Abstract
Congenital malformations are a prevalent cause of infant mortality in the United States and their induction has been linked to a variety of factors, including exposure to teratogens. However, the molecular mechanisms of teratogenicity are not fully understood. MicroRNAs are an important group of small, non-coding RNAs that regulate mRNA expression. MicroRNA roles in early embryonic development are well established, and their disruption during development can cause abnormalities. We hypothesized that developmental exposure to teratogens such as valproic acid alters microRNA expression profiles in developing embryos. Valproic acid is an anticonvulsant and mood-stabilizing drug used to treat epilepsy, bipolar disorder and migraines. To examine the effects of valproic acid on microRNA expression during development, we used zebrafish embryos as a model vertebrate developmental system. Zebrafish embryos were continuously exposed to valproic acid (1 mM) or vehicle control (ethanol) starting from 4 hours post-fertilization (hpf) and sampled at 48 and 96 hpf to determine the miRNA expression profiles prior to and after the onset of developmental defects. At 96 hpf, 95% of the larvae showed skeletal deformities, abnormal swimming behavior, and pericardial effusion. Microarray expression profiling was done using Agilent zebrafish miRNA microarrays. Microarray results revealed changes in miRNA expression at both time points. Thirteen miRNAs were differentially expressed at 48 hpf and 22 miRNAs were altered at 96 hpf. Among them, six miRNAs (miR-16a, 18c, 122, 132, 457b, and 724) were common to both time points. Bioinformatic target prediction and examination of published literature revealed that these miRNAs target several genes involved in the normal functioning of the central nervous system. These results suggest that the teratogenic effects of valproic acid could involve altered miRNA expression.
Keywords: microRNA, teratogens, microarrays, development, zebrafish
1. Introduction
MicroRNAs (miRNAs) are a group of small non-coding RNAs of 18–25 nucleotides in length that play important roles in embryonic development, physiology and disease. MiRNAs act post-transcriptionally to regulate gene expression by inhibiting translation and sometimes also inducing degradation of target messenger RNAs. Dysregulation of miRNA biogenesis and/or expression has been implicated in the pathology of various cancers and other metabolic, cardiovascular and neurodegenerative diseases (Calin and Croce, 2006; Delay et al., 2012; Esteller, 2011; Lorenzen et al., 2012; Salta and De Strooper, 2012; Small and Olson, 2011). Biogenesis of miRNAs begins with the transcription of miRNA genes into primary miRNAs (pri-miRNAs) of approximately 300–1000 bp in length (Bartel, 2004; Krol et al., 2010). In the nucleus, these pri-miRNAs are processed into 70–80 bp long transcripts called precursor miRNAs (pre-miRNAs), which form distinctive hairpin loop structures by folding back to themselves. The pre-miRNAs are exported into the cytoplasm, where they are further processed by the endonuclease dicer, leading to the generation of 18–25 bp long double-stranded miRNA. One of the strands in the duplex is incorporated into RNA-induced silencing complex (RISC). The miRNA in the RISC complex binds to target messenger RNA with extensive sequence complementarity. Each target messenger RNA contains specific sites in the 3’ untranslated region to which the miRNA binds and directs post-translational repression.
Although the mechanisms of miRNA action in physiology and disease are becoming well understood, few studies have focused on understanding how environmental contaminants affect miRNA expression during early development and whether altered expression of miRNAs may be involved in the developmental toxicity and teratogenicity of chemicals. Early embryogenesis is a sensitive phase in the life history of an organism during which cellular proliferation, tissue differentiation, and other processes involved in morphogenesis occur. MiRNAs have been shown to play an essential role in the regulation of these processes by fine-tuning the gene expression patterns both spatially and temporally during development (Wienholds and Plasterk, 2005). For example, members of the miRNA-430 family play an important role during maternal-zygotic transition by promoting clearance of maternal RNAs at the onset of zygotic transcription (Giraldez et al., 2005). Similarly, miRNA-9, 124, 132, 212 are important for normal development of the nervous system (Coolen and Bally-Cuif, 2009; Wanet et al., 2012; Zeng, 2009). While the developmental roles of individual miRNAs are well understood, the effects of developmental toxicants and teratogens on miRNA expression have not been thoroughly investigated and are not well understood. In addition, most of the studies conducted so far have been done using in vitro cell culture systems; only a few studies have utilized in vivo developmental model systems to study the effect of exposure to teratogens on miRNA expression (Gueta et al., 2010; Jenny et al., 2012; Soares et al., 2012; Tal et al., 2012; Zhang et al., 2011; Zhao et al., 2011).
Exposure to teratogens leads to a wide variety of morphological and physiological defects that are often characterized by defects in developmental signaling pathways and changes in gene expression patterns. Recently, there is increasing evidence suggesting that miRNAs play a role in mediating teratogenicity. For example, exposure of zebrafish embryos to ethanol during early development (4–24 hours post-fertilization) altered expression of miRNAs involved in cell cycle regulation and apoptosis (Soares et al., 2012). Furthermore, developmental exposure to ethanol was shown to affect larval behavior by altering miRNA expression patterns (Tal et al., 2012). Similarly, we recently demonstrated that exposure of zebrafish embryos to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), a well-established teratogen, altered expression of miRNAs that target genes involved in hematopoiesis and cardiovascular development (Jenny et al., 2012). Other studies have explored embryonic miRNA expression after exposure to microcystins (Zhao et al., 2011) and perfluorooctanosulfate (PFOS) (Zhang et al., 2011). All of these studies suggest that perturbing miRNA expression is a potential mechanism of teratogenic action. Because miRNAs are major regulators of gene expression, any perturbation to the biosynthesis of miRNAs could directly affect gene expression and subsequently impact embryonic growth and differentiation.
In this study, we tested the hypothesis that valproic acid (VPA; 2-propylpentanoic acid), a widely used anti-epileptic drug and a well-known teratogen, alters miRNA expression patterns in developing embryos. Although first developed as an anti-epileptic drug, VPA is now used in the treatment of variety of other disease states such as bipolar disorder, migraine, cancer, and Alzheimer’s disease (Terbach and Williams, 2009; Wiltse, 2005). One of the side effects of VPA is teratogenicity; administration of VPA during pregnancy induces birth defects such as spina bifida and other neural tube defects (Ornoy, 2009; Terbach and Williams, 2009). The teratogenicity of VPA has been shown to be due, at least in part, to direct inhibition of histone deacetylases (HDACs)(Gurvich et al., 2005; Gurvich et al., 2004). HDACs are enzymes important in chromatin remodeling (Gurvich et al., 2004; Marks et al., 2001a; Marks et al., 2001b), deacetylating the amino terminal ends of histones and thereby repressing gene expression. Recent studies have demonstrated that VPA and other HDAC inhibitors alter miRNA expression in neuronal cells in vitro (Agostini et al., 2011a; Agostini et al., 2011b; Hunsberger et al., 2013). However, there are no published studies reporting the VPA effects on miRNA expression in in vivo developmental model systems. To test the hypothesis that VPA can alter miRNA expression during development, we used zebrafish embryos, a well established model for studying vertebrate development and developmental toxicity (de Esch et al., 2012; Levin et al., 2003; Levin et al., 2011; Linney et al., 2004; Scalzo and Levin, 2004; Selderslaghs et al., 2013; Sylvain et al., 2011; Tal and Tanguay, 2012). We exposed zebrafish embryos to VPA and quantified the miRNA expression patterns at two different time points, prior to and after the onset of developmental phenotypes. MicroRNA expression was profiled using Agilent zebrafish miRNA microarrays. We demonstrate that developmental exposure to VPA altered the expression of several miRNAs that are critical for normal functioning of the central nervous system. Altered expression occurred prior to the onset of observable developmental defects.
2. Materials and Methods
2.1. Experimental animals
The wild-type TL (Tupfel/Long fin mutations) strain of zebrafish was used in this study. Procedures used to conduct these experiments were approved by the Animal Care and Use Committee of the Woods Hole Oceanographic Institution. Freshly fertilized eggs were obtained from breeding of multiple tanks with 30 female and 30 male fish.
2.2. Exposure of zebrafish embryos to VPA
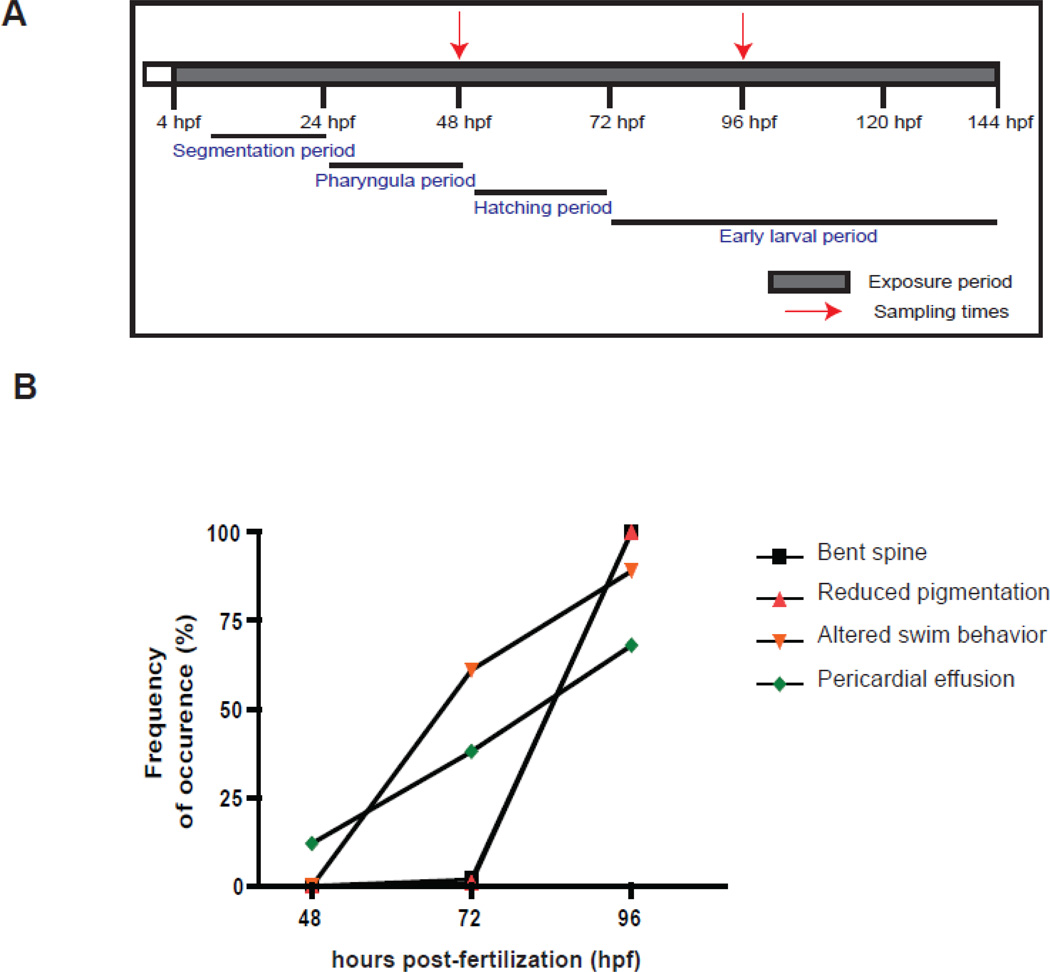
Several previous studies have documented effects of VPA on zebrafish development (Brannen et al., 2010; Cowden et al., 2012; Gurvich et al., 2005; Herrmann, 1993; Li et al., 2009; Selderslaghs et al., 2009; Teixido et al., 2013; Terbach et al., 2011)(Supplemental Table 1). The concentrations required to cause teratogenic effects vary among studies, but generally have been in the range of 0.1 – 3.0 mM. We therefore performed an initial concentration-response experiment to determine the sensitivity of zebrafish embryos and timing of VPA effects in our hands. Zebrafish embryos were exposed to final concentrations of 0.02, 0.05, or 1.0 mM VPA sodium salt (Sigma-Aldrich, St.Louis, MO) or carrier control (ethanol, 0.001% v/v), starting at 4 hours post-fertilization (hpf) continuously until 144 hpf (Fig. 1). Embryos were maintained at 28.5°C at a density of 3 embryos per mL in 0.3 × Danieau's medium (pH 7.2). Water was exchanged daily and fresh vehicle- or VPA-containing medium was added. Embryos were observed daily for any phenotypic abnormalities. No overt developmental phenotypes were observed in embryos exposed to 0.02 and 0.05 mM VPA. In 1 mM VPA exposed embryos, we observed developmental phenotypes such as pericardial effusion, spinal curvature, decreased pigmentation starting from 72 hpf. Based on these results, 1 mM was selected as the exposure concentration for assessing the effect of VPA on miRNA expression. Embryos were exposed to 1 mM VPA as described above. Each treatment consisted of four biological replicates with 50 embryos per replicate. During exposure, VPA- and vehicle exposed zebrafish embryos were observed under the microscope for any morphologic or behavioral abnormalities, as described below. Embryos were sampled for RNA at 48 and 96 hpf, before and after the onset of developmental deformities, by quickly euthanizing and flash freezing in liquid nitrogen. Samples were stored at −80 °C until RNA isolation.
