Abstract
In our in vitro model for HPV16-mediated transformation, HPV16-immortalized human keratinocytes (HKc/HPV16) give rise to differentiation resistant, premalignant cells (HKc/DR). HKc/DR, but not HKc/HPV16, are resistant to growth inhibition by transforming growth factor beta (TGF-β), due to a partial loss of TGF-β receptor type I. We show that TGF-β activates a Smad-responsive reporter construct in HKc/DR to about 50% of the maximum levels of activation observed in HKc/HPV16. To investigate the functional significance of residual TGF-β signaling in HKc/DR, we compared gene expression profiles elicited by TGF-β treatment of HKc/HPV16 and HKc/DR on Agilent 44k human whole genome microarrays. TGF-β altered the expression of cell cycle and MAP kinase pathway genes in HKc/HPV16, but not in HKc/DR. However, epithelial-mesenchymal transition (EMT) responses to TGF-β were comparable in HKc/HPV16 and HKc/DR, indicating that the signaling pathways through which TGF-β elicits growth inhibition diverge from those that induce EMT in HPV16-transformed cells.
Keywords: HPV, TGF-beta, Smad, Ski, EMT, Human Keratinocytes
Introduction
Cervical cancer ranks second as a leading cause of cancer-related deaths among women worldwide (Vaccarella et al., 2013). High-risk human papillomaviruses (HR-HPV), the etiologic agent of cervical cancer (zur Hausen, 2000), immortalize human keratinocytes (HKc) in culture (Pirisi et al., 1987) (Pirisi et al., 1988) (Kaur and McDougall, 1988). Both in vitro and in vivo, HPV-mediated transformation is elicited by the HR-HPV E6 and E7 oncoproteins, which bind to and functionally inactivate tumor suppressor proteins p53 and pRb, respectively (Scheffner et al., 1990) (Munger et al., 1989). Infection with HR-HPV is necessary but not sufficient to produce cervical cancer, suggesting that additional factors contribute to cervical cancer progression (zur Hausen, 2000). To study the molecular and cellular changes accompanying HPV-mediated transformation, our laboratory has established an in vitro model using HPV16 DNA and normal foreskin HKc. HKc immortalized by transfection with HPV16 DNA (HKc/HPV16) progress towards malignancy via a series of phenotypically well-defined and reproducible steps, including growth factor independent (HKc/GFI) and differentiation resistant (HKc/DR) stages (Pirisi et al., 1988). Interestingly, HKc/DR but not HKc/HPV16 or HKc/GFI undergo malignant conversion upon transfection with v-ras or herpes simplex virus 2 (HSV2) DNA (DiPaolo et al., 1990) (DiPaolo et al., 1989).
Transforming growth factor-beta (TGF-β) is a multifunctional cytokine that regulates numerous biological processes, including wound healing, embryogenesis and immune cell functions in a variety of conditions including fibrotic diseases and cancer (Chuang et al., 2013) (Massague, 2008). In cancer, the biological responses elicted by TGF-β depend on the cell type and surrounding microenvironment. For example, TGF-β inhibits cell proliferation in normal tissue and at early stages of carcinogenesis. However, during progression, tumor cells become partly or completely resistant to growth inhibition by TGF-β, which instead stimulates tumor cell growth and cell survival (see (Wendt et al., 2012) for a recent review). Accordingly, we have previously shown that while HKc/HPV16 are sensitive to the growth inhibitory effects of TGF-β, HKc/DR are resistant (Creek et al., 1994) (Mi et al., 2000) (Hypes et al., 2009).
TGF-β elicits intracellular signals through cell surface receptors of the serine/threonine kinase family and via the Smad proteins, downstream of the receptors, although non-Smad pathways of TGF-β signaling also exist (Massague et al., 2000; Seoane, 2006; Ten Dijke et al., 2002; Zhang, 2009). HKc/DR's complete resistance to growth inhibition by TGF-β is associated with a partial loss of TGF-β receptor type I (TGFBRI) expression (Mi et al., 2000). However, TGF-β signaling is not completely lost in HKc/DR, as TGF-β treatment of these cells partially represses E7 expression (Borger et al., 2000) and decreases Ski protein levels (Baldwin et al., 2004; Chen et al., 2013). Importantly, re-expression of exogenous TGFBRI in HKc/DR fully restores sensitivity to growth inhibition by TGF-β (Mi et al., 2000).
The primary goal of this study was to explore the functional significance of the TGF-β signaling still present in HKc/DR, and the possible differences in gene expression responses elicited by TGF-β in HKc/HPV16 and HKc/DR. Hence, we further explored Smad signaling in HKc/DR, and then compared the gene expression profiles of HKc/HPV16 and HKc/DR treated with or without TGF-β1 on Agilent 4x44k human oligonucleotide microarrays. We found that a 50% reduction in Smad signaling in HKc/DR is associated with a complete loss of gene expression responses to TGF-β related to growth inhibition. In addition, and most interestingly, TGF-β elicits the same EMT and cell motility gene expression profiles in both HKc/HPV16 and HKc/DR. Therefore, in this model of HPV16-mediated transformation, the functional activities of TGF-β that predominate when this cytokine's role “switches” from tumor suppressor to tumor promoter, during cancer progression, are present from early stages of transformation. However, in HKc/HPV16 the growth inhibitory effects of TGF-β predominate, while in HKc/DR TGF-β elicits only EMT and cell motility responses.
Materials and Methods
Cell Culture and TGF-β1 Treatment
The four HKc/HPV16 lines and their corresponding HKc/DR lines used in this study, and the methods for their establishment and culture have been previously described (Pirisi et al., 1988; Pirisi et al., 1987).
For microarray studies, cells were cultured to ~70% confluence and then fed fresh medium without or with 40 pM TGF-β1 (R&D Systems, Inc.) at time 0 and then again at 24 h. Cells were harvested for RNA extraction after 48 h of TGF-β1 treatment. By this time, cells were ~90% confluent.
TGF-β Induction of a Smad-Responsive Reporter Construct
A firefly luciferase reporter construct containing six tandem Smad-binding elements (SBE) cloned upstream of the SV40 promoter (p6SBE-Luc) and a control plasmid, identical to p6SBE-Luc in overall structure, but carrying six mutated inactive SBE (p6SME-Luc; both a gift of Dr. Scott Kern) (Dai et al., 1998) were co-transfected with the Renilla luciferase expression vector pLR-SV40 (Promega) into HKc/HPV16 and HKc/DR in six-well plates. Cells were treated with TGF-β1 or an equivalent amount of 4 mM HCl-1 mg/mL BSA vehicle (control) at the indicated doses 24 h after plating, and harvested for Dual Luciferase Assay (Promega) after 22 h of TGF-β treatment. Firefly luciferase values were normalized for Renilla luciferase and expressed as relative light units (RLU). Results were expressed as fold-change of RLU values of TGF-β treated cells over untreated controls.
Microarray and Data Analysis
RNA samples were obtained from HKc/HPV16d-1, d-2, d-4, and d-5 and their corresponding HKc/DR lines (Pirisi et al., 1988). Comparisons between control and TGF-β treated groups for each HKc/HPV16 and HKc/DR line were replicated using a dye-swap design, thereby giving 16 sets of hybridizations for both the HKc/HPV16 stage and the HKc/DR stage of the in vitro model system. Total RNA was extracted from control and TGF-β1 treated cells using the Total RNA Isolation Mini Kit (Agilent Technologies, Santa Clara, CA). RNA quality and quantity were assessed on an Agilent 2100 Bioanalyzer. Only RNA samples with RNA Integrity Number (RIN) 9.8 or above were used. Total RNA (500 ng per sample) was amplified and labeled using the Agilent Low RNA Input Linear Amplification Kit. RNA Spike-Ins (Invitrogen) was used as an internal control. Hybridization was performed on Whole Human Genome 4x44k 60mer oligonucleotide microarrays (Agilent Technologies, Santa Clara, CA). Images were quantified and normalized using ImaGene v7.5 software (BioDiscovery, Hawthorne, CA). Data were analyzed using GeneSifter software (Geospiza, Seattle, WA) and subjected to KEGG Pathway and Gene Ontology (GO) Analysis. The pathway and ontology analysis are based on a z-score report that identifies significantly changed gene-lists based on the total number of genes on the array. A z-score greater than 2 or less than −2 is considered significant.
Quantitative Real Time PCR
To validate microarray results, real-time RT-PCR was performed for 5 genes (FN1, VIM, ITGB6, INHBA and COL4A1). Reverse transcription was performed with 1 μg total RNA using the iScript™ cDNA synthesis kit (Bio-Rad, Hercules, CA) following the manufacturer's protocol. Serial dilutions of a cDNA sample were used to estimate primer efficiency for each gene. RT-PCR was carried out using gene-specific primers with the iQ-SYBR green kit (Bio-Rad) using the same total RNA used in the microarray study, but without performing whole-RNA amplification. Samples were assayed either in duplicate or triplicate. The Pfaffel quantification method (Pfaffl, 2001) was implemented to normalize all values against those for the reference gene beta-D-glucuronidase (GUSB) to determine the difference in the expression of the gene of interest. The primers were chosen using Primer3 software and their specificity for the selected genes was verified by performing a Blast analysis. The sequences of the primers used are listed in Supplementary Table 1.
Phase Contrast Microscopy
HKc/HPV16 and HKc/DR were grown in 100-mm tissue culture dishes. Cells were treated for 4 days with or without 40 pM TGF-β1 with fresh TGF-β1 administered every 24 h in fresh medium. After 4 days, control and TGF-β1 treated HKc/HPV16 and HKc/DR were examined for changes in morphology under phase contrast using a Zeiss Axiovert 200 inverted microscope (Carl Zeiss, Munich, Germany).
