into the wound was quantified as the percentage of wound closed at each time point; mean ± SD; *P < .0001. (B) Transwell migration assay of U87 and U251 cells stably expressing NFIA or vector; n = 3; *P < .005, **P = .001. (C) Transwell migration of U87-vector cells in conditioned media harvested from U87-vector or -NFIA stably expressing cells. Migration was measured as in B; *P < .005. See also Figure S7B. (D) NFIA represses PAI1 expression. Top: U87 and U251 cells stably expressing NFIA show lower PAI1 protein level. A representative immunoblotting of whole cell lysates; immunodensitometry from 3 independent experiments is shown in Figure S7C; *P < .0001. Bottom: relative PAI1 mRNA normalized to GAPDH in NFIA versus empty vector expressing cells in 3 experiments; quantification is shown in Figure S7C; *P < .001. (E) PAI1 secretion is suppressed in U87 cells overexpressing NFIA. Conditioned media from equal number of cells (2 × 105) stably expressing NFIA or vector were harvested at the indicated times, analyzed by immunoblotting, and quantified by densitometry (see also Fig. S7E); *P < .05, **P = .0005. (F) Conditioned medium (24 h) from U87 cells stably expressing NFIA or vector from equal cell numbers was analyzed for expression of MMP2 by immunoblotting. Upper and lower arrows indicate pro and mature MMP2, respectively. Right: densitometry of MMP2 mature/total in 3 experiments; *P = .0001. (G) NFIA represses the PAI1 promoter. Top: NFIA DNA binding site (-540 bp) in the pGL3-PAI1 promoter reporter plasmid: wild-type or mutated to destroy the PAI1 binding site. Left: wild-type PAI1 reporter luciferase activity in U87 cells stably expressing NFIA or -vector, or 3 days post lentiviral transduction of shNFIA, or controls; *P < .05, **P < .0005. Right: relative luciferase activity of pGL3 wild-type (wt) or mutant (mt) PAI1 promoter reporter in U87 cells expressing shNFIA or shCont; *P < .005, **P = .01. (H and I) Addition of recombinant human PAI1 (rhPAI1) inhibits NFIA-induced migration in glioma cells. (H) Transwell migration of U87-NFIA or vector expressing cells with/without rhPAI1 (1 ng/mL). Left: representative field of 5. Right: mean ± SD migrated cells from 3 experiments; *P < .0001, **P < .005. (I) The effect of addition of rhPAI1 (1 ng/mL) in U251-NFIA or -vector stably expressing cells was measured by wound-healing assay. Percentage of wound closure; *P < .0005, **P < .0001. See also Figure S7F in Supplementary information.
Interestingly, conditioned medium from NFIA-expressing cells was also able to enhance migration of U87 vector-control cells across the membranes of Transwells (Figs 5C and S7B), suggesting an effect by a secreted factor. The plasminogen activation (PA) system and its inhibitor PAI1 have been implicated in invasion, migration, and metastasis of cancers, including gliomas,38 through activation of matrix metalloproteinases (MMPs) and degradation of extracellular matrix proteins.39 In addition, PAI1 was found to harbor the consensus NFI-binding motif within its promoter,40 suggesting that PAI1 may mediate NFIA-induced migration. We therefore first examined whether NFIA could regulate PAI1 expression in glioma cells. Overexpression of NFIA in U87 and U251 cells significantly reduced PAI1 protein levels and PAI1 mRNA expression (Figs 5D and S7C). The effect of NFIA on PAI1 mRNA was specific, as the mRNA levels of 2 other components of the system, the urokinase-type plasminogen activator (uPA) and the uPA receptor (uPAR), were unchanged (Supplementary material, Fig. S7D). Furthermore, NFIA also decreased secretion of PAI1 and increased processing of MMP2 to mature MMP2 in conditioned media of NFIA-expressing U87 cells (Figs 5E, F and S7E). To determine whether the effect of NFIA on PAI1 repression was also through transcriptional regulation, we measured PAI1 promoter activity using luciferase reporter constructs. PAI1 promoter activity was repressed by overexpressing NFIA in U87 cells and increased in cells treated with shNFIA (Figs 5G, left panel, and S4C). Mutation of the NFIA binding site in the promoter of PAI in the reporter (Fig. 5G, top panel, pGL3-PAI1-Luc mt) increased PAI1 promoter activity independently of native NFIA knockdown by shNFIA (Fig. 5G, right panel). To demonstrate that downregulation of PAI1 was the cause of the promigratory effect of NFIA in glioma cells, we tested if recombinant human PAI1 (rhPAI1) could reverse the effect of NFIA on migration. Indeed, addition of rhPAI1 to U87 cells overexpressing cells abolished their NFIA-induced migration in Transwells (Fig. 5H). Consistent with this, addition of rhPAI1 to the medium also blocked NFIA-enhanced wound healing (Figs 5I and S7F). Together, these findings suggest that NFIA promotes glioma cell migration and that this is, at least in part, by transcriptional repression of PAI1.
