Summary
Stem cell niches in mammalian tissues are often heterogeneous and compartmentalized, however whether distinct niche locations determine different stem cell fates remains unclear. To test this hypothesis, we utilized the mouse hair follicle niche and devised a novel approach by combining intravital microscopy with genetic lineage tracing to re-visit the same stem cell lineages, from their exact place of origin, throughout regeneration in live mice. Using this method, we show directly that the position of a stem cell within the hair follicle niche can predict whether it is likely to remain uncommitted, generate precursors or commit to a differentiated fate. Furthermore, using laser ablation we demonstrate that hair follicle stem cells are dispensable for regeneration and that epithelial cells, which do not normally participate in hair growth, re-populate the lost stem cell compartment and sustain hair regeneration. This study provides a general paradigm for niche-induced fate determination in adult tissues.
Keywords: Stem Cells, Niche, Hair Follicle, Regeneration, Intravital Microscopy
In adult tissues, stem cell niches constitute a spatially distinct microenvironment, including neighboring cells, signals and extracellular material 1,2. Anatomical and molecular heterogeneity appears to be a common feature between mammalian stem cell niches across different tissues 3-6; however, it is unclear whether the specific location that a stem cell occupies within the niche can influence its function.
Hematopoietic stem cells with divergent roles in homeostasis and pathophysiology are proposed to associate with distinct niche compartments, such as the endosteum or the vasculature in the central bone marrow; which may affect their behavior and possibly long-term fate 7-13. Moreover in the intestinal crypt, fast-cycling stem cell and quiescent progenitor populations reside in distinct positions at the bottom of the crypt, while the transient amplified pool of precursors line the walls of the crypt, progressively differentiating as they reach the surface of the villi 14-17. A neutral competition model has been proposed for long-term homeostasis of the intestinal niche 18,19; however, the short-term behavior of individual stem cells in different positions at the bottom of the crypt has not been determined. The hair follicle in the skin represents another highly compartmentalized niche where stem cells reside in the bulge while a pool of progenitors, called the hair germ, is clustered in a different niche location directly below 20-24. This common theme of niche compartmentalization in the above examples raises the question whether stem cells within their compartments are functionally equivalent. Specifically, it is not clear if each stem cell can stochastically generate every lineage in a tissue, or whether the precise position within the niche can impose a distinct fate.
The murine hair follicle is a self-contained mini-organ that represents a unique system for monitoring niche behavior in vivo, since the location of stem cells and differentiated cell types is anatomically distinct and molecularly well defined 20-22,25-31 (Extended Data Fig. 1a, b). Hair follicles normally undergo stereotypic cycles of regeneration, which in the young mouse are highly synchronized across large areas of the skin and therefore the exact timing of rest, growth and regression phases can be accurately predicted 32 (Extended Data Fig. 1c). During hair growth, mesenchymal-epithelial crosstalk at the bottom of the hair follicle niche induces the formation and upward expansion of seven concentric differentiated layers. These inner layers make up the hair shaft and supportive Inner Root Sheath (IRS), while a relatively undifferentiated outer cell layer called Outer Root Sheath (ORS) grows downward to fully envelop the elongating hair follicle 29,31,33-37 (Extended Data Fig. 1b). Taking advantage of the accessibility of the skin hair follicle we previously established the ability to visualize these processes non-invasively, in vivo 24. Here, we have developed a novel approach to mark single stem cells in different positions within the niche and re-visit the same lineages over the period of several weeks to months, in live mice. Furthermore, we use laser-induced cell ablation to test whether hair follicle stem cells are required for hair regeneration and to address how injury-induced cell mobility between different niches affects their fate.
Niche location predicts stem cell fate
To explore the significance of specific niche positioning to stem cell fate we implemented an in vivo lineage tracing approach, at single-cell resolution by live imaging (Extended Data Fig. 2). To genetically mark hair follicle stem cells in the bulge and hair germ compartments we used mice harboring either K19-CreER or Lgr5-CreER, in addition to Rosa-stop-tdTomato reporter alleles 16,38-41 (Extended Data Fig. 2a). K14H2BGFP and Lef1RFP were used as general epithelial and mesenchymal fluorescent reporters as described previously 24 (Extended Data Fig. 1a, 3a). Mice were induced in the first rest phase of the hair cycle (1st Telogen, ∼ P20) and stem cells were visualized in vivo three days later (∼ P23), while the hair follicles were still quiescent (Extended Data Fig. 3a). We verified that marked cells did not translocate from their initial location within the niche and that no additional ectopic expression of the Cre reporter occurred due to Cre recombinase leakage while hair follicles remained quiescent (Extended Data Fig. 3b). As hair regeneration commenced, we re-visited the same follicles in separate imaging sessions and the lineage progression of previously identified single stem cells was documented (Fig. 1).
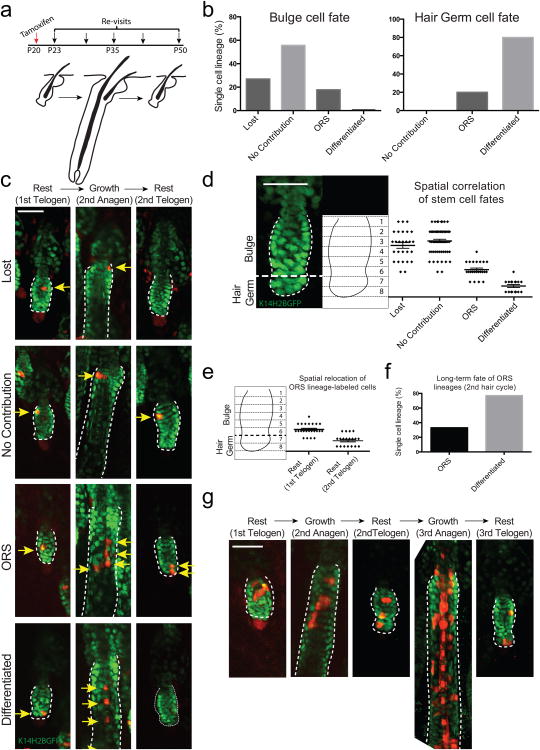
Figure 1. Niche location can predict the fate of hair follicle stem cells.
