Abstract
A pool of PTEN localizes to the nucleus. However, the exact mechanism of action of nuclear PTEN remains poorly understood. We have investigated PTEN’s role during DNA damage response. Here we report that PTEN undergoes chromatin translocation after DNA damage, and that its translocation is closely associated with its phosphorylation on S366/T370 but not on S380. Deletional analysis reveals that the C2 domain of PTEN is responsible for its nuclear translocation after exposure to genotoxin. Both casein kinase 2 and GSK3β are involved in the phosphorylation of the S366/T370 epitope, as well as PTEN’s association with chromatin after DNA damage. Significantly, PTEN specifically interacts with Rad52 and colocalizes with Rad52, as well as γH2AX, after genotoxic stress. Moreover, PTEN is involved in regulating Rad52 sumoylation. Combined, our studies strongly suggest that nuclear/chromatin PTEN mediates DNA damage repair through interacting with and modulating the activity of Rad52.
Keywords: DNA damage, PTEN, Rad52, chromatin, genotoxic stress, phosphorylation
Introduction
PTEN is a tumor suppressor gene whose importance in protection of mammalian cells from malignant transformation is second only to that of p53.1,2 The PTEN tumor suppressor is frequently mutated in cancer cells, and inherited PTEN mutations cause cancer-susceptibility conditions.1,2 The PTEN level as well as its activity in an organism profoundly influences tumor susceptibility, because PTEN+/‒ mice also develop tumors in multiple organs.3,4
PTEN functions as a lipid and protein phosphatase.1,2 The PTEN protein consists of an N-terminal phosphatase domain, a lipid-binding C2 domain, and a C-terminal “tail” domain. The N terminus contains a PIP2-binding motif spanning from residues 6–15; the C tail contains PEST motifs which commonly serve as molecular signals for rapid proteolytic cleavage.5 Biochemically, PTEN dephosphorylates the lipid second-messenger phosphatidylinositol 3, 4, 5-trisphosphate (PIP3) to generate phosphatidylinositol 3, 4-bisphosphate (PIP2) and, by doing so, antagonizes the PI3K/Akt signaling pathway. PTEN also has nuclear functions, since a fraction of PTEN is consistently detected in the nucleus.6,7 Several independent studies have shown that nuclear PTEN plays a significant role in the maintenance of genomic stability through the modulation of DNA repair, chromosomal segregation, and cell cycle arrest.8-12
PTEN is extensively phosphorylated at the C-tail region by several protein kinases. Phosphorylation of several Ser/Thr residues, including S370 in the C-tail region of PTEN by casein kinase 2 (CK2), is essential for the tail-dependent regulation of stability as phospho-defective mutant proteins exhibit decreased stability.13-15 GSK3β phosphorylates PTEN at S362 and T366.13 PTEN can be modified by mono- or polyubiquitination, which regulates its nuclear localization and stability, respectively.5 NEDD4-1 and WWP2 and RFP are 3 reported ubiquitin E3 ligases that mediate PTEN ubiquitination,16-18 although NEDD4-1 appears to be dispensable for the regulation of its subcellular localization and stability.19 PTEN also contains atypical nuclear localization sequences.20,21
Rad52 is a key component in DNA double-strand break repair and homologous recombination in yeast.22,23 Rad52 homologs in mammals are also identified and characterized as having similar biochemical activities to those of the yeast counterpart.22,23 Intriguingly, mammalian Rad52 plays a more modest role in vivo as Rad52 knockout in mice results in neither lethality, nor overt adverse phenotypes. However, Rad52 inactivation is synthetically lethal with BRCA2 deficiency,24 strongly suggesting that BRCA2 perform functions similar to those of Rad52 in vivo. Rad52 is regulated by sumoylation, which is induced by DNA damage and involved in regulating Rad52 stability and activity.25,26
In the current study, we report that a significant fraction of PTEN underwent time-dependent nuclear translocation after DNA damage induced by reactive oxygen species, and that the accumulation of chromatin PTEN was associated with its phosphorylation on S366/T370. Deletional analysis coupled with ectopic expression shows that the C2 domain was responsible for chromatin translocation of PTEN. PTEN specifically interacted with Rad52, and the interaction was enhanced after treatment with H2O2. Our further study revealed that nuclear PTEN was involved in regulating sumoylation of Rad52, a process essential for Rad52 function in DNA homologous recombination.
