Abstract
The role of the Forkhead box class O (FoxO)3a transcription factor in breast cancer migration and invasion is controversial. Here we show that FoxO3a overexpression decreases motility, invasiveness, and anchorage-independent growth in estrogen receptor α-positive (ERα+) cancer cells while eliciting opposite effects in ERα-silenced cells and in ERα-negative (ERα−) cell lines, demonstrating that the nuclear receptor represents a crucial switch in FoxO3a control of breast cancer cell aggressiveness. In ERα+ cells, FoxO3a-mediated events were paralleled by a significant induction of Caveolin-1 (Cav1), an essential constituent of caveolae negatively associated to tumor invasion and metastasis. Cav1 induction occurs at the transcriptional level through FoxO3a binding to a Forkhead responsive core sequence located at position −305/−299 of the Cav1 promoter. 17β-estradiol (E2) strongly emphasized FoxO3a effects on cell migration and invasion, while ERα and Cav1 silencing were able to reverse them, demonstrating that both proteins are pivotal mediators of these FoxO3a controlled processes. In vivo, an immunohistochemical analysis on tissue sections from patients with ERα+ or ERα− invasive breast cancers or in situ ductal carcinoma showed that nuclear FoxO3a inversely (ERα+) or directly (ERα−) correlated with the invasive phenotype of breast tumors. In conclusion, FoxO3a role in breast cancer motility and invasion depends on ERα status, disclosing a novel aspect of the well-established FoxO3a/ERα interplay. Therefore FoxO3a might become a pursuable target to be suitably exploited in combination therapies either in ERα+ or ERα− breast tumors.
Keywords: forkhead transcription factors, estrogen receptor, motility, invasion, breast cancer
Introduction
The forkhead box class O3a (FoxO3a) is one of the four members (FoxO1a, FoxO3a, FoxO4, and FoxO6) belonging to the subfamily of winged-helix forkhead transcription factors (FoxOs), whose functions are negatively regulated by the insulin-phosphatidylinositol 3-kinase (PI3K)-protein kinase B (PKB) signaling.1 In the absence of insulin or growth factors, FoxOs are mainly located within the nuclei and regulate a set of target genes, thereby promoting cell cycle arrest, stress resistance, apoptosis, DNA damage repair, and metabolism.2 In presence of insulin or growth factors, FoxOs undergo phosphorylation, bind to the chaperone proteins 14-3-3 and are exported into the cytoplasm, where they are degraded via the ubiquitin–proteasome pathway.1
An increasing interest in FoxOs factors has been lately observed in the oncologic research field. In particular, in breast cancer, its role is still controversial, in fact, FoxO3a overexpression has been shown to inhibit tumor growth in vitro and tumor size in vivo,3-5 and cytoplasmic location of FoxO3a seems to correlate with patients poor survival.3 Moreover, genetic deletion of the FoxOs alleles (FoxO1a, FoxO3a, and FoxO4) generates progressive cancerous phenotypes, such as thymic lymphomas and hemangiomas. These data elucidate FoxOs as bona fide tumor suppressor genes.6 Additionally, FoxO members seem to be important mediators of the well-established functional cross-talk between estrogens and growth factors, which play a pivotal role in breast cancer development and progression.7 In fact, growth factors are known to influence the expression and activity of estrogen receptor α (ERα) and its transcriptional cofactors; conversely, ERα regulates the expression of growth factor receptors and their ligands and signaling intermediates.8 In this context, several reports have recently suggested a functional interaction between ERα and FoxO members. 17β-estradiol (E2) has been noted to determine ERα binding to FoxO1a, FoxO3a, and FoxO4, which, in turn, showed either coactivator or corepressor functions on estrogen-responsive element (ERE) sites, depending on the cellular model.5,9,10 Moreover, we introduced the importance of Akt2/FoxO3a axis in the control of ERα-mediated transcription in ERα-positive (ERα+) breast cancer cells. Our results indicate that Akt2 inhibition reduces ERα transcriptional activity through FoxO3a activation, suggesting that FoxO3a, acting as a co-repressor for ERα, could exert a protective role in ERα+ breast tumors.11
In line with this assumption, Belguise et al. showed that ectopic expression of a constitutively active FoxO3a overrode transforming growth factor-B1-mediated invasive phenotype and induced a more epithelial phenotype in ERα+ mouse mammary tumors.12 However, more recently, FoxO3a has been described to behave in an opposite fashion in several other cancer cell lines, which, interestingly, were all ERα-negative (ERα-); in fact, Storz et al. reported that, in tested cells, nuclear retention of FoxO3a resulted in greatly increased invasion, through the induction of matrix metalloproteinase 9 (MMP-9) and MMP-13.13 Due to the inconsistency of the data available from ERα+ and ERα− breast cancer cells, the interplay between ERα and FoxO3a in tumor metastasis needs further investigations and is the goal of the present study. Since it is well documented that, in breast cancer, ERα signaling strongly correlates with a lower invasiveness and reduced metastatic potential,14 we assume that FoxO3a/ERα interplay could be responsible for the reduction of the migrating and invasive phenotype only in ERα+ cells, while, in ERα− cells, the lack of the α isoform of the receptor might enable FoxO3a to act in an opposite fashion. Thus, the present work was aimed to undertake an accurate study on the molecular mechanisms through which FoxO3a regulates migration and invasion in ERα+ breast cancer cells. Our results offer new interesting insights on FoxO3a activity, elucidating additional mechanisms that could represent novel targets in breast cancer therapy.
Results
Cell motility, invasion, and anchorage-independent growth are inhibited in ERα+ breast cancer cells overexpressing FoxO3a
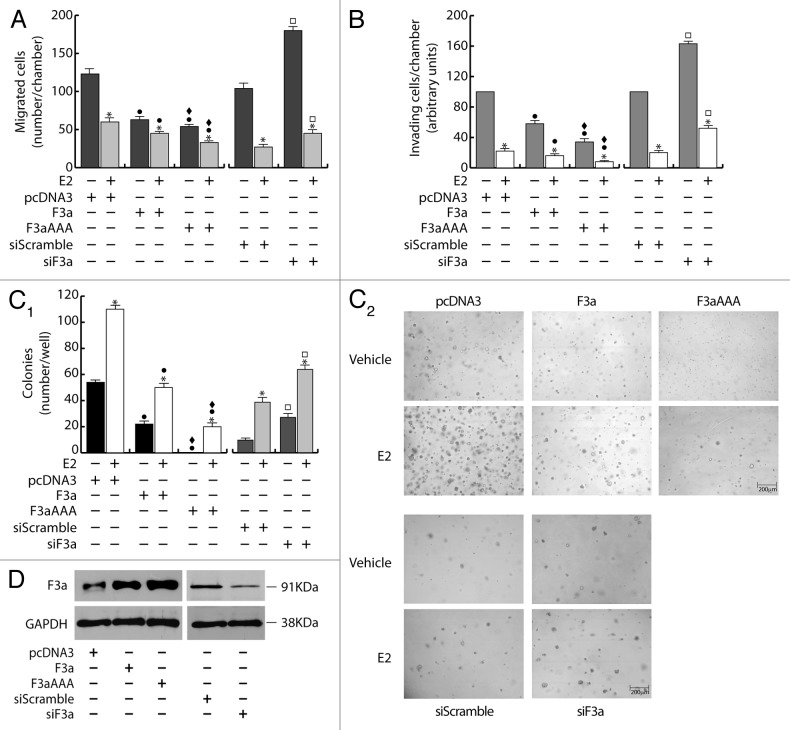
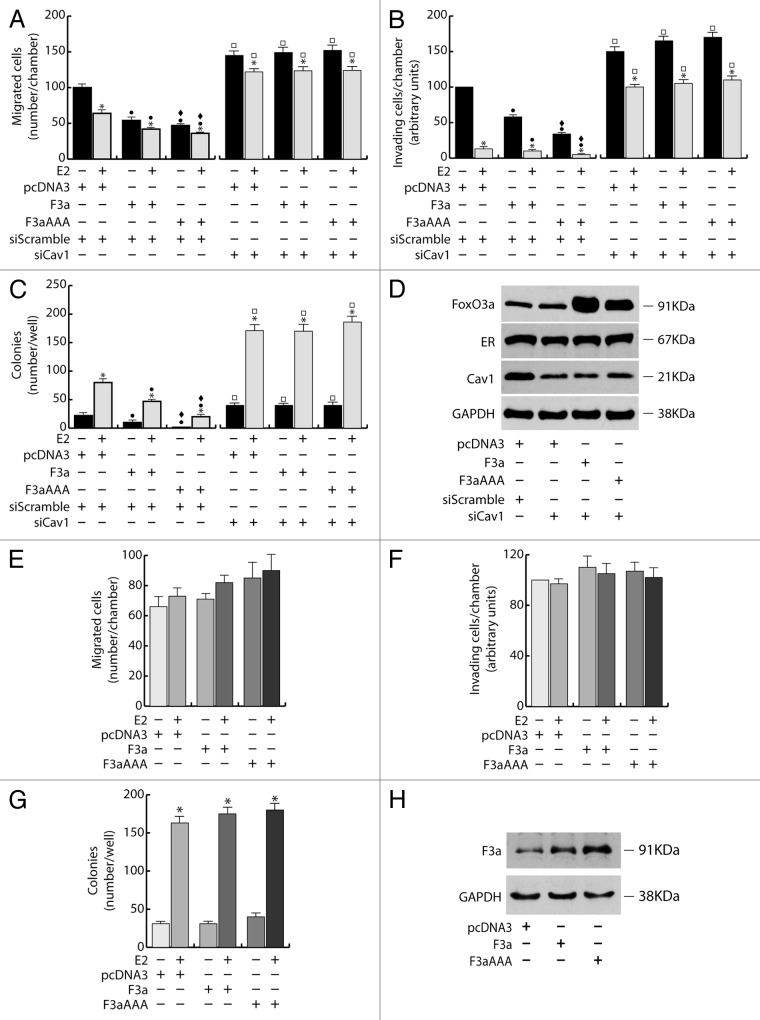
To assess the role of FoxO3a in the metastatic and invading potential of breast cancer cells, wild-type FoxO3a (F3a) was overexpressed in ERα+ MCF-7. Our results show a significant reduction of migrating and invading MCF-7/F3a cells (Fig. 1A and B), compared with control samples. Ectopic expression of the constitutively active triple mutant of FoxO3a (F3aAAA), where the 3 known PKB phosphorylation sites have been mutated to alanine, so that FoxO3a can no longer be inhibited by PKB-mediated phosphorylation, emphasized the phenomenon (Fig. 1A and B), suggesting that FoxO3a modulation of the migrating and the invading potential could involve the transcriptional induction of Forkhead responsive genes. FoxO3a silencing (siF3a) confirmed these data, since it led to a substantial increase in cell migration and invasion (Fig. 1A and B). Moreover, in agreement with our previous observations,15 E2 treatment strongly reduced motility and invasion, and the effect was additive in F3a- and F3aAAA-overexpressing samples, while siF3a only in part was able to counteract E2-mediated effects (Fig. 1A and B).