Figure 1.
(A). Outline of the experimental design. Zebrafish embryos were exposed to VPA (1 mM) and ethanol (solvent carrier control; 0.001% v/v) starting from 4 hours post-fertilization (hpf) continuously until the end of the experiment (144 hpf). Embryos were observed daily for developmental abnormalities and sampled at 48 and 96 hpf for microarray analysis. The exposure period is shown in shaded colors and the red arrows represent the two sampling time points. The key stages of zebrafish development are highlighted (Westerfield, 2000). (B). VPA-induced phenotypic defects during the course of exposure. Embryos were observed under the microscope for any phenotypic abnormalities every 24 hours until the end of the experiment. The different phenotypic defects observed include bent spine, reduced pigmentation, altered swimming behavior and pericardial effusion. The percentage of embryos with each defect was calculated by dividing the total number of embryos with the deformity by the total number in each replicate.
2.3. Assessment of phenotypes
Embryos exposed to VPA or vehicle were observed for morphological and behavioral abnormalities at 24, 48, 72, and 96 hpf. Embryos were scored individually for several phenotypes, including pericardial effusion, spinal curvature, reduced pigmentation, altered swimming behavior, twitching, and startle reaction. Results are expressed as the percentage of embryos exhibiting a phenotype at a specific time of observation. Not all phenotypes were scored at each observation time.
2.4. Total RNA isolation
Total RNA was isolated using the TRIzol method (Invitrogen, CA) following manufacturer’s instructions. Total RNA was quantified using a Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE) and RNA quality was assessed using an Agilent 2100 Bioanalyzer. Only the samples with RNA integrity numbers between 9.5–10 were used in microarray analysis.
2.5. Agilent microarray analysis
Agilent microRNA microarray hybridization and analysis was conducted as described previously (Jenny et al., 2012). Briefly, Agilent oligonucleotide microRNA microarrays were custom designed based on zebrafish mature microRNA sequences from miRbase v.16. Each individual array contained a total of 548 unique probes representing 259 mature microRNAs from zebrafish (245), Fugu rubripes (11) and Tetraodon nigriviridis (3). Total RNA labeling and hybridization were carried out using Agilent's microRNA complete labeling and hybridization kit (Agilent Technologies). The hybridized slides were washed using Agilent microarray wash buffers and scanned using the Genepix 4100A scanner (Molecular Devices Corporation, Sunnyvale, CA). Agilent Feature Extraction software (version 9.5.3.) was used for image analysis and data extraction. Quality control (QC) reports generated by AFE software were used to assess data quality for each microarray and to identify outliers. The raw data were used in statistical analysis for determining differential gene expression patterns. Statistical analysis was carried out using AgiMicroRna, a Bioconductor package (Lopez-Romero, 2011). Differential expression of microRNAs was determined by fitting a linear model using an empirical Bayes approach (Smyth, 2004). We controlled for multiple testing by estimating false discovery rate (FDR) using the Benjamini and Hochberg method (Benjamini and Hochberg, 1995). In addition, B-statistic (log-odds that the gene is differentially expressed) was also used as a cut-off for determining differentially expressed genes. Only microRNAs with adjusted p-values less than 0.05 and B-statistic above zero were considered to be statistically significant. All microarray data presented in this manuscript were collected in accordance with MIAME guidelines and have been deposited in the NCBI GEO database (GEO accession number GSE48054).
2.6. Quantitative real-time PCR
We selected seven miRNAs for confirmation using qPCR; they include miR-451, 122, 124, 132/212, 141 and 144. Complementary cDNA synthesis for mature miRNA was done using the stem-loop reverse transcription (RT) primer method as we described earlier (Jenny et al., 2012). Mature miRNA-132 and 212 differ by only one nucleotide and thus were quantified together. The reverse transcription reaction was carried out using the TaqMan microRNA reverse transcription kit (Applied Biosystems, CA) following manufacturer's instructions. Each reverse transcription reaction contained 100 ng of total RNA, 50 nM stem-loop primer, 1 × RT buffer, 0.25 mM each of dNTPs, 0.25 U/µL RNase inhibitor and 3.33 U/µL of multi-scribe reverse transcriptase in a total volume of 15 µL.
Real-time RT-PCR was performed using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) as described previously (Jenny et al., 2012). Each 25 µL PCR reaction consisted of 1 µL of RT product, 1 × PCR master mix, 1.5 µM each of microRNA specific forward primer and reverse primer. U6 snRNA was used as a reference gene. The RT and qPCR primers are given in Table 1. The PCR reaction conditions used were 95 °C for 3 min followed by 40 cycles of 95 °C for 15 s and Tm for 1 min, followed by melt curve analysis. All samples were run in triplicate. A no-template control reaction was run on each PCR plate. The specificity of these primers was confirmed by cloning and sequencing the qPCR products (Jenny et al., 2012).
Table 1.
cDNA synthesis (RT primer) and qRT-PCR primers used in this study.
| Small RNA | Primer (5’ – 3’) | |
|---|---|---|
| miR-141 | RT | GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACG CAT CG |
| miR-144 | RT | GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACA GTA CA |
| miR-451 | RT | GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACA ACT CA |
| miR-122 | RT | GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACC AAA CAC |
| miR-124 | RT | GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACT TGG CAT |
| miR-132/212 | RT | GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACC GAC CA |
| U6 | Forward | TCG CTA CGG TGG CAC ATA |
| Reverse | TAT GGA GCG CTT CAC GG | |
| miR-141 | Forward | GCC GCT AAC ACT GTC TGG TA |
| miR-144 | Forward | GCC CGG CCT ACA GTA TAG ATG |
| miR-451 | Forward | GGC CCA AAC CGT TAC CA |
| miR-122 | Forward | CCG CTG GAG TGT GAC AAT GG |
| miR-124 | Forward | GCT AAG GCA CGC GGT GAA TG |
| miR-132/212 | Forward | CCG CGC TAA CAG TCT ACA GC |
| miR | Reverse | GTG CAG GGT CCG AGG TAT TC |
| β-actin | Forward | CAA CAG AGA GAA GAT GAC ACA GAT CA |
| Reverse | GTC ACA CCA TCA CCA GAG TCC ATC AC | |
| amot | Forward | ACG AAA GCA GCT ACA TCA GCC ACA |
| Reverse | GGC ACG ATC CAC ACT CCG TCT | |
| bdnf | Forward | ACG ACT GGA CAG TAA AGT TCC AAC A |
| Reverse | CTG CAC GCC CGG GAT CTC TC | |
| ncor2 | Forward | AAG CAC CGC AGC CTT GTC CA |
| Reverse | ACA AAG GCA GCT CTA CTC GGG G | |
| paip2 | Forward | GCG CGT TAC AAT GCC GGA ACC |
| Reverse | TTC CCT TGT CCA CTG CCG CC | |
| mmp9 | Forward | CAC ATA CAG GAT TTT GAA CTA TTC G |
| Reverse | GAT CAC CGT GAT CTG CTT TCC |
Relative levels of miRNA abundance was calculated using the 2−ΔΔCt method (where ΔΔCt = [Ct(miRNA)-Ct(U6)] VPA - [Ct(miRNA)-Ct(U6)] DMSO) (Livak and Schmittgen, 2001). Statistical analysis of the qPCR data was conducted using the Prism 4 software package (GraphPad Software Inc., San Diego, CA). Logarithmic-transformed relative expression values were used in analysis of variance (ANOVA). Two-way ANOVA was used to determine the effect of time and treatment. Bonferoni’s post-hoc test was used to determine the statistical significance. P-values of less than or equal to 0.05 are considered statistically significant.
2.7. MicroRNA target prediction and functional annotation
The target prediction for differentially expressed miRNAs was done using their human ortholog sequences. The list of human orthologs is provided in Table 2. Putative mRNA targets of candidate miRNAs were predicted using three prediction algorithms: MicroCosm (http://www.ebi.ac.uk/enright-srv/microcosm/cgi-bin/targets/v5/search.pl), Target Scan (http://targetscan.org/), and microrna.org (http://www.microrna.org/microrna/home.do). We compared the predicted mRNA targets from the three algorithms and only the targets commonly predicted by all three algorithms were used in the downstream analyses. In order to identify transcripts that are targeted by multiple differentially expressed miRNAs, we searched the 3’ untranslated region of the mRNA targets for seed sequences of these miRNAs. We conducted this analysis separately for upregulated and downregulated miRNAs. Once the mRNAs with multiple miRNA targets were identified, we searched the zebrafish database (http://www.targetscan.org/fish_62/) to determine if orthologous zebrafish transcripts are targeted by the same miRNAs. From this list, we selected four genes for validation using qPCR.
Table 2.