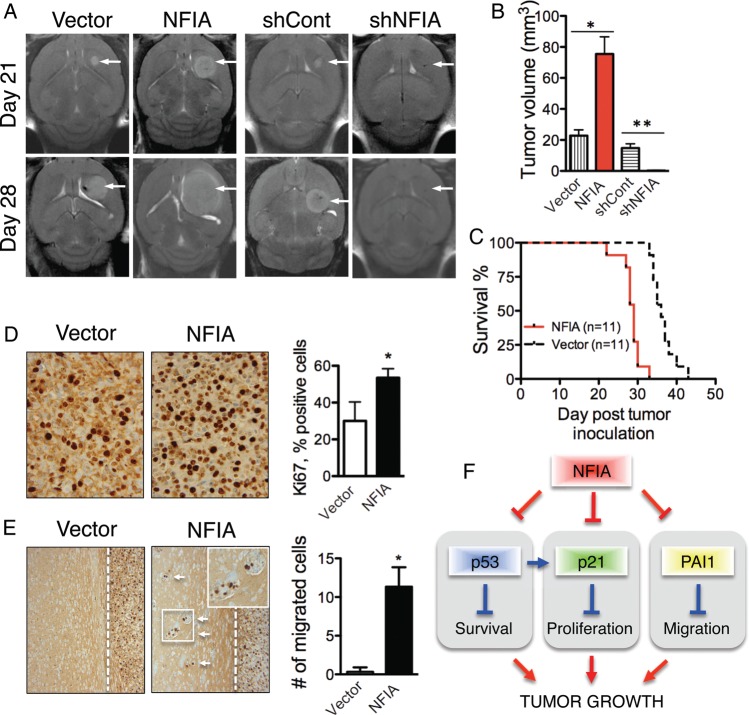
NFIA Is Necessary and Sufficient for Promoting Growth and Invasion of Gliomas in Mouse Brain
Lastly, we evaluated the effect of NFIA expression on growth of intracranial gliomas in vivo. Pools of U87 cells stably expressing NFIA, NFIA-shRNA (shNFIA), or their controls (empty vector or shCont, respectively) were implanted orthotopically into the forebrains of nude mice, and tumor growth was followed by MRI (Fig. 6A). Tumors from NFIA-overexpressing cells were larger on day 21 and day 28 compared with empty vector controls (Fig. 6A and B). Complementing this, mice injected with shNFIA-transduced cells did not form any tumors, whereas mice injected with shCont-transduced cells exhibited tumors comparable with those injected with empty vector controls (Fig. 6A and B). It is possible that they were not capable of growing in vivo due to their low proliferative capacity, ongoing senescence, and apoptosis, although shNFIA cells were viable and healthy-appearing when injected intracranially on day 3 after their transduction. Similar results were obtained using U251 cells expressing NFIA and vector controls (Supplementary material, Fig S8). All mice implanted with U87 cells overexpressing NFIA showed tumor-related symptoms by 33 days (Fig. 6C). Furthermore, the NFIA-overexpressing tumors were more proliferative, as reflected by their higher fraction of Ki-67-positive cells (Fig. 6D). Moreover, NFIA-overexpressing U87 tumors, but not vector controls, demonstrated invasion of tumor cells beyond the tumor border (Fig. 6E). These findings are consistent with our in vitro results and suggest that NFIA is necessary and sufficient for promoting growth and migration of gliomas in vivo (Fig. 6F).