Figure 1. FoxO3a inhibits migration, invasion and anchorage independent growth in ERα+ MCF-7 breast cancer cells. A double set of MCF-7 cells was transiently transfected with 1 μg/35 mm dish of F3a, F3aAAA, or pcDNA3 as control. Another double set was silenced for FoxO3a expression (siF3a), using a siScramble as control (60 pmol siRNAs/35 mm dish). After 5 h cells were switched to PRF-SFM, and the next day one of each set of cells was harvested and subjected to migration (A), invasion (B), and soft agar assay (C1 and C2). Migration and invasion assays were conducted as described in “Materials and Methods”, adding 100 nM E2 in the bottom of the wells where indicated. Migrated and invading cells were evaluated after 24 h and 72 h of incubation, respectively. In soft agar assay, colonies >50 μm diameter formed after 14 d from plating were photographed at 4× magnification (C2) and counted under the microscope (C1). The second set of either transfected or silenced MCF-7 cells was used for total protein extractions and WB analysis to assess transfections efficiency; GAPDH was evaluated as a loading control (D). Results are reported as the mean ± s.d. of at least 3 independent experiments. In all experiments, significance values were as follows: *, P < 0.01 vs. untreated; ●, P < 0.01 vs. corresponding pcDNA3; ♦, P < 0.05 vs. corresponding F3a; □, P < 0.01 vs. corresponding siScramble.
In addition, anchorage independence, a characteristic of malignancy and tumor progression, was also investigated in F3a-overexpressing and silenced MCF-7 cells through soft agar colony-formation assay. We observed a dramatic decrease of the number as well as of the dimensions of the colonies in MCF-7/F3a samples, reaching almost completely the condition of single cells in F3aAAA-expressing cells (Fig. 1C1 and C2). The same trend was evidenced in E2-treated samples, showing how FoxO3a, especially in its active form, is able to counteract the well-known positive effect of the nuclear hormone on the colony formation of MCF-7 cells.16 As expected, an increase in the number of colonies was observed following siF3a, and such increase became more evident in presence of E2 (Fig. 1C1 and C2). Transfections and silencing efficiency were assessed on total protein lysates (Fig. 1D).
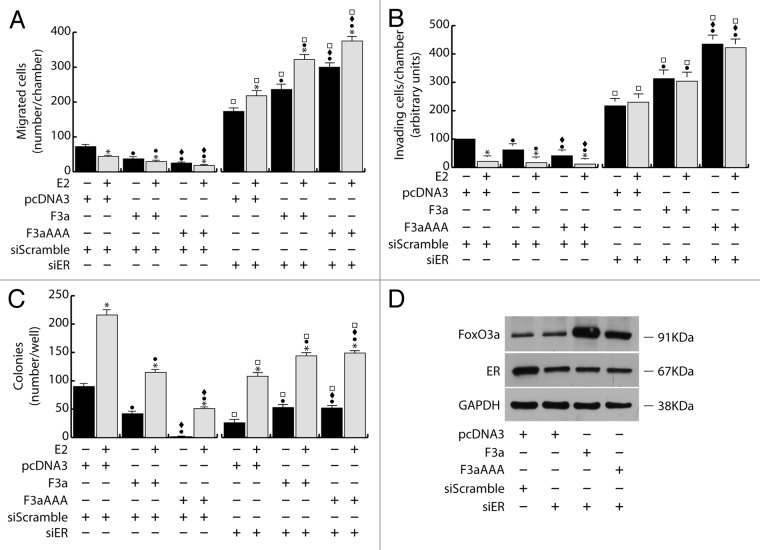
Figure 2. FoxO3a mediated inhibition of breast cancer cell migration, invasion and growth in suspension depends on ERα Two double sets of MCF-7 cells were silenced either for ERα (siER), using siScramble as control. After 5 h cells were switched to PRF-SFM and transiently transfected with F3a, F3aAAA, or pcDNA3. Next day cells were harvested and one set of each experiment was subjected to migration, invasion, and soft agar assay in the presence or in the absence of E2. Migrated (A) and invading (B) cells were evaluated after 24 h and 72 h of incubation, respectively. In soft agar assay, colonies ≥50 μm diameter formed after 14 d from plating were counted under the microscope (C). The second set of each experiment was used for total protein extraction to evaluate transfections efficiency by WB analysis; GAPDH was used as loading control (D). Results are the mean ± s.d. of at least three independent experiments. *, P < 0.05 vs. untreated; ●, P < 0.01 vs. corresponding pcDNA3; ♦, P < 0.01 vs. corresponding F3a; □, P < 0.01 vs. corresponding siScramble.
Interestingly, F3a and F3aAAA overexpression in other ERα-positive cell lines, ZR-75 (breast cancer) and Ishikawa (endometrial cancer), led to results that were comparable to those obtained from MCF-7, both in presence or absence of E2 (Fig. S1, upper panels)
The lack of ERα reverses FoxO3a-mediated inhibition of migration, invasion, and colonies formation
To assess if the effects of FoxO3a on motility, invasiveness, and colony formation could depend on ERα, silencing experiments were conducted in MCF-7, using specific siRNAs against ERα (siER) (Fig. 2). Interestingly, ERα silencing was able to counteract FoxO3a-mediated inhibition of the above-mentioned pathological features.
In particular, compared with control (siScramble), siER led to an increase in cell migration and invasion, which became even more evident in F3a and, especially, in F3aAAA-expressing cells (Fig. 2A and B), confirming that ERα is a hallmark of a less motile and invading phenotype,15,17 and that FoxO3a’s effect on cell motility and invasiveness can switch from inhibitory to stimulatory, depending on the presence or absence of ERα, respectively. Moreover, in siER samples, reasonably due to the lack of the receptor, E2 treatment no longer caused the reduction of the invading potential of MCF-7 (Fig. 2B) and even showed the opposite effect on cell motility, which rather increased over the respective controls (Fig. 2A). These evidences suggest that, in absence of a functional ERα, E2 could trigger some other pathway that stimulates cell migration (although not invasion), and that FoxO3a can somehow cooperate with the hormone in this process.
As expected, ERα silencing was able to inhibit both basal and E2 induced MCF-7 growth in soft agar by strongly reducing the number and the dimensions of colonies compared with non-treated and E2-treated siScramble samples, respectively (Fig. 2C). However, as in migration and invasion experiments, the inactivation of the nuclear receptor reversed the effect of ectopic F3a and F3aAAA, which, either in absence or presence of E2 treatment, induced an increase in the number of colonies, instead of the decrease observed in siScramble samples (Fig. 3C).
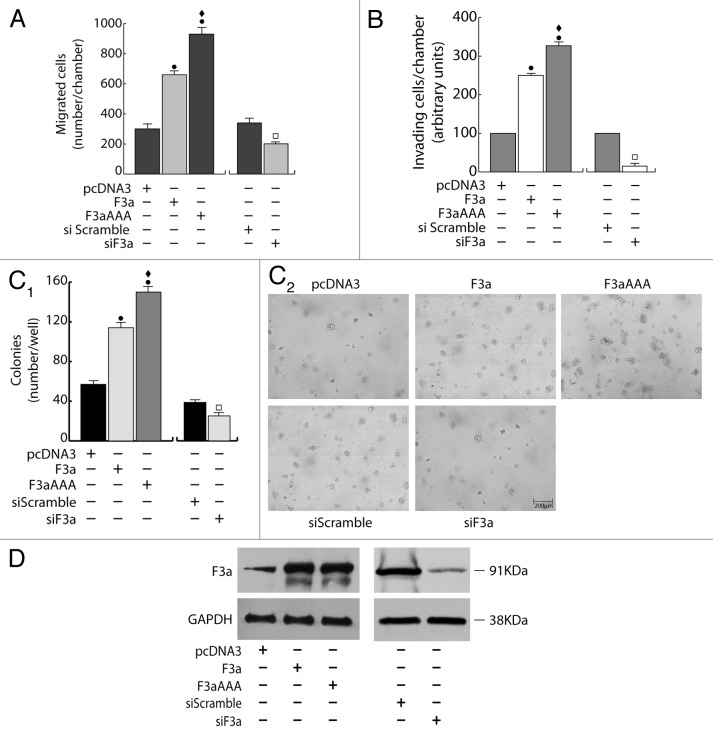
Figure 3. FoxO3a promotes migration, invasion, and anchorage-independent growth in ERα− MDA-MB-231 breast cancer cells. A double set of MDA-MB-231 cells were transiently transfected with 1 μg/35 mm dish of F3a, F3aAAA, or pcDNA3 or silenced for FoxO3a expression (siF3a) using a siScramble as control (60 pmol siRNAs/35 mm dish). Both transfection and silencing were made on cells in suspended PRF-GM. After 5 h cells were serum starved and, 24 h later, harvested. One set was subjected to migration (A), invasion (B), or soft agar assay (C1 and C2). Migrated and invading cells were evaluated after 16 h and 48 h of incubation, respectively. In soft agar assay, colonies > 50 μm diameter formed after 14 d from plating were photographed at 4× magnification (C2) and counted under the microscope (C1). The second set of either transfected or silenced MCF-7 cells was used to assess transfections efficiency by WB analysis on total protein extracts; GAPDH was evaluated as a loading control (D). Results are reported as the mean ± s.d. of at least 3 independent experiments. ●, P < 0.01 vs. pcDNA3; ♦, P < 0.01 vs. F3a; □, P < 0.05 vs. siScramble.