Differentially expressed zebrafish miRNAs and their human orthologs based on sequence conservation. The miRNAs listed in parenthesis are the IDs from the latest version of miRbase.
| microRNA | Human ortholog | |
|---|---|---|
| dre-miR-124 | hsa-miR-124-3p | |
| dre-miR-22a | hsa-miR-22-3p | |
| dre-miR-96 | hsa-miR-95-5p | |
| dre-miR-135a | hsa-miR-135a-5p | |
| dre-miR-182 | (dre-miR-182-5p) | hsa-miR-182-5p |
| dre-miR-182* | (dre-miR-182-3p) | hsa-miR-182-3p |
| dre-miR-122 | hsa-miR-122-5p | |
| dre-miR-140 | (dre-miR-140-5p) | hsa-miR-140-5p |
| dre-miR-16a | hsa-miR-16-5p | |
| dre-miR-18c | hsa-miR-18b-5p | |
| dre-miR-457b | hsa-miR-16-5p | |
| dre-miR-132 | (dre-miR-132-3p) | hsa-miR-132-3p |
| dre-miR-10b | hsa-miR-10b-5p | |
| dre-miR-16b | hsa-miR-16-5p | |
| dre-miR-210* | (dre-miR-210-5p) | hsa-miR-210 |
| dre-miR-451 | hsa-miR-451a | |
| dre-miR-21 | hsa-miR-21-5p | |
| dre-miR-29a | hsa-miR-29c-3p | |
| dre-miR-34 | (dre-miR-34a) | hsa-miR-34a-5p |
| dre-miR-194a | hsa-miR-194-5p | |
| dre-miR-10c | hsa-miR-10a-5p | |
| dre-miR-22b | hsa-miR-22-3p | |
| dre-miR-217 | hsa-miR-217 | |
| dre-miR-455 | (dre-miR-455a) | hsa-miR-455-5p |
| dre-miR-27c | (dre-miR-27c-3p) | hsa-miR-27b-3p |
| dre-miR-29b | hsa-miR-29b-3p | |
| dre-miR-192 | hsa-miR-192-5p | |
| dre-miR-212 | hsa-miR-212 | |
| dre-miR-724 | - |
For functional annotation of predicted mRNA targets, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was conducted on the predicted mRNA targets to identify signaling pathways that are over-representated upon VPA exposure. KEGG pathway analysis was carried out using DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/).
2.8. Validation of predicted mRNA targets using Quantitative real-time PCR
We selected four genes (Angiomotin (amot), nuclear receptor corepressor 2 (ncor2), matrix metalloproteinase 9 (mmp9) and poly (A) binding protein interacting protein 2 (paip2) for validation using qPCR. In addition, we also quantified the expression of brain-derived neurotrophic factor (bdnf) because this gene has been found to be involved in the regulation of miR-132/212. Complementary cDNA was synthesized from 1 µg of total RNA using an iScript cDNA synthesis kit (BioRad, Herculus, CA) and qPCR was performed as described in section 2.6. The primers used for amplifying the gene products are provided in Table 1. Relative expression levels were calculated using the 2−ΔΔCt method (where ΔΔCt = [Ct(GOI)-Ct(β-actin)] VPA - [Ct(GOI)-Ct(β-actin)] DMSO; GOI – gene of interest). Statistical analysis was performed as described in section 2.6.
3. Results
3.1. VPA-induced phenotypic changes
Zebrafish embryos exposed to VPA displayed a number of morphological and behavioral abnormalities. At 48 hpf, most VPA-exposed embryos appeared to be developing normally. A small percentage (5%) of VPA-exposed embryos displayed yolk sac effusion at this time, but other abnormalities were not observed. By 72 hpf, pericardial effusion was observed in approximately 30% of the embryos. In addition, the embryos also exhibited altered swimming behavior. VPA-exposed embryos showed circular swimming behavior in comparison to the forward motion exhibited by vehicle-exposed embryos. By 96 hpf, a large proportion of the embryos displayed a number of other abnormalities such as pericardial effusion, reduced pigmentation, spinal curvature, twitching, altered swimming behavior, and lack of startle response (Fig. 1b). These phenotypes are similar to those reported in previous studies of VPA effects in zebrafish embryos (Brannen et al., 2010; Cowden et al., 2012; Gurvich et al., 2005; Herrmann, 1993; Li et al., 2009; Selderslaghs et al., 2009; Teixido et al., 2013; Terbach et al., 2011). The list of developmental phenotypes observed and their frequency of occurrence is provided in Supplemental Table 2.
3.2. MicroRNA expression profiling by microarray
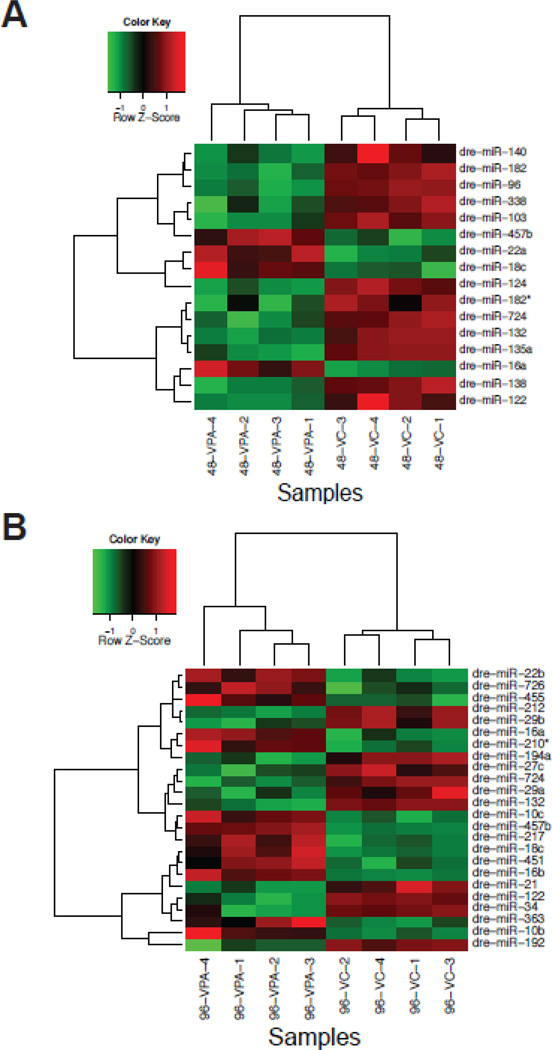
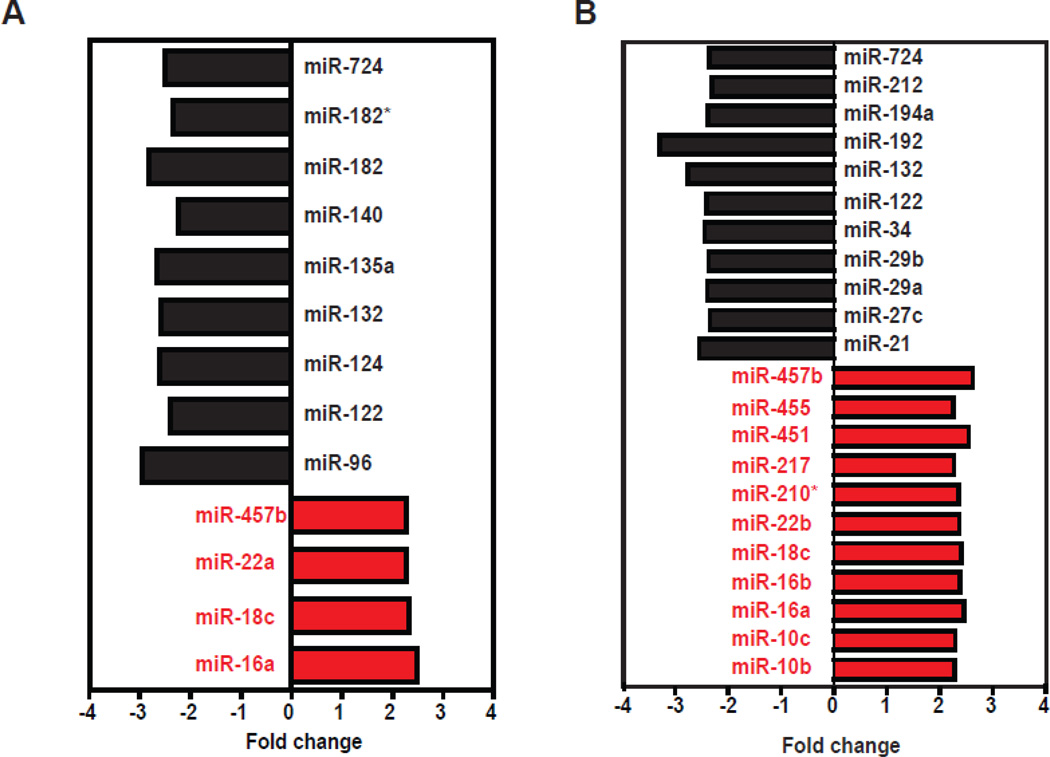
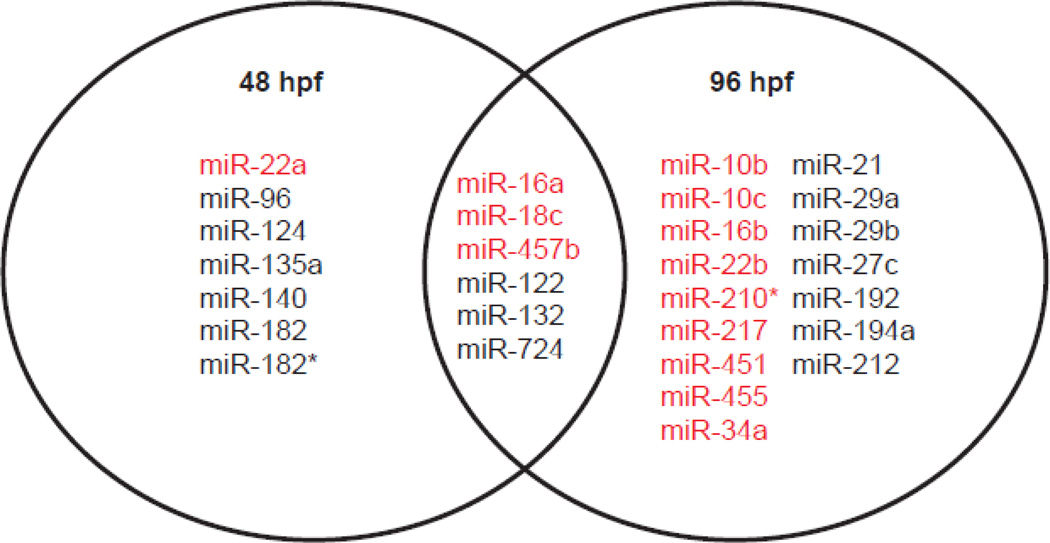
MicroRNA expression was measured at 48 hpf (prior to the onset of developmental abnormalities) and at 96 hpf (after the appearance of several abnormalities). Overall, VPA exposure significantly altered 35 miRNAs during development. Thirteen miRNAs were differentially expressed at 48 hpf. Of these, four miRNAs (miR-16a, 18c, 22a, 457b) were up-regulated and nine (miR-96, 122, 124, 132, 135a, 140, 182, 182* and 724) were down-regulated (Fig. 2A). At 96 hpf, 22 miRNAs were differentially expressed, with equal numbers that were up-regulated (miR-10b, 10c, 16a, 16b, 18c, 22b, 210*, 217, 451, 455 and 457b) and down-regulated (miR-21, 27c, 29a, 29b, 34, 122, 132, 192, 194a, 212 and 724) (Fig. 2B). The magnitude of differential expression ranged from +2.6-fold to −3.3-fold (Fig. 3). Six miRNAs were differentially expressed at both time points: miRNA-16a, 18c, 122, 132, 457b and 724 (Fig. 4). Three were up-regulated and three were downregulated; in each case the changes seen at 48 and 96-hpf were in the same direction.