Fig. 6.
NFIA is sufficient and necessary for glioma growth and invasion in mouse brain. U87 cells stably expressing NFIA or vector, or 3 days post lentiviral transduction of shNFIA, or control were orthotopically implanted into the brains of nude mice and followed. (A) MRI of intracranial tumors (arrows) in representative mice. Each brain is shown on day 21 and day 28 post tumor inoculation. (B) Mean ± SD of tumor volumes on day 28 MRI, measured using Metamorph. *P < .005 between NFIA versus control (n = 11 mice in each group); **P = .0001 between shNFIA vs shCont;n = 10 mice in each group. Performed in 2 separate identical experiments. (C) Kaplan–Meier survival curve of mice implanted with U87 cells overexpressing NFIA and vector control; n = 11 in each group; P < .001). (D) Ki67 immunohistochemistry of the orthotopic U87 tumors overexpressing NFIA or vector control. Mice were euthanized on day 28 post inoculation, and Ki67-positive cells were counted in 2 high-powered fields (x400) from 3 tumors for each condition. Bar graph shows means ± SD; *P = .0005. (E) Ectopic expression of NFIA in U87 enhances migration in vivo. Small groups of satellite tumor cells were found away from the main tumor mass in orthotopic U87 tumors overexpressing NFIA (examined at day 28). The inset of migrated cells is shown in magnification. Empty vector-expressing tumors revealed well-circumscribed tumor border (examined at days 35–38). Migrated cells were counted in 2 fields (x200) from 3 tumors for each condition; *P < .005. (F) NFIA promotes GBM growth by regulation of p53, p21, and PAI1. NFIA suppresses p53, p21, and PAI1 expression, resulting in decreased apoptosis and increased proliferation and migration, which leads to the enhanced tumor growth.
Discussion
The NFI family of transcription factors are key regulators in development and in central nervous system processes.9 We have reported that NFIA, a member of this family involved in glial development, is highly expressed in astrocytomas.16 In this work, using gain-of-function and loss-of-function experiments, we demonstrated that NFIA inhibited cell death and enhanced cell survival, proliferation, and migration in GBM through negative regulation of p53, p21, and PAI1. These newly described tumor-promoting activities of NFIA establish a mechanistic link between this glial fate determinant and GBM aggressiveness.
NFIA is critical in lineage decision during development: it determines glial cell fate in the CNS and is also involved in erythroid/granulocytic lineage decision of hematopoietic progenitors. In cancer cells, however, NFIA may have differential roles. While our study demonstrated a tumor-promoting role for NFIA in glioma, overexpression of NFIA in chronic myelogenous leukemia (K562) cells restores normal erythroid program,41 and knockdown of NFIA in acute promyelocytic leukemia cells enhances myeloid differentiation.42 In addition, NFIA gene silencing allowed myeloid progenitors to enter terminal granulocytic commitment through epigenetic modulation.43 This suggests that the function and effect of NFIA may differ depending on the context and the cell type.
TP53, a potent tumor suppressor that regulates cell cycle progression and induces apoptosis, is frequently mutated in GBM26,44 and is mainly regulated by posttranscriptional or posttranslational mechanisms. Our finding that part of the prosurvival effect of NFIA is mediated via transcriptional repression of the p53 promoter uncovers a novel and important molecular mechanism by which p53 signaling can be silenced in otherwise TP53-wild-type GBMs. This may be particularly relevant in the classical GBM subgroup tumors, which typically have wild-type TP5326 yet express high NFIA mRNA (Supplementary material, Fig. S1), which may suppress their wild-type-p53 and thus contribute to tumor progression.