The fact that ERα exerts a pivotal role in determining FoxO3a behavior was confirmed by the results obtained in ERα− cells. Indeed, overexpression of FoxO3a in ERα− breast cancer MDA-MB-231 cells was able to induce an evident increase (rather than a decrease, as in ERα+ cells) of the migrating and invading potential (Fig. 3A and B), as well as, when grown in soft agar, F3a-overexpressing cells formed many more and larger colonies compared with control vector (Fig. 3C1 and C2). Once again, in all experiments, F3aAAA was more effective than F3a, while an evident reduction of migration, invasion and number and dimensions of colonies was observed in F3a silenced samples (Fig. 3A–C2). Transfections and silencing efficiency were determined concomitantly (Fig. 3D).
Noteworthy, as in MDA-MB-231, F3a and F3aAAA overexpression led to comparable results in other ERα− breast cancer cell lines (MDA-MB-468 and MDA-MB-435) as well as in ERα− cervical cancer HeLa cells, indicating that FoxO3a functions trough mechanisms that are not tissue-specific (Fig. S1, lower panels and data not shown).
FoxO3a and E2 synergistically induce caveolin-1 expression in ERα+ cancer cells
To the aim of identifying the mechanism through which FoxO3a modulates cell motility and invasiveness, we focused our attention on caveolin-1 (Cav1), a protein that has been reported to be induced by both Forkhead transcription factors18 and E2.19,20 Since, in breast cancer, Cav1 has been negatively21 and positively22 linked to tumor progression, motility, and invasiveness, we questioned if FoxO3a could control migration and invasion of breast cancer cells through the modulation of Cav1 expression.
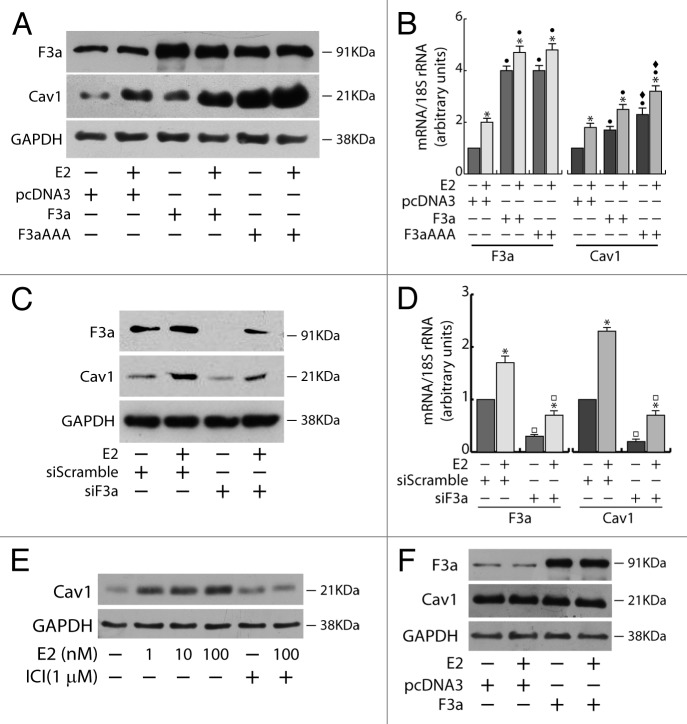
In ERα+ MCF-7 cells, the ectopic expression of FoxO3a caused a strong upregulation of Cav1 protein and mRNA, which was even more evident in F3aAAA transfectants, suggesting that FoxO3a induction of Cav1 expression could occur at the transcriptional level. As expected, E2 treatment increased Cav1 levels, and the effect was additive to that exerted by F3a or F3aAAA (Fig. 4A and B). Silencing experiments confirmed FoxO3a involvement in Cav1 transcription, leading to a decrease in Cav1 content and attenuating the E2-dependent Cav1 induction (Fig. 4C and D). Notably, Cav1 undergoes similar regulation by E2 and FoxO3a in the other 2 tested ERα+ cell lines, ZR-75 and Ishikawa (Fig. S2). In particular, the induction of Cav1 by E2 is ERα-dependent, since (1) the pure antiestrogen ICI 172.780 was able to abrogate the effect of E2 on Cav1 expression in ERα+ MCF-7 cells (Fig. 4E); and (2) the hormone did not increase Cav1 expression in ERα−, although ERβ+, MDA-MB-231 cells (Fig. 4F).
Figure 4. Cav1 expression depends on E2 and FoxO3a in ERα+ MCF-7 breast cancer cells. A double set of MCF-7 cells were either transiently transfected with F3a, F3aAAA, or pcDNA3 or silenced for FoxO3a, serum starved after 5 h and treated the next day with 100 nM E2 for 24 h. Cells were then harvested and total proteins and RNA were extracted, and subjected to WB (A and C) and RT-PCR analysis (B and D), respectively, for F3a and Cav1 expression assessment. (E) MCF-7 cells were seeded in growing medium, serum starved the next day for 24 h, pre-treated or not for 1 h with the pure antiestrogen ICI 182.780 and then treated with increasing concentrations of E2 (0, 1, 10, and 100 nM). (F) MDA-MB-231 cells were transiently transfected with F3a or pcDNA3 as control, serum starved for 24 h and then treated or not with 100 nM E2. After 24 h of E2 treatment, total proteins were extracted and subjected to WB analysis. GAPDH was analyzed as loading control in WB assays. For RT-PCR assays, each sample was normalized to its 18S rRNA content. Results are reported as the mean ± s.d. of at least 3 independent experiment. *, P < 0.01 vs. untreated; ●, P < 0.01 vs. pcDNA3; ♦, P < 0.01 vs. F3a; □, P < 0.05 vs. siScramble.
In light of these evidences we could hypothesize that, in ERα+ cells, FoxO3a might promote a less aggressive phenotype by cooperating with the hormone receptor in CAV1 gene induction.
Cav1 is a mediator of FoxO3a-dependent inhibition of migration, invasion, and growth in suspension in ERα+ breast cancer cells
Cav1 involvement in FoxO3a-mediated inhibition of motility, invasiveness, and colonies formation was assessed by silencing experiments using specific siRNAs against Cav1 (siCav1) in ERα+ breast cancer cells, (Fig. 5A–D). Cav1 silencing was able to counteract FoxO3a effects, leading to an overall increase of cell migration and invasion in MCF-7 cells, although F3a and F3aAAA overexpression did not contribute to such increase, nor was siCav1 sufficient to completely reverse the inhibitory effect exerted by E2 treatment (Fig. 5A and B). A similar trend was observed in soft agar experiments, where the number of colonies was much greater in siCav1 samples, especially under E2 treatment (note that ERα protein content was not affected by siCav1, Fig. 5D), compared with the respective controls (siScramble) (Fig. 5C). Again, F3a and F3aAAA did not have any additive effect on colony growth (Fig. 5C).
Figure 5. Cav1 is a mediator of FoxO3a dependent inhibition of migration, invasion and growth in suspension of ERα+ breast cancer cells. (A–D) Two double sets of MCF-7 cells were silenced for Caveolin-1 (siCav1), using siScramble as control. After 5 h cells were switched to PRF-SFM and transiently transfected with F3a, F3aAAA, or pcDNA3. Next day cells were harvested and one set of each experiment was subjected to migration, invasion, and soft agar assay, in the presence or in the absence of E2. Migrated (A) and invading (B) cells were evaluated after 24 h and 72 h of incubation, respectively. In soft agar assay, colonies ≥50 μm diameter formed after 14 d from plating were counted under the microscope (C). Transfection efficiency was evaluated by WB analysis on total protein extracted by the second set of cells; GAPDH was used as loading control (D). Results are the mean ± s.d. of at least 3 independent experiments. *, P < 0.05 vs. untreated; ●, P < 0.01 vs. corresponding pcDNA3; ♦, P < 0.01 vs. corresponding F3a; □, P < 0.01 vs. corresponding siScramble. (E–H) A double set of T47D cells were transiently transfected with F3a, F3aAAA or pcDNA3. After 5h cells were switched to PRF-SFM and the next day one set of cells was harvested and subjected to migration (E), invasion (F), or soft agar assay (G), with or without 100 nM E2. Migrated and invading cells were counted after 24 h and 72 h of incubation, respectively. In soft agar assay, colonies formed after 14 d from plating were exposed to MTT and counted under the microscope. The second set of cells was lysed, and total protein was used for WB analysis to assess transfections efficiency; GAPDH was used as loading control (H). Results are the mean ± s.d. of at least 3 independent experiments. *, P < 0.01 vs. untreated.
These results show how, in MCF-7, FoxO3a control of cell migration, invasion, and anchorage-independent cell growth depends, in part, on Cav1, while it is strictly linked to ERα expression (Fig. 2). Indeed, in Cav1-negative T47D cells, which, in addition, bear a very low content of ERα, F3a, and F3aAAA overexpression did not lead to any significant decrease in motility, invading potential and colony formation in soft agar, reflecting a sort of compromise between the results observed following either Cav1 or ERα silencing in MCF-7 cells (Figs. 2 and 5E–G), thus indicating that these 2 proteins are mediators of both E2 and FoxO3a activity.