Figure 2.
Hierarchical clustering heat map of fold changes (log2) depicting VPA-induced differential expression of miRNAs at (A) 48 and (B) 96 hpf. Each row represents a miRNA and columns represent biological replicate samples of control and VPA-treated embryos. The legend represents the color mapping of row-wise Z-scores used to display the up-(red) and down (green)-regulated genes. Row-wise Z-scores were calculated by subtracting the mean from each cell and dividing the value by the standard deviation of the row.
Figure 3.
Magnitude of fold change in miRNA expression in response to VPA exposure at (A) 48 hpf and (B) 96 hpf. The magnitude change in expression varied from −3.2 to + 2.6.
Figure 4.
Venn diagram showing the overlap in differentially expressed miRNA at 48 and 96 hpf in VPA-exposed embryos. A total of 6 miRNAs were differentially expressed at both time points. Of these, three miRNAs were down-regulated and three were up-regulated. Red = up-regulated; black = down-regulated.
In addition to VPA-induced changes in miRNA expression, we also observed developmental changes in miRNA expression in vehicle controls between 48 and 96 hpf. Comparison of the two time points revealed that 93 miRNAs were differentially expressed (Fig. 5); the fold change in expression ranged from +9 to −8. None of these miRNAs showed differences in response to VPA exposure. The complete list of differentially expressed miRNAs is provided in the supplemental information (Supplemental Table 3: VPAarray-supplemental Information.xlsx).
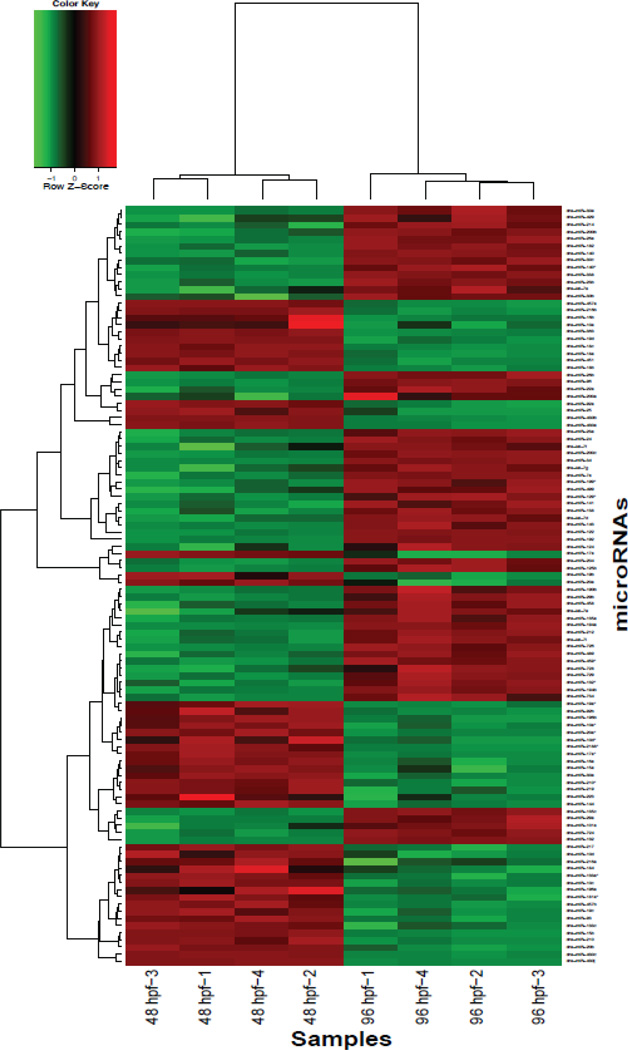
Figure 5.
Heat map representation of fold changes (log2) showing differential expression of miRNAs during development. Fold change comparisons were done between 48 hpf and 96 hpf vehicle-treated groups and analyzed using hierarchical clustering. A total of 93 miRNAs were differentially expressed.
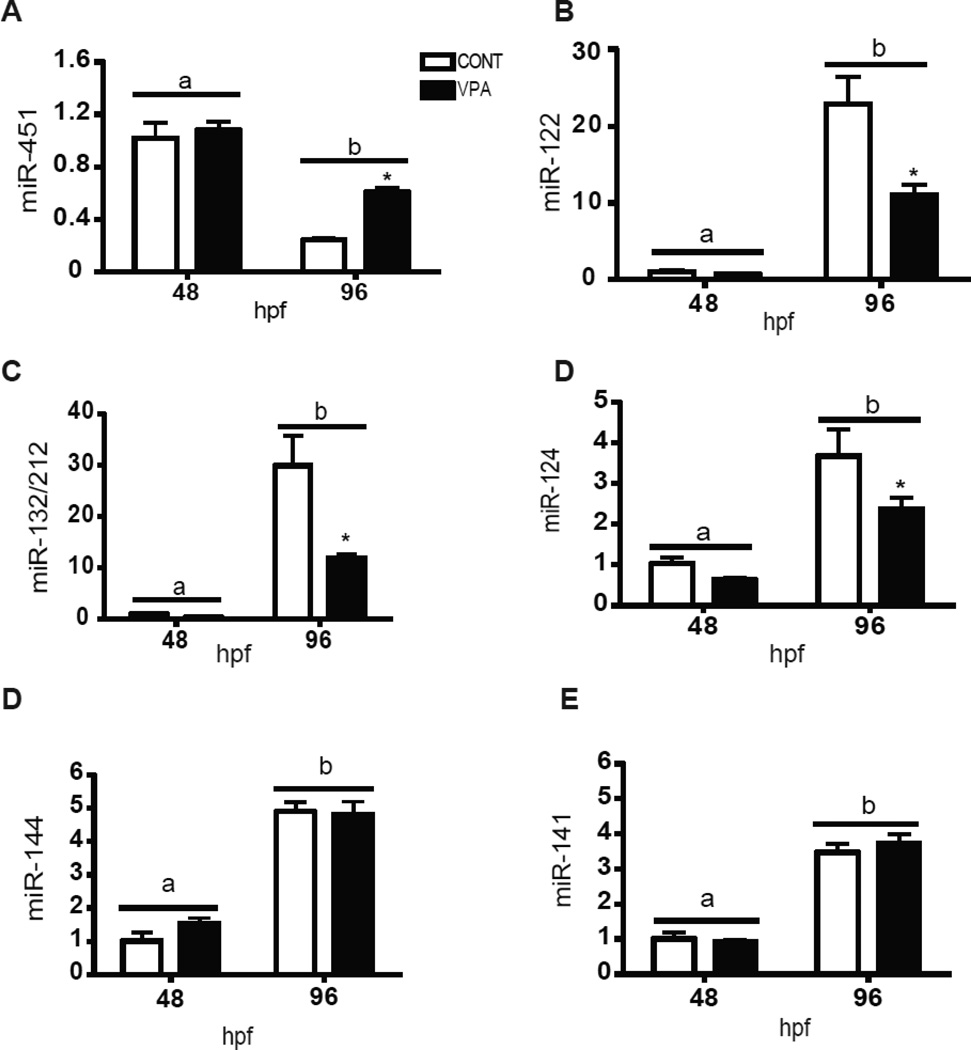
3.3. MicroRNA measurement by qPCR
We selected seven miRNAs that showed differential expression with either VPA exposure (miR-451, 122, 124, 132/212) or during development (miR-141, 144). (Fig. 6). MiRNA-451 was up-regulated with VPA exposure at 96 hpf, confirming microarray results. MiRNA-122, 124 and 132/212 showed apparent down-regulation at both time points but the results were statistically significant only at 96 hpf. This is somewhat different from the microarray results, in which miRNA-122 and 132 were down-regulated at both time points and miRNA-124 was down-regulated only at 48 hpf. Consistent with the array data on developmental changes, qPCR showed that miRNA-451 was significantly down-regulated between 48 and 96 hpf, whereas the other miRNAs were significantly up-regulated at 96 vs. 48 hpf.
Figure 6.
Quantitative RT-PCR confirmation of microarray results. Seven microRNAs (MiRNA-451, 122, 132/212, 124, 141 and 144) were selected to be quantified using qRT-PCR. U6 was used as a housekeeping gene. MiRNA expression was calculated using delta-delta Ct method. Data are expressed as fold change from 48 hpf control group. Values represent mean+S.D. (Two-way ANOVA; n=4). Different letters denotes significant differences between the two time-points. Statistically significant effect of VPA on miRNA expression is denoted by asterisk (*).
3.4. MicroRNA target prediction and functional annotation
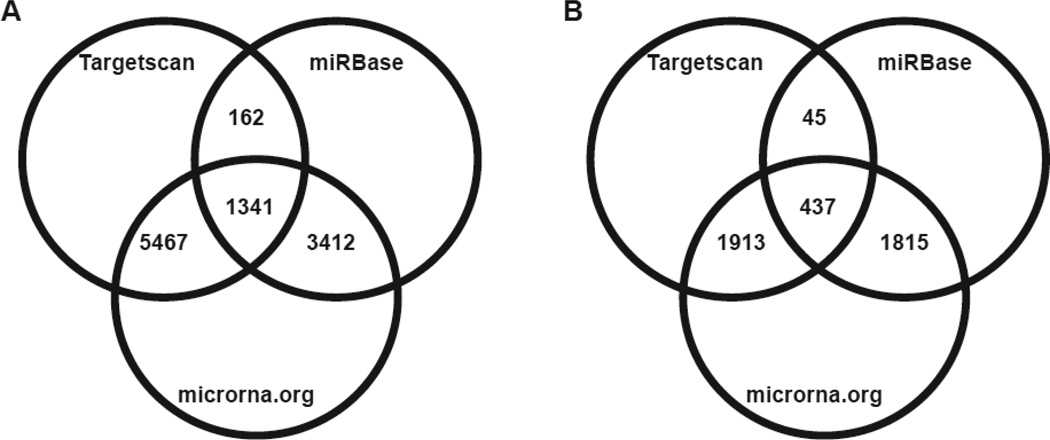
Using Targetscan, microcosm and mirna.org target prediction software, we identified the predicted mRNA targets of differentially expressed miRNAs (Fig. 7). Only predicted targets that were identified by all three algorithms were considered. Using this criterion, a total of 437 and 1341 transcripts were predicted to be the targets of up-regulated and down-regulated miRNAs, respectively. Among the targets of up-regulated miRNAs, three transcripts (teneurin transmembrane protein 2 (ODZ2), nuclear receptor corepressor 2 (NCOR2) and CUB and Sushi multiple domains 1 (CSMD1)) were found to be commonly targeted by 3 miRNAs (Supplemental Table 4). Similarly, four transcripts are common targets of several downregulated miRNAs. They include poly (A) binding protein interacting protein 2 (PAIP2), a disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type1 motif, 6 (ADAMTS6), RAS p21 protein activator (GTPase activating protein) 1 (RASA1) and angiomotin (AMOT). As the target prediction analysis was conducted using human miRNAs, we searched for the presence of these genes in zebrafish and for the conservation of miRNA seed sequences. The list of miRNA targets and the presence of these targets in zebrafish 3’UTR is provided in Supplemental Table 4.