Western Blot Analysis Immunofluorescence
Cell lysates (15 μg protein) were resolved on an 8% SDS-PAGE gel, transferred to PVDF membranes (BioRad) and probed with anti-E-cadherin (Santa Cruz BioTech, Santa Cruz, CA, sc-71008), anti- Fibronectin (Sigma, St. Louis, F3648) or anti-Tubulin (Sigma) antibodies. Proteins were detected using the ECL Western Blotting detection kit (Amersham Biosciences, GE Health Care Life Science, Pittsburg, PA).
Immunofluorescence Studies
HKc/HPV16 and HKc/DR were grown on coverslips pre-coated with Poly-L-lysine and collagen type I (Sigma) overnight at 37° C. The cells were treated for 4 days without or with 40 pM TGF-β1 with administration of fresh TGF-β1 every 24 h. After 4 days, coverslips were fixed with 4% paraformaldehyde (PFA), probed with primary anti-fibronectin antibody (Sigma, F3648) at 1:100 dilution in 2% blocking solution, followed by incubation with Alexa fluor-546 labeled secondary antibody (Invitrogen, Life Technologies Corporation, Grand Island, NY) at 1:250 dilution in 2% blocking solution. Cells were also stained with Alexa Fluor 488-Phalloidin to detect F-actin (Invitrogen, A12379) and nuclei were stained with DAPI (Invitrogen). Coverslips were mounted using Vectashield mounting media (VectorLabs, Burlingame, CA). Images were captured using a LSM Meta 510 Confocal Microscope (Carl Zeiss) and a Zeiss Axiovert 200 inverted fluorescence microscope.
Cell Migration Assay
HKc/HPV16 and HKc/DR were grown to confluence and treated with or without TGF-β1 (40 pM) for 12 h. A wound was then incised by scratching the cell monolayers using a pipette tip. Images were captured under phase contrast microscopy immediately after the incision (time 0) and at 6 h intervals until the wound closed. At least six different regions on each tissue culture dish for each condition were examined and imaged. Image analysis to quantify the assay was conducted using ImageJ (NIH).
Results
TGF-β Activation of a Smad-Responsive Reporter Construct in HKc/HPV16 and HKc/DR
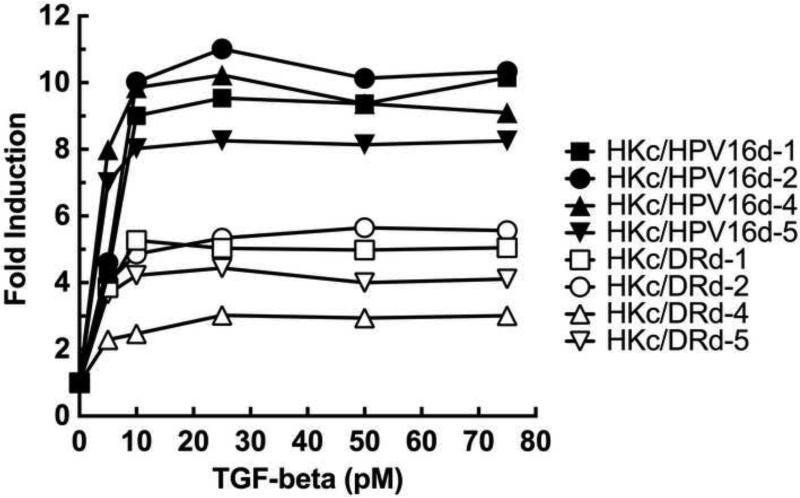
To compare the extent of Smad-dependent signaling in HKc/HPV16 and HKc/DR, we performed dose-response curves of TGF-β1 induction of p6SBE-Luc activity in four independently-derived HKc/HPV16 lines and their corresponding HKc/DR lines. We determined that p6SBE-Luc activity is induced maximally in both HKc/HPV16 and HKc/DR by about 10 pM TGF-β, with no further induction of the reporter by TGF-β1 concentrations as high as 75 pM (Fig. 1). However, while TGF-β1 induced p6SBE-Luc activity by 8.7 to 12.2-fold in the HKc/HPV16 lines, the reporter was only activated by 3.2 to 5.9-fold in the HKc/DR lines (Fig. 1). As expected, the p6SME control plasmid did not show induction by TGF-β1 at any concentration in either HKc/HPV16 or HKc/DR (data not shown).
Figure 1. TGF-β activation of a Smad-responsive luciferase reporter construct in HKc/HPV16 and HKc/DR.
Four HKc/HPV16 and their corresponding HKc/DR lines (d-1, d-2, d-4 and d-5) were transiently transfected in triplicate wells per experimental condition with the p6SBE-Luc reporter construct and pRL-SV40. Cells were treated without or with the indicated concentrations of TGF-β1 24 h after transfection, and harvested for dual luciferase assay after 22 h of TGF-β treatment. Firefly luciferase values, measured in Relative Light Units (RLU), were normalized against Renilla luciferase values, and the results expressed as fold induction over control (without TGF-β).
Identification of Differentially Regulated Genes in HKc/HPV16 and HKc/DR Treated With and Without TGF-β
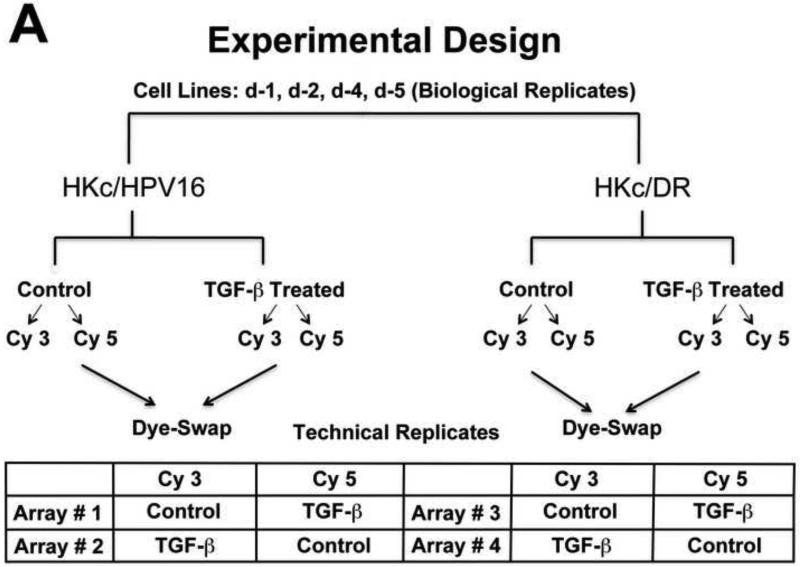
To investigate the functional significance of the TGF-β signaling remaining in HKc/DR, we compared the gene expression profiles of HKc/HPV16 and HKc/DR treated with and without 40 pM TGF-β1 for 48 h. A schematic representation of the microarray study design is presented in Figure 2A. To identify genes that exhibited statistically significant expression changes in TGF-β-treated cells versus controls, we performed a pairwise t test analysis of the microarray data at p <0.05, with a cut-off of two-fold (up or down). All four HKc/HPV16 and HKc/DR dye-swap microarray pairs were selected for analysis, thus n=8 for all culture conditions and cell lines.
Figure 2. Experimental design and initial analysis of microarray experiments.
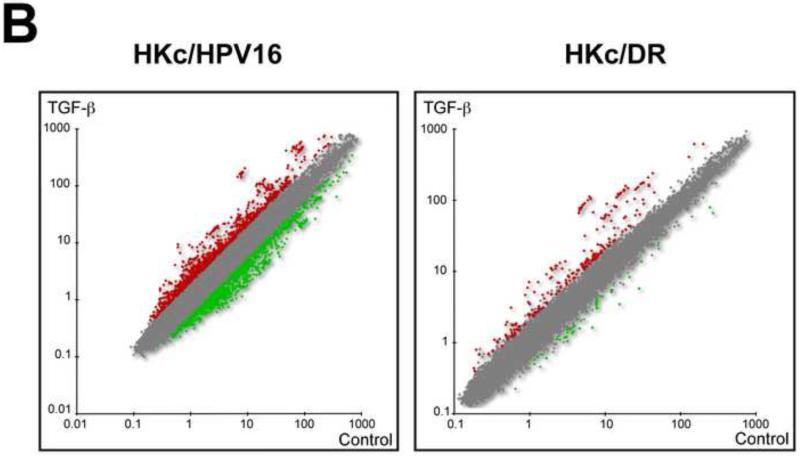
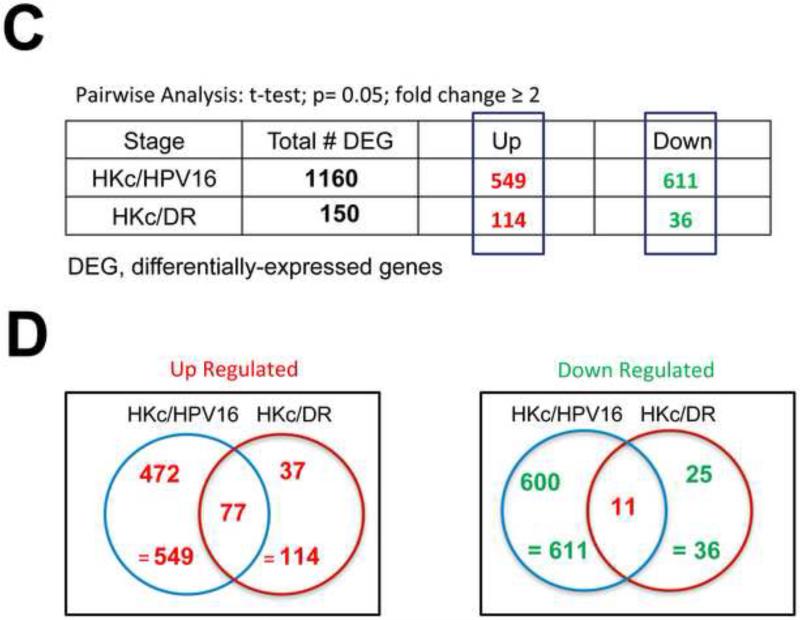
(A) Experimental design: Four separate HKc/HPV16 lines and their respective HKc/DR counterparts were treated without or with 40 pM TGF-β for 48 h before harvesting. Labeled RNA samples, using a dye-swap design, were hybridized to Agilent 4x44k whole human genome microarrays for each of the HKc/HPV16 and HKc/DR lines, resulting in eight microarrays per transformation stage of the model system. RNA amplification, hybridization, washing and scanning for each individual donor (HKc/HPV16 and HKc/DR) were performed at the same time on separate dates. (B) Scatter plot of gene expression levels detected by microarray analysis described in A. Red: genes up-regulated by ≥ 2- fold; green: genes down-regulated by ≥ 2- fold; gray: genes exhibiting no statistically significant change, or < 2-fold change in expression between TGF-β treated and control cells. (C) Numbers of genes significantly changed in HKc/HPV16 and HKc/DR as a consequence of TGF-β treatment. (D) Venn diagrams showing how many genes are up- or down-regulated (≥ 2 fold) in all four HKc/HPV16 and HKc/DR lines after TGF-β treatment, and how many of the TGF-β regulated genes are common between HKc/HPV16 and HKc/DR. red: ≥ 2-fold up-regulated, green ≥ 2-fold down-regulated.