FoxO3a binds to and trans-activates the Cav1 promoter in MCF-7 cells
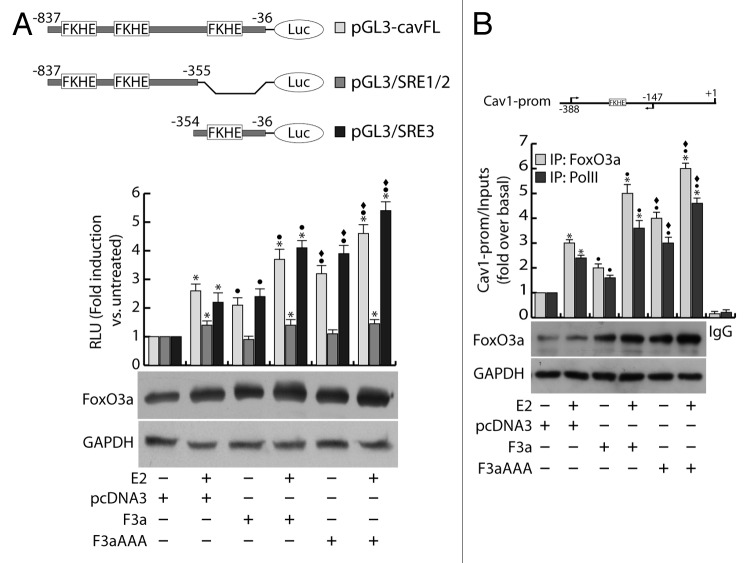
To deepen the understanding of the mechanism underlying the FoxO3a/ERα interplay in Cav1 induction, through an accurate analysis of the Cav1 promoter (GenBank accession #AF095591.1), we verified the presence of several Forkhead core sequences (FKHE), and we questioned if any of the identified regions may be involved in the FoxO3a/ERα-mediated regulation of Cav1 gene expression in ERα+ breast cancer cells. To this aim, a vector bearing the luciferase gene under the control of the -837/-36 region of Cav1 promoter (pGL3-cavFL) was co-transfected with F3a or F3aAAA in MCF-7 cells and exposed or not to E2 treatment. In line with the results reported in Figure 4A and B, E2 stimulation significantly induced the Cav1 promoter activity, and such effect was increasingly higher in F3a- and F3aAAA-transfected cells (Fig. 6A). Interestingly, the construct pGL3/SRE1/2 (nt −837/−355), although containing FKHE core sequences, failed to be induced by FoxO3a but still weakly responded to hormone stimulation, most likely for the presence of Sp1 and AP-1 sites; on the contrary, the construct pGL3/SRE3 (nt −354/−36), bearing only one FKHE motif (nt −305/−299) and several Sp1 and AP-1 sites, was induced by both E2 and overexpressed FoxO3a, with a trend comparable to that observed with the pGL3-cavFL construct (Fig. 6A).
Figure 6. FoxO3a binds to and transactivates the Cav1 promoter. (A) MCF-7 were seeded in culture medium on 24-well plates, serum starved for 24 h, co-transfected in PRF-CT with pGL3-cavFL, or pGL3/SRE1/2, or pGL3/SRE3 and pRL-Tk, in presence of either pcDNA3 or F3a or F3aAAA vectors. After 6 h, E2 (100 nM) was added to the medium, where opportune, and the next day cells were harvested, and luciferase activity was evaluated. Cell extracts were also processed by WB analysis to assess F3a and F3aAAA transfection efficiency; GAPDH was used as loading control. (B) ChIP analysis was performed on the nuclear extracts from subconfluent MCF-7 cells seeded in 15 cm dish diameter, switched to PRF-SFM, and transfected with pcDNA3, F3a, or F3aAAA vectors. Twenty-four hours after transfection, the cells were treated with 100 nM E2 for 30 min or left untreated. The FKHE-containing Cav1 promoter region, precipitated with either anti-FoxO3a or anti-PolII pAbs were amplified using a specific pair of primers reported in “Materials and Methods”. E2-treated samples were also precipitated with normal rabbit IgG and used as negative control. FoxO3a expression in transfected samples was analyzed by WB on Cytosolic lysates from the same set of cells. Data represents the mean ± s.d. of 3 independent experiments. *, P < 0.05 vs. untreated; ●, P < 0.05 vs. corresponding pcDNA3; ♦, P < 0.05 vs. corresponding F3a.
The involvement of E2 and FoxO3a in the transcriptional activation of the Cav1 promoter was corroborated by chromatin immunoprecipitation (ChIP) experiments, which evidenced a significant recruitment of FoxO3a on the region containing the −305/−299 FKHE sequence. Once again, E2 treatment strongly increased FoxO3a occupancy of the promoter, especially in F3a- and F3aAAA-overexpressing samples (Fig. 6B). A similar pattern was observed in Polymerase II (PolII) precipitates, confirming that E2 and FoxO3a, both independently and synergistically, are able to induce Cav1 gene transcription (Fig. 6B).
Nuclear FoxO3a correlates in an opposite way with the tumor grade and the invasive phenotype in ERα+ and ERα− breast tumors
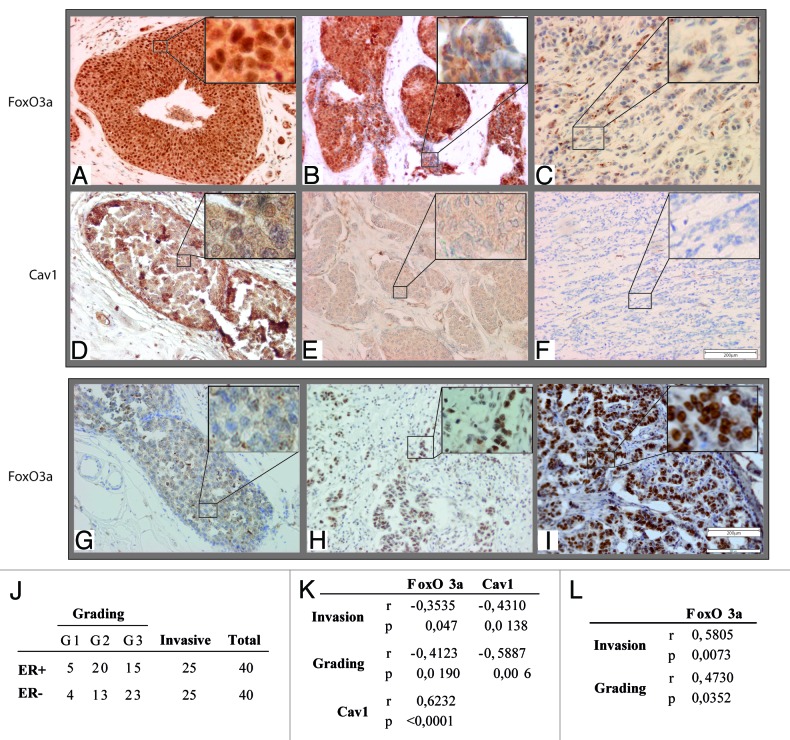
Tissue specimens from ductal carcinomas in situ (DCIS) and invading ductal carcinomas (IDC) (Fig. 7J) were analyzed to investigate if FoxO3a expression could correlate with the tumor grade and the invasive potential in ERα+ and ERα− breast tumors, as well as with Cav1 expression (in ERα+ tumors only).
Figure 7. Nuclear FoxO3a is highly expressed in non-invasive ERα+, and in invasive ERα− breast tumors. FoxO3a (A–C) and Cav1 (D–F) expression in ERα+ breast tumors and FoxO3a (G–I) in ERα− breast tumor samples. IHC was conducted on tissue sections deriving from biopsies diagnosed as DCIS (A and D), microinvasive DCIS (B and E), DCIS with contiguous IDC areas (G and H) and highly aggressive IDC (C, F, and I). Representative fields were photographed at 20× magnification. Insets, showing details of proteins subcellular localization, were taken at 100× magnification. (J) Samples descriptions and classification; (K) correlation between nuclear FoxO3a or Cav1 content and the tumor grading and invasive potential in ERα+ breast cancer samples; (L) correlation between nuclear FoxO3a content and the tumor grading and invasive potential in ERα− breast cancer samples. The correlation coefficient (r) and the statistical significance (P) are reported.
In all sections, tumor cells were clearly distinguishable from either infiltrating immune cells or stromal cells. In non-invading, well-differentiated ERα+ tumors, FoxO3a was strongly expressed, showing a very high nuclear localization (Fig. 7A). Strikingly nuclear FoxO3a positivity was gradually lost in invading and less differentiated cells (see insets in Fig. 7B), while cytoplasmic localization was not as indicative. Concomitantly, Cav1 expression tended to decrease from tumors with positive to negative FoxO3a nuclear staining, and was completely lost in highly invading ERα+ tumors (Fig. 7D–F). Statistical analysis of these samples showed that both FoxO3a nuclear expression and Cav1 were inversely correlated with tumor grade and the invasive potential, while cytosolic FoxO3a did not result to be significantly correlated with any clinicopathological feature (Fig. 7K); moreover, Cav1 expression resulted directly correlated with FoxO3a nuclear content (Fig. 7K).
On the contrary, a very weak or even absent FoxO3a nuclear localization was observed in intraductal, well delimited areas of ERα− tumors (Fig. 7G), while a very strong nuclear staining was detected in invading areas of the same samples (Fig. 7H) and in clearly invasive carcinomas (Fig. 7I). This observation was confirmed by statistical analysis that evidenced a direct correlation between FoxO3a expression and both tumor grading and the invasive potential of ERα− breast cancer tissues (Fig. 7L).