Loss of NFIA-mediated repression of p53 by functional mutation of the NFIA binding site in the p53 promoter suggests that this repression is highly sequence-specific and indicates that NFIA may target the p53 promoter in human GBM. Interestingly, Johannson et al reported that NFIA has much lower affinity than another isoform, NFIC2, to the NFI binding site in the mouse p53 promoter and that it is not involved in p53 gene regulation in mammary epithelial cells.45 This suggests that the effects of NFI family members may be isoform-specific and/or cell-type specific, as previously suggested.35,46
Our findings that NFIA knockdown caused apoptosis in both p53-wild-type and p53-mutant GBM (Figs 2 and 3) indicate that the anti-apoptotic effect of NFIA may have both p53-dependent and p53-independent components. As p21 is a downstream target of p53 and an inhibitor of cell cycle progression,47 our finding that p21 was downregulated by NFIA in both p53-wild-type and p53-mutated GBM cells may explain the p53-independent effects of NFIA. NFIA repressed p21 luciferase reporter activity in GBM cells via the NFIA-recognition site in the p21 promoter (Figs 4F and S4B), further suggesting that p53-independent repression of p21 by NFIA may contribute to the growth advantage in p53-mutated GBM cells. The clinical relevance of this is further supported by TCGA analysis,26 which shows high NFIA in proneural GBMs, a subgroup in which TP53 mutations are common (Supplementary material, Fig S1). This suggests that the p53-independent effects of NFIA may further contribute to growth of TP53-mutated proneural GBMs.
Levels of cellular p21 are mainly controlled by transcriptional regulation,48 in particular by wild-type p53. p53 binds to distal regions on the p21 promoter at −2.26 kb and −1.37 kb,49 which are both included in the p21 reporter construct used here. In our studies, NFIA repressed p21 transcription via a proximal region at -161 bp on the p21 promoter, a site that is distinct from the p53 recognition site (Fig. 4F). The proximal interaction of NFIA with the p21 promoter seems to be a central inhibitory mechanism, as other NFI family members also bind to this site and repress p21 transcription in other cell types.37,46 Considering that NFIA and p53 may bind to distinct sites in the p21 promoter, it is unlikely that their binding would be competitive. This is supported by our observation that the p21-Luc-mt reporter activity was higher than that of the p21-Luc-wt in U87 control cells (p53 wild-type) (Figs 4F and S6C). The increase in p21 promoter activity in U87 cells harboring the mutated p21 promoter reporter (and without knockdown of NFIA) was likely due to inability of the native NFIA to bind to this mutated p21 promoter reporter (Supplementary material, Fig. S6C). When U87 cells expressing the mutated p21 luciferase reporter were additionally treated with shNFIA, p21 promoter activity was further increased, likely due to increase in endogenous p53 brought on by the decreased endogenous NFIA. This interpretation is consistent with the maximal increase of the p21 promoter mutated at the NFIA binding site in the p53-mutated line, LN18 (Fig. 4F bottom; schema in S6C), which showed no further increase by NFIA knockdown. Thus, our report indicates that NFIA is a critical regulator of p21 and cell proliferation in GBMs.
The invasive nature of GBM is a major clinical challenge. PAI1 is a critical regulator of invasion, angiogenesis, and metastasis in cancer.50 While PAI1 impairs migration/invasion of many cell types, including glioma cells (D54MG),38,51 its expression is associated with poor survival in gliomas.52–54 These seemingly conflicting results could be partly explained by dose-dependent effects of the uPA/PAI1 system55 or non-cell autonomous effect on other cells such as endothelial cells.56 However, the mechanism by which PAI1 contributes to the malignant phenotype of glioma is still largely unknown. We demonstrate here that NFIA promotes migration of glioma cells by inhibiting PAI1 expression and thereby activating MMP2, whichb indicates a novel NFIA effect on a protease-dependent pathway mediated by PAI1 (Fig. 5). The specific repression of PAI1 by NFIA in GBM and the role of NFIA in GBM migration were previously unrecognized, and thus our results also contribute to the understanding of GBM invasiveness. A TGFβ-responsive CTF/NFI binding site in the PAI1 promoter has been described in Hep3B and NIH3T3 cells.40 Under those conditions, however, the binding site mediated TGFβ-induced increase in PAI1 transcription rather than repression, as we found here. During development, NFI family members have also been implicated in migration of cortical granular neurons and astrocyte precursor cells in mouse brains through regulation of cell adhesion molecules (ephrin B1 and N-cadherin) and Apcdd1 (adenomatosis polyposis coli downregulated 1), respectively.14,57 It is currently unknown if these targets are also involved in NFIA-induced migration in GBM or if there is a functional relationship between PAI1 and these targets in NFIA-induced migration.
