Abstract
Multiprotein complexes have three-dimensional shapes and dynamic functions that impact almost every aspect of biochemistry. Despite this, our ability to rapidly assess the structures of such macromolecules lags significantly behind high-throughput efforts to identify their function, especially in the context of human disease. Here, we describe results obtained by coupling ion mobility-mass spectrometry with automated robotic sampling of different solvent compositions. This combination of technologies has allowed us to explore an extensive set of solution conditions for a group of eight protein homo-tetramers, representing a broad sample of protein structure and stability. We find that altering solution ionic strength in concert with dimethylsulfoxide content is sufficient to disrupt the protein-protein interfaces of all of the complexes studied here. Ion mobility measurements captured for both intact assemblies and subcomplexes matched expected values from available X-ray structures in all cases save two. For these exceptions, we find that distorted subcomplexes result from extreme disruption conditions, and are accompanied by small shifts in intact tetramers size, thus enabling the removal of distorted subcomplex data in downstream models. Furthermore, we find strong correlations between the relative intensities of disrupted protein tetramers and the relative number and type of interactions present at interfaces as a function of disrupting agent added. In most cases, this correlation appears strong enough to quantify various types of protein interfacial interactions within unknown proteins following appropriate calibration.
Introduction
Proteins form most of the central macromolecular machines that support cellular life through the creation of dynamic multiprotein complexes.1 By the same token, such complexes comprise most of the important drug targets sought in treatment efforts for human disease.2,3 Due to their biochemical importance, broad efforts are currently underway to determine the structures of protein complexes on a proteome-wide scale. These efforts typically rely upon high throughput X-ray crystallography and NMR protocols, with many other technologies operating in a supporting role to ensure high protein purity, stability, and concentrations.4,5 While this has been a very successful strategy, leading to the determination of many structures for large protein complexes, continued application of these basic approaches has also served to highlight their limitations. For example, such experiments typically require the availability of a sufficient quantity of homogeneous material and significant method development efforts for success.6 These conditions are often difficult to meet and, thus, the number of structures of multi-subunit protein complexes deposited in structural databases remains relatively lowwhen compared to monomeric proteins.7
Recently, technologies based on mass spectrometry (MS) have provided dramatic insights into the composition and structure of multiprotein complexes.8,9 Growing significantly from previous yeast two-hybrid screens,10 affinity-based purification coupled to MS detection has provided some of clearest depictions of macromolecular protein networks in terms of their connectivity,11 leading eventually to include sophisticated quantitative proteomics protocols.11 Chemical cross linking (CXL) in conjunction with MS experiments also have expanded our knowledge of both global protein networks and individual macromolecular protein complexes.11,12 Hydrogen-deuterium exchange (HDX) and oxidative labeling are being used with increasing frequency prior to MS in order to assess dynamic structural changes within a host of multiprotein systems.13,15 While the implementation of most MS experiments involve the proteolytic digestion of denatured proteins following the steps taken within the protocol to capture native protein-protein contact or structure information,16 MS experiments that introduce intact protein complexes directly into the instrument can also be used to provide, in many cases, a greater amount of native structure information.17 Despite the challenges inherent in analyzing large protein ions by MS, such experiments have aided in the development of protein structure models for a range of complexes in advance of more-conventional structural biology approaches.17,19
While such MS measurements rely principally upon preserving an entire multiprotein assembly during both sample preparation and detection,20 recent experiments in this area have highlighted the utility of partially disrupting the protein-protein contacts within complexes so that various subcomplexes are also detected during the experiment.19,21,22 Such disruption steps enable the determination of protein complex connectivity and stoichiometry to a level of detail that few other approaches can match.23,24 Typically, two separate and complementary approaches for subcomplex formation are pursued: gas-phase activation and solution-phase disruption.21,25,28 The former most-often utilizes collisional activation of gas-phase ions to initiate dissociation, forming individual protein subunit and stripped protein complex product ions.25,29,30 While an indispensible tool for verifying protein connectivity and stoichiometry, larger subcomplexes are typically not formed in the unimolecular decay reactions that proceed during collision induced dissociation (CID) of multiprotein complex ions, thus somewhat limiting the overall utility of the technique in the context of protein structure modeling efforts.19 Charge manipulation in combination with collisional activation, as well as alterative activation methods, have shown promise in altering the overall mechanism of protein complex CID to realize increased information content.31 Current practice, however, dictates significant solution phase protein complex disruption efforts in order to generate sufficient information to fully-define protein topology and connectivity by intact MS methods.32 Such disruption is often initiated by altering solvent composition, pH and ionic strength to favor protein subcomplex formation.32 The subcomplexes formed in such experiments have been shown to reflect the known organizational substructure of complexes,21,22,33 and when combined with CID, can be used to deduce a complete organizational diagram of protein complexes of previously unknown structure.34 However, conditions that optimize the ion signal recorded for protein subcomplexes are often found through a time consuming, trial-and-error based process that can substantially limit the throughput of MS experiments.