Discussion
FoxO transcription factors are crucial for regulating a myriad of physiological processes, including proliferation, metabolism, cell differentiation, cell cycle arrest, DNA repair, and apoptosis. FoxOs also play important roles in tumorigenesis, since they have been shown to be deregulated in many types of human cancers, and restoring their expression/activity has been shown to be effective in tumor suppression.2
The involvement of FoxOs in tumor metastasis is controversial, e.g., FoxO3a has been reported to have either a protective or a promoting role on cell motility and invasion.12,13 Our hypothesis was that such a difference might be ascribed to ERα status, since activated FoxO3a was able to reverse the invasive phenotype of ERα+ breast cancer cells12 while promoting tumor cell invasion in other cancer cell lines, which, notably, were all ERα−.13 Thus, the present study was aimed to verify if the effect exerted by FoxO3a on the metastatic potential of ERα+ breast cancer could derive from a general mechanism through which FoxO3a cooperates with the nuclear receptor in reducing motility and invasiveness of ERα+ tumors, while in absence of the receptor FoxO3a favors a more migrating and invasive phenotype. Indeed, since ERα signaling is well known to strongly correlate with a lower invasiveness and reduced motility of breast cancer cells,15 and considering that increasing evidences recognize Forkhead factors as important modulators of ERα transcriptional activity,9-11 it won’t surprise to ascertain that, in ERα+ tumors, FoxO3a could reduce cell migration and invasion through a functional interaction with ERα. On the other hand, in ERα− tumors, the absence of the receptor could enable FoxO3a to trigger some different pathway that leads to an opposite outcome.
To prove our hypothesis, minimally motile and invasive ERα+ MCF-7 and ZR-75 breast cancer cell lines have been transfected with wild-type F3a and constitutively active F3aAAA mutant, and the effects on cell migration, invasion, and colony formation in soft agar were observed. The results presented here show that FoxO3a overexpression reduces the migratory and invasive potential, as well as anchorage-independent growth (a hallmark of tumor progression), in ERα+ tested cells. It is worth noting that, in all experiments, the constitutively active mutant F3aAAA was always more effective than the wild-type FoxO3a, suggesting that the regulation of the above-mentioned features could occur at the transcriptional level, through the induction of Forkhead-responsive genes. Moreover, the expected reduced motility and invasiveness of ERα+ cells upon E2 stimulation15 was more evident in F3a and, especially, in F3aAAA-overexpressing cells, providing evidence that E2 and FoxO3a act synergistically on these 2 features (Fig. 1A and B; Fig. S1, upper panels). On the contrary, E2 stimulation does not show an anti-metastatic behavior in presence of growth factors, since it favors the anchorage-independent growth,16 suggesting that other growth factors regulated pathways do prevail on that of ERα in the control of this feature. However, in line with our previous observations,11 FoxO3a overexpression was able to counteract the proliferative effect of E2, and its silencing led to an increase in basal as well as in E2-dependent cell growth (Fig. 1C1 and C2). Taken together, these results suggest, once again, that FoxO3a might act as a co-repressor (e.g., by quenching E2/ERα dependent proliferative signals11) or a co-activator (e.g., by potentiating E2/ERα mediated inhibition of cell motility and invasion15) for ERα.10
More importantly, ERα is the key regulator of FoxO3a function, as evidenced by the opposite behavior of overexpressed F3a (and F3aAAA) in ERα-silenced cells if compared with the corresponding ERα-expressing samples (Fig. 2). Thus, the lack of the hormone receptor is responsible for the switch of FoxO3a biological function, which shifts from inhibitory (when ERα is present) to stimulatory (when ERα is absent) on cell motility, invasion, and growth in suspension.
This is confirmed by the fact that FoxO3a overexpression exhibits a stimulating (rather than inhibitory as in ERα+ cells) effect on the same features in ERα− MDA-MB-231, MDA-MB-468, and MDA-MB-435S breast cancer cells. Notably, since the results observed in ERα+ and ERα− breast cancer cells following F3a and F3aaAAA ectopic expression, were similar to those obtained in non-breast cancer Ishikawa (ERα+ human endometrial adenocarcinoma) and HeLa (ERα− human cervical cancer) cell lines, respectively, we could assume that FoxO3a controls cell migration, invasion, and growth in suspension with a general, not tissue-specific, mechanism, which seems to depend on ERα expression (Fig. 3; Fig. S1).
Our results also show how Cav1 represents the ultimate downstream target through which FoxO3a modulates the metastatic potential of ERα+ cells. Cav1 is a multifunctional scaffolding protein that is associated with cell surface caveolae and the regulation of lipid raft domains. Cav1 regulates multiple cancer-associated processes, including cellular transformation, tumor growth, cell migration and metastasis, cell death and survival, multidrug resistance, and angiogenesis. In breast cancer, Cav1 seems to function as a tumor suppressor.23 In fact, Cav1 mRNA and protein are downregulated or absent in primary human cancers as well as in several mouse and human breast cancer cell lines. Forced re-expression of Cav1 in transformed mammary cell lines abrogates numerous of their tumorigenic properties, including anchorage-independent growth and invasiveness24 and suppresses growth of breast cancer cell-derived xenografts in nude mice.25 Moreover, Cav1−/− mice showed an accelerated onset of mammary tumors and lung metastases.26 In accordance, Cav1 expression has been inversely related to the grade of the primary breast tumors and its upregulation was found to reduce metastasis to distant organs.21
In light of this evidence, we questioned if FoxO3a could exert a protective role in ERα+ breast cancer cells through the induction of Cav1 expression. Indeed, in all ERα+ cells tested, FoxO3a overexpression increased the RNA and protein amounts of Cav1, and such increase was additive to that observed under E2 treatment, suggesting that ERα is also involved in the transcriptional induction of Cav1 (Fig. 4), which, in turn, seems to be the effector of a less aggressive phenotype, as evidenced by Cav1-silencing experiments (Fig. 5A–D) and by the fact that F3a and F3aAAA overexpression failed to inhibit migration, invasion, and growth in suspension in Cav1-negative T47D cells, despite the presence of a low, but still functional, content of ERα (Fig. 5E–H).
Since the highest induction of Cav1 has always been observed in F3aAAA-transfected cells, Cav1 regulation by FoxO3a and estrogens at the transcriptional level was investigated. In fact, the 5′-flanking region of the CAV1 gene, including the promoter region, bear several perfect and predicted forkhead consensus sequences, one of which (at position −1814, located above the promoter sequence) has been reported to be responsible for forkhead dependent CAV1 gene regulation.18 However, as the same authors stated, it is possible that other FKHE, also present within the 5′-flanking region, may play a role in Cav1 transcriptional activation by FoxO as well. Indeed, the data presented here clearly show how FoxO3a is able to induce Cav1 transcription by binding to a FKHE motif, mapping nt −305/−299 of its promoter; in addition, the FoxO3a-dependent Pol II recruitment confirms the occurrence of a transcriptional event (Fig. 6). To explain the induction exerted by E2, alone or in combination with FoxO3a, on Cav1 expression, we exclude, at the present stage, the direct involvement of ERα in the transcriptional process, since an integrated analysis of ERα binding sites upstream of the Cav1 gene, through Myles Brown lab data sets (http://research.dfci.harvard.edu/brownlab/datasets/index.php?dir=ER_whole_human_genome/)27 and Cistrome-web application (http://cistrome.dfci.harvard.edu/ap/), evidenced that ERα recruitment to the chromatin occurs at a very large distance from the promoter, on 3 distinct positions around 80–100 Kb upstream of the transcription start site. No ERα binding is reported in the data sets at the promoter level or in its close proximity, as also confirmed by ChIP experiments conducted on several predicted estrogen-responsive motifs identified within the +1/−5000bp region (data not shown). Additionally, neither Sp1 nor AP-1 transcription factors, 2 well-established mediators of the ERα “non-classical” genomic pathway28 that have been reported to transcriptionally cooperate with FoxO3a,29,30 resulted to be involved in Cav1 regulation. In fact, both Sp1 silencing and c-Jun inhibition achieved through the dominant-negative (DN)/c-fos plasmid31 did not lead to any significant decrease in FoxO3a/E2-dependent Cav1 promoter activation, nor to a reduction of Cav1 protein content (data not shown). Despite these observations, the evidence that liganded ERα induces Cav1 expression, and that E2 and FoxO3a, separately or synergistically, lead to a significant increase of Pol II recruitment on the Cav1 promoter region (Fig. 6), suggests that it would be interesting to investigate, by means of the recent and fascinating techniques Chromosome conformation capture (3C) technology and detection of loops in DNA-picked chromatin (DPC),32,33 if the combined effect of E2 and FoxO3a on Cav1 expression could be ascribed to the interaction of at least one of the 3 above mentioned ERα binding sites, at 80–100 Kb upstream of the transcription start site, where FoxO3a is recruited to the CAV1 gene promoter (ongoing experiments). In fact, recent studies using tiled microarrays to identify the ERα interacting sites of estrogen responsive genes, showed that EREs can function as enhancer elements far away (up to 100 Kb) from gene promoters, and that other cooperating transcription factors (e.g., FoxA1, AP1 and Sp1) can participate with ERα to regulate the expression of E2-induced genes.27,34
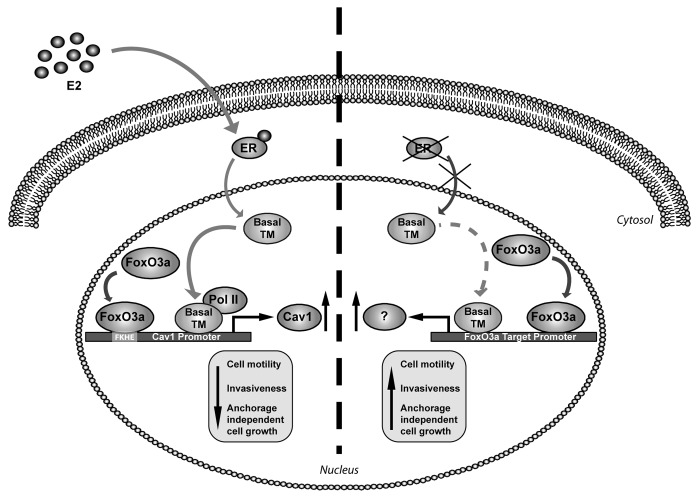
Taken together, the results obtained in ERα+ cancer cells show that FoxO3a-dependent decrease of migration, invasion, and colony formation is mediated by both ERα and Cav1, as confirmed by knockout experiments of these two factors (Figs. 2, 4, and 5). In particular, ERα cooperates with FoxO3a in the transcriptional induction of Cav1, which, in turn, is responsible of the reduced aggressive phenotype of FoxO3- overexpressing ERα+ cells (Fig. 8).