a) Scheme of single stem cell lineage tracing in live mice. b) Statistical analysis of the fate of stem cells originating from the bulge or hair germ (n = 108 and 20 lineages, respectively in 8 mice). c) Representative examples of single stem cell lineages (arrows) traced during a full hair cycle. Each sequence represents a different fate that correlates with a specific niche location. d) Graphical correlation between the original location of a single stem cell and its fate after a full hair cycle (n = 128; error bars represent S.E.M.). e) Spatial relocation of ORS lineages after a full hair cycle (n = 23). f) Quantification of the fate of ORS lineages in the 2nd hair cycle (n = 9). g) In vivo lineage tracing of bulge stem cells over two consecutive hair cycles. Scale bars: 50μm.
Analysis of the in vivo lineage tracing data showed that during this process the fate of individual stem cells followed highly stereotypic patterns, which correlated with their original location within the niche at the onset of a regeneration cycle (Fig. 1b - d). Specifically, the majority of the stem cells located within the bulge did not contribute to the subsequent hair cycle or were lost while a smaller fraction of bulge stem cells produced lineages only in the relatively undifferentiated outer layer (ORS) (Fig. 1b, c; Extended Data Fig. 4, 5; Video S1). Conversely, cells located in the hair germ consistently contributed to hair follicle growth by generating differentiated lineages (Fig. 1b, c; Extended Data Fig. 4, 6; Video S1). Even within each niche compartment the precise location dictated different stem cell behaviors. For example, within the bulge, stem cells situated in the lower half of the compartment were more likely to proliferate and generate ORS lineages compared to stem cells situated in the upper half, which were either quiescent or generated limited clones that remained in the bulge (Fig. 1d). These data show a direct correlation between a specific niche location and stem cell fate.
To test the long-term fate of hair follicle stem cells we traced bulge lineages over two consecutive hair cycles. Bulge stem cells that persisted in the upper portion of the bulge compartment after the first cycle remained there during the second cycle (Extended Data Fig. 7). However, lower bulge descendants that acquired an ORS fate were often found in the hair germ after hair follicle regression and entry into the next rest phase (2nd Telogen; Fig. 1e; Extended Data Fig. 8). These ORS clones more frequently gave rise to differentiated lineages in the second hair cycle, consistent with their new position in the hair germ (Fig. 1f). Thus, bulge stem cells can contribute to hair growth by following a step-wise transition to differentiation through an intermediate ORS fate (Fig. 1g). These data also enforce the notion that fate is established at the onset of a new regeneration cycle depending on the specific location that a stem cell occupies in the niche.
ORS expansion is spatially regulated
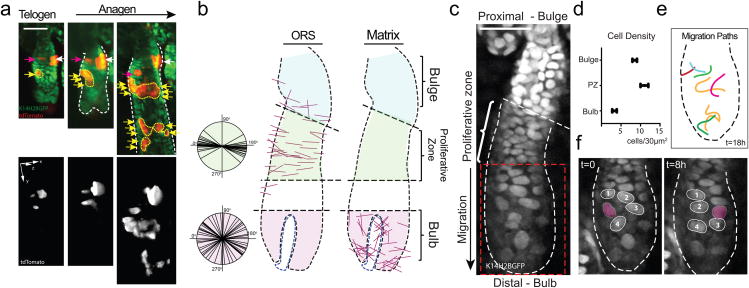
Our data suggest that the ORS represents an intermediate stage between quiescent bulge stem cells and hair germ cells. Interestingly, lineage tracing indicated that ORS clones often expanded discontinuously toward the bulb (Fig. 2a; Extended Data Fig. 8; Video S2). To understand how the niche influences this mode of ORS expansion we collected a number of time-lapse recordings of hair follicles in advanced growth stages (Anagen III - IV; Video S3 and S4). Analysis of cell behavior at this stage of growth revealed that ORS undergoes a spatially regulated mode of expansion. Specifically, cell proliferation was restricted to a narrow zone between the lower bulge and the bulb (Fig. 2b; Video S3 and S4). Cell divisions in this “proliferative zone” were highly oriented, with a mitotic spindle perpendicular to the long axis of growth (Fig. 2b). These oriented cell divisions may not contribute directly to the longitudinal expansion of the ORS and as a result this “proliferative zone” displayed higher cell density compared to other areas (Fig. 2c, d; Video S3 and S4). Further analysis of the time-lapse videos revealed that cells at the distal border of the “proliferative zone”, became mobile and migrated rapidly towards the bulb, thus directly contributing to the downward expansion of the ORS. This previously uncharacterized mode of cell migration indicates highly dynamic cell-cell contacts and may partially explain the discontinuous appearance of the ORS clones observed by lineage tracing (Fig. 2e, f; Video S5). This bimodal type of ORS growth and the spatially defined areas of proliferation and migration highlight the regional control that the niche exerts during growth.
Figure 2. Mode of ORS growth.
a) In vivo lineage tracing sequence (top) and corresponding 3D renderings of the Cre reporter (bottom) showing ORS expansion during hair growth. Arrows denote different lineages. b) Graphical representation of the location and axis of cell divisions in the ORS and Matrix (Bulb) in advanced hair follicle growth (Anagen III-IV). c) ORS cell distribution during active hair growth. d) Quantification of cell density in different regions of the outer hair follicle epithelial layer (n = 9 per region; error bars represent S.E.M.). e) Individual traces of migrating ORS nuclei (see also Video S1). f) Traces of ORS nuclei depicting their relative positions at two time-points eight hours apart (see also Video S3). Scale bars: 50μm.