Figure 7.
Venn diagram showing the target prediction results of (A) down-regulated and (B) up-regulated miRNAs. Only the number of transcripts that are commonly identified by the prediction software are represented.
Functional annotation of these targets based on KEGG pathway analysis revealed that the up-regulated miRNAs regulate genes involved in p53 signaling, cancer, cell cycle and Wnt signaling (Table 3), whereas the down-regulated miRNAs have target genes involved in focal adhesion, MAP kinase signaling, insulin signaling, ECM-receptor interaction, cancer and axon guidance (Table 4).
Table 3.
KEGG pathway annotation of mRNA targets of up-regulated microRNAs.
| KEGG Term | Count | % | P-Value | Genes | FDR (%) |
|---|---|---|---|---|---|
| p53 signaling pathway | 10 | 2.50 | 4.57E-05 | RFWD2, CCNE1, CDKN1A, PPM1D, SERPINE1, TP53, SIAH1, CHEK1, SESN1, CDK2 | 0.05 |
| Prostate cancer | 9 | 2.25 | 0.0017 | FGFR1, CCNE1, CDKN1A, SOS1, SOS2, TP53, PIK3CA, TCF7L2, CDK2 | 1.96 |
| Cell cycle | 10 | 2.50 | 0.0042 | CCNE1, CDKN1A, CDKN2D, YWHAQ, TP53, CDC23, CHEK1, CDK2, WEE1, STAG2 | 4.68 |
| Wnt signaling pathway | 10 | 2.50 | 0.0139 | CTNNBIP1, PPP2R1A, NKD1, DKK1, LRP6, TP53, SIAH1, NFATC3, WNT7A, TCF7L2 | 14.81 |
| Pathways in cancer | 16 | 4.01 | 0.0181 | FGFR1, FLT3, TP53, TCF7L2, CDK2, TPM3, CCNE1, MAX, CUL2, CDKN1A, CRKL, SOS1, SOS2, PIK3CA, WNT7A, CSF1R | 18.83 |
Only KEGG terms with significant overrepresentation (p-value less than 0.05) are reported. Count and % columns represent the number and percentage of up-regulated miRNA targets represented in each KEGG term. False discovery rate (FDR) was used as a multiple testing correction method.
Table 4.
KEGG pathway annotation of mRNA targets of down-regulated microRNAs.
| KEGG Term | Count | % | P-Value | Genes | FDR (%) |
|---|---|---|---|---|---|
| Focal adhesion | 30 | 2.90 | 3.71E-05 | TLN2, COL3A1, COL2A1, ITGB1, IGF1R, PTK2, COL6A3, COL6A2, PDGFC, THBS1, FIGF, RAPGEF1, COL11A1, COL4A1, ACTN4, MET, MYLK2, MAPK10, COL5A3, PPP1CC, COL4A6, LAMA2, VEGFC, CRKL, VEGFA, ITGA7, MAPK3, COL1A2, RELN, CRK | 0.045 |
| MAPK signaling pathway | 33 | 3.19 | 5.22E-04 | FGF14, FGF9, MRAS, FASLG, CACNB3, HSPA1B, FOS, TNFRSF1A, BDNF, MAP3K4, RRAS, TRAF6, RASA1, NTF3, TAOK1, RELA, NF1, NR4A1, MAPK10, CACNA2D3, RPS6KA5, DUSP5, CRKL, DUSP2, RPS6KA4, MAPK13, MAPK3, MAPK7, DUSP9, CRK, CACNA1D, MAP3K12, CACNA1A | 0.632 |
| Pathways in cancer | 37 | 3.57 | 0.0012 | FGF14, FGF9, PPARG, FASLG, ITGB1, TCF7L1, RBX1, FOS, IGF1R, PTK2, SLC2A1, RARA, HHIP, TRAF6, FIGF, TRAF4, AXIN1, DVL2, RET, COL4A1, MSH2, RELA, MET, FZD3, MAPK10, RALGDS, COL4A6, RAD51, LAMA2, VEGFC, CRKL, PIAS4, ARAF, VEGFA, MAPK3, PTCH1, CRK | 1.495 |
| Insulin signaling pathway | 19 | 1.83 | 0.0028 | FLOT2, PHKA1, FLOT1, PDE3B, PDE3A, MAPK10, PPP1CC, CRKL, ARAF, PRKAR1A, MAPK3, GYS1, RHEB, PTPN1, TRIP10, CRK, RAPGEF1, LIPE, EIF4E2 | 3.362 |
| ECM-receptor interaction | 14 | 1.35 | 0.0028 | COL4A1, COL3A1, COL2A1, COL5A3, ITGB1, COL4A6, LAMA2, ITGA7, COL6A3, COL6A2, COL1A2, RELN, THBS1, COL11A1 | 3.467 |
| Axon guidance | 18 | 1.74 | 0.0041 | PLXNA3, GNAI2, PLXNB2, MET, NTNG1, L1CAM, DPYSL2, ITGB1, EPHA2, PTK2, SEMA4G, ROBO1, SEMA7A, CFL1, MAPK3, SEMA4C, EFNA5, RASA1 | 4.899 |
Only KEGG terms with significant overrepresentation (p-value less than 0.05) are reported. Count and % columns represent the number and percentage of down-regulated miRNA targets represented in each KEGG term. False discovery rate (FDR) was used as a multiple testing correction method.
3.5. Validation of target prediction results using qPCR
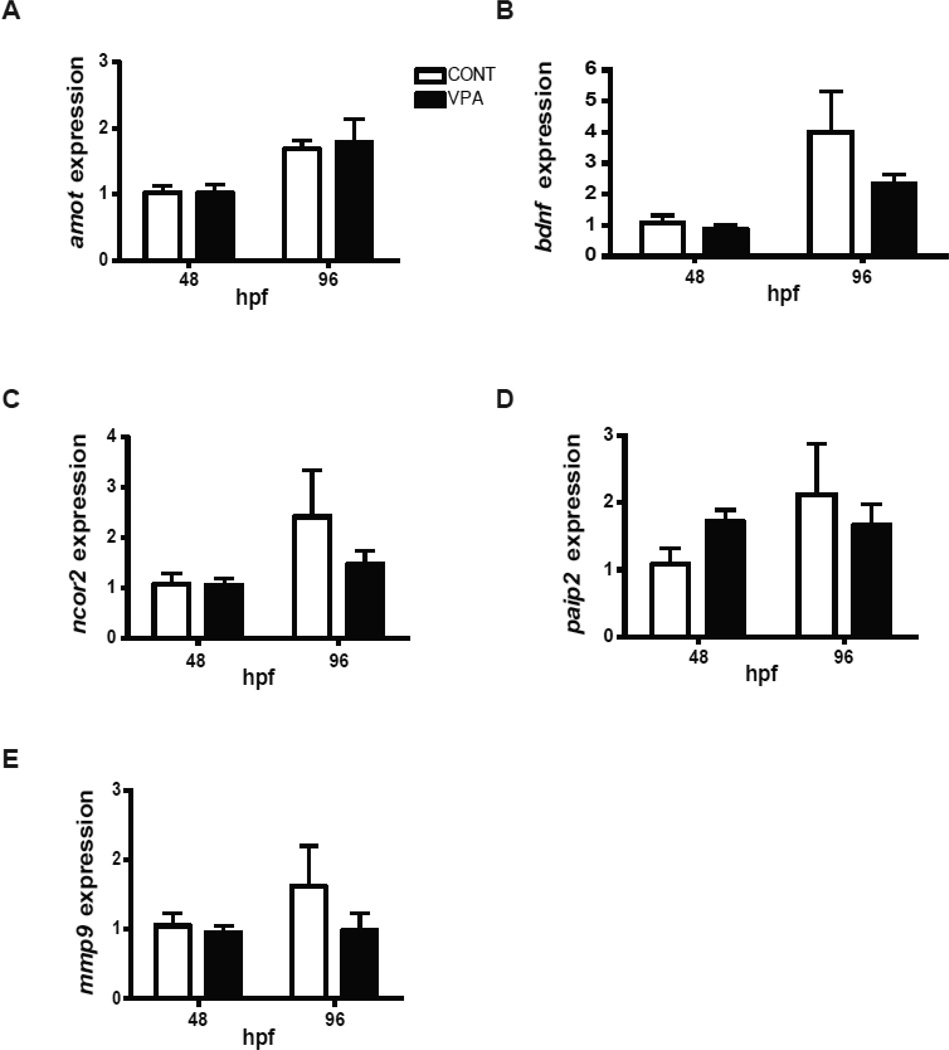
To determine whether altered microRNA expression was reflected in altered transcript levels, we measured mRNA for four predicted microRNA targets, selected as described above. Quantitative PCR results show no statistically significant effect of VPA exposure on the relative expression of any of the four transcripts (amot, ncor2, mmp9 and paip2) (Fig. 8). In addition, VPA exposure did not significantly affect bdnf expression. However, there was a decreasing trend in bdnf, ncor2 and mmp9 expression in VPA-exposed embryos at 96 hpf (Fig. 8).
Figure 8.
Quantitative RT-PCR confirmation of putative mRNA targets. Five mRNAs (amot, bdnf, ncor2, paip2 and mmp9) were selected for quantification using qRT-PCR. β-actin was used as a housekeeping gene. Relative mRNA expression was calculated using delta-delta Ct method. Data are expressed as fold change from 48 hpf control group. Values represent mean + S.D. (Two-way ANOVA; n=4).
4. Discussion
In this study we demonstrated that VPA, widely used as a psychotherapeutic drug and a well-known teratogen in vertebrates including humans (Ornoy, 2009), alters microRNA expression during development in zebrafish. Altered miRNA expression was seen prior to the onset of morphological and behavioral abnormalities. Most of the miRNAs that were altered by VPA exposure are enriched in the central nervous system and essential for proper neurodevelopment (Fiore et al., 2008; Gao, 2008). Furthermore, dysregulation of some of these miRNAs has been shown to be responsible for a variety of neurological and neurodegenerative diseases such as schizophrenia, bipolar disorder, autism, Alzheimer’s and Parkinson’s diseases (Delay et al., 2012; Mellios and Sur, 2012; Miller et al., 2012; Minones-Moyano et al., 2011; Schonrock et al., 2010; Wanet et al., 2012).