Figure 8. Proposed model for FoxO3a-mediated control of cell motility and invasiveness in presence or absence of ERα. F3a and ERα synergistically induce the expression of Cav1, which, in turn, reduces cell motility and invasiveness of ERα+ breast cancer cells. Transcriptionally active F3a binds to a FKHE located on the Cav1 proximal promoter and increases the recruitment of RNA Polymerase II, which is enhanced upon E2 stimulation. The lack of the hormone receptor enables active F3a to behave in an opposite fashion, thus increasing cell motility and invasion. Basal TM, basal transcriptional machinery.
On the other hand, several reports called into question Cav1 role as a tumor suppressor, since it has been found overexpressed in highly aggressive inflammatory breast cancer (IBC) human specimens and cell lines35 as well as in invasive human breast cancers samples, where its expression was significantly associated with basal-like phenotype, high histological grade, shorter disease-free and overall survival, and, more interestingly, lack of steroid hormone receptors positivity.36,37 Moreover, in ERα− cancer cells, Cav1 has been found in membrane protrusions, where it promotes tumor cell migration and invasion by regulating either the function of membrane type 1 matrix metalloproteinase (MT1-MMP),38 or, when phosphorylated (pY14Cav1), the focal adhesion turnover.22 Therefore, we investigated if the more aggressive phenotype of FoxO3a overexpressing ERα− cells could depend, also in this case, on Cav1 induction. However, no differences in Cav1 levels or phosphorylation status have been detected in ERα− cells following FoxO3a overexpression, nor E2 treatment, possibly through ERβ, has been able to induce Cav1 expression (Fig. 4, and data not shown).
Although MMP-9 and MMP-13 induction has been proposed as the mechanism through which FoxO3a increases invasion of cells lacking the hormone receptor,13 not all the ERα− cell lines tested do express these MMPs, or do express negligible levels. Moreover we failed to detect a reproducible increase in MMP-9 transcripts and in MMP-13 mRNA and protein in FoxO3a-overexpressing cells (data not shown), thus other markers are currently being investigated in our laboratory to justify the higher motility and greater invading ability induced by FoxO3a in ERα− cells. However, it is worth to underline that ERα silencing is a sufficient condition to reverse the effect of FoxO3a on migration, invasion and colony formation in ERα+ cells (Fig. 2), thus ERα seems to be a pivotal regulator of FoxO3a function, which switches from protective to malignant depending, respectively, on the presence or absence of the hormone receptor. A schematic representation of our findings is shown in Figure 8.
Finally, an immunohistochemical study from Yoshino’s research group showed that nuclear FoxO3a associates with IDC and lymph node metastasis, and the same authors speculated that, in some cases, aberrant activation of FoxO3a may cause the recruitment of metastasis-related molecules, instead of inducing apoptotic genes.39 Since no association with ERα status has been considered in this study, it might be possible that nuclear FoxO3a could correlate to a more metastatic phenotype only in the subset of ERα− IDC. In line with this hypothesis, nuclear FoxO3a has been recently proposed as a good prognostic factor in luminal-like breast cancer, which contain principally ERα+ cases,40 where it directly correlates with biomarkers of good prognosis and inversely with mitotic counts and tumor grade. Moreover, with respect to patient outcome, FoxO3a nuclear localization was associated with longer breast cancer specific survival and longer distant metastasis-free interval, independently of the well-established breast cancer prognostic factors.41
The screening of nuclear FoxO3a on opportunely selected ERα+ and ERα− tissue samples from patients with breast cancer of ductal origin gave results that perfectly fit with the above-mentioned reports and also confirm the in vitro studies presented in this work. Moreover, the co-expression of Cav1 and FoxO3a in ERα+ tumors, together with the functional link provided by our in vitro data, supports a potentially important role for these 2 proteins in predicting a better tumor prognosis. However, a more systematic evaluation within various subtypes of ERα+ and ERα− non-invasive and invasive breast cancers, in absence or in presence of lymph node and/or long distance metastasis, would help to better clarify the biological and prognostic role of FoxO3a protein expression, also with respect to its subcellular localization. For instance, since no correlation has been found between FoxO3a and ERα 41, the loss of an active (nuclear) FoxO3a might be predictive of a worse phenotype in the subset of ERα+ breast cancers that do not respond to therapy. At the same time, a more accurate immunohistochemical analysis on the biological link between FoxO3a and Cav1 in hormone-positive tumors needs to be addressed. In fact, although Cav1 expression has been associated with lack of the steroid hormone receptor,37 its positivity in luminal-like tumors could represent a good prognostic factor when associated to a FoxO3a nuclear prevalence.
In conclusion, the results presented here give new insights on the functional role of nuclear FoxO3a, whose overexpression seems to be associated to a low motile phenotype in ERα+ breast cancers and to a more metastatic potential in those lacking the hormone receptor, harboring the idea that ERα may represent the molecular switch determining FoxO3a biological behavior. These evidences clearly suggest that FoxO3a has the potential to become a relevant prognostic factor and a suitable pharmacological target to be exploited in combination therapies for both ERα+ (through FoxO3a activation) and ERα− (through FoxO3a disruption) breast cancer patients.
Materials and Methods
Cell culture, conditions, and treatments
The human breast cancer epithelial cell lines MCF-7, ZR75, T47D, MDA-MB-231, and MDA-MB-468 and the cervical epithelial cell line, HeLa, were purchased from Interlab Cell Line Collection, ICLC, Italy. Ishikawa human endometrial cancer cell line was obtained from D Picard (University of Geneva). MCF-7 and ZR75 were maintained in DMEM/Ham F-12 medium (1:1) (DMEM/F-12) supplemented with 5% FBS. Ishikawa and HeLa cells were grown in MEM containing 10% FBS and 1% non-essential amino acids. MDA-MB-231 and MDA-MB-468 cells were cultured in 10% FBS DMEM. T47D cells were cultured in RPMI containing 10% FBS, 2.5 g/ml glucose, 1% Na-Pyruvate, 10 nM Hepes, and 0.2 U/ml insulin. Additionally, culture media were supplemented with 100 IU/ml penicillin, 100 ng/ml streptomycin, and 0.2 mM L-glutamine. For experimental purposes, cells were synchronized in phenol red-free and serum-free media (PRF-SFM) for 24 h and then, where opportune, switched to PRF-media containing 5% charcoal-treated FBS (PRF-CT) or FBS (ERα+ and ERα− cells, respectively), in presence or absence of 17β-estradiol (E2, Sigma-Aldrich). All media and reagents were purchased from Invitrogen.
Plasmids and transfections assays
The following plasmids were used: pcDNA3 empty vector (Invitrogen); 1038 pcDNA3 flag FKHRL1 (F3a) encoding full-length FoxO3a and 1319 pcDNA3 flag FKHRL1 AAA (F3aAAA), encoding the constitutively active triple mutant of FoxO3a (provided by William Sellers, Addgene plasmids 10708 and 10709,42 respectively). MCF-7, ZR75, and MDA-MB-231 and MDA-MB-468 cells were resuspended in PRF-growing medium (PRF-GM) and transfected with Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions, while transfection of T47D, Ishikawa and HeLa cells were conducted with FuGENE HD (Promega). Six hours after transfections, cells were synchronized for 24 h and then subjected either to migration, invasion, and soft agar assays or switched to FBS (ERα− cells) or PRF-CT, in presence or absence of E2 (ERα+ cells), for protein and RNA extraction purposes.
For luciferase assays, the following constructs of the Cav1 promoter43 were used: pGL3-cavFL, driving the expression of firefly luciferase under the control of the Cav1 promoter full-length (nt −837/−36 from the ATG), pGL3/SRE1/2 (nt −837/−355) and pGL3/SRE3 (nt −354/−36).
Transfections were performed using FuGENE HD. Luciferase activity was measured using the dual-luciferase assay system, normalized to pRL-Tk activity (both from Promega), and expressed as fold-induction over the control.
siRNA-mediated RNA interference
Custom-synthesized siRNA-annealed duplexes (25 bp double-stranded RNA [dsRNA]) were used for effective depletion of FoxO3a (siF3a) and Caveolin-1 (siCav1) transcripts. A scramble siRNA (siScramble) lacking identity with known gene targets was used as a negative control. Cells were transfected in suspension with Lipofectamine 2000 in PRF-GM, using the appropriate amounts of siRNA duplexes (Life Technologies). ERα silencing was conducted according to manufacturer’s instructions using siER and the appropriate transfection reagent HiPerFect HTS Reagent purchased from Qiagen. For each silenced gene, at least 2 different siRNAs have been employed with comparable outcome.
Migration and invasion assays
Migration assays were performed as previously described.15 Briefly, 6 h after transfection or silencing, cells were serum starved for 24 h, resuspended in PRF-SFM, and seeded (104 cells/insert) on the upper face of 24-well modified Boyden chambers (8 μm) (Corning); 500 μl of 5% PRF-CT with or without 100 nM E2 (for ERα+ cells) or PRF-GM (for ERα− cells) were added to the bottom of the wells. After opportune incubation, migrated cells were stained with Coomassie brilliant blue and counted under the microscope.
For invasion experiments, 30 μl of Matrigel™ Basement Membrane Matrix (BD Biosciences) (1:3 dilution in PRF-SFM) were coated on the internal surfaces of the Boyden chambers and let solidify at RT for 30 min. The lower chambers were loaded as described for migration assays. Cells suspended in 200 μl of 1% PRF-CT (ERα+ cells) or 1% FBS (ERα− cells), respectively, were plated into the upper chambers (105 cells/insert). After the appropriate times of incubation, cells in the upper chamber were removed by a cotton tip; membranes were then mixed in methanol for 10 min at −20 °C, rinsed with PBS, stained with DAPI (Sigma Aldrich, Italy) for 5 min, rinsed again in PBS and dried. The filters were then detached from the chamber, and mounted onto slides using Fluoromount mounting medium (Sigma Aldrich) and observed under a fluorescence microscope (Olympus BX51 fluorescence microscope, Olympus Italia srl). Invading cells were photographed at 10× magnification using ViewFinder™ Software, through an Olympus camera system dp50 and then counted using ImageJ software (NIH).