Ion mobility (IM) separation,35 when coupled to MS, enables the collection of protein complex size information, and when this is combined with the connectivity information described above, coarse-grained (CG) or atomic models of the assemblies can be constructed.32,36 In addition, IM-MS provides signal-to-noise, spectral deconvolution, and tandem MS benefits unique to the analysis of the multiply-charged protein complex ions produced by nESI.37 Specifically, IM-MS was used to refine a connectivity model for the eukaryotic ribosomal initiation factor 3 complex,22 provided key structural data for models of rotary adenosine triphosphatases/synthases,38 pallindromic repeat-associated protein complexes,39 and utilized to deduce complete models for αB-crystallin40 and DNA replisome assemblies.41 In addition, models for the highly-dynamic olgiomers involved in the amyloid-β,42 norwalk virus,43 and β-2 microglobulin44 aggregation/assembly pathways have been constructed using IM-MS. In every case, IM measurements of many inter-related protein complexes, connected either through assembly, equilibrium, or intentional disruption, were used to construct the resultant models. Key assumptions surrounding such IM-MS derived models include: a strong relationship between subcomplex and higher-order assembly quaternary structure, a high degree of structural correlation between gas-phase and native state proteins, and a lack of significant protein subunit conformation changes upon higher-order complex formation or disruption. While most IM-MS models have been cross-validated using other structural biology tools and control samples,22,39,40 there are many foundational questions that still surround the use subcomplex information as a constraint for larger related oligomers, especially when ideal disruption conditions introduce significant amounts of potential chemical denaturants in solution.
Here, we couple automated titration and nESI of protein complex samples to IM-MS in order to deduce the optimum protocols and analysis guidelines necessary for high-throughput protein topology mapping experiments. Our total dataset contains ca. 400 individual IM-MS datasets, thus necessitating an automated approach. We discover that IM-MS measurements of intact multiprotein complexes and subcomplexes can be readily mapped onto known solution structures, with few exceptions. For those exceptions, the distortion of subcomplexes is mirrored in distorted intact complexes, which are also detected by IM-MS. We find strong correlations between the disruption of intact complexes with specific reagents and quantitative values for the different classes of interactions that form the protein-protein interfaces within the intact assembly. We propose that such information can be used in the future to deduce the critical contacts within unknown complexes, and project the utility of IM-MS in structural proteomics and genomics efforts broadly in light of the data presented.
Experimental
Sample Preparation
Homo-tetrameric protein complexes studied included transthyretin (TTR, human, Sigma P1742), avidin (AVD, egg white, Sigma A9275), concanavalin A (CON, jack bean, Sigma C2010), alcohol dehydrogenase (ADH, Saccharomyces cerevisiae, Sigma A7011), aldolase (ALD, rabbit muscle, Sigma A2714), pyruvate kinase (PKI, rabbit muscle, Sigma P9136), catalase (CAT, bovine liver, CalBioChem 219001) and β-galactosidase (βGL, E.coli, Sigma G5635). Standards used to construct collision cross-section (CCS) calibration curves include: a mixture of peptides (Waters, product code: 186002337), monomeric cytochrome C (Sigma, C2506), and the protein complexes AVD, CON, ADH, and glutamate dehydrogenase (GDH) (Sigma, G7882). All protein complexes were solubilized in 200 mM aqueous ammonium acetate (NH4Ac) solutions and stored at −80°C. Stock samples (50 μM with respect to the complex, diluted 10 fold afterwards) were then buffer-exchanged just prior to disruption experiments and IM-MS analysis, using a Micro Bio-Spin 30 column (Bio-Rad, Hercules, CA) equilibrated with 200 mM aqueous NH4Ac. While the final concentration of protein complexes is unknown, control experiments performed with the protein complexes studied here over a range of concentrations (as low as 3μM) did not alter the oligomeric states detected. The peptide mixture (~2 μM for each peptide) was prepared in a water/methanol/acetic acid (49/49/2) solution. Stock cytochrome C (5 μM) was prepared in 200 mM aqueous NH4Ac, and stored at −80°C.