In summary, this is the first report demonstrating that NFIA promotes the malignant behavior of GBM, namely inhibition of cell death and promotion of proliferation and migration, and that these NFIA effects are mediated through specific transcriptional regulation of p53, p21, and PAI1. Our report provides new insights into the role of NFIA in glioma growth and may prove important in development of novel therapeutic approaches for glioblastomas.
Supplementary Material
Funding
This work was supported in part by grants from National Institutes of Health (R01NS071153 to B.D.); Grayson's Gift, Nautica Malibu Triathlon Fund, and Bogart Pediatric Cancer Research Program (A.E.); Child Neurology Foundation Shields Award, Children's Cancer Research Fund, Hyundai Hope on Wheels, National Institutes of Health (K12HD052954, and K08NS064297 to H.S.); and NYU Cancer Center Support Grant P30CA016087.
Supplementary Material
Acknowledgments
We thank Valina Dawson for the NFIA antibody, Hong Zhou for technical assistance, and Moses Chao and Martine Torres for critical review of the manuscript.
Conflict of interest statement. None declared.
References
- 1.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Siebzehnrubl FA, Reynolds BA, Vescovi A, Steindler DA, Deleyrolle LP. The origins of glioma: E Pluribus Unum? Glia. 2011;59(8):1135–1147. doi: 10.1002/glia.21143. [DOI] [PubMed] [Google Scholar]
- 4.Ligon KL, Huillard E, Mehta S, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53(4):503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Y, Meijer DH, Alberta JA, et al. Phosphorylation state of Olig2 regulates proliferation of neural progenitors. Neuron. 2011;69(5):906–917. doi: 10.1016/j.neuron.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahaman SO, Harbor PC, Chernova O, Barnett GH, Vogelbaum MA, Haque SJ. Inhibition of constitutively active Stat3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. Oncogene. 2002;21(55):8404–8413. doi: 10.1038/sj.onc.1206047. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Alizadeh D, Van Handel M, Kortylewski M, Yu H, Badie B. Stat3 inhibition activates tumor macrophages and abrogates glioma growth in mice. Glia. 2009;57(13):1458–1467. doi: 10.1002/glia.20863. [DOI] [PubMed] [Google Scholar]
- 8.Konnikova L, Kotecki M, Kruger MM, Cochran BH. Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer. 2003;3:23. doi: 10.1186/1471-2407-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gronostajski RM. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249(1–2):31–45. doi: 10.1016/s0378-1119(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 10.Sun P, Dong P, Dai K, Hannon GJ, Beach D. p53-independent role of MDM2 in TGF-beta1 resistance. Science. 1998;282(5397):2270–2272. doi: 10.1126/science.282.5397.2270. [DOI] [PubMed] [Google Scholar]
- 11.das Neves L, Duchala CS, Tolentino-Silva F, et al. Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc Natl Acad Sci U S A. 1999;96(21):11946–11951. doi: 10.1073/pnas.96.21.11946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shu T, Butz KG, Plachez C, Gronostajski RM, Richards LJ. Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice. J Neurosci. 2003;23(1):203–212. doi: 10.1523/JNEUROSCI.23-01-00203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52(6):953–968. doi: 10.1016/j.neuron.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 14.Kang P, Lee HK, Glasgow SM, et al. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 2012;74(1):79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scrideli CA, Carlotti CG, Jr, Okamoto OK, et al. Gene expression profile analysis of primary glioblastomas and non-neoplastic brain tissue: identification of potential target genes by oligonucleotide microarray and real-time quantitative PCR. J Neurooncol. 2008;88(3):281–291. doi: 10.1007/s11060-008-9579-4. [DOI] [PubMed] [Google Scholar]