Anchorage-independent growth assay
Transfected or silenced ERα+ cells were seeded in 1 mL of 0.3% GellyPhor® HR agarose (Euroclone S.p.A.) on top a base of 0.6% agarose in 12-multiwell plates in PRF-CT (2 × 104 cells/well) and treated with 100nM E2 or left untreated; ERα− cells were seeded in PRF-GM (3 × 104 cells/well). On day 14, the colonies (>50 μm) were exposed to 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) for 2 h, photographed at 4× magnification and counted under the microscope (Olympus BX51 microscope).
RNA extraction, reverse transcription, and real-time (RT)-PCR
Total RNA was isolated using TRI-reagent (Ambion) and treated with DNase I (Life Technologies). Two micrograms of total RNA were reverse transcribed with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s instructions. cDNA was diluted 1:3 in nuclease-free water, and 5 μl were analyzed in triplicate by RT-PCR in a iCycler iQ Detection System (Bio-Rad) using SYBR green Universal PCR Master Mix (Bio-Rad) and the following pairs of primers: FoxO3a forward 5′- CAAACCCAGG GCGCTCTT-3′ and reverse 5′- CTCACTCAAG CCCATGTTGC T-3′ (68 bp); Cav1 forward 5′- CAGTTTTCAT CCAGCCACGG-3′ and reverse 5′- CGGATGGGAA CGGTGTAGAG-3′ (82 bp).
Negative controls contained water instead of first-strand cDNA. Each sample was normalized on its 18S rRNA content. The relative gene expression levels were normalized to a calibrator that was chosen to be the basal, untreated sample. The final results were expressed as n-fold differences in gene expression relative to 18S rRNA and the calibrator, calculated using the ΔΔCT method as follows: n-fold = 2−(ΔCTsample − ΔCTcalibrator), where the ΔCT values of the sample and calibrator were determined by subtracting the average CT value of the 18S rRNA reference gene from the average CT value of the different genes analyzed.
Western blotting (WB) assays
Protein expression was assessed by WB assay as previously described.44 Total lysates were extracted using RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.25% Na deoxycholate) plus inhibitors (0.1 mmol/liter Na3VO4, 1% PMSF, and 20 mg/ml aprotinin). The protein content was determined using Bradford dye reagent (Bio-Rad). Fifty μg of lysates were separated on an 11% polyacrylamide denaturing gel and transferred to nitrocellulose membranes. Proteins of interest were detected with specific polyclonal (p) or monoclonal (m) antibodies (Abs), recognized by peroxidase-coupled secondary Abs, and developed using the ECL Plus Western Blotting Detection System (Amersham Pharmacia Biotech). The following Abs were used: anti-FoxO3a (75D8) pAb (Cell Signaling), anti-Cav1 (N-20) pAb, anti-ERα (F-10) mAb, and anti-GAPDH (FL-335) pAb (Santa Cruz Biotechnology). Images were acquired by using an Epson Perfection scanner (Epson).
Chromatin immunoprecipitation (ChIP)
ChIP assay was performed as previously described.11 The immuno-cleared chromatin was precipitated with anti-FoxO3a pAb (Abcam, USA) and anti-Polymerase II (N-20) pAb (Santa Cruz Biotechnology). Normal rabbit IgG (Santa Cruz Biotechnology) was used instead of primary Abs as negative controls. Immunoprecipitated DNA was analyzed by RT-PCR, as described above. A pair of primers (5′-GAGATGATGC ACTGCGAAAA-3′ and reverse 5′-GCCAAAGGTT TGTTCTGCTC -3′) (242 bp) mapping the FKHE-containing Cav1 promoter region forward was used.
Tissue collection, immunohistochemistry (IHC), and data analysis
Formalin-fixed paraffin-embedded tissue sections were prepared from primary operable breast cancer cases (15 DCIS and 25 IDC from ERα+ tumors and an equal number from ERα− tumors) from patients under age 80 who underwent mastectomy at the Cosenza Hospital (Cosenza Hospital Authority) between 2011 and 2012. FoxO3a, ERα and Cav1 expression were assessed by IHC. The rabbit anti-FoxO3a pAb (cat. PA1-14171, Thermo Scientific) and the rabbit anti-Caveolin-1 pAb (N-20) (sc-894, Santa Cruz Biotechnology) were optimized at a working dilution of 1:200 in Dako Real antibody diluent (DAKO); the mouse anti-ERα (Clone 1D5, DAKO) was ready to use. Deparaffinization, rehydration, and antigen unmasking was obtained by incubation in tris-phospahte buffer (Envision Flex target retrieval solution) in a Pre-Treatment Module for Tissue Specimens (PTLINK), according to the manufacturer’s instructions (DAKO). The staining was performed in a Dako Autostainer Link48 immunostainer, using a linked streptavidin biotin technique (Envision Flex kit High pH, DAKO) in accordance with the manufacturer’s instructions. Sections were counterstained in hematoxylin and coverslipped using DPX mounting medium (both from Sigma-Aldrich).
The expression and subcellular localization of FoxO3a and Cav1 were evaluated microscopically. Pictures of representative fields were taken at opportune magnification using ViewFinder™ Software, through an Olympus camera system dp50.
Ethical statement
The clinical investigation has been conducted in accordance with the ethical standards and according to the Declaration of Helsinki of 1975 and to national and international guidelines and has been approved by the Research Ethics Committee of Cosenza Hospital Authority. The informed consent was not requested, since the study was retrospective and the data were analyzed anonymously.
Statistical analysis
All data were expressed as the mean ± s.d. of at least 3 independent experiments. Statistical significances were evaluated using Student t test. The correlations between nuclear and cytoplasmic FoxO3a and Cav1 with respect to tumor grading and invasiveness were examined with Pearson correlation test.
Supplementary Material
Acknowledgments
We thank Cao Sheng and Vijay Shah from the Gastroenterology Research Unit and Tumor Biology Program, Mayo Clinic College of Medicine (Rochester, Minnesota) for have kindly provided the human Cav1 promoter constructs. This study was supported by Grant IG 11595/2012 and IG 12849/2012 from Associazione Italiana Ricerca sul Cancro (AIRC), and MIUR EX 60%.
Glossary
Abbreviations:
- Cav1
caveolin-1
- E2
17β-estradiol
- ERα−
estrogen receptor alpha negative
- ERα+
estrogen receptor alpha positive
- FoxO3a
Forkhead box class O 3a
- F3a
1038 pcDNA3 flag FKHRL1 (Addgene) encoding full-length FoxO3a
- F3aAAA
1319 pcDNA3 flag FKHRL1 AAA (Addgene) encoding the constitutively active triple mutant of FoxO3a
- IDC
invading ductal carcinomas
- IHC
immunohistochemistry
- DCIS
ductal carcinomas in situ
- MMPs
matrix metalloproteinases
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PKB
protein kinase B
- PRF-CT
phenol red-free medium containing charcoal-treated FBS
- PRF-GM
PRF-growing medium
- PRF-SFM
PRF and serum-free media
- siCav1
siRNA for effective depletion of Caveolin-1 transcripts
- siER
siRNA for effective depletion of ERα transcripts
- siF3a
siRNA for effective depletion of FoxO3a transcripts
- WB
western blotting assay
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/26421
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26421
References
- 1.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–25. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 2.Yang JY, Hung MC. Deciphering the role of forkhead transcription factors in cancer therapy. Curr Drug Targets. 2011;12:1284–90. doi: 10.2174/138945011796150299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–37. doi: 10.1016/S0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 4.Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, Lang JY, Lai CC, Chang CJ, Huang WC, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008;10:138–48. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou Y, Tsai WB, Cheng CJ, Hsu C, Chung YM, Li PC, Lin SH, Hu MC. Forkhead box transcription factor FOXO3a suppresses estrogen-dependent breast cancer cell proliferation and tumorigenesis. Breast Cancer Res. 2008;10:R21. doi: 10.1186/bcr1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–23. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sisci D, Surmacz E. Crosstalk between IGF signaling and steroid hormone receptors in breast cancer. Curr Pharm Des. 2007;13:705–17. doi: 10.2174/138161207780249182. [DOI] [PubMed] [Google Scholar]
- 8.Lanzino M, Morelli C, Garofalo C, Panno ML, Mauro L, Andò S, Sisci D. Interaction between estrogen receptor alpha and insulin/IGF signaling in breast cancer. Curr Cancer Drug Targets. 2008;8:597–610. doi: 10.2174/156800908786241104. [DOI] [PubMed] [Google Scholar]
- 9.Schuur ER, Loktev AV, Sharma M, Sun Z, Roth RA, Weigel RJ. Ligand-dependent interaction of estrogen receptor-alpha with members of the forkhead transcription factor family. J Biol Chem. 2001;276:33554–60. doi: 10.1074/jbc.M105555200. [DOI] [PubMed] [Google Scholar]
- 10.Zhao HH, Herrera RE, Coronado-Heinsohn E, Yang MC, Ludes-Meyers JH, Seybold-Tilson KJ, Nawaz Z, Yee D, Barr FG, Diab SG, et al. Forkhead homologue in rhabdomyosarcoma functions as a bifunctional nuclear receptor-interacting protein with both coactivator and corepressor functions. J Biol Chem. 2001;276:27907–12. doi: 10.1074/jbc.M104278200. [DOI] [PubMed] [Google Scholar]