The scatter plots in Figure 2B show a graphical view of the expression levels of all genes on the microarrays, from all four HKc/HPV16 lines and all four HKc/DR lines. Red corresponds to spots with increased signal (TGF-β treated vs control) on the microarray; spots with decreased signal are shown in green, and the gray area represents spots that do not meet the two-fold change cutoff criteria in either direction. For HKc/HPV16 treated with TGF-β1, the scatter plot shows a balanced distribution of up-regulated and down-regulated genes (Fig. 2B, HKc/HPV16). However, many more spots had an increased signal than a decreased signal in HKc/DR following TGF-β1 treatment (Fig. 2B, HKc/DR). Furthermore, it was visually evident that the number of genes whose expression changed by more than 2-fold in TGF-β1–treated cells was much higher in HKc/HPV16 than in HKc/DR (Fig. 2B).
Pairwise comparisons identified 1160 differentially expressed genes (DEG) that had changed by at least two-fold after treatment with TGF-β in HKc/HPV16: 549 genes were up-regulated and 611 genes were down-regulated. In contrast, only 150 DEG (about 8-fold less than found in HKc/HPV16) were identified following TGF-β treatment of HKc/DR: 114 genes were up-regulated and only 36 genes were down-regulated (Fig. 2C). We generated Venn diagrams by combining the separate gene lists obtained from the Genesifter analysis, and then manually identified the common genes between the two stages (Fig. 2D). We found 77 genes that were up-regulated (listed in Table S2), and 11 genes down-regulated (listed in Table S3) by TGF-β in both HKc/HPV16 and HKc/DR (Fig. 2D). In addition, 62 genes (37 up-regulated and 25 down-regulated) were regulated by TGF-β1 in HKc/DR but not in HKc/HPV16 (Table S4).
Validation of TGF-β Induced Genes by Real-Time PCR
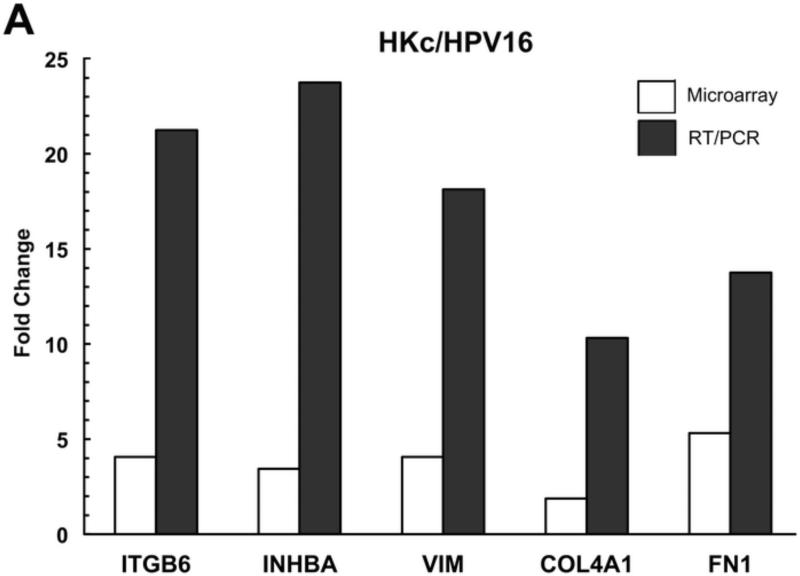
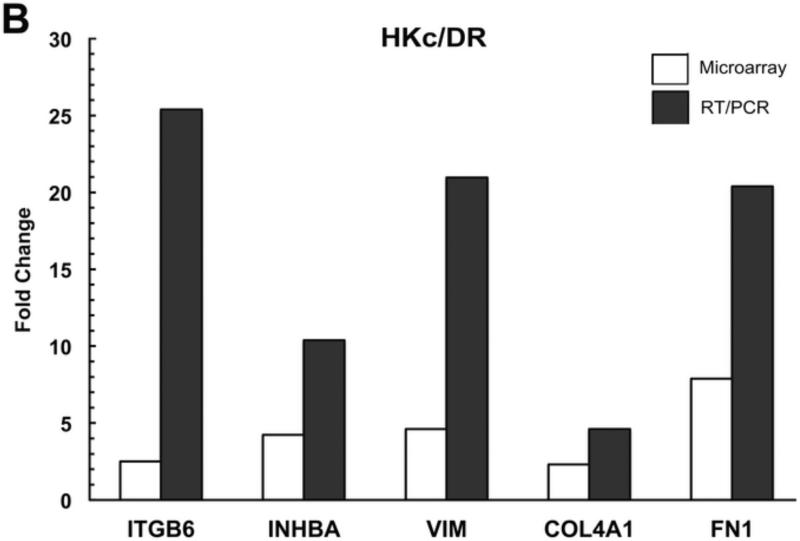
To validate the array data, we performed quantitative RT-PCR for five genes (ITGB6, INHBA, VIM, COL4A1, and FN1) whose expression was increased by TGF-β in both HKc/HPV16 and HKc/DR. We confirmed by RT-PCR that TGF-β up-regulated the expression of these five genes in comparison with untreated controls in both HKc/HPV16 (Fig. 3A) and HKc/DR (Fig. 3B). A comparison of the fold induction of each of these five genes as determined by microarray and RT-PCR found that induction of gene expression as determined by microarrays was consistently several fold less than that determined by RT-PCR (Fig. 3).
Figure 3. RT/PCR validation of microarray results.
Fold-change induced by TGF-β of the expression of a panel of genes, determined by microarray analysis (open bars) and RT-PCR (solid bars) in HKc/HPV16 (A) and HKc/DR (B).
Pathways and Biological Processes Affected by TGF-β in HKc/HPV16 and HKc/DR
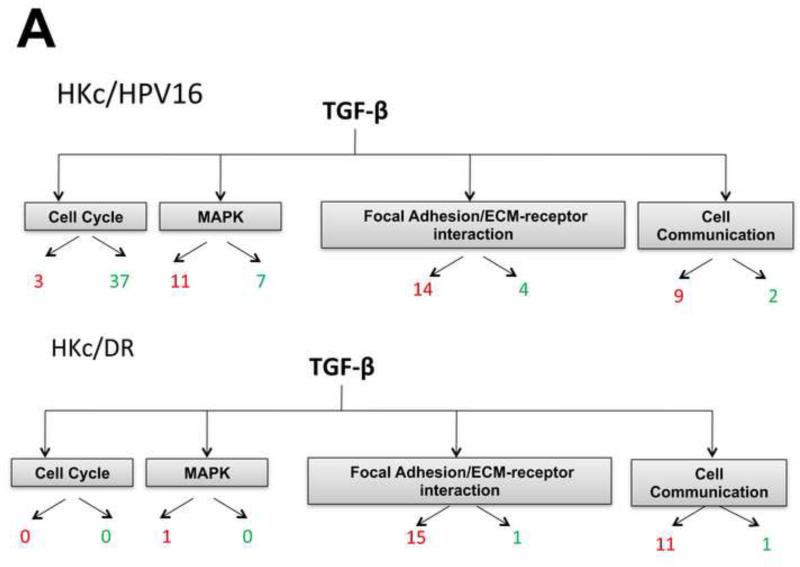
To explore which pathways were affected by TGF-β1 treatment, we categorized the significant DEG using the KEGG pathway analysis tool within the GeneSifter software. Pathways with z-scores >2 or < −2 in either HKc/HPV16 or HKc/DR were compared. In HKc/HPV16, pathways altered by TGF-β1 included the cell cycle and mitogen-activated protein kinase (MAPK) pathways, with many cell cycle genes being down-regulated, as expected, by TGF-β1 (Fig. 4A, HKc/HPV16). However, the same pathways were virtually non-responsive to TGF-β1 treatment in HKc/DR (Fig. 4A, HKc/DR). This was not surprising since previous studies from our laboratory demonstrated that HKc/HPV16 are as sensitive to the growth inhibitory effects of TGF-β as normal HKc, while HKc/DR are completely resistant (Creek et al., 1995; Mi et al., 2000). Tables S5 and S6 present a list of the cell-cycle and MAPK genes that were significantly up- or down- regulated in response to TGF-β1 in HKc/HPV16, but not in HKc/DR.