Bulge stem cells are dispensable
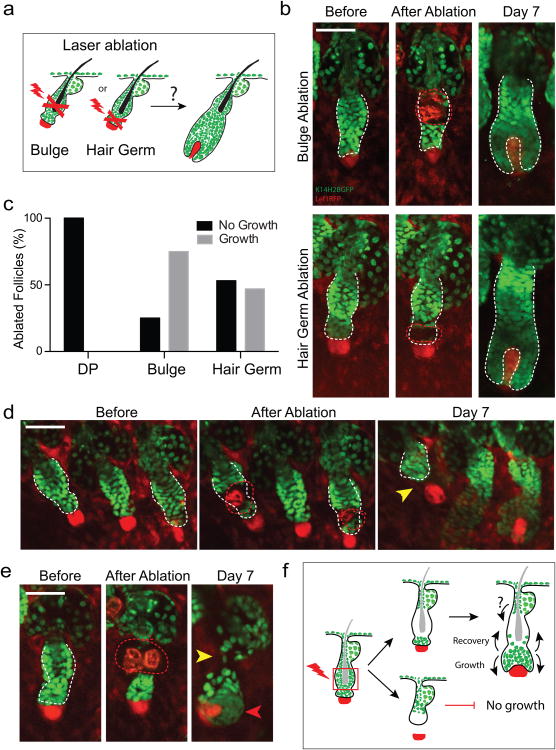
Our lineage tracing experiments suggest a functional compartmentalization of the niche, where stem cells positioned in the lower bulge may specify the ORS and those in the hair germ the differentiated hair lineages. To test the stringency of niche-imposed fates towards hair regeneration we employed laser-induced cell ablation to specifically remove either the bulge or the hair germ at the onset of hair growth (1st Telogen, ∼ P20; Fig. 3a). To recognize each targeted compartment we used reliable anatomical features of the niche 24 (Extended Data Fig. 1a), since available genetic markers label overlapping populations that extend across both the bulge and hair germ 28,40,41. Remarkably, following ablation of either the bulge or hair germ, the niche consistently recovered the lost cell population, regained its anatomical features and proceeded with hair regeneration (Fig. 3b, c). To verify the efficiency of the ablation process we re-visited the ablated follicles shortly after ablation. Instances where bulge or hair germ ablation impaired hair regeneration were the result of extensive damage that affected the entire niche and/or the mesenchyme (DP), consistent with previous reports 24,29,42 (Fig. 3c). However, some such examples provided critical information on the dynamics between the epithelium and the mesenchyme. For instance, when the epithelium and the mesenchyme were physically separated as the result of laser ablation, both the full recovery of the niche and hair regeneration was impaired, even though the mesenchymal DP lingered a few micrometers below (Fig. 3d). Conversely, in some follicles where the bulge was ablated the hair germ was able to initiate growth, encompassing the DP before the full recovery of the bulge (Fig. 3e). Overall, our data show that the bulge and hair germ populations are mutually dispensable for hair regeneration as long as a functional interaction between the epithelium and the mesenchymal dermal papilla is maintained (Fig. 3f). Furthermore, they suggest that the ability of the hair germ to initiate hair growth may occur independently of bulge input.
Figure 3. Bulge and hair germ are mutually dispensable for hair regeneration.
a) Scheme of the laser ablation experiment. b) Sequential snapshots of hair follicles immediately or a week after bulge and hair germ ablation. The red dotted line represents the ablated region. c) Quantification of the regenerative capacity of follicles with ablated dermal papilla, bulge and hair germ, respectively (n = 30, 32, 28 follicles, respectively in 9 mice). d) Example of hair growth impairment due to the physical separation of the epithelium from the mesenchymal dermal papilla after ablation. e) Example of hair follicle growth initiation before full niche recovery. f) Scheme of hair follicle niche responses after laser ablation. Scale bars: 50μm.
Cell fate changes upon niche injury
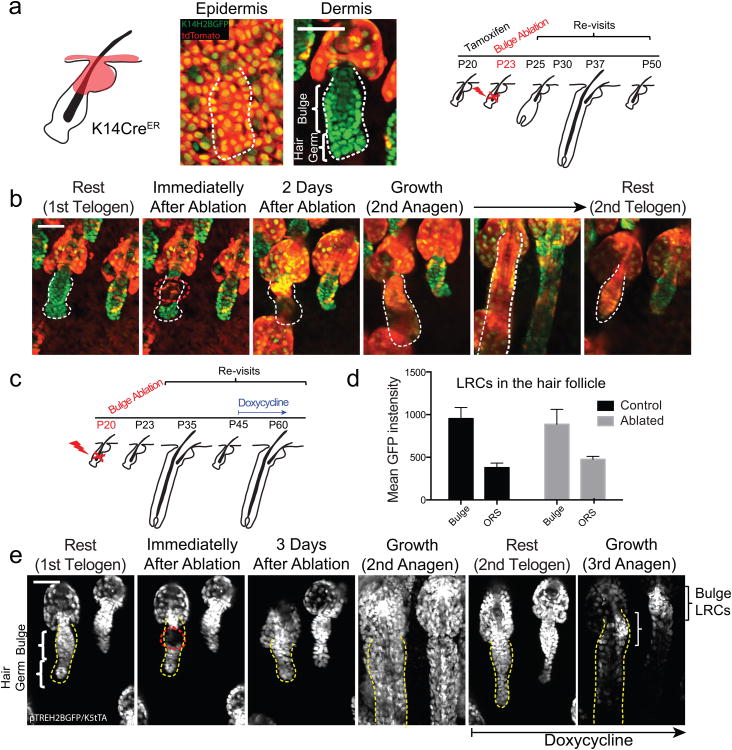
To explore the cellular mechanisms of niche recovery, we performed time-lapse recordings shortly after bulge laser ablation. Hair germ became proliferative consistent with previous experiments that show hair germ contribution to the niche upon stem cell depletion due to plucking 23. Surprisingly, distant epithelial cells, above the bulge (Infundibulum) were also observed to become proliferative and some cells descended rapidly into the niche (Video S6 and S7). These findings raised the possibility that neighboring epithelial cells situated above the bulge may contribute to the recovery of the niche. To test this hypothesis, we implemented our in vivo lineage tracing approach to monitor the behavior of cells outside the hair follicle niche after bulge ablation. To exclusively mark the outermost epithelial layers located above the bulge we took advantage of the particular expression profile of K14-CreER/Rosa-stop-tdTomato mice, in which labeling is strongly biased towards the interfollicular epidermis (IFE), infundibulum and sebaceous glands (Fig. 4a; Video S8). After induction follicles that did not contain any labeled cells within the niche were targeted for bulge ablation (Fig. 4a, b). In the days following the ablation there was a significant influx of labeled epithelial cells into the niche, in contrast to neighboring non-ablated follicles where no additional tdTomato+ cells appeared to enter the hair follicle (Fig. 4b).