Ion Mobility-Mass Spectrometry
Solution composition screens for subcomplex formation were performed using an automatable, chip-based nano electrospray (nESI) robot, (TriVersa NanoMate, Advion, Ithaca NY), capable of both rapid sampling and mixing. Solution parameters were screened in a two-dimensional fashion to be compatible with robotic sampling of a 96-well plate. An Advanced User Interface (AUI) module in the control software allowed programmed sequential aspiration, and allowed for protein samples to be quickly dispensed into individual wells and mixed with disrupting agents under highly-controlled temperatures (18 °C). After mixing, chip-based nESI using nano machined emitters (5μm) was initiated through the robotic sampling of individual solutions with conductive pipette tips. In general, samples were sprayed immediately after mixing, however, various incubation times were tested for selected protein complexes (e.g., AVD), ranging from minutes to hours, and resulted in no significant change in disruption behavior. Chip based emitters were maintained at 1.3 kV to 1.9 kV. Samples prepared at high ionic strengths or solutions containing large amounts of organic co-solvent generally required lower nESI voltages and higher backing pressures (0.1-1 psi) to avoid salt cluster-related chemical noise and generate stable spray conditions respectively.
IM-MS data were acquired on a Synapt G2 (Waters, Milford, MA) 45 optimized for the study of large multiprotein complexes, as described previously.46 For the purposes of topology construction, the accuracy of CCS measurements for both intact protein complexes and dissociated products was optimized through highly linear CCS calibration curves (R2 > 0.98), resulting from high pressure (3-4 mbar), low wave height (12-15 V) and low wave velocity (150-200 m/s) travelling wave IM conditions.46 Initial IM-MS Data processing was performed using Masslynx v4.1, Driftscope v2.0 (both from Waters, Milford, MA), as well as previously-described IM calibration tools.37,47
X-ray and IM Data Comparisons and Computational Analysis
Coarse-grained (CG) topology models for the protein tetramers discussed in this report were constructed either using a simple symmetry analysis of X-ray structures using in-house python scripts and visualized in PyMol, or using IM-derived CCSs as described below. Protein data bank (PDB) IDs for the X-ray structures used here are given in on-line Supporting Information. All model structures were evaluated in terms of their agreement with IM data using a version of the program Mobcal modified for the analysis of coarse grained structures.37 For all model CCS values, a scaled version of the projection approximation (PA) found in Mobcal was used,48 as such methods have exhibited strong, broad agreement with experimental IM-MS data.49 For model topologies constructed using IM data, the size of a single sphere was constrained using monomer CCS data. Then, the distance between monomer units was constrained using protein dimer CCS values. Finally, a dimer-dimer distance was constrained using tetramer CCS measurements. The final IM-based model was then compared to the same CG values derived from the X-ray structure. Note that not all tetramers studied in this report are planar, and that the above protocol does not evaluate the relative angles between protein subunits.33,41 To resolve a planar tetramer geometry from that of a tetrahedral structure, an IM CCS resolution in excess of 100 has been projected to be necessary for protein complexes comprised of >10 kDa subunits, whereas the maximum CCS resolution recorded to date on the Synapt G2 is 50-60.37,46 A number of quantitative values related to protein structure were extracted from X-ray and IM-MS data, and compared. These values are defined in detail in on-line Supporting Information.