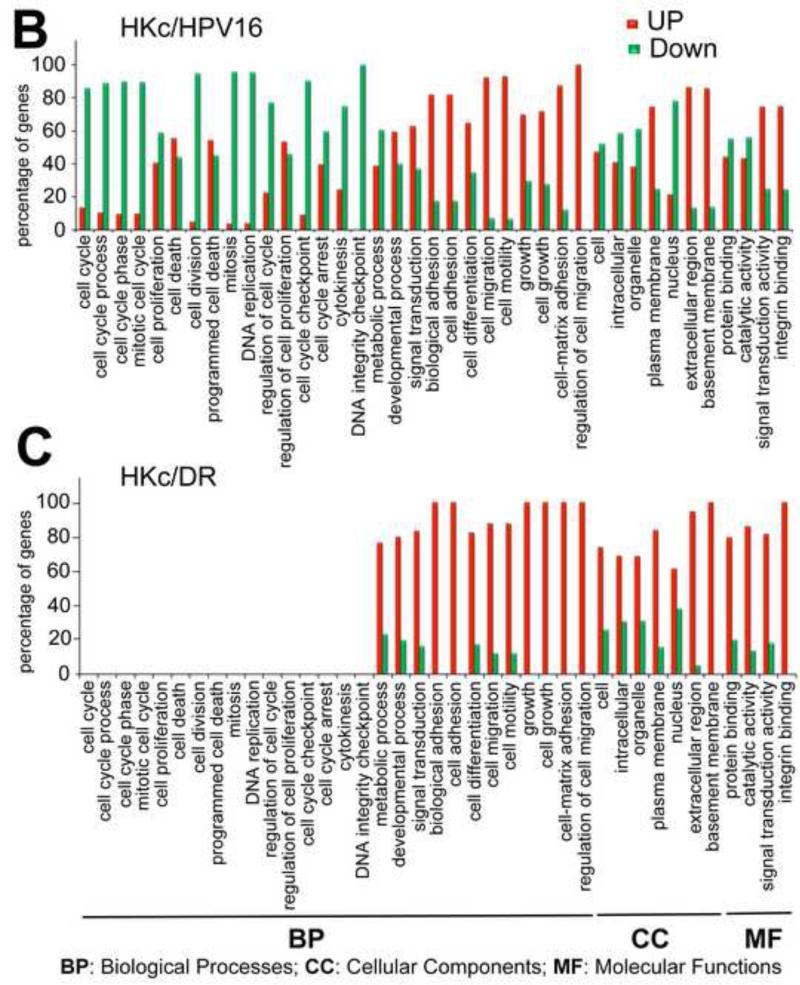
Figure 4. KEGG pathway analysis of differentially expressed genes.
(A) Summary of KEGG analysis results for genes belonging to the cell cycle, MAPK, focal adhesion/ECM-receptor interaction and cell communication pathways found changed by TGF-β treatment in HKc/HPV16 (upper panel) and HKc/DR (lower panel). Red: ≥ 2-fold up-regulated; green β 2-fold down-regulated by TGF-β. (B, C) Gene ontology analysis of differentially expressed genes as determined using the GeneSifter software. The y-axis represents the percentage of genes changed up or down, within each group of genes (in each cellular process) that changed in response to 48 h of TGF-β treatment (40 pM) in HKc/HPV16 (B) and HKc/DR (C) by β 2-fold. Data are grouped into different ontology groups (x-axis) and each is further separated into increased (red bars) and decreased (green bars) expression.
Interestingly, pathways related to EMT (focal adhesion/ECM-receptor interaction and cell communication pathways) were induced by TGF-β to a similar extent in both HKc/HPV16 and HKc/DR, based upon the number of significantly changed genes (Fig. 4A, compare HKc/HPV16 with HKc/DR). Table S7 shows a list of the genes changed in these pathways that were up- or down- regulated by TGF-β1 in HKc/HPV16 and HKc/DR.
GO analysis confirmed and expanded upon the results of the KEGG pathway analysis: genes down-regulated by TGF-β1 in HKc/HPV16 were related to the cell cycle (Fig. 4B) and many genes belonging to processes such as cell adhesion, cell-matrix adhesion or migration and other related processes were up-regulated in HKc/HPV16 (Fig. 4B). In HKc/DR, genes belonging to cell cycle-related processes were not responsive to TGF-β treatment, while a majority of the genes belonging to the cell-adhesion, cell-matrix adhesion or migration, and other related processes were up-regulated to a similar extent in HKc/DR, compared to HKc/HPV16 (Fig. 4C). Many of the specific gene targets in the TGFβ induced pathways (Fig. 4A-C, Table S7) such as FN1, VIM, COL4A1, and ITGB6 participate in cell-cell/cell-matrix adhesion and migration processes involved in EMT. Therefore, the microarray data suggest that TGF-β1 inhibits cell proliferation and promotes EMT in HKc/HPV16, but promotes only EMT in HKc/DR.
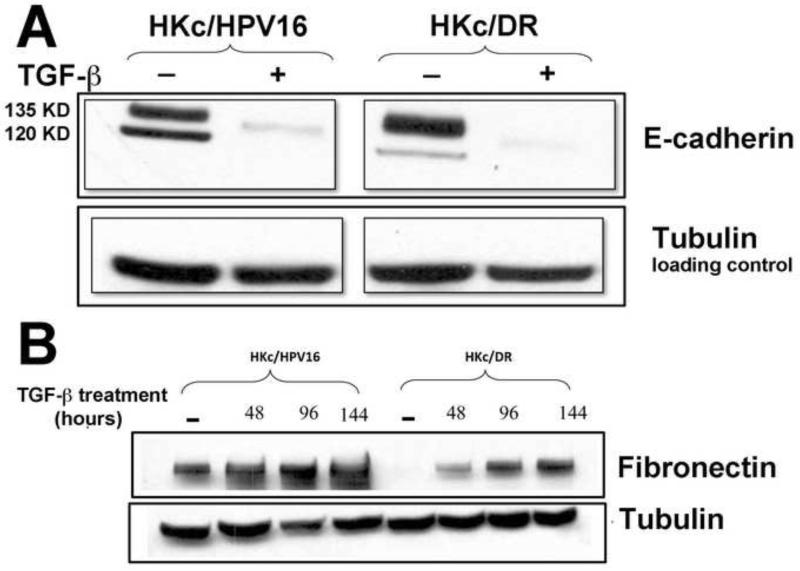
Decreased Protein Levels of E-cadherin and Increased Protein Levels of Fibronectin in both HKc/HPV16 and HKc/DR treated with TGF-β
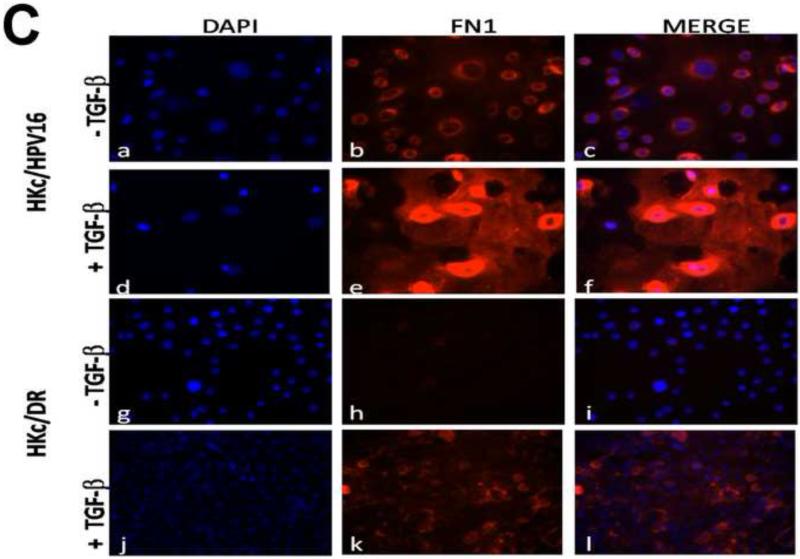
To functionally validate the role of TGF-β as an inducer of EMT at both early and late stages of HPV-mediated transformation in our in vitro model, we studied events associated with EMT in HKc/HPV16 and HKc/DR treated without and with TGF-β1. At the protein level, E-cadherin, an epithelial cell marker, was significantly reduced in HKc/HPV16 and HKc/DR following TGF-β1 treatment (40 pM for 48 h) (Fig. 5A). In addition, the mesenchymal marker fibronectin (FN1) was induced by TGF-β1 in a time-dependent manner in HKc/HPV16 and HKc/DR (Fig. 5B). Interestingly, the basal cellular levels of fibronectin were much lower in HKc/DR than in HKc/HPV16, so that following 96 h of TGF-β1 treatment, fibronectin levels in HKc/DR are about the same as in HKc/HPV16 in the absence of TGF-β (Fig. 5B). These changes in fibronectin expression determined by Western blot analysis were also confirmed by immunofluorescence confocal analysis of HKc/HPV16 and HKc/DR treated without or with 40 pM TGF-β1 for 96 h (Fig. 5C). These findings show that TGF-β1 treatment of both HKc/HPV16 and HKc/DR induces markers of EMT.