Figure 4. Functional reconstitution of the stem cell niche from non-hair epithelial populations.
a) Scheme of in vivo lineage tracing of non-hair epithelial populations following bulge laser ablation. b) Example of bulge-ablated hair follicle showing the influx of labeled non-hair epithelial cell populations into the niche and their contribution to hair growth. c) Scheme of label retention experiment following bulge ablation. d) Quantification of label retention in control and bulge-ablated hair follicles (n = 15 and 12 follicles; error bars represent S.E.M.). e) Example of label retention in the niche following bulge ablation and recovery. Scale bars: 50μm.
Strikingly, we found that these “new” niche cells not only contributed to re-establishing the lost bulge compartment but also participated in the subsequent hair growth, suggesting that they acquired a different fate upon assuming their new position in the hair follicle niche (Fig. 4b). To further test if the niche influences the same type of behavior on the epithelial cells that re-populated the bulge we used label retention to analyze for quiescence, a hallmark of bulge stem cells20,21, during the second growth cycle following bulge ablation (Fig. 4c). At full growth (3rd Anagen) the bulge of ablated hair follicles displayed significant label retention compared to the lower growing portion of the follicle, but similar to the bulge of non-ablated neighboring follicles (Fig. 4d, e). Thus, these data provide direct evidence that loss of a stem cell pool due to injury can induce neighboring epithelial cell populations that do not normally have a hair follicle fate to be mobilized and contribute to re-establishing the niche anatomically as well as functionally. Most importantly, once these cells enter the niche they display characteristics consistent with a hair follicle fate enacted on them in their new location.
Discussion
The relationship between niche position and stem cell fate is a fundamental question in mammalian stem cell biology that has remained unanswered. Current approaches to address this problem involve the use of genetic lineage tracing tools based on inducible Cre recombinase, driven by stem cell-specific promoters 43. However, the mosaic expression of the Cre reporter within the stem cell pool and the inability to follow individual stem cells over time has greatly limited our understanding of the fate of individual cells at precise locations within the niche. To overcome these limitations we have devised a novel system that combines genetic lineage tracing with intravital microscopy to monitor the progression of single stem cell lineages from their initial position, by re-visiting the same undisturbed niche in separate experiments in live mice.
Using this approach we found evidence that establish a strong link between a specific niche location and stem cell fate. While a cell-autonomous model is plausible, our data support a model for fate determination in the hair follicle that is based on the spatial organization of the niche (Extended Data Fig. 9). According to this model, a cell in the upper half of the bulge is favored to remain uncommitted to a specific fate and therefore more likely to remain quiescent or self-renew. In contrast, a cell situated in the lower bulge will be subject to activating stimuli from the niche driving it to undergo limited amplification as part of the still relatively undifferentiated ORS. The fraction of the ORS pool that survives the regression phase of the hair cycle will now be situated in the compartment that becomes the new hair germ. Once in that part of the niche these cells receive different stimuli pushing them to commit towards a differentiation pathway in order to support the subsequent hair cycle.
Our model is consistent with previous data from the hair follicle and other stem cell niches 22,31,35,36,44,45 but directly demonstrates the significance of the niche for stem cell fate determination. Our results from the laser ablation experiments further support this notion, highlighting the fact that niche stem cells can be dispensable for tissue regeneration; provided that the overall integrity of the niche is maintained. In this context, injury can induce cell mobility between different tissue compartments but the overall structure and function of the tissue is maintained because cells are capable of adopting new fates as dictated by their new niche microenvironment (Extended Data Fig. 9). This may also explain how certain hierarchies that exist between different stem cell pools under homeostatic conditions can be re-shuffled and new ones established following injury, as part of a wound healing process 46. Identifying the extrinsic factors that make up a particular niche microenvironment is paramount for understanding the mechanism of stem cell fate determination and our ability to manipulate stem cells for therapeutic purposes.
Methods Summary
K19-CreER mice were created and obtained from Guoqiang Gu's Laboratory38. Lgr5-CreER were created by Hans Clevers's Laboratory16 and obtained from JAX.K14-CreER mice were created by Elaine Fuchs's Laboratory47 and obtained from JAX. Rosa-stop-tdTomato mice were created by Hongkui Zeng's Laboratory39 and obtained from JAX. All studies and procedures involving animal subjects were approved by the Institutional Animal Care and Use Committee at Yale School of Medicine and conducted in accordance with the approved animal handling protocol. Expression of the Cre fluorescent reporter for the lineage tracing experiments was induced with a single intraperitoneal injection of Tamoxifen (20μg/g and 1μg/g in corn oil for K19-CreER and Lgr5-CreER, respectively) at ∼ P20 or times specified. For lineage tracing of epithelial populations above the hair follicle, K14-CreER/Rosa-stop-tdTomato mice were given a single intraperitoneal injection of Tamoxifen (0.2mg/g in corn oil). For the label retention experiment, K5tTA/pTREH2BGFP mice were given Doxycycline (1mg/ml) in potable water at times specified. Intravital microscopy and laser ablation procedures were carried out as described previously24. All lineage tracing and ablation experiments were repeated at least in triplicates or otherwise indicated.