4.1. VPA-induced developmental phenotypes and potential mechanisms of action
It is well demonstrated that in utero exposure to VPA causes neural tube defects such as spina bifida in mammals, including humans (Ornoy, 2009; Wiltse, 2005). Similarly, developmental exposure of zebrafish embryos to VPA has been shown to cause developmental defects such as bent spine, edema, and brain deformities (Brannen et al., 2010; Cowden et al., 2012; Gurvich et al., 2005; Herrmann, 1993; Li et al., 2009; Selderslaghs et al., 2009; Teixido et al., 2013; Terbach et al., 2011). The VPA concentration (1 mM) used in this study is higher than the concentrations used in some previous studies (Supplemental Table 1). We did not observe any developmental defects at lower concentrations. The differences in responsiveness could be related to the uptake of VPA by the embryos. VPA uptake is pH-dependent, with higher uptake under acidic conditions (Terbach et al., 2011). The pH of our exposure medium was 7.2 throughout the exposure period and that could be responsible for lower VPA uptake by the embryos, requiring a higher nominal exposure concentration. Nevertheless, the teratogenic effects that we observed are similar to those reported previously in fish and other vertebrate species.
The mechanism of action of VPA in causing these teratogenic effects is unclear. Of the several mechanisms that have been proposed (Wiltse, 2005), the most well understood is the inhibition of chromatin remodeling proteins, the histone deacetylases (HDACs) (Gottlicher et al., 2001; Gurvich et al., 2005; Gurvich et al., 2004). HDAC inhibitors such as VPA prevent the deacetylation of histones and thus induce a more open chromatin configuration, leading to misexpression of genes. In addition to HDAC inhibition, VPA is known to directly target the nervous system by modulating the enzymes involved in the synthesis and degradation of the neurotransmitter gamma aminobutyric acid (GABA), thereby affecting its levels in the brain (Laeng et al., 2004). VPA is also known to affect intracellular signal transduction processes by altering protein kinase C (PKC) (Hahn et al., 2005), ERK/MAP (Yuan et al., 2001) and Wnt/β-catenin pathways (Blaheta et al., 2002; Phiel et al., 2001). Recent studies have determined that VPA alters the expression of miRNAs in the nervous system (Fayyad-Kazan et al., 2010; Lee et al., 2011a; Zhou et al., 2009). Because miRNAs are particularly important for regulating brain and nervous system development, it is possible that some of the teratogenic effects of VPA could be mediated by altered expression of these miRNAs. To our knowledge, this is the first study to investigate the effect of VPA on miRNA expression patterns in vertebrate embryos in vivo.
We determined the VPA effects on miRNA expression at two developmental time points (48 and 96 hpf), prior to and after the onset of phenotypic changes. At 48 hpf, VPA exposure did not cause any phenotypic abnormalities and the changes in miRNA expression (13 differentially expressed miRNAs) observed at this time point could be considered direct or primary effects of VPA exposure. At 96 hpf, VPA-exposed embryos showed phenotypic abnormalities such as spinal defects (100% embryos), altered swimming behavior, reduced pigmentation and pericardial effusion. The behavioral phenotypes could reflect direct effects of VPA exposure on neurodevelopment or could be secondary to other phenotypes. Microarray expression profiling at 96 hpf resulted in differential expression of 22 miRNAs. Some of these changes could be indirect effects of VPA, a result of the VPA-induced abnormalities rather than a direct effect of VPA. Comparison of the known and predicted functions of miRNAs with altered expression at these two times may provide insight into the mechanism by which VPA disrupts embryonic development. There is a need for detailed dose-response studies to determine the VPA exposures associated with altered microRNA expression and the relationship with levels of exposure resulting in the observed phenotypic changes.
4.2. VPA exposure altered expression of neuromiRs
Among the miRNAs that are differentially expressed in VPA-exposed embryos, most are essential for the development of the nervous system, and deregulation of these miRNAs is associated with neurodegenerative disorders. Overall, VPA exposure caused downregulation of 17 miRNAs, most of which are highly conserved among vertebrates. For instance, miR-132 and 212, down-regulated with VPA exposure, are highly expressed in the neurons and are critical for neuronal morphogenesis (Wanet et al., 2012). These two miRNAs are induced by cAMP-response element binding (CREB) protein and brain-derived neurotrophic factor (BDNF) (Remenyi et al., 2010; Vo et al., 2005). We observed a decreasing trend in bdnf transcript levels, suggesting that VPA may downregulate miR-132/212 expression by blocking their induction by BDNF. This suggestion needs additional study, including measurements of BDNF protein expression. In concert with these BDNF and CREB, miR-212 and 132 play an important role in neuronal differentiation, maturation and survival. Both miR-132 and 212 are intergenic miRNAs that are localized in tandem in most vertebrates including humans (chromosome 17), mice (chr. 11), rats (chr. 10) and zebrafish (chr. 15). Similar to humans and rodents, zebrafish miR-132 and 212 share the same primary transcript and have highly similar mature miRNAs, thereby targeting the same mRNAs. Some of the targets of these miRNAs include genes associated with neurodevelopment, synaptic transmission, angiogenesis and inflammation (Wanet et al., 2012). In addition, deficiency in the function of these miRNAs has been shown to be responsible for neurodegenerative disorders such as Alzheimer’s disease, Huntington’s disease, schizophrenia and bipolar disorders (Lee et al., 2011b; Wang et al., 2011).
Our target prediction analysis also revealed that miR-212 and 132 target genes associated with a number of physiological pathways that could indirectly influence neurodevelopment. We quantified the expression of two genes, amot and paip2, that were computationally predicted to be targeted by miR-132/212. Angiomotin is an important player in angiogenesis during early development (Bratt et al., 2005) and its knock-down has been shown to disrupt normal vascular development (Aase et al., 2007). Although VPA exposure has been shown to affect angiogenesis during zebrafish development (Farooq et al., 2008), we did not observe any change in amot transcript levels. In addition, we quantified paip2, an important player in the initiation of protein translation and experimentally validated target of miR-132 (Alvarez-Saavedra et al., 2011). Knock-down of miR-132 increased paip2 protein expression in the Neuro-2a cell line (Alvarez-Saavedra et al., 2011). We observed a moderate but not statistically significant increase in paip2 transcript levels at 48 hpf in response to VPA exposure. The lack of significant effect on the levels of amot and paip2 transcripts is not surprising given the fact that miRNAs generally inhibit translation, often without affecting transcript levels. However, the bioinformatic target prediction results support previous observations that miR-132 and miR-212 target genes involved in the regulation of number of signaling pathways, including neuro and vascular development. These results, together with multiple phenotypic defects observed, suggest that VPA exposure affects normal development either directly or indirectly. Further studies are needed to characterize the direct and indirect effects of VPA on zebrafish development.
Similar to miR-212 and 132, miR-124 was down-regulated in VPA-exposed embryos at 48 hpf. MiR-124 is a CNS-specific miRNA that is expressed at very high levels in the brain (25–45% of all miRNAs) (Krichevsky et al., 2006; Lagos-Quintana et al., 2002). miR-124 is highly expressed during neuronal differentiation in embryonic brain development in mammals (Sempere et al., 2004). In zebrafish embryos, miR-124 is ubiquitously expressed between 12–18 hpf, and after that expression is restricted to brain, spinal cord and the eye as observed by in situ hybridization (Weinholds et al., 2005). In larvae, miR-124 is expressed in all differentiating neuronal cells in the brain and retina (Wienholds et al., 2005). Overexpression of miR-124 in HeLa cells caused brain-type patterns of gene expression, suggesting that miR-124 is very important for maintaining neuronal identity (Lim et al., 2005). In mice, miR-124 knockdown caused abnormalities in adult neurogenesis (Cheng et al., 2009). All these observations clearly suggest that miR-124 is essential for normal development of the nervous system in vertebrates; further studies are needed to characterize the effects of VPA on nervous system development.
Another miRNA that was down-regulated by VPA exposure is miR-34a. This miRNA is ubiquitously expressed in the brain and its function is well characterized. One recent study demonstrated that exposure of hippocampal neurons to VPA down-regulated miR-34a expression and elevated the levels of metabotropic glutamate receptor 7 (GRM7) transcripts (Zhou et al., 2009). GRM7 is an important player in glutaminergic neurotransmission and perturbation in glutamate signaling is associated with neuropathological conditions such as bipolar disorder (Alliey-Rodriguez et al., 2011). Another important miRNA family that was down-regulated by VPA exposure is miR-29. The zebrafish miRNA-29 family is comprised of three mature miRNAs (miR-29a, b and c). Of these, miR-29a (ortholog of human miR-29c) and 29b were down-regulated by VPA exposure (96 hpf) in our studies. Among the main targets of miR-29 are extracellular matrix (ECM) genes, including a large number of collagen isoforms, laminin, fibrilin, elastin, matrix metalloproteinases and integrin. Matrix metalloproteinase 9 (mmp9) was found to be a putative target of miR-29 in zebrafish, but we did not find any significant change in expression of mmp9 mRNA in response to VPA. In addition, the miR-29 family is involved in the regulation of cell proliferation, differentiation and apoptosis. These two functions of the miR-29 family are important determinants of long-term neuronal survival, and defects in their function are associated with development of Alzheimer's disease (AD) (Hebert et al., 2008). In addition, miR-29b has been shown to be an inhibitor of apoptosis in neurons (Kole et al., 2011). These observations suggest a neuroprotective function of miR-29. All these studies are consistent with the idea that VPA may impair normal development of the nervous system by altering miRNA expression. KEGG pathway analysis of predicted mRNA targets suggest that most of the down-regulated miRNAs target transcripts associated with functions known to be perturbed by VPA exposure. For example, VPA has been shown to interfere with cytoskeletal organization (Walmod et al., 1998; Walmod et al., 1999), alter Wnt signaling pathways (Gould and Manji, 2002), arrest cell growth in colon, lung and prostrate carcinoma cell lines (Gurvich et al., 2004; Marks et al., 2001a), and disrupt neurodevelopment.
VPA also down-regulated a liver-specific miRNA, miR-122. These results are interesting given the previous observation that VPA inhibits liver development by inhibiting HDAC activity (Farooq et al., 2008). Further studies are necessary to determine if the downregulation of miR-122 by VPA is the cause or consequence of HDAC inhibition. In addition, miR-122 is considered as a potential tumor suppressor miRNA in hepatic carcinogenesis and genes that promote tumorigenesis are some of its validated targets (Fornari et al., 2009; Lin et al., 2008; Saito et al., 2013; Tsai et al., 2009).
4.3. VPA-up-regulated miRNAs
VPA exposure up-regulated several microRNAs involved in the regulation of apoptosis, tumorigenesis, senescence, and metabolism. These include members of miR-15/16 cluster, miR-22a, 22b, 10b, 10c, 217, 210*, 451 and 455. Three members of the miR-15/16 cluster— miR-16a, 16b and 457b—were up-regulated at both 48 and 96 hpf. Human orthologs of miR-16a and 16b exist, but miR-457b is specific to zebrafish. miRNAs belonging to this cluster have been demonstrated to inhibit cell proliferation, promote apoptosis, and suppress tumorigenicity (Aqeilan et al., 2010). Some of the validated targets of this cluster include multiple oncogenes and the genes that control cell-cycle progression. Similarly, miR-22, which was up-regulated with VPA exposure, is also known for tumor suppressor effects and one well known target is HDAC4, a critical player in the development of cancer (Jovicic et al., 2013).