Figure 5. TGF-β decreases E-cadherin and increases fibronectin protein levels in HKc/HPV16 and HKc/DR.
Whole-protein lysates were prepared from HKc/HPV16 and HKc/DR treated without or with TGF-β (40 pM) for 48 h (A) or for the indicated times (B) and Western blot analysis was performed for E-cadherin (A), fibronectin (B) or tubulin (A, B) as a loading control. (C) Control and TGF-β treated (40 pM for 96 h) HKc/HPV16 and HKc/DR were fixed in 4% paraformaldehyde. Slides were stained with DAPI to visualize nuclei (blue; panels a, d, g, and j) and with anti-fibronectin antibody (FN1) (red; panels b, e, h, and k). Merged images are presented in panels c, f, i, and l.
Effect of TGF-β on Cell Morphology in HKc/HPV16 and HKc/DR
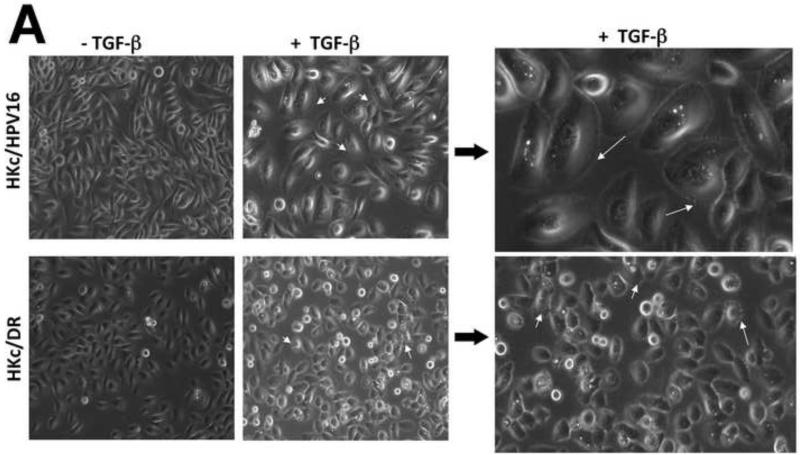
We next compared the morphological changes induced by treatment of HKc/HPV16 and HKc/DR with TGF-β1. We cultured HKc/HPV16 and HKc/DR in the absence and presence of TGF-β1 (40 pM) for 4 days and examined the cells under phase contrast light microscopy (Fig. 6A). After TGF-β treatment, HKc/HPV16 exhibited a significant increase in cell size compared to the control. Furthermore, HKc/HPV16 also showed the presence of “ruffles” along the cell membrane (Fig. 6A, top panel). In contrast, treatment of HKc/DR with TGF-β did not result in any marked changes in cell size. However, more prominent cell surface protrusions were observed in HKc/DR treated with TGF-β1 (Fig. 6A, lower panel) compared to their respective controls and to TGF-β1 treated HKc/HPV16.
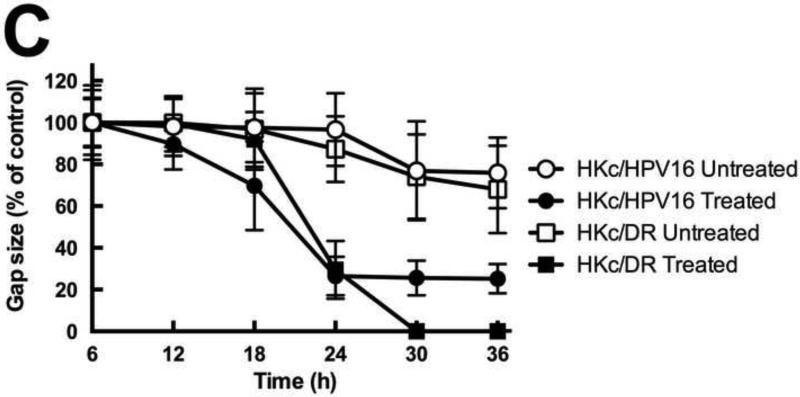
Figure 6. TGF-β induces morphological changes, re-organizes actin cytoskeleton, and enhances cell migration in HKc/HPV16 and HKc/DR.
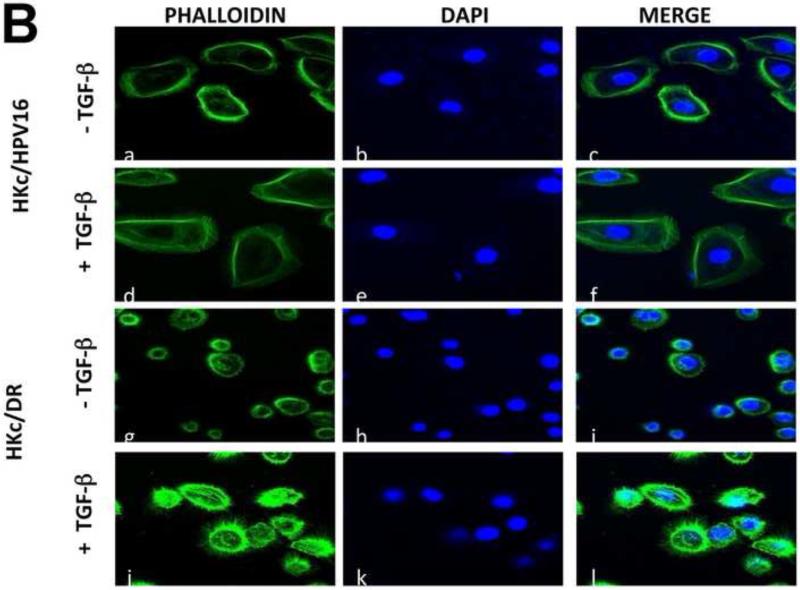
(A) Cells were treated with or without TGF-β (40 pM for 4 days) and examined under phase contrast using a Zeiss Axiovert 200 inverted microscope. White arrows show projections protruding out from the cell surface of HKc/HPV16 and HKc/DR. (B) F-actin was stained with Alexa fluor-488 labeled Phalloidin and nuclei with DAPI in HKc/HPV16 and HKc/DR, which were then visualized under a confocal microscope. F-actin, green: panels a, d, g and j; nuclei, blue: panels b, e, h and k. Merged images are presented in panels c, f, i, and l. (C) Quantification of scratch assay. HKc/HPV16 and HKc/DR were grown to confluence and treated with or without 40 pM TGF-β1 for 12 h. Wounds were then incised by scratching the cell monolayer with a pipet tip, and incubation with or without TGF-β1 continued for 36 h. Images were captured under phase-contrast microscopy at 6 h intervals after the incision and quantified with ImageJ. At least ten measurements of the width of the gap were assessed for each time point, starting at the 6 h time point and continuing until 36 h after the scratch.
TGF-β Causes the Actin Cytoskeleton to Reorganize in HKc/HPV16 and HKc/DR
During EMT, the actin cytoskeleton is re-arranged. Therefore, we sought to determine whether TGF-β causes re-arrangements of the actin cytoskeleton in HKc/HPV16 and HKc/DR. We stained the cells for F-actin using Alexa fluor 488-phalloidin, and examined the effect of TGF-β1 on the actin cytoskeleton by confocal microscopy. In the absence of TGF-β1, HKc/HPV16 showed an orderly organization of cortical F-actin filaments (Fig. 6B). Treatment of HKc/HPV16 with TGF-β resulted in filopodia-like protrusions seen as slender “brushlike” projections on the cell surface (Fig. 6B). Some cortical actin filaments were also detected in TGF-β–treated HKc/HPV16, although to a lesser extent than in controls. HKc/DR were characterized by a radial network of F-actin filaments at the plasma membrane, in the absence of TGF-β. Upon treatment with TGF-β1, HKc/DR displayed intense cortical actin staining, as well as a marked increase in the number and density of lamellipodia or actin-rich microspikes (Fig. 6B), indicative of EMT. The results show that TGF-β modulates cell morphology and reorganizes the actin cytoskeleton in both HKc/HPV16 and HKc/DR.
TGF-β Increases Cell Migration in both HKc/HPV16 and HKc/DR
An important property of cells undergoing EMT is their ability to migrate. To determine whether TGF-β increased cell migration in HKc/HPV16 and HKc/DR, we performed an in vitro scratch assay. Confluent HKc/HPV16 and HKc/DR were treated with or without 40 pM TGF-β for 12 h, then cell monolayers were scratched to produce wounds, and images were captured under phase contrast microscopy immediately after the scratch (time 0) and at 6 h intervals until the wound closed. The gap width was measured by ImageJ on images collected at each time point, starting at 6 h after the scratch and at 6 h intervals thereafter, up to 36 h of observation. TGF-β treatment led to increased migratory behavior and wound closure in both HKc/HPV16 and HKc/DR compared to their respective controls (Fig. 6C). The scratch closed with approximately the same dynamics in HKc/HPV16 and HKc/DR treated with TGF-β, HKc/DR closed the gap somewhat faster than HKc/HPV16 and more completely (Fig. 6C). Untreated cells also behaved in comparable ways, failing to close the gap during the time of observation (Fig. 6C).