Methods
Mice
K19-CreER38 mice were provided by G. Gu.K14H2BGFP 21, Lef1RFP 48 and pTREH2BGFP 21 mice were obtained from E. Fuchs. K5tTA 49 mice were provided by A. Glick. Lgr5-CreER 16, K14-CreER 47 and Rosa-stop-tdTomato 39 mice were obtained from Jackson Laboratories. All studies and procedures involving animal subjects were approved by the Institutional Animal Care and Use Committee at Yale School of Medicine and conducted in accordance with the approved animal handling protocol.
In vivo imaging
Three-week old mice were anesthetized with intraperitoneal injection of ketamine/xylazine (15mg/ml and 1mg/ml, respectively in PBS). Anesthesia was maintained throughout the course of the experiment with vaporized isofluorane delivered by a nose cone (1% in air). The area of the skin around the head region shaved using an electrical shaver and depilatory cream (Nair). The skin connecting the ear with the head was then mounted on a custom-made stage and a glass coverslip was placed directly against it. Image stacks were acquired with a LaVision TriM Scope II (LaVision Biotec, Germany) microscope equipped with a Chameleon Vision II (Coherent, USA) 2-Photon laser. For collection of serial optical sections a laser beam (940nm for GFP and 1040nm for RFP and tdTomato, respectively) was focused through a 20× water immersion lens (N.A. 1.0; Olympus ,USA) and scanned with a field of view of 0.5mm2 at 600Hz. Z-stacks were acquired in 2-3μm steps to image a total depth of 100μm within the tissue. Laser power intensity was increased accordingly for each wavelength to normalize exposure and counteract the loss of signal in higher tissue depths.
In vivo lineage tracing
In order to genetically mark hair follicle stem cells we utilized an inducible Cre-mediated system, using mice harboring either K19-CreER/Rosa-stop-tdTomato or Lgr5-CreER /Rosa-stop-tdTomato alleles. Keratin 19 and Lgr5 are expressed in distinct but overlapping cell populations located in the bulge and hair germ, respectively 40,41. To mark epithelial cells above the bulge we used the K14-CreER which displays a biased expression toward the interfollicular epidermis (IFE), infundibulum and sebaceous glands. K14H2BGFP and Lef1RFP were used as general epithelial and mesenchymal fluorescent reporters as described previously. For induction mice were given a single intraperitoneal injection of Tamoxifen (20μg/g, 1μg/g, and 0.2mg/g in corn oil for K19-CreER, Lgr5-CreER and K14-CreER, respectively) at ∼ P20 or times specified. Three days later (∼ P23), expression of the Cre reporter was assessed by in vivo imaging. Serial optical sections with a volume that included the epidermis and the entire volume of hair follicles were acquired, in a pattern of successive and overlapping XY fields-of-view that spanned a skin area of ∼2mm2. In each of these serial sections individual hair follicles were evaluated and single labeled stem cells were identified. For re-visiting the same hair follicles in separate experiments, distinctive inherent landmarks of the skin were used to navigate back to the original region; including the vasculature and clustering of hair follicles in uniquely arranged groups. For time-lapse recordings serial optical sections were obtained at 5-minute intervals.
Label retention
Mice hemizygous for the pTREH2BGFP and K5tTTA alleles were used. This combination confers expression of H2BGFP in all the Keratin 5 positive epithelial populations, which can be repressed with the administration of Doxycycline. Mice were given Doxycycline (1mg/ml) in potable water at times specified.
Laser ablation
Laser ablation was carried out with the same optics as for acquisition. A 900nm laser beam was used to scan the target area (10-50μm2) and ablation was achieved using 20-40 % laser power for ∼ 1-5 sec. Ablation parameters were adjusted according to the depth of the target (30-80μm).
Image Analysis
Raw image stacks were imported into ImageJ (NIH, USA) for further analysis. For visualizing single-labeled stem cells that expressing the tdTomato Cre reporter the brightness and contrast was adjusted accordingly for the green (GFP) and red (RFP/tdTomato) channels and composite serial image sequences were assembled. In the red channel the signal from the tdTomato Cre reporter was significantly higher and could easily be distinguished from the Lef1RFP. To account for every tdTomato labeled cell in the hair follicle maximum intensity z-projections of sequential optical sections that included the entire volume of the hair follicle were obtained. For quantifying H2BGFP signal intensities for the label retention experiment 16-bit serial optical sections were acquired for the green channel only. Pixel intensity levels were equalized and z-projections of the entire volume of the hair follicles were obtained. Mean grey values were measured in 20μm2 areas within the bulge and ORS regions of the follicles, respectively using ImageJ.
Statistical Analysis
Data are expressed as percentages or mean ± SEM. Statistical calculations and graphical representation of the data were performed using the Prism software package (GraphPad,USA).
Supplementary Material
Extended Data Figure 1 - Hair follicle anatomy and physiology. a) Scheme of a murine hair follicle in quiescence. Different cell populations reside in defined anatomical compartments. b-c) In homeostasis the hair follicle undergoes repeated cycles of regeneration. b) Hair growth is fueled by stem cells in the niche that proliferate and differentiate to form the seven concentric layers of the mature hair shaft and Inner Root Sheath (IRS), while a basal epithelial layer called Outer Root Sheath (ORS) surrounds the entire structure. Notice that the seven inner layers expand from the matrix, at the interphase with the mesenchymal Dermal Papilla where they are generated toward the surface of the skin, while the ORS has a different mode of growth and expands in the opposite direction. c) A complete hair cycle alternates between phases of rest (Telogen), growth (Anagen) and regression (Catagen).