Results
PTEN has been implicated in DNA damage responses.27 To understand how PTEN is regulated during DNA damage responses, we first examined PTEN subcellular localization after treatment with adriamycin, etoposide, and H2O2, chemical compounds that cause DNA double-strand breaks. We observed that at high concentrations, both adriamycin and H2O2, but not etoposide, induced chromatin association of PTEN (Fig. 1). The presence of chromatin-PTEN was inversely correlated with the amount of cytoplasmic PTEN, especially after H2O2 treatment, suggesting its subcellular translocation under severe genotoxic stress. Both adriamycin and etoposide induced cleavage of PARP-1 at high concentrations, indicating apoptosis caused by these agents. On the other hand, H2O2-induced chromatin translocation was not associated with PARP-1 cleavage. Moreover, chromatin-associated PTEN was phosphorylated at the epitope of T366/S370, as revealed by probing with a phospho-specific antibody.
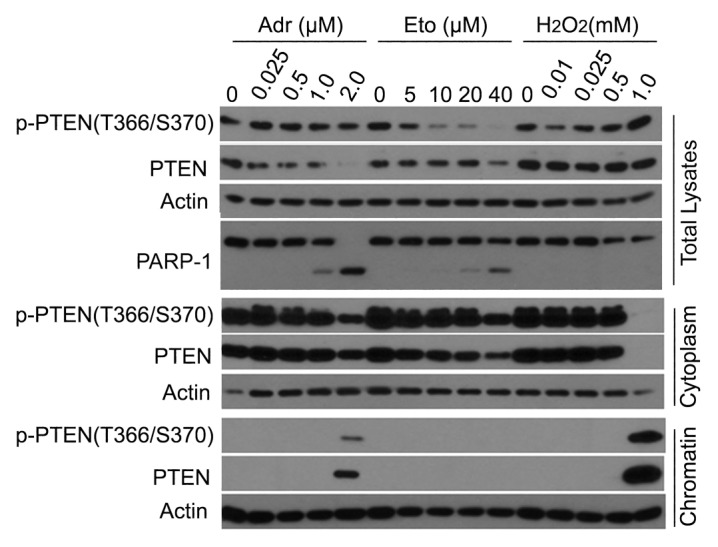
Figure 1. Chromatin localization of PTEN in response to genotoxic stresses. HeLa cells were treated with adriamycin (Adr), etoposide (Eto), and hydrogen peroxide (H2O2) at various concentrations for 24 h. Cells were then collected at the end of the treatment. Both cytoplasmic and chromatin-bound fractions of cell lysates, as well as total cell lysates, were obtained. Equal amounts of the lysates were blotted for phospho-PTEN(T366/S370), total PTEN, PARP-1, and/or actin.
We next analyzed the time course of PTEN subcellular translocation after treatment with H2O2, given its strong effect on inducing chromatin association (Fig. 1). Immunoblotting analysis revealed that PTEN underwent a time-dependent decrease in the cytoplasm with a concomitant increase on chromatin (Fig. 2). PTEN phosphorylation on epitope T366/S370, but not epitope S380, was increased after exposure to H2O2, and its level was closely associated with chromatin translocation. Moreover, DNA double-strand breaks as revealed by phospho-H2AX signals peaked at about 2 h after H2O2 treatment. The p-H2AX signals were inversely correlated with the increase of chromatin PTEN.
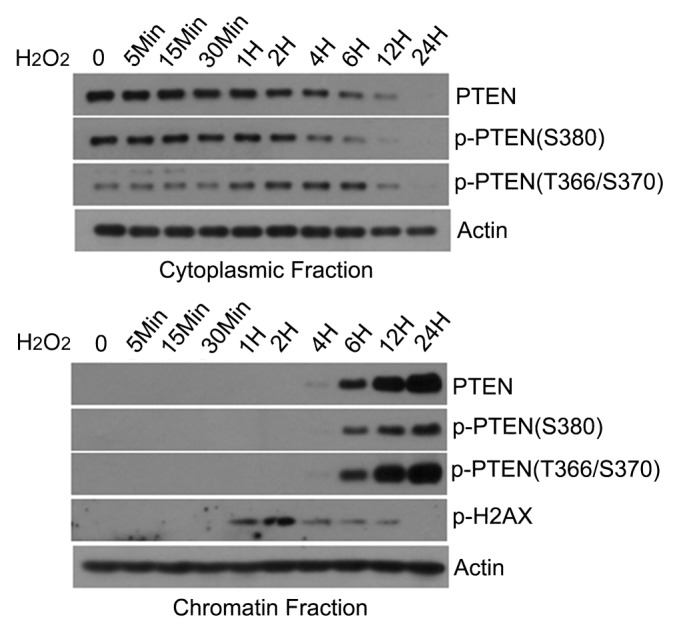
Figure 2. Time-dependent increase of chromatin-associated PTEN. HeLa cells were treated with 1 mM H2O2 for various times as indicated. Cells were collected and cytoplasmic and chromatin-bound fractions were obtained. Equal amounts of lysates from each fraction were blotted for phospho-PTEN (epitopes T366/S370 and S380, respectively), total PTEN, phospho-H2AX, and actin.