Discussion
Relative roles of E7 and TGF-β in the control of proliferation of HPV16-transformed cells
A major advantage of our model for HPV16-mediated transformation of human cells is that it allows for studies of the interplay of HPV16 oncoproteins with cellular pathways key to growth control, transformation, and progression. The results we present here, combined with those we published over the years about the role of TGF-β in this model (Creek et al., 1995) (Mi et al., 2000) (Baldwin et al., 2004; Hypes et al., 2009) (Chen et al., 2013) allow us to begin describing how the interplay of HPV oncoproteins and TGF-β drives in vitro progression of HPV16-transformed cells. In normal HKc, TGF-β causes marked (but reversible) growth inhibition (Shipley et al., 1986) (Batova et al., 1992) and promotes EMT (Fukawa et al., 2012). In HPV16-immortalized cells, E7-mediated degradation of RB (Munger et al., 1989) and E7 interference with p300 (Bernat et al., 2003) (Fera and Marmorstein, 2012) (Pouponnot et al., 1998) and with the Smads (Habig et al., 2006) (Lee et al., 2002) may be expected to produce resistance to growth inhibition by TGF-β from the very beginning of the transformation process. However, we have shown that early passage HKc/HPV16 are just as sensitive to growth inhibition by TGF-β as normal HKc (Creek et al., 1995) (Mi et al., 2000) but progressively become resistant to growth inhibition by TGF-β during in vitro progression. The explanation for this apparent paradox may reside in the observation that TGF-β inhibits E6/E7 expression by a mechanism that involves decreased binding of NF1/Ski complexes with the HPV16 upstream regulatory region (URR) (Baldwin et al., 2004). This decrease is due primarily to a dramatic loss of Ski protein produced by TGF-β treatment of HKc, including HKc/HPV16 and HKc/DR (Chen et al., 2013). Ski complexes with NF1 and increases its transcriptional activity on the HPV16 URR, therefore a loss of Ski translates into decreased E6/E7 transcription (Baldwin et al., 2004). Our data thus far support the interpretation that the activities of HPV16 E7 and TGF-β exist in a balance: E7 interferes with TGF-β inhibition of growth; TGF-β, in turn, decreases E7 expression. Therefore, TGF-β treatment of HKc/HPV16 not only increases the expression of CDK inhibitors, but also inhibits E6/E7 expression, lessening the impact of the HPV oncoproteins on Rb and p53. We postulate that this balance of activities in HKc/HPV16 leads to growth inhibition by TGF-β that is comparable in magnitude to that of normal HKc, although elicited through somewhat different mechanisms.
As HKc/HPV16 progress toward the HKc/DR phenotype, E6/E7 levels rise (our unpublished observation). We postulate that this increase in E6/E7 expression may be due to the fact that Ski is much more abundant in HKc/DR than in HKc/HPV16 (Baldwin et al., 2004) (Chen et al., 2013) and consequently NF1/Ski-mediated activation of the URR may be more robust. Even though TGF-β still causes Ski degradation in HKc/DR, the levels of Ski in TGF-β treated HKc/DR are comparable to those of untreated normal HKc (Chen et al., 2013). Accordingly, TGF-β dramatically suppresses E7 expression in HKc/HPV16, but not that much in HKc/DR (Borger et al., 2000). During progression to HKc/DR, as E6 levels rise, p53 protein levels decline. We and others have shown that a loss of p53 causes YY1 protein levels to increase (Sui et al., 2004) (Bheda et al., 2008). This leads to a global shift in cell responses co-regulated by YY1, which has been shown to interfere with TGF-β and BMP signaling, preventing TGF-β induction of differentiation in HKc (Kurisaki et al., 2003).
During in vitro progression from HKc/HPV16 to HKc/DR, HKc/HPV16 progressively lose TGFBRI by mechanisms directly dependent upon E6/E7 expression (Hypes et al., 2009; Mi et al., 2000). Consequently, Smad signaling decreases to about 50% of normal (shown here); Ski levels rise; E6/E7 levels increase; and sensitivity to growth control by TGF-β decreases until, at the HKc/DR stage, TGF-β no longer suppresses growth and only partially inhibits HPV16 E7 expression (Borger et al., 2000; Creek et al., 1995). Ski's role in this process is relatively clear. Whether and how YY1 participates in these processes, and what other factors are involved remain to be investigated.
During progression of HKc/HPV16 to HKc/DR we observed a marked decrease in the basal, steady-state levels of fibronectin protein, although TGF-β stimulated fibronectin expression at both stages of in vitro progression. Although initially puzzling, this observation can be probably viewed in the same context as those discussed above, as a reversible change depending on the changing levels of E6/E7 and their interactions with cellular proteins. It has been reported that the HPV oncoproteins induce EMT markers (Hellner et al., 2009) therefore a robust expression of fibronectin in HKc/HPV16 is not surprising. In HKc/DR, which express E7 at higher levels, fibronectin expression in the absence of TGF-β is very low. This apparently contradictory behavior finds support in the literature, as it has been reported that transient expression of E7 results in inhibition of fibronectin transcription, and that fibronectin promoter activity is lower in HPV-positive cell lines than in HPV-negative ones (Rey et al., 2000). We postulate that the apparent contradiction between these two published reports may result from different levels of E7 being expressed in the different models utilized in these two studies. While this conclusion remains to be confirmed in a single experimental system, the observations summarized here above bring forth the idea that changing levels of E7 (and E6, as well) during progression of HPV-transformed cells lead to different gene/protein expression landscapes that, in turn, profoundly affect cell behavior.
Importantly, all of these interactions are based upon the balance of activities of cellular factors with HPV oncoproteins, and therefore are generally reversible, as opposed to a permanent loss of tumor suppressor gene activity due to gene deletion or mutation (as for example with p53 in most solid tumors) or increased oncogene activity due to mutation, gene amplification or insertional activation.
Differential effects of TGF-β on cyclins in HKc/HPV16 and HKc/DR
As expected, gene expression analysis shows that TGF-β down-regulates the expression of cyclins A1, A2, B1 and B2 and up-regulates the expression of cyclin-dependent kinase inhibitors p21, p57 and p15 in HKc/HPV16, but not in HKc/DR (Table S5). In normal epithelial cells, TGF-β mediates G1-arrest by the Smad-dependent induction of cyclin-dependent kinase inhibitors p21 and p15 (reviewed in (Massague and Gomis, 2006)). Previous studies have demonstrated that TGF-β decreases the expression of c-myc in normal murine mammary gland epithelial cells as well as NIH3T3 cells, also by Smad-dependent mechanisms (Hsu et al., 1994). Consistent with our growth data, our microarray results show inhibition of myc expression by TGF-β in HKc/HPV16, but not in HKc/DR. In HKc/HPV16, TGF-β also suppressed the expression of cdc2, a well-established cell cycle regulator important for cell division, by approximately 5-fold. Binding of cdc2 to cyclin B1 is required for its activity. Cyclin B1 was also down-regulated by 5-fold by TGF-β in HKc/HPV16 (Table S5). Up-regulation of cell cycle control genes such as CDKN1A (p21), CDKN2B (p15) and down-regulation of myc by TGF-β are known to be mediated via Smad activation (reviewed in (Massague and Gomis, 2006)).
Stimulation of growth and EMT by TGF-β in tumor cells
Originally reported as a tumor suppressor, TGF-β can also stimulate growth of tumor cells depending on the cell type. During progression tumor cells break through the basement membrane, migrate and establish themselves at new sites where they lay down an extracellular matrix (ECM) (Dawes et al., 2007; Garamszegi et al., 2009). TGF-β can rearrange the cytoskeleton and stimulate cell migration and invasion in vitro and in vivo in human breast cancer, colon cancer and pancreatic cancer (Garamszegi et al., 2009; Kalluri and Neilson, 2003; Kang and Massague, 2004; Lee et al., 2008; Thiery, 2003; Wu and Zhou, 2008; Yang and Weinberg, 2008). Matrix metalloproteases (MMPs) play a role in degrading the basement membrane (Kalluri and Neilson, 2003; Thiery, 2003). The expression of MMP9 was found to be upregulated by TGF-β at both the HKc/HPV16 and HKc/DR stages, implicating TGF-β in the processes of tumor progression in our model. Data from several microarray studies have demonstrated induction of genes involved in ECM by TGF-β (Dawes et al., 2007; Kalluri and Neilson, 2003). We show here that treatment of HKc/HPV16 and HKc/DR with TGF-β up-regulated the expression of genes involved in ECM and cell adhesion processes including: collagens (COL4A1), integrins (ITGAV, ITBG6), and laminins (LAMA3, LAMC2). Integrins serve as cell surface receptors for ECM components, by which the cells attach to and migrate across the ECM (Dawes et al., 2007). In addition, the expression of vimentin (intermediate filament), N-cadherin (mesenchymal marker) and fibronectin (ECM component and mesenchymal marker) was also induced in both HKc/HPV16 and HKc/DR (Table S2). Increased expression of TGF-β1 has been observed in fibrotic diseases as well as human cancers (Hsu et al., 1994; Khalil et al., 2001; Oft et al., 1998) and endogenous TGF-β1 production after exogenous stimulation by TGF-β1 has been noted to support EMT (Yao et al., 2004). Our microarray results also demonstrate increased levels of TGF-β1 mRNA following TGF-β1 treatment of HKc/HPV16 and HKc/DR (Table S2) suggesting a self-induction of TGF-β expression. It is possible that for the maintenance of EMT, HPV16 transformed cells may require continuous TGF-β production which is stimulated in an autocrine or paracrine manner.