Extended Data Figure 2 - Method for single stem cell lineage tracing in live mice. a) Single hair follicle stem cell labeling is achieved using a combination of either K19-CreER/Rosa-stop-tdTomato or Lgr5-CreER /Rosa-stop-tdTomato alleles and administration of a single low dose of Tamoxifen to achieve low frequency of Cre-mediated loxP recombination and mosaic expression of the fluorescent tdTomato reporter within the stem cell niche. b) The lineage of single labeled stem cells is traced in vivo during hair growth. c) Using two-photon laser scanning microscopy we can re-visit the same hair follicles, non-invasively in live mice at different stages of hair regeneration. Each panel depicts low (top) and high (bottom) magnification images of live hair follicles captured in 1st Telogen, 2nd Anagen and 2nd Telogen, respectively.
Extended Data Figure 3 - Fluorescent proteins and kinetics of the inducible tdTomato Cre reporter. a) Panels depicting the green (left) and red (middle) channel as well as a composite image (right) of a group of follicles in rest phase (Telogen). K14H2BGFP marks all the epithelial cells in the skin including the hair follicles. Lef1RFP marks mesenchymal cells in the dermis including the Dermal Papilla at the bottom of the hair follicles. The tdTomato Cre reporter (K19- or Lgr5- driven) displays mosaic expression in the stem cell niche after administering a low dose of Tamoxifen. Notice that the fluorescent intensity of the tdTomato is several-fold higher and easily distinguishable from the RFP in the red channel. b) Individual channels and composite images of a group of follicles three (P23) and five days (P25) after administering a low dose of Tamoxifen showing a non-leaky expression of the Cre reporter (tdTomato) and a quiescent niche between these time-points. Scale bar: 100μm.
Extended Data Figure 4 – Classification of hair follicle cell types in vivo. a) Panel showing single optical sections (top) or 3D volume renderings (bottom) of the outer (ORS; left) or inner hair follicle cell layers (right) as seen in vivo using a K14H2BGFP fluorescent reporter. b) Single optical sections showing cells marked with the tdTomato Cre reporter (in addition to K14H2BGFP) in the outer (ORS; left) and inner hair follicle layers (right). Notice the differences in morphology between cells located in different layers within the hair follicle.
Extended Data Figure 5 - Relocation of bulge stem cells and progeny over a hair cycle. Examples of in vivo lineage tracing of bulge cells in rest and growth phases of a full hair cycle. Arrows point to the location of the original cell and that of its progeny occupying different positions in the niche after a full hair cycle. Scale bar: 50μm.
Extended Data Figure 6 - A single hair germ cell generates a spatially restricted differentiated lineage. In vivo lineage tracing of a single cell located in the hair germ in rest and growth phases over a full hair cycle. In advanced hair growth (Anagen) an Inner Root Sheath differentiated lineage can be visualized and it is restricted to one side of the follicle as the original founder cell. Scale bar: 50μm.
Extended Data Figure 7 - Long-term fate of bulge stem cells. Examples of in vivo lineage tracing of a single bulge cell in rest and growth phases over two consecutive hair cycles. Arrows point to the location of the stem cell that remains uncommitted in the niche during both hair cycles. Scale bar: 50μm.
Extended Data Figure 8 - Origin of the hair germ. Examples of in vivo lineage tracing of a single bulge cell lineage in rest and growth phases over two consecutive hair cycles. A bulge cell positioned in the lower bulge undergoes limited and more extended amplification in the ORS over two consecutive hair cycles. After regression of the follicle at the end of the second hair cycle some surviving ORS clones form part of the hair germ before the third hair cycle commences. Arrows depict the clonal expansion and contraction of the bulge stem cell lineage during the two hair cycles. Scale bar: 50μm.
Extended Data Figure 9 – Model for determining stem cell fate based on the spatial organization of the hair follicle niche in homeostasis and after injury. In homeostasis a stem cell located in the upper bulge does not commit to a specific fate and is likely to remain quiescent, self-renew or become lost. In contrast, a cell positioned in the lower bulge is more likely to become activated and undergo limited amplification as part of the Outer Root Sheath (ORS), while still remaining relatively undifferentiated. ORS cells that survive the regression phase in one hair cycle can be situated in the niche compartment that becomes the new hair germ. Once positioned within the hair germ a cell will commit towards a differentiation pathway generating the cell types necessary to support hair growth. Loss of a stem cell pool due to injury can induce an epithelial cell to enter the niche and contribute to its recovery. However, once a cell enters the niche it is subject to the same inputs as previous resident cells and as a result it will acquire a hair fate and actively contribute to hair regeneration.
Acknowledgments
We are grateful to Stephen King, Shanquin Guo and Arthur Horwich for critical feedback on the manuscript. We thank Elaine Fuchs for the K14H2BGFP; Lef1RFP, pTREH2BGFP mice, Adam Glick for the K5tTA mice and Guoqiang Gu for the K19CreER mice. We thank David Gonzalez and Ann Haberman for technical support with intravital microscopy. P.R. is a New York Stem Cell Foundation - Druckenmiller Fellow. This work was supported by grants to V.G. from the American Cancer Society (RGS-12-059-01-DCC) and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (1RO1AR063663-01).
Footnotes
Reprints and permissions information is available in www.nature.com/reprints. Readers are welcome to comment on the online version of the paper.
Author Contributions: P.R. and V.G. designed experiments and wrote the manuscript; P.R. performed the experiments and analyzed the data. K.M. assisted with the revisions.
Author Information: The authors declare no competing financial interests.