To determine which domain of PTEN mediated chromatin translocation, we made a series of deletion mutants as depicted in Figure 2. PTEN mutants, as well as the wild-type counterpart, were expressed as GFP-tagged fusion proteins. After transfection, HeLa cells were treated with or without H2O2 for 12 h. Immunoblotting revealed that deletion constructs C and D with an intact C2 domain underwent nuclear translocation in a manner similar to that of wild-type PTEN (Fig. 3). Phosphatase domain appeared to suppress the chromatin translocation, because mutants with the phosphatase domain alone or with the C2 domain failed to be associated with chromatin with or without H2O2 treatment. C tail domain was constitutively present in both cytoplasmic and chromatin fractions, and the genotoxic stress did not significantly change the amount of the mutant peptide on chromatin. This may be due to fact that the fragment is small, which could freely diffuse between compartments.
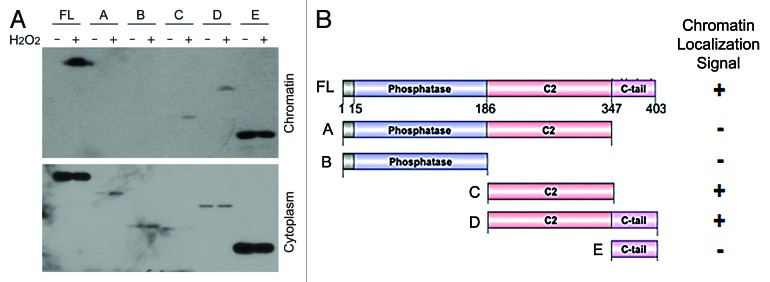
Figure 3. C2 domain of PTEN contains a signal for chromatin localization. (A) HEK293T cells were transfected with plasmids expressing GST-PTEN or individual deletion fragments, as indicated, for 24 h. Cells were then treated 1 mM H2O2 for 24 h. Cytoplasmic and chromatin-bound proteins were obtained at the end of treatment. Equal amounts of lysates from both fractions were blotted with the anti-GST antibody. (B) Schematic presentations of PTEN domains and various mutant constructs, as well as the summary of their chromatin localization. Plus (+) or minus (−) signs denote the presence and absence of a chromatin localization signal, respectively.
Given that T366/S370 phosphorylation was associated with chromatin translocation of PTEN, we next determined potential protein kinases that might mediate the translocation. We treated cells with chemicals that specifically inhibited CK2 or GSK3β, because it is known that GSK3β and CK2 phosphorylates T366 and S370, respectively.13 Immunoblotting revealed that both CK2 and GSK3β inhibitors, but not CK1 inhibitor, significantly reduced phosphorylation of the T366/S370 epitope, which was correlated with a reduced amount of PTEN on chromatin after treatment with H2O2 (Fig. 4A and B), strongly implicating the phosphorylation of the epitope in the positive regulation of PTEN chromatin association.
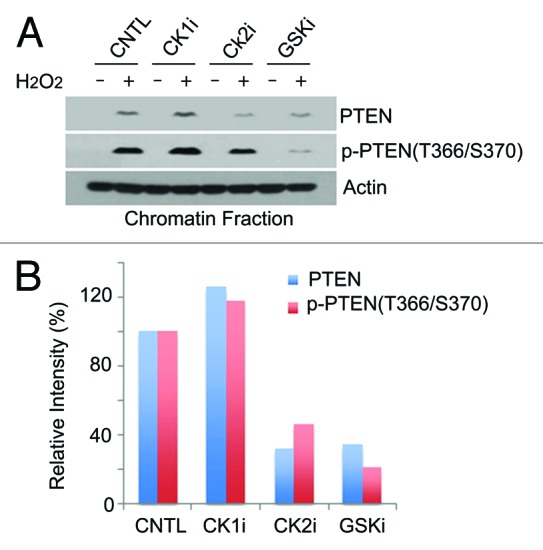
Figure 4. Regulation of PTEN chromatin localization by CK2 and GSK3β. (A) HeLa cells were treated with specific inhibitors for CK1 (CK1i), CK2 (CK2i), or GSK3β (GSKi) for 30 min, and then either left untreated or treated with H2O2 for 24 h. Cells were then lysed and chromatin-bound proteins were extracted. Equal amounts of cell lysates were blotted for phospho-PTEN(T366/S370), phospho-PTEN(S380), total PTEN, and actin. (B) Quantification of relative amounts of chromatin association of PTEN (blue) or phospho-PTEN (red).