TGF-β modulation of E-cadherin expression
Epithelial cells are sealed to one another by adherent junctions, as well as tight junctions (Thiery, 2003; Yang and Weinberg, 2008). E-cadherin is a hallmark epithelial marker, which forms tight cell-cell associations with E-cadherin of neighboring epithelial cells, thus preventing the dissociation and movement of epithelial cells from their origin. An early event in EMT is the disruption of epithelial junctions. An increasing body of literature documents that E-cadherin repression in several carcinomas correlates well with tumor progression (Ewing et al., 1995; Kasai et al., 2005). In vitro studies show that treatment of human epithelial cells of various origins with TGF-β results in the down-regulation of E-cadherin expression (Bhowmick et al., 2001; Kasai et al., 2005; Miettinen et al., 1994). Consistent with previous reports (Bhowmick et al., 2001; Kasai et al., 2005; Miettinen et al., 1994), we show that 48 h treatment with TGF-β induces loss of E-cadherin protein in both HKc/HPV16 to HKc/DR. However, our microarray data did not reveal any of the known E-cadherin repressors such as Snail, Slug, and Twist (Wu and Zhou, 2008) as significantly changed (below 2-fold cut-off) at the mRNA level, following TGF-β treatment of HKc/HPV16 and HKc/DR. Therefore, the mechanism by which TGF-β mediates down-regulation of E-cadherin in our model remains to be determined. It has been shown that TGF-β can disrupt the stability of E-cadherin junctions via Rho-dependent mechanisms (Bruewer et al., 2004). Furthermore, studies using siRNA against Smad2 have shown that Smad2 inhibition suppresses TGF-β-mediated EMT (Kasai et al., 2005). Concomitant with the loss of E-cadherin expression, HKc/HPV16 and HKc/DR expressed increased levels of the mesenchymal marker fibronectin after treatment with TGF-β. It has also been demonstrated that overexpressing Smad3 increases fibronectin expression (Uemura et al., 2005). These results are in accordance with several published reports on the effects of TGF-β during EMT (Frolik et al., 1984; Hsu et al., 1994; Thiery, 2003; Zavadil and Bottinger, 2005).
TGF-β-mediated reorganization of the cytoskeleton
Beyond changes in phenotypic markers, we found that TGF-β treatment of HKc/HPV16 and HKc/DR resulted in the reorganization of the actin cytoskeleton from cortical actin to actin microspikes, and induced cell migration, which is dependent on the dynamics of the actin cytoskeleton. In line with its ability to destabilize adherens junctions, TGF-β mediates actin re-organization via the Rho-GTPases (Edlund et al., 2002; Vardouli et al., 2008). Therefore it is possible that TGF-β-induced cell migration in HKc/HPV16 and HKc/DR may be the result of TGF-β mediated Rho-dependent mechanisms that reduce E-cadherin expression and re-organize the actin cytoskeleton.
Conclusions
This study integrates microarray results with morphological and biochemical observations to show that TGF-β switches from a growth suppressor and inducer of EMT to solely a stimulator of EMT during in vitro progression of HPV16-transformed HKc. This switch occurs in the presence of considerable residual SMAD signaling which, however, is no longer sufficient to cause growth inhibition, known to occur through canonical TGF-β signaling pathways. Of particular importance is the finding that TGF-β not only inhibits cell proliferation, but also induces several features of EMT at early stages of HPV16-mediated transformation. Hence, the gene expression patterns demonstrated by our microarray analysis suggest that branches of the same signaling pathways are cut-off, rather than entirely different pathways being activated by TGF-β in HKc/DR. TGF-β signaling may branch within the Smad dependent route, because many of the gene expression changes identified in HKc/DR are known to be Smad-mediated, at least in other systems (Massague and Gomis, 2006). Alternatively, non-canonical signaling pathways may take over at the HKc/DR stage, with the residual Smad signaling playing at best an accessory role in the process of EMT induction. While the overall patterns of gene expression we observed in response to TGF-β recapitulate mechanisms of TGF-β action that have been identified in other systems, here the interactions of TGF-β signaling pathways with responses elicited by E6 and E7 provide unique circumstances that dictate functional effects specific to HPV-mediated cancers. In this context, it will be important to dissect at which branch point/s in the TGF-β signaling cascade, TGF-β activities bifurcate to exert antiproliferative and proEMT actions in HPV-transformed cells.
Supplementary Material
Research Highlights.
TGF-β inhibits growth of early HPV16-transformed human keratinocytes (HKc/HPV16) but not premalignant HKc/DR
Smad signaling in HKc/DR in response to TGF-β is about 50% of that observed in HKc/HPV16
TGF-β elicits gene expression changes indicative of EMT in both HKc/HPV16 and HKc/DR
All cell cycle-related gene expression responses to TGF-β are abolished in HKc/DR
Acknowledgements
We thank Christian A. Graves for critical reading of the manuscript, and Scott Kern for the pSBE and pSME plasmids. Grant Support: This work was funded by grants 1R01CA89502 and 1P20MD001770 from the National Institutes of Health. Quality control of microarray data analysis was provided by Dr. Pierre Rivaillier, of the Bioinformatics Core of SC INBRE (P20GM103499) from which the South Carolina College of Pharmacy/USC DNA Microarray Facility also receives partial support. The sponsors had no role in the design or execution of the experiments described in this paper, or in the interpretation of the results.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: No potential conflicts of interest were disclosed.
References
- Baldwin A, Pirisi L, Creek KE. NFI-Ski interactions mediate transforming growth factor beta modulation of human papillomavirus type 16 early gene expression. Journal of virology. 2004;78:3953–3964. doi: 10.1128/JVI.78.8.3953-3964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batova A, Danielpour D, Pirisi L, Creek KE. Retinoic acid induces secretion of latent transforming growth factor beta 1 and beta 2 in normal and human papillomavirus type 16-immortalized human keratinocytes. Cell growth & differentiation : the molecular biology journal of the American Association for Cancer Research. 1992;3:763–772. [PubMed] [Google Scholar]
- Bernat A, Avvakumov N, Mymryk JS, Banks L. Interaction between the HPV E7 oncoprotein and the transcriptional coactivator p300. Oncogene. 2003;22:7871–7881. doi: 10.1038/sj.onc.1206896. [DOI] [PubMed] [Google Scholar]
- Bheda A, Creek KE, Pirisi L. Loss of p53 induces epidermal growth factor receptor promoter activity in normal human keratinocytes. Oncogene. 2008;27:4315–4323. doi: 10.1038/onc.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME, Arteaga CL, Moses HL. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol Biol Cell. 2001;12:27–36. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borger DR, Mi Y, Geslani G, Zyzak LL, Batova A, Engin TS, Pirisi L, Creek KE. Retinoic acid resistance at late stages of human papillomavirus type 16-mediated transformation of human keratinocytes arises despite intact retinoid signaling and is due to a loss of sensitivity to transforming growth factor-beta. Virology. 2000;270:397–407. doi: 10.1006/viro.2000.0282. [DOI] [PubMed] [Google Scholar]
- Bruewer M, Hopkins AM, Hobert ME, Nusrat A, Madara JL. RhoA, Rac1, and Cdc42 exert distinct effects on epithelial barrier via selective structural and biochemical modulation of junctional proteins and F-actin. Am J Physiol Cell Physiol. 2004;287:C327–335. doi: 10.1152/ajpcell.00087.2004. [DOI] [PubMed] [Google Scholar]
- Chen Y, Pirisi L, Creek KE. Ski protein levels increase during in vitro progression of HPV16-immortalized human keratinocytes and in cervical cancer. Virology. 2013 doi: 10.1016/j.virol.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang PY, Menon MC, He JC. Molecular targets for treatment of kidney fibrosis. Journal of molecular medicine. 2013;91:549–559. doi: 10.1007/s00109-012-0983-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creek KE, Geslani G, Batova A, Pirisi L. Progressive loss of sensitivity to growth control by retinoic acid and transforming growth factor-beta at late stages of human papillomavirus type 16-initiated transformation of human keratinocytes. Advances in experimental medicine and biology. 1995;375:117–135. doi: 10.1007/978-1-4899-0949-7_11. [DOI] [PubMed] [Google Scholar]
- Creek KE, Jenkins GR, Khan MA, Batova A, Hodam JR, Tolleson WH, Pirisi L. Retinoic acid suppresses human papillomavirus type 16 (HPV16)-mediated transformation of human keratinocytes and inhibits the expression of the HPV16 oncogenes. Advances in experimental medicine and biology. 1994;354:19–35. doi: 10.1007/978-1-4899-0939-8_2. [DOI] [PubMed] [Google Scholar]
- Dai JL, Turnacioglu KK, Schutte M, Sugar AY, Kern SE. Dpc4 transcriptional activation and dysfunction in cancer cells. Cancer research. 1998;58:4592–4597. [PubMed] [Google Scholar]
- Dawes LJ, Elliott RM, Reddan JR, Wormstone YM, Wormstone IM. Oligonucleotide microarray analysis of human lens epithelial cells: TGFbeta regulated gene expression. Mol Vis. 2007;13:1181–1197. [PubMed] [Google Scholar]
- DiPaolo JA, Woodworth CD, Popescu NC, Koval DL, Lopez JV, Doniger J. HSV-2-induced tumorigenicity in HPV16-immortalized human genital keratinocytes. Virology. 1990;177:777–779. doi: 10.1016/0042-6822(90)90548-6. [DOI] [PubMed] [Google Scholar]