Results and Discussion
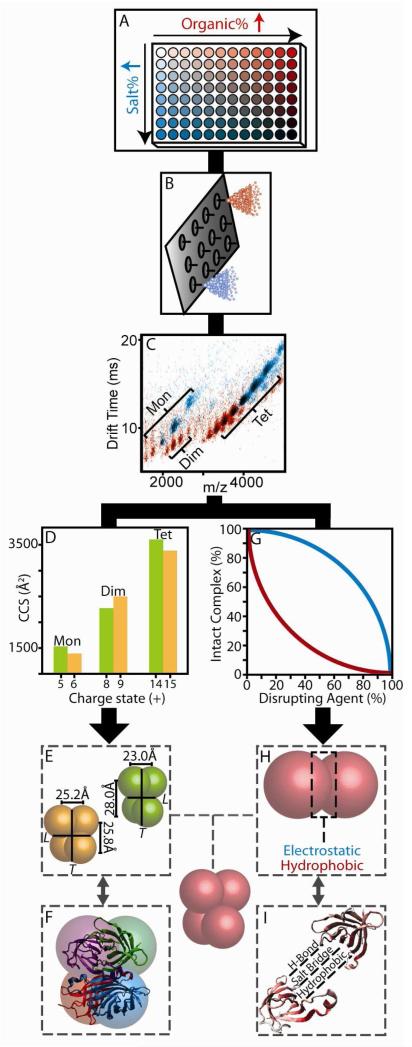
Figure 1 illustrates the experimental protocol used in this study. Protein complex sample was partitioned into a 96-well plate where different amounts of disrupting agents were robotically added into the existing aqueous solutions (Figure 1A). The amounts of these agents were then modified in a stepwise fashion across one axis of the plate. For the complexes studied here, we found that a two-component screen, that varied the DMSO (red wells, Figure 1A) and NH4Ac (blue wells, Figure 1A) content of the protein complex containing solutions was sufficient to disrupt and interrogate all eight protein tetramers, while keeping the total protein concentration constant (5μM). Solutions contained, at most, 67% DMSO by volume and 4M NH4Ac, beyond which significant protein secondary structure changes may occur.50 Following the addition of disruption agents, samples were then immediately sprayed through one of the nozzles on a nESI chip (Figure 1B). IM-MS data were then recorded (Figure 1C), and subtle changes in protein oligomeric state, CCS, and charge state detected. As observed previously,18,23 the sub-assemblies observed under disruption conditions reflect the known sub-structure of the intact complexes analyzed. All complexes studied here are tetramers comprised of dimeric subcomplexes, possessed of non covalent bonds of varying strengths. By quantifying the sizes of the subcomplexes observed by IM-MS (Figure 1D) over a range of charge states, CG models of the relative interface sizes within these tetramers were constructed (Figure 1E) and compared against X-ray datasets (Figure 1F) to detect correlations. In parallel with this analysis track, the relative intensities of the subcomplexes were recorded (Figure 1G) and correlated to the influence of specific disrupting agents likely to disproportionately influence selected sub-classes of protein-protein contacts (Figure 1H). These data were then also correlated with X-ray data where specific interactions were precisely measured (Figure 1I), allowing us to evaluate the ability of IM-MS to quantify the type of interactions present within unknown protein-protein interfaces.
Figure 1.
IM-MS data collection and analysis procedures. (A) Disrupting agents and protein complexes are mixed in solvents containing either increased amounts of organic (DMSO, red) or salt (NH4Ac, blue). (B) Each sample is sprayed using a chip based nESI emitter array. (C) An example overlay of two IM-MS plots under high organic (red) and high salt (blue) conditions. (D) IM based size measurements recovered for both intact complexes and subcomplexes as a function of charge state (E & F). Protein sizes are converted to distance constraints, and tested against those calculated from X-ray. (G) Signal intensities recorded for intact complex ions are plotted against the amount of disrupting agent added in solution. (H & I) These data are analyzed with respect to the separate interaction types found within the protein-protein interfaces from X-ray.
Evaluating the Fidelity of IM-MS Derived Protein Topology
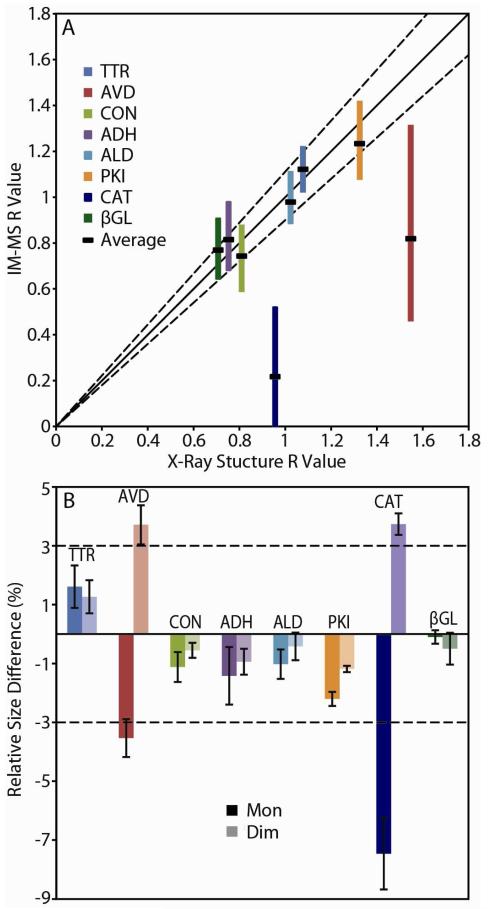
In order to evaluate the agreement between IM-MS derived protein topology measurements and those from X-ray datasets, we generated R values from both classes of data, which track the relative distance between dimers and monomers within the complex (see Supporting Information). A plot of IM-MS R values against those extracted from X-ray structures reveals a tight correlation between the two for all complexes save two: AVD and CAT (Figure 2A). For the other six protein tetramers, average topologies, as well as much of the total spread of possible topologies created if all charge state based CCS combinations are considered with equal weight, fall within a 90% confidence prediction interval. Conversely, the entire range of R values generated from IM-MS for AVD and CAT fall outside of the same prediction interval in Figure 2A. Note that AVD and CAT present larger R value ranges than the other protein complexes studied here, and that such data on its own could be used in broader IM-MS topology construction efforts to identify-distortion prone complexes.