- 16.Song HR, Gonzalez-Gomez I, Suh GS, et al. Nuclear factor IA is expressed in astrocytomas and is associated with improved survival. Neuro Oncol. 2010;12(2):122–132. doi: 10.1093/neuonc/nop044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng S, Eacker SM, Hong SJ, Gronostajski RM, Dawson TM, Dawson VL. NMDA-induced neuronal survival is mediated through nuclear factor I-A in mice. J Clin Invest. 2010;120(7):2446–2456. doi: 10.1172/JCI33144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plachez C, Lindwall C, Sunn N, et al. Nuclear factor I gene expression in the developing forebrain. J Comp Neurol. 2008;508(3):385–401. doi: 10.1002/cne.21645. [DOI] [PubMed] [Google Scholar]
- 19.Ishii N, Maier D, Merlo A, et al. Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol. 1999;9(3):469–479. doi: 10.1111/j.1750-3639.1999.tb00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong G, Husseiny MI, Song L, et al. Novel cancer vaccine based on genes of Salmonella pathogenicity island 2. Int J Cancer. 2010;126(11):2622–2634. doi: 10.1002/ijc.24957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, Erdreich-Epstein A, Gonzalez-Gomez I, et al. Novel cell lines established from pediatric brain tumors. J Neurooncol. 2012;107(2):269–280. doi: 10.1007/s11060-011-0756-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosol M, Harutyunyan I, Xu J, et al. Metabolism of orthotopic mouse brain tumor models. Mol Imaging. 2009;8(4):199–208. [PubMed] [Google Scholar]
- 23.Sun L, Hui AM, Su Q, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9(4):287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Bredel M, Bredel C, Juric D, et al. Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res. 2005;65(19):8679–8689. doi: 10.1158/0008-5472.CAN-05-1204. [DOI] [PubMed] [Google Scholar]
- 25.Murat A, Migliavacca E, Gorlia T, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26(18):3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 26.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okada H, Mak TW. Pathways of apoptotic and non-apoptotic death in tumour cells. Nat Rev Cancer. 2004;4(8):592–603. doi: 10.1038/nrc1412. [DOI] [PubMed] [Google Scholar]
- 28.Lee HK, Xiang C, Cazacu S, et al. GRP78 is overexpressed in glioblastomas and regulates glioma cell growth and apoptosis. Neuro Oncol. 2008;10(3):236–243. doi: 10.1215/15228517-2008-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawada M, Nakashima S, Banno Y, et al. Ordering of ceramide formation, caspase activation, and Bax/Bcl-2 expression during etoposide-induced apoptosis in C6 glioma cells. Cell Death Differ. 2000;7(9):761–772. doi: 10.1038/sj.cdd.4400711. [DOI] [PubMed] [Google Scholar]
- 30.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 31.Blagosklonny MV. Loss of function and p53 protein stabilization. Oncogene. 1997;15(16):1889–1893. doi: 10.1038/sj.onc.1201374. [DOI] [PubMed] [Google Scholar]
- 32.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26(12):1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Zhu S, Cloughesy TF, Liau LM, Mischel PS. p53 disruption profoundly alters the response of human glioblastoma cells to DNA topoisomerase I inhibition. Oncogene. 2004;23(6):1283–1290. doi: 10.1038/sj.onc.1207244. [DOI] [PubMed] [Google Scholar]
- 34.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2(2):a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furlong EE, Rein T, Martin F. YY1 and NF1 both activate the human p53 promoter by alternatively binding to a composite element, and YY1 and E1A cooperate to amplify p53 promoter activity. Mol Cell Biol. 1996;16(10):5933–5945. doi: 10.1128/mcb.16.10.5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johansson EM, Kannius-Janson M, Gritli-Linde A, Bjursell G, Nilsson J. Nuclear factor 1-C2 is regulated by prolactin and shows a distinct expression pattern in the mouse mammary epithelial cells during development. Mol Endocrinol. 2005;19(4):992–1003. doi: 10.1210/me.2004-0359. [DOI] [PubMed] [Google Scholar]
- 37.Ouellet S, Vigneault F, Lessard M, Leclerc S, Drouin R, Guerin SL. Transcriptional regulation of the cyclin-dependent kinase inhibitor 1A (p21) gene by NFI in proliferating human cells. Nucleic Acids Res. 2006;34(22):6472–6487. doi: 10.1093/nar/gkl861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hjortland GO, Bjornland K, Pettersen S, et al. Modulation of glioma cell invasion and motility by adenoviral gene transfer of PAI-1. Clin Exp Metastasis. 2003;20(4):301–309. doi: 10.1023/a:1024040718238. [DOI] [PubMed] [Google Scholar]