References
- 1.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 2.Spradling AC, et al. Stem cells and their niches: integrated units that maintain Drosophila tissues. Cold Spring Harb Symp Quant Biol. 2008;73:49–57. doi: 10.1101/sqb.2008.73.023. [DOI] [PubMed] [Google Scholar]
- 3.Fuchs E. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell. 2009;137:811–819. doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greco V, Guo S. Compartmentalized organization: a common and required feature of stem cell niches? Development. 2010;137:1586–1594. doi: 10.1242/dev.041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Copley MR, Beer PA, Eaves CJ. Hematopoietic stem cell heterogeneity takes center stage. Cell Stem Cell. 2012;10:690–697. doi: 10.1016/j.stem.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Wilson A, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 8.Sato T, et al. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat Med. 2009;15:696–700. doi: 10.1038/nm.1973. [DOI] [PubMed] [Google Scholar]
- 9.Kopp HG, Avecilla ST, Hooper AT, Rafii S. The bone marrow vascular niche: home of HSC differentiation and mobilization. Physiology (Bethesda) 2005;20:349–356. doi: 10.1152/physiol.00025.2005. [DOI] [PubMed] [Google Scholar]
- 10.Lo Celso C, et al. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature. 2009;457:92–96. doi: 10.1038/nature07434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie Y, et al. Detection of functional haematopoietic stem cell niche using real-time imaging. Nature. 2009;457:97–101. doi: 10.1038/nature07639. [DOI] [PubMed] [Google Scholar]
- 12.Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495:231–235. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenbaum A, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeda N, et al. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 17.Buczacki SJA, et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 18.Lopez-Garcia C, Klein AM, Simons BD, Winton DJ. Intestinal stem cell replacement follows a pattern of neutral drift. Science. 2010;330:822–825. doi: 10.1126/science.1196236. [DOI] [PubMed] [Google Scholar]
- 19.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 20.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 21.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greco V, et al. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito M, Kizawa K, Hamada K, Cotsarelis G. Hair follicle stem cells in the lower bulge form the secondary germ, a biochemically distinct but functionally equivalent progenitor cell population, at the termination of catagen. Differentiation. 2004;72:548–557. doi: 10.1111/j.1432-0436.2004.07209008.x. [DOI] [PubMed] [Google Scholar]
- 24.Rompolas P, et al. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature. 2012;487:496–499. doi: 10.1038/nature11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trempus C, et al. Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. J Invest Dermatol. 2003;120:501–511. doi: 10.1046/j.1523-1747.2003.12088.x. [DOI] [PubMed] [Google Scholar]
- 26.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Claudinot S, Nicolas M, Oshima H, Rochat A, Barrandon Y. Long-term renewal of hair follicles from clonogenic multipotent stem cells. Proc Natl Acad Sci U S A. 2005;102:14677–14682. doi: 10.1073/pnas.0507250102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Lyle S, Yang Z, Cotsarelis G. Keratin 15 promoter targets putative epithelial stem cells in the hair follicle bulge. J Invest Dermatol. 2003;121:963–968. doi: 10.1046/j.1523-1747.2003.12600.x. [DOI] [PubMed] [Google Scholar]
- 29.Ito M, et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- 30.Morris RJ, et al. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 31.Zhang YV, Cheong J, Ciapurin N, McDermitt DJ, Tumbar T. Distinct self-renewal and differentiation phases in the niche of infrequently dividing hair follicle stem cells. Cell Stem Cell. 2009;5:267–278. doi: 10.1016/j.stem.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller-Röver S, et al. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol. 2001;117:3–15. doi: 10.1046/j.0022-202x.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- 33.Jahoda CA, Horne KA, Oliver RF. Induction of hair growth by implantation of cultured dermal papilla cells. Nature. 1984;311:560–562. doi: 10.1038/311560a0. [DOI] [PubMed] [Google Scholar]
- 34.Kaufman CK, et al. GATA-3: an unexpected regulator of cell lineage determination in skin. Genes Dev. 2003;17:2108–2122. doi: 10.1101/gad.1115203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Legué E, Nicolas JF. Hair follicle renewal: organization of stem cells in the matrix and the role of stereotyped lineages and behaviors. Development. 2005;132:4143–4154. doi: 10.1242/dev.01975. [DOI] [PubMed] [Google Scholar]
- 36.Hsu YC, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sequeira I, Nicolas JF. Redefining the structure of the hair follicle by 3D clonal analysis. Development. 2012;139:3741–3751. doi: 10.1242/dev.081091. [DOI] [PubMed] [Google Scholar]
- 38.Means AL, Xu Y, Zhao A, Ray KC, Gu G. A CK19(CreERT) knockin mouse line allows for conditional DNA recombination in epithelial cells in multiple endodermal organs. Genesis. 2008;46:318–323. doi: 10.1002/dvg.20397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Youssef KK, et al. Identification of the cell lineage at the origin of basal cell carcinoma. Nat Cell Biol. 2010;12:299–305. doi: 10.1038/ncb2031. [DOI] [PubMed] [Google Scholar]
- 41.Jaks V, et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 42.Chi W, Wu E, Morgan BA. Dermal papilla cell number specifies hair size, shape and cycling and its reduction causes follicular decline. Development. 2013;140:1676–1683. doi: 10.1242/dev.090662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Keymeulen A, Blanpain C. Tracing epithelial stem cells during development, homeostasis, and repair. J Cell Biol. 2012;197:575–584. doi: 10.1083/jcb.201201041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barroca V, et al. Mouse differentiating spermatogonia can generate germinal stem cells in vivo. Nat Cell Biol. 2009;11:190–196. doi: 10.1038/ncb1826. [DOI] [PubMed] [Google Scholar]
- 45.Nystul T, Spradling A. An epithelial niche in the Drosophila ovary undergoes long-range stem cell replacement. Cell Stem Cell. 2007;1:277–285. doi: 10.1016/j.stem.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 46.Plikus MV, et al. Epithelial stem cells and implications for wound repair. Semin Cell Dev Biol. 2012;23:946–953. doi: 10.1016/j.semcdb.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci U S A. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rendl M, Lewis L, Fuchs E. Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol. 2005;3:e331. doi: 10.1371/journal.pbio.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diamond I, Owolabi T, Marco M, Lam C, Glick A. Conditional gene expression in the epidermis of transgenic mice using the tetracycline-regulated transactivators tTA and rTA linked to the keratin 5 promoter. J Invest Dermatol. 2000;115:788–794. doi: 10.1046/j.1523-1747.2000.00144.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended Data Figure 1 - Hair follicle anatomy and physiology. a) Scheme of a murine hair follicle in quiescence. Different cell populations reside in defined anatomical compartments. b-c) In homeostasis the hair follicle undergoes repeated cycles of regeneration. b) Hair growth is fueled by stem cells in the niche that proliferate and differentiate to form the seven concentric layers of the mature hair shaft and Inner Root Sheath (IRS), while a basal epithelial layer called Outer Root Sheath (ORS) surrounds the entire structure. Notice that the seven inner layers expand from the matrix, at the interphase with the mesenchymal Dermal Papilla where they are generated toward the surface of the skin, while the ORS has a different mode of growth and expands in the opposite direction. c) A complete hair cycle alternates between phases of rest (Telogen), growth (Anagen) and regression (Catagen).