To elucidate the function of chromatin PTEN, we next sought a potential binding partner(s) of chromatin PTEN. HeLa cells transfected with Flag-tagged PTEN or vector alone were lysed, and equal amounts of lysates were immunoprecipitated with the anti-Flag antibody. Immunoblotting showed that Rad52, but not ATR or Rad50, was present and enriched in Flag immunoprecipitates (Fig. 5A), suggesting a physical interaction between PTEN and Rad52. To determine whether PTEN co-localized with Rad52 during DNA damage responses, HeLa cells treated with or without H2O2 were fixed and stained with antibodies to PTEN and Rad52 and examined under a fluorescence microscope. PTEN signals were detected primarily in the cytoplasm and the cell surface membrane before H2O2 treatment (Fig. 5B). After the genotoxic stress, strong nuclear, punctuate signals of PTEN, as well as Rad52, were detected. PTEN signals significantly co-localized with Rad52 in cells treated with H2O2. Moreover, nuclear signals of PTEN co-localized with γH2AX signals after genotoxic stress (Fig. 5C), strongly suggesting a role of nuclear/chromatin PTEN in DNA damage responses.
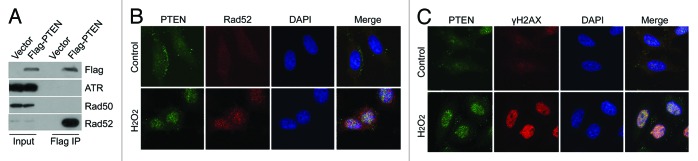
Figure 5. PTEN specifically interacts with Rad52. (A) HEK293T cells were transfected with plasmid expressing Flag-tagged PTEN for 48 h. Equal amounts of cell lysates prepared from various transfection samples were immunoprecipitated with the antibody to Flag. Immunoprecipitates, along with lysate inputs, were then blotted for Flag. ATR, Rad50, and Rad52. (B) HeLa Cells were treated 1 mM H2O2 for 24 h and then fixed and stained with antibodies against PTEN (green) and Rad52 (red). DNA was stained with DAPI. Representative images are shown. (C) HeLa Cells were treated 1 mM H2O2 for 24 h and then fixed and stained with antibodies against PTEN (green) and phospho-H2AX (γH2AX, red). DNA was stained with DAPI. Representative images are shown.
Rad52 is a sumoylated protein, and the sumoylation is essential for mediating its activity in DNA damage responses.25,28 To determine whether PTEN was involved in regulating Rad52, we downregulated endogenous PTEN via transfection of PTEN siRNAs. Immunoblotting analysis revealed that Rad52 was sumoylated in a time-dependent manner after treatment with H2O2 (Fig. 6A). Significantly, downregulation of PTEN compromised sumoylation of Rad52. Given the data described in this work, we propose the following model to explain the mode of action of chromatin-associated PTEN during the DNA damage response (Fig. 6B). PTEN is phosphorylated by both CK2 and GSK3β after genotoxic stress, and the phosphorylation facilitates chromatin translocation of PTEN. In the nucleus, PTEN interacts with Rad52 and regulates its sumoylation, which, in turn, is essential for mediating homologous recombination during DNA damage repair.
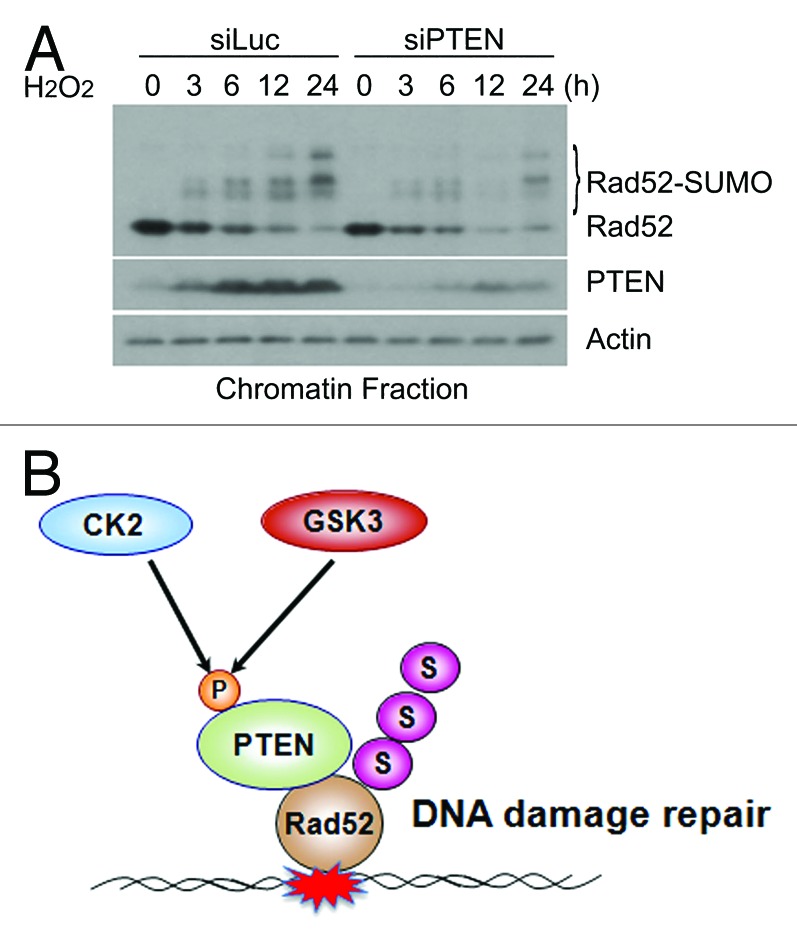
Figure 6. PTEN mediated regulation of Rad52 sumoylation. (A) HeLa cells were transfected with luciferase siRNAs (siLuc) or PTEN siRNAs (siPTEN) for 24 h and then treated with 1 mM H2O2 for various times as indicated. Cells were collected and chromatin-bound proteins were obtained. Equal amounts of protein lysates were then blotted for Rad52, PTEN, and actin. (B) A model of PTEN regulation in response to DNA damage induced by genotoxic stresses.