- 11.Morelli C, Lanzino M, Garofalo C, Maris P, Brunelli E, Casaburi I, Catalano S, Bruno R, Sisci D, Andò S. Akt2 inhibition enables the forkhead transcription factor FoxO3a to have a repressive role in estrogen receptor alpha transcriptional activity in breast cancer cells. Mol Cell Biol. 2010;30:857–70. doi: 10.1128/MCB.00824-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belguise K, Guo S, Sonenshein GE. Activation of FOXO3a by the green tea polyphenol epigallocatechin-3-gallate induces estrogen receptor alpha expression reversing invasive phenotype of breast cancer cells. Cancer Res. 2007;67:5763–70. doi: 10.1158/0008-5472.CAN-06-4327. [DOI] [PubMed] [Google Scholar]
- 13.Storz P, Döppler H, Copland JA, Simpson KJ, Toker A. FOXO3a promotes tumor cell invasion through the induction of matrix metalloproteinases. Mol Cell Biol. 2009;29:4906–17. doi: 10.1128/MCB.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rochefort H, Platet N, Hayashido Y, Derocq D, Lucas A, Cunat S, Garcia M. Estrogen receptor mediated inhibition of cancer cell invasion and motility: an overview. J Steroid Biochem Mol Biol. 1998;65:163–8. doi: 10.1016/S0960-0760(98)00010-7. [DOI] [PubMed] [Google Scholar]
- 15.Sisci D, Middea E, Morelli C, Lanzino M, Aquila S, Rizza P, Catalano S, Casaburi I, Maggiolini M, Andò S. 17β-estradiol enhances α(5) integrin subunit gene expression through ERα-Sp1 interaction and reduces cell motility and invasion of ERα-positive breast cancer cells. Breast Cancer Res Treat. 2010;124:63–77. doi: 10.1007/s10549-009-0713-6. [DOI] [PubMed] [Google Scholar]
- 16.Manni A, Wright C, Buck H. Growth factor involvement in the multihormonal regulation of MCF-7 breast cancer cell growth in soft agar. Breast Cancer Res Treat. 1991;20:43–52. doi: 10.1007/BF01833356. [DOI] [PubMed] [Google Scholar]
- 17.Platet N, Cathiard AM, Gleizes M, Garcia M. Estrogens and their receptors in breast cancer progression: a dual role in cancer proliferation and invasion. Crit Rev Oncol Hematol. 2004;51:55–67. doi: 10.1016/j.critrevonc.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 18.van den Heuvel AP, Schulze A, Burgering BM. Direct control of caveolin-1 expression by FOXO transcription factors. Biochem J. 2005;385:795–802. doi: 10.1042/BJ20041449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charpentier AH, Bednarek AK, Daniel RL, Hawkins KA, Laflin KJ, Gaddis S, MacLeod MC, Aldaz CM. Effects of estrogen on global gene expression: identification of novel targets of estrogen action. Cancer Res. 2000;60:5977–83. [PubMed] [Google Scholar]
- 20.Razandi M, Oh P, Pedram A, Schnitzer J, Levin ER. ERs associate with and regulate the production of caveolin: implications for signaling and cellular actions. Mol Endocrinol. 2002;16:100–15. doi: 10.1210/me.16.1.100. [DOI] [PubMed] [Google Scholar]
- 21.Sloan EK, Stanley KL, Anderson RL. Caveolin-1 inhibits breast cancer growth and metastasis. Oncogene. 2004;23:7893–7. doi: 10.1038/sj.onc.1208062. [DOI] [PubMed] [Google Scholar]
- 22.Joshi B, Strugnell SS, Goetz JG, Kojic LD, Cox ME, Griffith OL, Chan SK, Jones SJ, Leung SP, Masoudi H, et al. Phosphorylated caveolin-1 regulates Rho/ROCK-dependent focal adhesion dynamics and tumor cell migration and invasion. Cancer Res. 2008;68:8210–20. doi: 10.1158/0008-5472.CAN-08-0343. [DOI] [PubMed] [Google Scholar]
- 23.Sotgia F, Rui H, Bonuccelli G, Mercier I, Pestell RG, Lisanti MP. Caveolin-1, mammary stem cells, and estrogen-dependent breast cancers. Cancer Res. 2006;66:10647–51. doi: 10.1158/0008-5472.CAN-06-2805. [DOI] [PubMed] [Google Scholar]
- 24.Fiucci G, Ravid D, Reich R, Liscovitch M. Caveolin-1 inhibits anchorage-independent growth, anoikis and invasiveness in MCF-7 human breast cancer cells. Oncogene. 2002;21:2365–75. doi: 10.1038/sj.onc.1205300. [DOI] [PubMed] [Google Scholar]
- 25.Wu P, Wang X, Li F, Qi B, Zhu H, Liu S, Cui Y, Chen J. Growth suppression of MCF-7 cancer cell-derived xenografts in nude mice by caveolin-1. Biochem Biophys Res Commun. 2008;376:215–20. doi: 10.1016/j.bbrc.2008.08.146. [DOI] [PubMed] [Google Scholar]
- 26.Williams TM, Medina F, Badano I, Hazan RB, Hutchinson J, Muller WJ, Chopra NG, Scherer PE, Pestell RG, Lisanti MP. Caveolin-1 gene disruption promotes mammary tumorigenesis and dramatically enhances lung metastasis in vivo. Role of Cav-1 in cell invasiveness and matrix metalloproteinase (MMP-2/9) secretion. J Biol Chem. 2004;279:51630–46. doi: 10.1074/jbc.M409214200. [DOI] [PubMed] [Google Scholar]
- 27.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–97. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 28.Safe S, Kim K. Non-classical genomic estrogen receptor (ER)/specificity protein and ER/activating protein-1 signaling pathways. J Mol Endocrinol. 2008;41:263–75. doi: 10.1677/JME-08-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lützner N, De-Castro Arce J, Rösl F. Gene expression of the tumour suppressor LKB1 is mediated by Sp1, NF-Y and FOXO transcription factors. PLoS One. 2012;7:e32590. doi: 10.1371/journal.pone.0032590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo X, Puig O, Hyun J, Bohmann D, Jasper H. Foxo and Fos regulate the decision between cell death and survival in response to UV irradiation. EMBO J. 2007;26:380–90. doi: 10.1038/sj.emboj.7601484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahn S, Olive M, Aggarwal S, Krylov D, Ginty DD, Vinson C. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998;18:967–77. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 33.Abbondanza C, De Rosa C, Ombra MN, Aceto F, Medici N, Altucci L, Moncharmont B, Puca GA, Porcellini A, Avvedimento EV, et al. Highlighting chromosome loops in DNA-picked chromatin (DPC) Epigenetics. 2011;6:979–86. doi: 10.4161/epi.6.8.16060. [DOI] [PubMed] [Google Scholar]
- 34.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 35.Van den Eynden GG, Van Laere SJ, Van der Auwera I, Merajver SD, Van Marck EA, van Dam P, Vermeulen PB, Dirix LY, van Golen KL. Overexpression of caveolin-1 and -2 in cell lines and in human samples of inflammatory breast cancer. Breast Cancer Res Treat. 2006;95:219–28. doi: 10.1007/s10549-005-9002-1. [DOI] [PubMed] [Google Scholar]
- 36.Savage K, Lambros MB, Robertson D, Jones RL, Jones C, Mackay A, James M, Hornick JL, Pereira EM, Milanezi F, et al. Caveolin 1 is overexpressed and amplified in a subset of basal-like and metaplastic breast carcinomas: a morphologic, ultrastructural, immunohistochemical, and in situ hybridization analysis. Clin Cancer Res. 2007;13:90–101. doi: 10.1158/1078-0432.CCR-06-1371. [DOI] [PubMed] [Google Scholar]
- 37.Elsheikh SE, Green AR, Rakha EA, Samaka RM, Ammar AA, Powe D, Reis-Filho JS, Ellis IO. Caveolin 1 and Caveolin 2 are associated with breast cancer basal-like and triple-negative immunophenotype. Br J Cancer. 2008;99:327–34. doi: 10.1038/sj.bjc.6604463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamaguchi H, Takeo Y, Yoshida S, Kouchi Z, Nakamura Y, Fukami K. Lipid rafts and caveolin-1 are required for invadopodia formation and extracellular matrix degradation by human breast cancer cells. Cancer Res. 2009;69:8594–602. doi: 10.1158/0008-5472.CAN-09-2305. [DOI] [PubMed] [Google Scholar]
- 39.Jin GS, Kondo E, Miyake T, Shibata M, Takashima T, Liu YX, Hayashi K, Akagi T, Yoshino T. Expression and intracellular localization of FKHRL1 in mammary gland neoplasms. Acta Med Okayama. 2004;58:197–205. doi: 10.18926/AMO/32088. [DOI] [PubMed] [Google Scholar]
- 40.Bertos NR, Park M. Breast cancer - one term, many entities? J Clin Invest. 2011;121:3789–96. doi: 10.1172/JCI57100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Habashy HO, Rakha EA, Aleskandarany M, Ahmed MA, Green AR, Ellis IO, Powe DG. FOXO3a nuclear localisation is associated with good prognosis in luminal-like breast cancer. Breast Cancer Res Treat. 2011;129:11–21. doi: 10.1007/s10549-010-1161-z. [DOI] [PubMed] [Google Scholar]
- 42.Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell. 2002;2:81–91. doi: 10.1016/S1535-6108(02)00086-7. [DOI] [PubMed] [Google Scholar]
- 43.Cao S, Fernandez-Zapico ME, Jin D, Puri V, Cook TA, Lerman LO, Zhu XY, Urrutia R, Shah V. KLF11-mediated repression antagonizes Sp1/sterol-responsive element-binding protein-induced transcriptional activation of caveolin-1 in response to cholesterol signaling. J Biol Chem. 2005;280:1901–10. doi: 10.1074/jbc.M407941200. [DOI] [PubMed] [Google Scholar]
- 44.Lanzino M, Sisci D, Morelli C, Garofalo C, Catalano S, Casaburi I, Capparelli C, Giordano C, Giordano F, Maggiolini M, et al. Inhibition of cyclin D1 expression by androgen receptor in breast cancer cells--identification of a novel androgen response element. Nucleic Acids Res. 2010;38:5351–65. doi: 10.1093/nar/gkq278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.