Members of the miR-10 family (miR-10b and 10c) also were up-regulated by VPA exposure. The miR-10 family is found within the Hox gene clusters in zebrafish and mammals. In zebrafish embryos, miR-10 regulates hox genes (HoxB1a and HoxB3a) that are important in anterior-posterior patterning during early embryonic development (Woltering and Durston, 2008). Furthermore, up-regulation of miR-10b has been found in glioblastoma, astrocytomas and pancreatic cancer (Lund, 2010). These results concur with our target prediction analysis, in which most of the genes targeted by up-regulated miRNAs are related to tumorigenesis, apoptosis and early embryonic development. One of the predicted mRNA targets of miR-10 is ncor2, an important player in hormone signaling during early embryonic development. NCOR2 acts as a transcriptional corepressor by recruiting histone-modifying proteins to the promoter regions of genes and thereby silencing their expression (Linney et al., 2011). It has been demonstrated recently that ectopic expression of miR-10a/10b regulates neuronal cell differentiation by downregulating NCOR2 in neuroblastoma cell lines (Foley et al., 2011). Our data suggested that there may be downregulation of ncor2 mRNA expression in VPA-exposed embryos and this could be due to post-transcriptional regulation of NCOR2 by miR-10a/b (Foley et al., 2011). These results provide an additional mechanism by which developmental exposure to VPA might impact various physiological processes during early development.
5. Conclusions
The results obtained in this study demonstrate that teratogenic effects of VPA are associated with dysregulation of miRNA expression. MicroRNA profiling results suggest that VPA specifically altered miRNAs involved in nervous system development, liver development, and tumorigenesis. Several of these miRNAs are known to be involved in various neuropatholgical conditions. Further mechanistic studies using antagomiRs against differentially expressed miRNAs are needed to determine the direct relationship between altered miRNA expression and the observed phenotypes.
Supplementary Material
Highlights.
Valproic acid exposure alters microRNA expression profiles in developing embryos.
miR-16a, 18c, 122, 132, 457b, 724 are among the differentially expressed miRNAs.
Most of these microRNAs are involved in the nervous system development.
Acknowledgements
This work was supported in part by National Institutes of Health (NIH) grants [R21ES017304 to MEH and MJJ, R00ES017044 to MJJ]. Additional support was provided by NIH grant P01ES021923 and National Science Foundation Grant OCE-1314642 through the Woods Hole Center for Oceans and Human Health. Financial support for KD was provided by a WHOI Summer Student Fellowship funded by The Carl and Pancha Peterson Endowed Fund for Support of Summer Fellows, WHOI Academic Programs funds, and the National Science Foundation Research Experience for Undergraduates (NSF-REU) Program. The U.S. Government is authorized to produce and distribute reprints for governmental purposes notwithstanding any copyright notation that may appear hereon. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The authors have no conflicts of interest concerning this manuscript.
References
- Aase K, Ernkvist M, Ebarasi L, Jakobsson L, Majumdar A, Yi C, Birot O, Ming Y, Kvanta A, Edholm D, Aspenstrom P, Kissil J, Claesson-Welsh L, Shimono A, Holmgren L. Angiomotin regulates endothelial cell migration during embryonic angiogenesis. Genes Dev. 2007;21:2055–2068. doi: 10.1101/gad.432007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M, Tucci P, Killick R, Candi E, Sayan BS, Rivetti di Val Cervo P, Nicotera P, McKeon F, Knight RA, Mak TW, Melino G. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proceedings of the National Academy of Sciences of the United States of America. 2011a;108:21093–21098. doi: 10.1073/pnas.1112061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M, Tucci P, Steinert JR, Shalom-Feuerstein R, Rouleau M, Aberdam D, Forsythe ID, Young KW, Ventura A, Concepcion CP, Han YC, Candi E, Knight RA, Mak TW, Melino G. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proceedings of the National Academy of Sciences of the United States of America. 2011b;108:21099–21104. doi: 10.1073/pnas.1112063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alliey-Rodriguez N, Zhang D, Badner JA, Lahey BB, Zhang X, Dinwiddie S, Romanos B, Plenys N, Liu C, Gershon ES. Genome-wide association study of personality traits in bipolar patients. Psychiatric genetics. 2011;21:190–194. doi: 10.1097/YPG.0b013e3283457a31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Saavedra M, Antoun G, Yanagiya A, Oliva-Hernandez R, Cornejo-Palma D, Perez-Iratxeta C, Sonenberg N, Cheng HY. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Human molecular genetics. 2011;20:731–751. doi: 10.1093/hmg/ddq519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell death and differentiation. 2010;17:215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Blaheta RA, Nau H, Michaelis M, Cinatl J., Jr Valproate and valproate-analogues: potent tools to fight against cancer. Curr Med Chem. 2002;9:1417–1433. doi: 10.2174/0929867023369763. [DOI] [PubMed] [Google Scholar]
- Brannen KC, Panzica-Kelly JM, Danberry TL, Augustine-Rauch KA. Development of a zebrafish embryo teratogenicity assay and quantitative prediction model. Birth Defects Res B Dev Reprod Toxicol. 2010;89:66–77. doi: 10.1002/bdrb.20223. [DOI] [PubMed] [Google Scholar]
- Bratt A, Birot O, Sinha I, Veitonmaki N, Aase K, Ernkvist M, Holmgren L. Angiomotin regulates endothelial cell-cell junctions and cell motility. J Biol Chem. 2005;280:34859–34869. doi: 10.1074/jbc.M503915200. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen M, Bally-Cuif L. MicroRNAs in brain development and physiology. Curr Opin Neurobiol. 2009;19:461–470. doi: 10.1016/j.conb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Cowden J, Padnos B, Hunter D, MacPhail R, Jensen K, Padilla S. Developmental exposure to valproate and ethanol alters locomotor activity and retino-tectal projection area in zebrafish embryos. Reprod Toxicol. 2012;33:165–173. doi: 10.1016/j.reprotox.2011.11.111. [DOI] [PubMed] [Google Scholar]
- de Esch C, Slieker R, Wolterbeek A, Woutersen R, de Groot D. Zebrafish as potential model for developmental neurotoxicity testing: a mini review. Neurotoxicology and teratology. 2012;34:545–553. doi: 10.1016/j.ntt.2012.08.006. [DOI] [PubMed] [Google Scholar]
- Delay C, Mandemakers W, Hebert SS. MicroRNAs in Alzheimer's disease. Neurobiology of disease. 2012;46:285–290. doi: 10.1016/j.nbd.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nature reviews Genetics. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Farooq M, Sulochana KN, Pan X, To J, Sheng D, Gong Z, Ge R. Histone deacetylase 3 (hdac3) is specifically required for liver development in zebrafish. Dev Biol. 2008;317:336–353. doi: 10.1016/j.ydbio.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Fayyad-Kazan H, Rouas R, Merimi M, El Zein N, Lewalle P, Jebbawi F, Mourtada M, Badran H, Ezzeddine M, Salaun B, Romero P, Burny A, Martiat P, Badran B. Valproate treatment of human cord blood CD4-positive effector T cells confers on them the molecular profile (microRNA signature and FOXP3 expression) of natural regulatory CD4-positive cells through inhibition of histone deacetylase. J Biol Chem. 2010;285:20481–20491. doi: 10.1074/jbc.M110.119628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochimica et biophysica acta. 2008;1779:471–478. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Foley NH, Bray I, Watters KM, Das S, Bryan K, Bernas T, Prehn JH, Stallings RL. MicroRNAs 10a and 10b are potent inducers of neuroblastoma cell differentiation through targeting of nuclear receptor corepressor 2. Cell death and differentiation. 2011;18:1089–1098. doi: 10.1038/cdd.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornari F, Gramantieri L, Giovannini C, Veronese A, Ferracin M, Sabbioni S, Calin GA, Grazi GL, Croce CM, Tavolari S, Chieco P, Negrini M, Bolondi L. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer research. 2009;69:5761–5767. doi: 10.1158/0008-5472.CAN-08-4797. [DOI] [PubMed] [Google Scholar]
- Gao FB. Posttranscriptional control of neuronal development by microRNA networks. Trends Neurosci. 2008;31:20–26. doi: 10.1016/j.tins.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Gottlicher M, Minucci S, Zhu P, Kramer OH, Schimpf A, Giavara S, Sleeman JP, Lo Coco F, Nervi C, Pelicci PG, Heinzel T. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould TD, Manji HK. The Wnt signaling pathway in bipolar disorder. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry. 2002;8:497–511. doi: 10.1177/107385802237176. [DOI] [PubMed] [Google Scholar]
- Gueta K, Molotski N, Gerchikov N, Mor E, Savion S, Fein A, Toder V, Shomron N, Torchinsky A. Teratogen-induced alterations in microRNA-34, microRNA-125b and microRNA-155 expression: correlation with embryonic p53 genotype and limb phenotype. BMC developmental biology. 2010;10:20. doi: 10.1186/1471-213X-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurvich N, Berman MG, Wittner BS, Gentleman RC, Klein PS, Green JB. Association of valproate-induced teratogenesis with histone deacetylase inhibition in vivo. Faseb J. 2005;19:1166–1168. doi: 10.1096/fj.04-3425fje. [DOI] [PubMed] [Google Scholar]
- Gurvich N, Tsygankova OM, Meinkoth JL, Klein PS. Histone deacetylase is a target of valproic acid-mediated cellular differentiation. Cancer research. 2004;64:1079–1086. doi: 10.1158/0008-5472.can-03-0799. [DOI] [PubMed] [Google Scholar]
- Hahn CG, Umapathy, Wang HY, Koneru R, Levinson DF, Friedman E. Lithium and valproic acid treatments reduce PKC activation and receptor-G protein coupling in platelets of bipolar manic patients. J Psychiatr Res. 2005;39:355–363. doi: 10.1016/j.jpsychires.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann K. Effects of the anticonvulsant drug valproic acid and related substances on the early development of the zebrafish (Brachydanio rerio) Toxicology in vitro : an international journal published in association with BIBRA. 1993;7:41–54. doi: 10.1016/0887-2333(93)90111-h. [DOI] [PubMed] [Google Scholar]
- Hunsberger JG, Fessler EB, Chibane FL, Leng Y, Maric D, Elkahloun AG, Chuang DM. Mood stabilizer-regulated miRNAs in neuropsychiatric and neurodegenerative diseases: identifying associations and functions. Am J Transl Res. 2013;5:450–464. [PMC free article] [PubMed] [Google Scholar]