Discussion
PTEN is arguably the second most commonly lost tumor suppressor in human cancer, behind only to p53.1,2,29 During tumor development and progression, PTEN functions are lost or compromised, which leads to uncontrolled cell proliferation. Thus, further elucidation of PTEN regulatory pathways will lead to the identification of new targets for cancer drug development. The PTEN tumor suppressor is a central negative regulator of the PI3K/PDK1/Akt signaling axis that controls multiple cellular functions, including cell growth, survival, proliferation, and angiogenesis.1,2,30 However, many independent studies have demonstrated that a pool of PTEN is nuclear-bound, and that nuclear/chromatin-associated PTEN plays a significant role in the maintenance of genomic stability through modulating DNA repair, chromosomal segregation, and cell cycle progression.6-9,11 Our current study reveals that PTEN chromatin translocation is regulated by phosphorylation after exposure to genotoxic stress, and that chromatin PTEN is involved in regulating Rad52, thus an integral component of the DNA damage response pathway. Supporting this, it has been recently shown that nuclear PTEN functions to control DNA damage repair and sensitivity to genotoxic stress.31
Rad52 is a key component in DNA double-strand break repair and homologous recombination in yeast.22,23 Rad52 homologs in mammals are also identified and characterized as having similar biochemical activities as those of the yeast counterpart.22,23 Intriguingly, mammalian Rad52 plays a more modest role in vivo, as Rad52 knockout in mice results in neither lethality nor overt adverse phenotypes. However, Rad52 inactivation is synthetically lethal with BRCA2 deficiency,24 strongly suggesting that BRCA2 performs functions similar to those of Rad52 in vivo. Rad52 is regulated by sumoylation, which is induced by DNA damage and involved in regulating Rad52 stability and activity.25,26 Our finding that PTEN is involved in Rad52 sumoylation opens up a new avenue of research in the elucidation of the function of PTEN in DNA damage responses.
The Smc5–Smc6 complex is essential for normal growth, as well as coping with genotoxic stress through regulating chromatid cohesion and homologous recombination.32 It has been shown that cells with an Smc6 mutation display an increased level of Rad52 foci at centromeres.33 Nse2/Mms21, a SUMO ligase, is a component of the Smc5/6 complex.32,34 Given that PTEN is reported to reside at centromeres,8 it is tempting to speculate that PTEN may affect the activity of Smc5–Smc6 complex, which, in turn, modulates sumoylation of Rad52.
Rad52 is also involved in mitotic recombination. Mitotic recombination is a frequent yet poorly understood event in eukaryotic cells. Mitotic recombination within the centromere has been documented in yeast and mammalian cells.35,36 Proteins that are key to homologous recombination, such as BRCA1 and BRCA2, also have mitotic functions,37-41 although available information about their molecular basis in mitosis is fragmentary. Independent studies have also shown that PTEN exhibits centromere localization.8,9 Given our recent observation that PTEN interacts with Rad52 (Fig. 4), we propose that PTEN may have a function in mitotic recombination through modulating the activity and/or stability of Rad52.
Materials and Methods
Cell culture and transfection
HeLa and HEK-293T cell lines obtained from the American Type Culture Collection were cultured in DMEM supplemented with 10% fetal bovine serum (FBS, Invitrogen) and antibiotics (100 μg/ml of penicillin and 50 μg/ml of streptomycin sulfate, Invitrogen) at 37 °C under 5% CO2. Transfection of 293T cells was achieved with Lipofectamine 2000 (Invitrogen) following the manufacturers’ protocol. Transfection efficiency was estimated to be between 80–100% in all cases through transfecting a GFP expressing plasmid.