- 39.Smith HW, Marshall CJ. Regulation of cell signalling by uPAR. Nat Rev Mol Cell Biol. 2010;11(1):23–36. doi: 10.1038/nrm2821. [DOI] [PubMed] [Google Scholar]
- 40.Riccio A, Pedone PV, Lund LR, Olesen T, Olsen HS, Andreasen PA. Transforming growth factor beta 1-responsive element: closely associated binding sites for USF and CCAAT-binding transcription factor-nuclear factor I in the type 1 plasminogen activator inhibitor gene. Mol Cell Biol. 1992;12(4):1846–1855. doi: 10.1128/mcb.12.4.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Starnes LM, Sorrentino A, Pelosi E, et al. NFI-A directs the fate of hematopoietic progenitors to the erythroid or granulocytic lineage and controls beta-globin and G-CSF receptor expression. Blood. 2009;114(9):1753–1763. doi: 10.1182/blood-2008-12-196196. [DOI] [PubMed] [Google Scholar]
- 42.Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123(5):819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 43.Zardo G, Ciolfi A, Vian L, et al. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood. 2012;119(17):4034–4046. doi: 10.1182/blood-2011-08-371344. [DOI] [PubMed] [Google Scholar]
- 44.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johansson EM, Kannius-Janson M, Bjursell G, Nilsson J. The p53 tumor suppressor gene is regulated in vivo by nuclear factor 1-C2 in the mouse mammary gland during pregnancy. Oncogene. 2003;22(38):6061–6070. doi: 10.1038/sj.onc.1206884. [DOI] [PubMed] [Google Scholar]
- 46.Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006;20(18):2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.el-Deiry WS, Tokino T, Velculescu VE, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 48.Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65(10):3980–3985. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- 49.Saramaki A, Banwell CM, Campbell MJ, Carlberg C. Regulation of the human p21(waf1/cip1) gene promoter via multiple binding sites for p53 and the vitamin D3 receptor. Nucleic Acids Res. 2006;34(2):543–554. doi: 10.1093/nar/gkj460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Czekay RP, Wilkins-Port CE, Higgins SP, et al. PAI-1: An Integrator of Cell Signaling and Migration. Int J Cell Biol. 2011;2011:562481. doi: 10.1155/2011/562481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stefansson S, Lawrence DA. The serpin PAI-1 inhibits cell migration by blocking integrin alpha V beta 3 binding to vitronectin. Nature. 1996;383(6599):441–443. doi: 10.1038/383441a0. [DOI] [PubMed] [Google Scholar]
- 52.Bindal AK, Hammoud M, Shi WM, Wu SZ, Sawaya R, Rao JS. Prognostic significance of proteolytic enzymes in human brain tumors. J Neurooncol. 1994;22(2):101–110. doi: 10.1007/BF01052886. [DOI] [PubMed] [Google Scholar]
- 53.Landau BJ, Kwaan HC, Verrusio EN, Brem SS. Elevated levels of urokinase-type plasminogen activator and plasminogen activator inhibitor type-1 in malignant human brain tumors. Cancer Res. 1994;54(4):1105–1108. [PubMed] [Google Scholar]
- 54.Yamamoto M, Sawaya R, Mohanam S, et al. Expression and cellular localization of messenger RNA for plasminogen activator inhibitor type 1 in human astrocytomas in vivo. Cancer Res. 1994;54(13):3329–3332. [PubMed] [Google Scholar]
- 55.Stefansson S, McMahon GA, Petitclerc E, Lawrence DA. Plasminogen activator inhibitor-1 in tumor growth, angiogenesis and vascular remodeling. Curr Pharm Des. 2003;9(19):1545–1564. doi: 10.2174/1381612033454621. [DOI] [PubMed] [Google Scholar]
- 56.Hjortland GO, Lillehammer T, Somme S, et al. Plasminogen activator inhibitor-1 increases the expression of VEGF in human glioma cells. Exp Cell Res. 2004;294(1):130–139. doi: 10.1016/j.yexcr.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 57.Wang W, Mullikin-Kilpatrick D, Crandall JE, Gronostajski RM, Litwack ED, Kilpatrick DL. Nuclear factor I coordinates multiple phases of cerebellar granule cell development via regulation of cell adhesion molecules. J Neurosci. 2007;27(23):6115–6127. doi: 10.1523/JNEUROSCI.0180-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.