- DiPaolo JA, Woodworth CD, Popescu NC, Notario V, Doniger J. Induction of human cervical squamous cell carcinoma by sequential transfection with human papillomavirus 16 DNA and viral Harvey ras. Oncogene. 1989;4:395–399. [PubMed] [Google Scholar]
- Edlund S, Landstrom M, Heldin CH, Aspenstrom P. Transforming growth factor-beta-induced mobilization of actin cytoskeleton requires signaling by small GTPases Cdc42 and RhoA. Mol Biol Cell. 2002;13:902–914. doi: 10.1091/mbc.01-08-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing CM, Ru N, Morton RA, Robinson JC, Wheelock MJ, Johnson KR, Barrett JC, Isaacs WB. Chromosome 5 suppresses tumorigenicity of PC3 prostate cancer cells: correlation with re-expression of alpha-catenin and restoration of E-cadherin function. Cancer Res. 1995;55:4813–4817. [PubMed] [Google Scholar]
- Fera D, Marmorstein R. Different regions of the HPV-E7 and Ad-E1A viral oncoproteins bind competitively but through distinct mechanisms to the CH1 transactivation domain of p300. Biochemistry. 2012;51:9524–9534. doi: 10.1021/bi3011863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolik CA, Wakefield LM, Smith DM, Sporn MB. Characterization of a membrane receptor for transforming growth factor-beta in normal rat kidney fibroblasts. J Biol Chem. 1984;259:10995–11000. [PubMed] [Google Scholar]
- Fukawa T, Kajiya H, Ozeki S, Ikebe T, Okabe K. Reactive oxygen species stimulates epithelial mesenchymal transition in normal human epidermal keratinocytes via TGF-beta secretion. Experimental cell research. 2012;318:1926–1932. doi: 10.1016/j.yexcr.2012.05.023. [DOI] [PubMed] [Google Scholar]
- Garamszegi N, Garamszegi SP, Shehadeh LA, Scully SP. Extracellular matrix-induced gene expression in human breast cancer cells. Mol Cancer Res. 2009;7:319–329. doi: 10.1158/1541-7786.MCR-08-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habig M, Smola H, Dole VS, Derynck R, Pfister H, Smola-Hess S. E7 proteins from high- and low-risk human papillomaviruses bind to TGF-beta-regulated Smad proteins and inhibit their transcriptional activity. Archives of virology. 2006;151:1961–1972. doi: 10.1007/s00705-006-0768-1. [DOI] [PubMed] [Google Scholar]
- Hellner K, Mar J, Fang F, Quackenbush J, Munger K. HPV16 E7 oncogene expression in normal human epithelial cells causes molecular changes indicative of an epithelial to mesenchymal transition. Virology. 2009;391:57–63. doi: 10.1016/j.virol.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S, Huang F, Hafez M, Winawer S, Friedman E. Colon carcinoma cells switch their response to transforming growth factor beta 1 with tumor progression. Cell Growth Differ. 1994;5:267–275. [PubMed] [Google Scholar]
- Hypes MK, Pirisi L, Creek KE. Mechanisms of decreased expression of transforming growth factor-beta receptor type I at late stages of HPV16-mediated transformation. Cancer letters. 2009;282:177–186. doi: 10.1016/j.canlet.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest. 2003;112:1776–1784. doi: 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Kasai H, Allen JT, Mason RM, Kamimura T, Zhang Z. TGF-beta1 induces human alveolar epithelial to mesenchymal cell transition (EMT). Respir Res. 2005;6:56. doi: 10.1186/1465-9921-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur P, McDougall JK. Characterization of primary human keratinocytes transformed by human papillomavirus type 18. Journal of virology. 1988;62:1917–1924. doi: 10.1128/jvi.62.6.1917-1924.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil N, Parekh TV, O'Connor R, Antman N, Kepron W, Yehaulaeshet T, Xu YD, Gold LI. Regulation of the effects of TGF-beta 1 by activation of latent TGF-beta 1 and differential expression of TGF-beta receptors (T beta R-I and T beta R-II) in idiopathic pulmonary fibrosis. Thorax. 2001;56:907–915. doi: 10.1136/thorax.56.12.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisaki K, Kurisaki A, Valcourt U, Terentiev AA, Pardali K, Ten Dijke P, Heldin CH, Ericsson J, Moustakas A. Nuclear factor YY1 inhibits transforming growth factor beta- and bone morphogenetic protein-induced cell differentiation. Molecular and cellular biology. 2003;23:4494–4510. doi: 10.1128/MCB.23.13.4494-4510.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DK, Kim BC, Kim IY, Cho EA, Satterwhite DJ, Kim SJ. The human papilloma virus E7 oncoprotein inhibits transforming growth factor-beta signaling by blocking binding of the Smad complex to its target sequence. The Journal of biological chemistry. 2002;277:38557–38564. doi: 10.1074/jbc.M206786200. [DOI] [PubMed] [Google Scholar]
- Lee MY, Chou CY, Tang MJ, Shen MR. Epithelial-mesenchymal transition in cervical cancer: correlation with tumor progression, epidermal growth factor receptor overexpression, and snail up-regulation. Clin Cancer Res. 2008;14:4743–4750. doi: 10.1158/1078-0432.CCR-08-0234. [DOI] [PubMed] [Google Scholar]
- Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Massague J, Gomis RR. The logic of TGFbeta signaling. FEBS letters. 2006;580:2811–2820. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Mi Y, Borger DR, Fernandes PR, Pirisi L, Creek KE. Loss of transforming growth factor-beta (TGF-beta) receptor type I mediates TGF-beta resistance in human papillomavirus type 16-transformed human keratinocytes at late stages of in vitro progression. Virology. 2000;270:408–416. doi: 10.1006/viro.2000.0283. [DOI] [PubMed] [Google Scholar]
- Miettinen PJ, Ebner R, Lopez AR, Derynck R. TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J Cell Biol. 1994;127:2021–2036. doi: 10.1083/jcb.127.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger K, Werness BA, Dyson N, Phelps WC, Harlow E, Howley PM. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. The EMBO journal. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oft M, Heider KH, Beug H. TGFbeta signaling is necessary for carcinoma cell invasiveness and metastasis. Curr Biol. 1998;8:1243–1252. doi: 10.1016/s0960-9822(07)00533-7. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic acids research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirisi L, Creek KE, Doniger J, DiPaolo JA. Continuous cell lines with altered growth and differentiation properties originate after transfection of human keratinocytes with human papillomavirus type 16 DNA. Carcinogenesis. 1988;9:1573–1579. doi: 10.1093/carcin/9.9.1573. [DOI] [PubMed] [Google Scholar]
- Pirisi L, Yasumoto S, Feller M, Doniger J, DiPaolo JA. Transformation of human fibroblasts and keratinocytes with human papillomavirus type 16 DNA. Journal of virology. 1987;61:1061–1066. doi: 10.1128/jvi.61.4.1061-1066.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouponnot C, Jayaraman L, Massague J. Physical and functional interaction of SMADs and p300/CBP. The Journal of biological chemistry. 1998;273:22865–22868. doi: 10.1074/jbc.273.36.22865. [DOI] [PubMed] [Google Scholar]
- Rey O, Lee S, Park NH. Human papillomavirus type 16 E7 oncoprotein represses transcription of human fibronectin. Journal of virology. 2000;74:4912–4918. doi: 10.1128/jvi.74.10.4912-4918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- Seoane J. Escaping from the TGFbeta anti-proliferative control. Carcinogenesis. 2006;27:2148–2156. doi: 10.1093/carcin/bgl068. [DOI] [PubMed] [Google Scholar]
- Shipley GD, Pittelkow MR, Wille JJ, Jr., Scott RE, Moses HL. Reversible inhibition of normal human prokeratinocyte proliferation by type beta transforming growth factor-growth inhibitor in serum-free medium. Cancer research. 1986;46:2068–2071. [PubMed] [Google Scholar]
- Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR, Shi Y. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Ten Dijke P, Goumans MJ, Itoh F, Itoh S. Regulation of cell proliferation by Smad proteins. J Cell Physiol. 2002;191:1–16. doi: 10.1002/jcp.10066. [DOI] [PubMed] [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Uemura M, Swenson ES, Gaca MD, Giordano FJ, Reiss M, Wells RG. Smad2 and Smad3 play different roles in rat hepatic stellate cell function and alpha-smooth muscle actin organization. Mol Biol Cell. 2005;16:4214–4224. doi: 10.1091/mbc.E05-02-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccarella S, Lortet-Tieulent J, Plummer M, Franceschi S, Bray F. Worldwide trends in cervical cancer incidence: Impact of screening against changes in disease risk factors. European journal of cancer. 2013 doi: 10.1016/j.ejca.2013.04.024. [DOI] [PubMed] [Google Scholar]
- Vardouli L, Vasilaki E, Papadimitriou E, Kardassis D, Stournaras C. A novel mechanism of TGFbeta-induced actin reorganization mediated by Smad proteins and Rho GTPases. Febs J. 2008;275:4074–4087. doi: 10.1111/j.1742-4658.2008.06549.x. [DOI] [PubMed] [Google Scholar]
- Wendt MK, Tian M, Schiemann WP. Deconstructing the mechanisms and consequences of TGF-beta-induced EMT during cancer progression. Cell and tissue research. 2012;347:85–101. doi: 10.1007/s00441-011-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhou BP. New insights of epithelial-mesenchymal transition in cancer metastasis. Acta Biochim Biophys Sin (Shanghai) 2008;40:643–650. doi: 10.1111/j.1745-7270.2008.00443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Yao HW, Xie QM, Chen JQ, Deng YM, Tang HF. TGF-beta1 induces alveolar epithelial to mesenchymal transition in vitro. Life Sci. 2004;76:29–37. doi: 10.1016/j.lfs.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. Journal of the National Cancer Institute. 2000;92:690–698. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.