Antibodies and reagents
Antibodies to PTEN p-PTENS380, β-actin, ATR, Rad50, Rad52, and Flag were purchased from Cell Signaling Technology. Antibody to GFP was purchased from Santa Cruz Biotechnology. Antibody to phospho-PTENT366/T370 (p-PTENT/S) was purchased from Epitomics. CK2 inhibitor (4,5,6,7-Tetrabromobenzimidazole abbreviated as TBBz) was purchased from Sigma Aldrich. CK1 (4-[4-{2,3-Dihydrobenzo[1,4]dioxin-6-yl}-5-pyridin-2-yl-1H-imidazol-2-yl] benzamide as D4476) and GSK3β inhibitor (6-[2-{4-(2,4-Dichlorophenyl)-5-(4-methyl-1H-imidazol-2-yl)-pyrimidin-2-ylamino}ethyl-amino]-nicotinonitrile as GSK-3 Inhibitor XVI) were purchased from Calbiochem. Adriamycin, etoposide, and hydrogen peroxide (H2O2) were purchased from Sigma.
Plasmids
Plasmid encoding Flag-tagged PTEN (pCMV Flag WT-PTEN) was purchased from Addgene. To make plasmids encoding GST-PTEN fusion proteins, PCR products encoding human PTEN residues of 1–403, 1–347, 1–186, 186–347, 186–403, or 347–403 were inserted into pGEX-3X (GE Healthcare Life Sciences). Individual deletion fragments of GST-PTEN cDNA, as well as wild-type GST-PTEN cDNA, were subcloned into pcDNA3 plasmid, a mammalian expression vector. All deletion constructs were confirmed by DNA sequencing.
Lysate preparation and immunoblotting
Total cell lysates were prepared in a buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% IGEPAL, 0.1% SDS, and 0.5% sodium deoxycholate) supplemented with a mixture of protease and phosphatase inhibitors. Chromatin and cytosolic/soluble extracts were obtained as previously described.42 In brief, cell extracts were prepared in the harvest buffer (10 mM HEPES [pH 8.0], 50 mM NaCl, 0.5 M sucrose, 0.1 M EDTA, 0.5% Triton X-100) containing both protease inhibitors (1 mM dithiothreitol [DTT], 2 mg/ml pepstatin, 4 mg/ml aprotinin, 100 mM PMSF) and phosphatase inhibitors [10 mM] tetrasodium pyrophosphate, 100 mM NaF, 17.5 mM β-glycerophosphate). The low-speed supernatant (500 g) containing cytoplasmic proteins was collected and nuclear extracts were made by vortexing the nuclei for 15 min at 4°C in a buffer containing 20 mM HEPES (pH 7.9), 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.1% IGEPAL CA-630, and protease inhibitors. For Flag-PTEN immunoprecipitation, cells were collected and lysed in a lysis buffer (LB: 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 50 mM NaF, 0.5% Triton X-100, plus protease and phosphatase inhibitors). Flag-PTEN was immunoprecipitated with anti-flag M2 agarose (Sigma), and the immunocomplexes were washed 5 times with LB. After washing, the flag-beads were eluted at 95 °C for 5 min in 1× laemmli buffer. Protein concentrations were measured using the bicinchoninic acid protein assay reagent kit (Pierce Chemical Co). Equal amounts of protein lysates from various samples were used for SDS–PAGE analysis followed by immunoblotting. Specific signals on immunoblots (polyvinylidene difluoride) were visualized using enhanced chemiluminescence (Super-Signal, Pierce Chemical Co).
Immunofluorescence microscopy
Fluorescence microscopy was performed essentially as described.43 Briefly, cells were fixed with 4% paraformaldehyde (w/v) for 30 min, washed 3 times in PBS-T (PBS-Tween 0.1%), and permeabilized with 0.1% Triton X-100 in PBS before blocking with 4% Bovine serum albumin (BSA) in PBS-T for 30 min. Primary antibodies were incubated for 1 h, and secondary antibodies conjugated to Alexa-Fluor 488 and 555 were incubated for 30 min at room temperature in 4% BSA. DAPI staining was performed for 10 min in PBS. After washing, coverslips were dried and mounted on glass slides. Fluorescence microscopy was performed using a Leica Confocal Laser Scanning Microscopes SP2 and SL2 using the 100× objective (1.4).
Statistical analysis
Each experiment was performed at least 3 times. The data were plotted as the mean ± SD. Student t test was used for all comparisons. A P value of less than 0.05 was considered statistically significant.