- Jenny MJ, Aluru N, Hahn ME. Effects of short-term exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin on microRNA expression in zebrafish embryos. Toxicology and applied pharmacology. 2012;264:262–273. doi: 10.1016/j.taap.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicic A, Zaldivar Jolissaint JF, Moser R, Silva Santos Mde F, Luthi-Carter R. MicroRNA-22 (miR-22) overexpression is neuroprotective via general anti-apoptotic effects and may also target specific Huntington's disease-related mechanisms. PloS one. 2013;8:e54222. doi: 10.1371/journal.pone.0054222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole AJ, Swahari V, Hammond SM, Deshmukh M. miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev. 2011;25:125–130. doi: 10.1101/gad.1975411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature reviews Genetics. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Laeng P, Pitts RL, Lemire AL, Drabik CE, Weiner A, Tang H, Thyagarajan R, Mallon BS, Altar CA. The mood stabilizer valproic acid stimulates GABA neurogenesis from rat forebrain stem cells. J Neurochem. 2004;91:238–251. doi: 10.1111/j.1471-4159.2004.02725.x. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Current biology : CB. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lee S, Jung JW, Park SB, Roh K, Lee SY, Kim JH, Kang SK, Kang KS. Histone deacetylase regulates high mobility group A2-targeting microRNAs in human cord blood-derived multipotent stem cell aging. Cell Mol Life Sci. 2011a;68:325–336. doi: 10.1007/s00018-010-0457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ST, Chu K, Im WS, Yoon HJ, Im JY, Park JE, Park KH, Jung KH, Lee SK, Kim M, Roh JK. Altered microRNA regulation in Huntington's disease models. Experimental neurology. 2011b;227:172–179. doi: 10.1016/j.expneurol.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Levin ED, Chrysanthis E, Yacisin K, Linney E. Chlorpyrifos exposure of developing zebrafish: effects on survival and long-term effects on response latency and spatial discrimination. Neurotoxicology and teratology. 2003;25:51–57. doi: 10.1016/s0892-0362(02)00322-7. [DOI] [PubMed] [Google Scholar]
- Levin ED, Sledge D, Roach S, Petro A, Donerly S, Linney E. Persistent behavioral impairment caused by embryonic methylphenidate exposure in zebrafish. Neurotoxicology and teratology. 2011;33:668–673. doi: 10.1016/j.ntt.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Jia S, Wang S, Wang Y, Meng A. Mta3-NuRD complex is a master regulator for initiation of primitive hematopoiesis in vertebrate embryos. Blood. 2009;114:5464–5472. doi: 10.1182/blood-2009-06-227777. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lin CJ, Gong HY, Tseng HC, Wang WL, Wu JL. miR-122 targets an anti-apoptotic gene, Bclw, in human hepatocellular carcinoma cell lines. Biochemical and biophysical research communications. 2008;375:315–320. doi: 10.1016/j.bbrc.2008.07.154. [DOI] [PubMed] [Google Scholar]
- Linney E, Perz-Edwards A, Kelley B. Identification and characterization of a functional zebrafish smrt corepressor (ncor2) Gene. 2011;486:31–36. doi: 10.1016/j.gene.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linney E, Upchurch L, Donerly S. Zebrafish as a neurotoxicological model. Neurotoxicology and teratology. 2004;26:709–718. doi: 10.1016/j.ntt.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Lorenzen J, Kumarswamy R, Dangwal S, Thum T. MicroRNAs in diabetes and diabetes-associated complications. RNA biology. 2012;9:820–827. doi: 10.4161/rna.20162. [DOI] [PubMed] [Google Scholar]
- Lund AH. miR-10 in development and cancer. Cell death and differentiation. 2010;17:209–214. doi: 10.1038/cdd.2009.58. [DOI] [PubMed] [Google Scholar]
- Marks PA, Richon VM, Breslow R, Rifkind RA. Histone deacetylase inhibitors as new cancer drugs. Curr Opin Oncol. 2001a;13:477–483. doi: 10.1097/00001622-200111000-00010. [DOI] [PubMed] [Google Scholar]
- Marks PA, Rifkind RA, Richon VM, Breslow R. Inhibitors of histone deacetylase are potentially effective anticancer agents. Clinical cancer research : an official journal of the American Association for Cancer Research. 2001b;7:759–760. [PubMed] [Google Scholar]
- Mellios N, Sur M. The Emerging Role of microRNAs in Schizophrenia and Autism Spectrum Disorders. Front Psychiatry. 2012;3:39. doi: 10.3389/fpsyt.2012.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, Willoughby D, Kenny PJ, Elsworth JD, Lawrence MS, Roth RH, Edbauer D, Kleiman RJ, Wahlestedt C. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3125–3130. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minones-Moyano E, Porta S, Escaramis G, Rabionet R, Iraola S, Kagerbauer B, Espinosa-Parrilla Y, Ferrer I, Estivill X, Marti E. MicroRNA profiling of Parkinson's disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Human molecular genetics. 2011;20:3067–3078. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- Ornoy A. Valproic acid in pregnancy: how much are we endangering the embryo and fetus? Reprod Toxicol. 2009;28:1–10. doi: 10.1016/j.reprotox.2009.02.014. [DOI] [PubMed] [Google Scholar]
- Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar MA, Klein PS. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J Biol Chem. 2001;276:36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- Remenyi J, Hunter CJ, Cole C, Ando H, Impey S, Monk CE, Martin KJ, Barton GJ, Hutvagner G, Arthur JS. Regulation of the miR-212/132 locus by MSK1 and CREB in response to neurotrophins. The Biochemical journal. 2010;428:281–291. doi: 10.1042/BJ20100024. [DOI] [PubMed] [Google Scholar]
- Saito Y, Hibino S, Saito H. Alterations of epigenetics and microRNAs in hepatocellular carcinoma. >Hepatology research : the official journal of the Japan Society of Hepatology. 2013 doi: 10.1111/hepr.12147. [DOI] [PubMed] [Google Scholar]
- Salta E, De Strooper B. Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet neurology. 2012;11:189–200. doi: 10.1016/S1474-4422(11)70286-1. [DOI] [PubMed] [Google Scholar]
- Scalzo FM, Levin ED. The use of zebrafish (Danio rerio) as a model system in neurobehavioral toxicology. Neurotoxicology and teratology. 2004;26:707–708. doi: 10.1016/j.ntt.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Schonrock N, Ke YD, Humphreys D, Staufenbiel M, Ittner LM, Preiss T, Gotz J. Neuronal microRNA deregulation in response to Alzheimer's disease amyloid-beta. PloS one. 2010;5:e11070. doi: 10.1371/journal.pone.0011070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selderslaghs IW, Hooyberghs J, Blust R, Witters HE. Assessment of the developmental neurotoxicity of compounds by measuring locomotor activity in zebrafish embryos and larvae. Neurotoxicology and teratology. 2013;37:44–56. doi: 10.1016/j.ntt.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Selderslaghs IW, Van Rompay AR, De Coen W, Witters HE. Development of a screening assay to identify teratogenic and embryotoxic chemicals using the zebrafish embryo. Reprod Toxicol. 2009;28:308–320. doi: 10.1016/j.reprotox.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares AR, Pereira PM, Ferreira V, Reverendo M, Simoes J, Bezerra AR, Moura GR, Santos MA. Ethanol exposure induces upregulation of specific microRNAs in zebrafish embryos. Toxicological sciences : an official journal of the Society of Toxicology. 2012;127:18–28. doi: 10.1093/toxsci/kfs068. [DOI] [PubMed] [Google Scholar]
- Sylvain NJ, Brewster DL, Ali DW. Embryonic ethanol exposure alters synaptic properties at zebrafish neuromuscular junctions. Neurotoxicology and teratology. 2011;33:313–321. doi: 10.1016/j.ntt.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Tal TL, Franzosa JA, Tilton SC, Philbrick KA, Iwaniec UT, Turner RT, Waters KM, Tanguay RL. MicroRNAs control neurobehavioral development and function in zebrafish. Faseb J. 2012;26:1452–1461. doi: 10.1096/fj.11-194464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal TL, Tanguay RL. Non-coding RNAs--novel targets in neurotoxicity. Neurotoxicology. 2012;33:530–544. doi: 10.1016/j.neuro.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixido E, Pique E, Gomez-Catalan J, Llobet JM. Assessment of developmental delay in the zebrafish embryo teratogenicity assay. Toxicology in vitro : an international journal published in association with BIBRA. 2013;27:469–478. doi: 10.1016/j.tiv.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Terbach N, Shah R, Kelemen R, Klein PS, Gordienko D, Brown NA, Wilkinson CJ, Williams RS. Identifying an uptake mechanism for the antiepileptic and bipolar disorder treatment valproic acid using the simple biomedical model Dictyostelium. Journal of cell science. 2011;124:2267–2276. doi: 10.1242/jcs.084285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terbach N, Williams RS. Structure-function studies for the panacea, valproic acid. Biochemical Society transactions. 2009;37:1126–1132. doi: 10.1042/BST0371126. [DOI] [PubMed] [Google Scholar]
- Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, Lin CD, Liao YL, Wang JL, Chau YP, Hsu MT, Hsiao M, Huang HD, Tsou AP. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walmod PS, Foley A, Berezin A, Ellerbeck U, Nau H, Bock E, Berezin V. Cell motility is inhibited by the antiepileptic compound, valproic acid and its teratogenic analogues. Cell motility and the cytoskeleton. 1998;40:220–237. doi: 10.1002/(SICI)1097-0169(1998)40:3<220::AID-CM2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Walmod PS, Skladchikova G, Kawa A, Berezin V, Bock E. Antiepileptic teratogen valproic acid (VPA) modulates organisation and dynamics of the actin cytoskeleton. Cell motility and the cytoskeleton. 1999;42:241–255. doi: 10.1002/(SICI)1097-0169(1999)42:3<241::AID-CM7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Wanet A, Tacheny A, Arnould T, Renard P. miR-212/132 expression and functions: within and beyond the neuronal compartment. Nucleic Acids Res. 2012;40:4742–4753. doi: 10.1093/nar/gks151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Huang Q, Hu Y, Stromberg AJ, Nelson PT. Patterns of microRNA expression in normal and early Alzheimer's disease human temporal cortex: white matter versus gray matter. Acta neuropathologica. 2011;121:193–205. doi: 10.1007/s00401-010-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS letters. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- Wiltse J. Mode of action: inhibition of histone deacetylase, altering WNT-dependent gene expression, and regulation of beta-catenin--developmental effects of valproic acid. Crit Rev Toxicol. 2005;35:727–738. doi: 10.1080/10408440591007403. [DOI] [PubMed] [Google Scholar]
- Woltering JM, Durston AJ. MiR-10 represses HoxB1a and HoxB3a in zebrafish. PloS one. 2008;3:e1396. doi: 10.1371/journal.pone.0001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan PX, Huang LD, Jiang YM, Gutkind JS, Manji HK, Chen G. The mood stabilizer valproic acid activates mitogen-activated protein kinases and promotes neurite growth. J Biol Chem. 2001;276:31674–31683. doi: 10.1074/jbc.M104309200. [DOI] [PubMed] [Google Scholar]
- Zeng Y. Regulation of the mammalian nervous system by microRNAs. Mol Pharmacol. 2009;75:259–264. doi: 10.1124/mol.108.052118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li YY, Zeng HC, Wei J, Wan YJ, Chen J, Xu SQ. MicroRNA expression changes during zebrafish development induced by perfluorooctane sulfonate. Journal of applied toxicology : JAT. 2011;31:210–222. doi: 10.1002/jat.1583. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Xiong Q, Xie P. Analysis of microRNA expression in embryonic developmental toxicity induced by MC-RR. PloS one. 2011;6:e22676. doi: 10.1371/journal.pone.0022676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yuan P, Wang Y, Hunsberger JG, Elkahloun A, Wei Y, Damschroder-Williams P, Du J, Chen G, Manji HK. Evidence for selective microRNAs and their effectors as common long-term targets for the actions of mood stabilizers. Neuropsychopharmacology. 2009;34:1395–1405. doi: 10.1038/npp.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.