Extended Data Figure 2 - Method for single stem cell lineage tracing in live mice. a) Single hair follicle stem cell labeling is achieved using a combination of either K19-CreER/Rosa-stop-tdTomato or Lgr5-CreER /Rosa-stop-tdTomato alleles and administration of a single low dose of Tamoxifen to achieve low frequency of Cre-mediated loxP recombination and mosaic expression of the fluorescent tdTomato reporter within the stem cell niche. b) The lineage of single labeled stem cells is traced in vivo during hair growth. c) Using two-photon laser scanning microscopy we can re-visit the same hair follicles, non-invasively in live mice at different stages of hair regeneration. Each panel depicts low (top) and high (bottom) magnification images of live hair follicles captured in 1st Telogen, 2nd Anagen and 2nd Telogen, respectively.
Extended Data Figure 3 - Fluorescent proteins and kinetics of the inducible tdTomato Cre reporter. a) Panels depicting the green (left) and red (middle) channel as well as a composite image (right) of a group of follicles in rest phase (Telogen). K14H2BGFP marks all the epithelial cells in the skin including the hair follicles. Lef1RFP marks mesenchymal cells in the dermis including the Dermal Papilla at the bottom of the hair follicles. The tdTomato Cre reporter (K19- or Lgr5- driven) displays mosaic expression in the stem cell niche after administering a low dose of Tamoxifen. Notice that the fluorescent intensity of the tdTomato is several-fold higher and easily distinguishable from the RFP in the red channel. b) Individual channels and composite images of a group of follicles three (P23) and five days (P25) after administering a low dose of Tamoxifen showing a non-leaky expression of the Cre reporter (tdTomato) and a quiescent niche between these time-points. Scale bar: 100μm.
Extended Data Figure 4 – Classification of hair follicle cell types in vivo. a) Panel showing single optical sections (top) or 3D volume renderings (bottom) of the outer (ORS; left) or inner hair follicle cell layers (right) as seen in vivo using a K14H2BGFP fluorescent reporter. b) Single optical sections showing cells marked with the tdTomato Cre reporter (in addition to K14H2BGFP) in the outer (ORS; left) and inner hair follicle layers (right). Notice the differences in morphology between cells located in different layers within the hair follicle.
Extended Data Figure 5 - Relocation of bulge stem cells and progeny over a hair cycle. Examples of in vivo lineage tracing of bulge cells in rest and growth phases of a full hair cycle. Arrows point to the location of the original cell and that of its progeny occupying different positions in the niche after a full hair cycle. Scale bar: 50μm.
Extended Data Figure 6 - A single hair germ cell generates a spatially restricted differentiated lineage. In vivo lineage tracing of a single cell located in the hair germ in rest and growth phases over a full hair cycle. In advanced hair growth (Anagen) an Inner Root Sheath differentiated lineage can be visualized and it is restricted to one side of the follicle as the original founder cell. Scale bar: 50μm.
Extended Data Figure 7 - Long-term fate of bulge stem cells. Examples of in vivo lineage tracing of a single bulge cell in rest and growth phases over two consecutive hair cycles. Arrows point to the location of the stem cell that remains uncommitted in the niche during both hair cycles. Scale bar: 50μm.
Extended Data Figure 8 - Origin of the hair germ. Examples of in vivo lineage tracing of a single bulge cell lineage in rest and growth phases over two consecutive hair cycles. A bulge cell positioned in the lower bulge undergoes limited and more extended amplification in the ORS over two consecutive hair cycles. After regression of the follicle at the end of the second hair cycle some surviving ORS clones form part of the hair germ before the third hair cycle commences. Arrows depict the clonal expansion and contraction of the bulge stem cell lineage during the two hair cycles. Scale bar: 50μm.
Extended Data Figure 9 – Model for determining stem cell fate based on the spatial organization of the hair follicle niche in homeostasis and after injury. In homeostasis a stem cell located in the upper bulge does not commit to a specific fate and is likely to remain quiescent, self-renew or become lost. In contrast, a cell positioned in the lower bulge is more likely to become activated and undergo limited amplification as part of the Outer Root Sheath (ORS), while still remaining relatively undifferentiated. ORS cells that survive the regression phase in one hair cycle can be situated in the niche compartment that becomes the new hair germ. Once positioned within the hair germ a cell will commit towards a differentiation pathway generating the cell types necessary to support hair growth. Loss of a stem cell pool due to injury can induce an epithelial cell to enter the niche and contribute to its recovery. However, once a cell enters the niche it is subject to the same inputs as previous resident cells and as a result it will acquire a hair fate and actively contribute to hair regeneration.