Acknowledgment
This study was supported in part by US Public Service Awards to WD (CA090658 and ES019929) an NIEHS Center grant (ES000260). This manuscript is dedicated to the fond memory of Victor Fung, Ph.D., a former Program Officer at NCI and former Scientific Review Officer of the Cancer Etiology study section of CSR, NIH, for his wisdom, compassion, integrity, his love of sciences and the arts, his incredible culinary skills, and, above all, his contributions to the career development of so many investigators during his own distinguished career.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26465
References
- 1.Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387–90. doi: 10.1016/S0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 2.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–14. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 3.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–55. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 4.Kwabi-Addo B, Giri D, Schmidt K, Podsypanina K, Parsons R, Greenberg N, Ittmann M. Haploinsufficiency of the Pten tumor suppressor gene promotes prostate cancer progression. Proc Natl Acad Sci U S A. 2001;98:11563–8. doi: 10.1073/pnas.201167798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Jiang X. PTEN: a default gate-keeping tumor suppressor with a versatile tail. Cell Res. 2008;18:807–16. doi: 10.1038/cr.2008.83. [DOI] [PubMed] [Google Scholar]
- 6.Planchon SM, Waite KA, Eng C. The nuclear affairs of PTEN. J Cell Sci. 2008;121:249–53. doi: 10.1242/jcs.022459. [DOI] [PubMed] [Google Scholar]
- 7.Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, Pavletich NP, Carver BS, Cordon-Cardo C, Erdjument-Bromage H, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–56. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, Yin Y. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–70. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 9.Song MS, Carracedo A, Salmena L, Song SJ, Egia A, Malumbres M, Pandolfi PP. Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell. 2011;144:187–99. doi: 10.1016/j.cell.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puc J, Parsons R. PTEN loss inhibits CHK1 to cause double stranded-DNA breaks in cells. Cell Cycle. 2005;4:927–9. doi: 10.4161/cc.4.7.1795. [DOI] [PubMed] [Google Scholar]
- 11.Gupta A, Yang Q, Pandita RK, Hunt CR, Xiang T, Misri S, Zeng S, Pagan J, Jeffery J, Puc J, et al. Cell cycle checkpoint defects contribute to genomic instability in PTEN deficient cells independent of DNA DSB repair. Cell Cycle. 2009;8:2198–210. doi: 10.4161/cc.8.14.8947. [DOI] [PubMed] [Google Scholar]
- 12.Leonard MK, Hill NT, Bubulya PA, Kadakia MP. The PTEN-Akt pathway impacts the integrity and composition of mitotic centrosomes. Cell Cycle. 2013;12:1406–15. doi: 10.4161/cc.24516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Khouri AM, Ma Y, Togo SH, Williams S, Mustelin T. Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3beta. J Biol Chem. 2005;280:35195–202. doi: 10.1074/jbc.M503045200. [DOI] [PubMed] [Google Scholar]
- 14.Torres J, Pulido R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J Biol Chem. 2001;276:993–8. doi: 10.1074/jbc.M009134200. [DOI] [PubMed] [Google Scholar]
- 15.Xu D, Yao Y, Jiang X, Lu L, Dai W. Regulation of PTEN stability and activity by Plk3. J Biol Chem. 2010;285:39935–42. doi: 10.1074/jbc.M110.166462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, Wang J, Erdjument-Bromage H, Tempst P, Cordon-Cardo C, et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–39. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maddika S, Kavela S, Rani N, Palicharla VR, Pokorny JL, Sarkaria JN, Chen J. WWP2 is an E3 ubiquitin ligase for PTEN. Nat Cell Biol. 2011;13:728–33. doi: 10.1038/ncb2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JT, Shan J, Zhong J, Li M, Zhou B, Zhou A, Parsons R, Gu W. RFP-mediated ubiquitination of PTEN modulates its effect on AKT activation. Cell Res. 2013;23:552–64. doi: 10.1038/cr.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fouladkou F, Landry T, Kawabe H, Neeb A, Lu C, Brose N, Stambolic V, Rotin D. The ubiquitin ligase Nedd4-1 is dispensable for the regulation of PTEN stability and localization. Proc Natl Acad Sci U S A. 2008;105:8585–90. doi: 10.1073/pnas.0803233105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lobo GP, Waite KA, Planchon SM, Romigh T, Nassif NT, Eng C. Germline and somatic cancer-associated mutations in the ATP-binding motifs of PTEN influence its subcellular localization and tumor suppressive function. Hum Mol Genet. 2009;18:2851–62. doi: 10.1093/hmg/ddp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gil A, Andrés-Pons A, Fernández E, Valiente M, Torres J, Cervera J, Pulido R. Nuclear localization of PTEN by a Ran-dependent mechanism enhances apoptosis: Involvement of an N-terminal nuclear localization domain and multiple nuclear exclusion motifs. Mol Biol Cell. 2006;17:4002–13. doi: 10.1091/mbc.E06-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–70. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiom K. Dna repair: Rad52 - the means to an end. Curr Biol. 1999;9:R446–8. doi: 10.1016/S0960-9822(99)80278-4. [DOI] [PubMed] [Google Scholar]
- 24.Lok BH, Carley AC, Tchang B, Powell SN. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene. 2013;32:3552–8. doi: 10.1038/onc.2012.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altmannova V, Eckert-Boulet N, Arneric M, Kolesar P, Chaloupkova R, Damborsky J, Sung P, Zhao X, Lisby M, Krejci L. Rad52 SUMOylation affects the efficiency of the DNA repair. Nucleic Acids Res. 2010;38:4708–21. doi: 10.1093/nar/gkq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sacher M, Pfander B, Hoege C, Jentsch S. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat Cell Biol. 2006;8:1284–90. doi: 10.1038/ncb1488. [DOI] [PubMed] [Google Scholar]
- 27.Ming M, He YY. PTEN in DNA damage repair. Cancer Lett. 2012;319:125–9. doi: 10.1016/j.canlet.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho JC, Warr NJ, Shimizu H, Watts FZ. SUMO modification of Rad22, the Schizosaccharomyces pombe homologue of the recombination protein Rad52. Nucleic Acids Res. 2001;29:4179–86. doi: 10.1093/nar/29.20.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker SJ. PTEN enters the nuclear age. Cell. 2007;128:25–8. doi: 10.1016/j.cell.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 30.Carracedo A, Pandolfi PP. The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene. 2008;27:5527–41. doi: 10.1038/onc.2008.247. [DOI] [PubMed] [Google Scholar]
- 31.Bassi C, Ho J, Srikumar T, Dowling RJ, Gorrini C, Miller SJ, Mak TW, Neel BG, Raught B, Stambolic V. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science. 2013;341:395–9. doi: 10.1126/science.1236188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kegel A, Sjögren C. The Smc5/6 complex: more than repair? Cold Spring Harb Symp Quant Biol. 2010;75:179–87. doi: 10.1101/sqb.2010.75.047. [DOI] [PubMed] [Google Scholar]
- 33.Yong-Gonzales V, Hang LE, Castellucci F, Branzei D, Zhao X. The Smc5-Smc6 complex regulates recombination at centromeric regions and affects kinetochore protein sumoylation during normal growth. PLoS One. 2012;7:e51540. doi: 10.1371/journal.pone.0051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stephan AK, Kliszczak M, Morrison CG. The Nse2/Mms21 SUMO ligase of the Smc5/6 complex in the maintenance of genome stability. FEBS Lett. 2011;585:2907–13. doi: 10.1016/j.febslet.2011.04.067. [DOI] [PubMed] [Google Scholar]
- 35.Jaco I, Canela A, Vera E, Blasco MA. Centromere mitotic recombination in mammalian cells. J Cell Biol. 2008;181:885–92. doi: 10.1083/jcb.200803042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liebman SW, Symington LS, Petes TD. Mitotic recombination within the centromere of a yeast chromosome. Science. 1988;241:1074–7. doi: 10.1126/science.3137657. [DOI] [PubMed] [Google Scholar]
- 37.Fukasawa K. Centrosome amplification, chromosome instability and cancer development. Cancer Lett. 2005;230:6–19. doi: 10.1016/j.canlet.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 38.Stolz A, Ertych N, Kienitz A, Vogel C, Schneider V, Fritz B, Jacob R, Dittmar G, Weichert W, Petersen I, et al. The CHK2-BRCA1 tumour suppressor pathway ensures chromosomal stability in human somatic cells. Nat Cell Biol. 2010;12:492–9. doi: 10.1038/ncb2051. [DOI] [PubMed] [Google Scholar]
- 39.Xiong B, Li S, Ai JS, Yin S, Ouyang YC, Sun SC, Chen DY, Sun QY. BRCA1 is required for meiotic spindle assembly and spindle assembly checkpoint activation in mouse oocytes. Biol Reprod. 2008;79:718–26. doi: 10.1095/biolreprod.108.069641. [DOI] [PubMed] [Google Scholar]
- 40.Choi E, Park PG, Lee HO, Lee YK, Kang GH, Lee JW, Han W, Lee HC, Noh DY, Lekomtsev S, et al. BRCA2 fine-tunes the spindle assembly checkpoint through reinforcement of BubR1 acetylation. Dev Cell. 2012;22:295–308. doi: 10.1016/j.devcel.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 41.Meng X, Fan J, Shen Z. Roles of BCCIP in chromosome stability and cytokinesis. Oncogene. 2007;26:6253–60. doi: 10.1038/sj.onc.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu PD, Jousse C, Marciniak SJ, Zhang Y, Novoa I, Scheuner D, Kaufman RJ, Ron D, Harding HP. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 2004;23:169–79. doi: 10.1038/sj.emboj.7600030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, Yang Y, Dai W. Differential subcellular localizations of two human Sgo1 isoforms: implications in regulation of sister chromatid cohesion and microtubule dynamics. Cell Cycle. 2006;5:635–40. doi: 10.4161/cc.5.6.2547. [DOI] [PubMed] [Google Scholar]