Figure 2.
(A) A plot of R values generated from IM-MS data vs. R values from X-ray structures for eight protein tetramers. Color-coded bands indicate the range of R values recovered from IM-MS data, and black bands represent the average. The solid line represents a 1:1 correlation between the IM-MS experiment and X-ray, with the dashed lines representing a 90% confidence interval. (B) Relative size differences observed between IM-MS and X-ray data for monomers (dark colors) and dimers (light colors) released during disruption. Dashed lines indicate the ±3% error for IM-MS CCS determinations.
If we evaluate protein CCS measurements used to generate the R values shown in Figure 2A in detail, the origins of the topological defects observed in our CAT and AVD datasets becomes apparent (Figure 2B). While CCS values recorded for the intact tetramer ions generated under control conditions (200 mM NH4Ac, pH 6.9) are all within 3% of the expected X-ray CCS values, the disrupted monomer and dimer CCS values determined experimentally vary considerably over the eight protein tetramers studied here. Notably, both CAT and AVD display compacted monomer and enlarged dimer CCS values. Specifically, AVD disruption generates monomers that are, on average, compressed by 3.5% and dimers that possess similarly inflated CCS values when compared to X-ray estimates. CAT dimers are also enlarged by 3.7%, but generate monomer CCS values upon disruption that are 7.5% compressed when compared to X-ray data. While the data shown in Figure 2B provides a clear empirical explanation for the topology defects detected in Figure 2A, it provides neither a structural explanation for the same observations nor a robust analytical methodology to avoid using such IM-MS data in the construction of CG structures for unknown protein complexes.
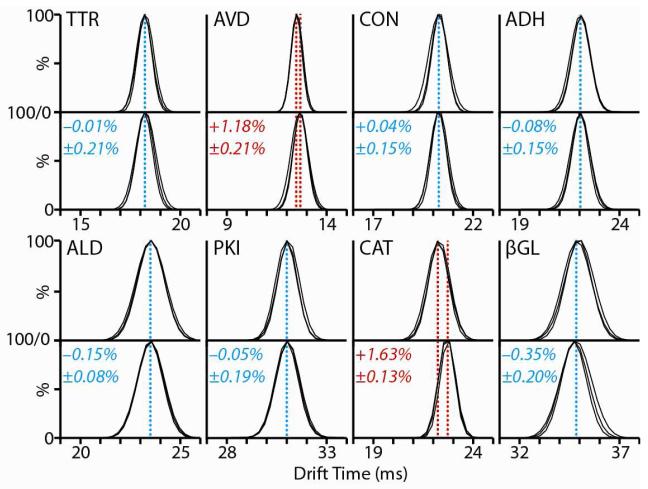
A broader analytical protocol for IM-MS based protein complex topology mapping can be derived from the data shown in Figure 3, which tracks the IM arrival time profiles for all eight protein tetramers under two separate conditions: those optimized for protein disruption and control measurements of ions formed from 200 mM NH4Ac. For six of the eight protein complexes, no significant differences are observed between the IM data recorded under either conditions (<1% deviation), both in terms of the centriods or peak widths of the IM distributions collected (three replicates shown for each dataset). In two cases, however, small yet significant increases (> 1%) are observed in the IM drift time centriod observed for intact tetramer ions. Since the intact masses of AVD and CAT are identical under both disruption and control solutions conditions (Figure S1, Supporting Information), the increase in drift time recorded is likely derived from conformational changes in the intact tetramers. These two complexes, AVD and CAT, correspond precisely to those that produced erroneous protein CG structures in our analysis shown in Figure 2. As such, the observation of similar shifts in IM data for intact complexes allows us to detect and remove distorted subcomplex CCS data from our modeling constraints. It is also notable that this strategy can, in theory, be extended to protein complexes of unknown structure, as the IM drift time shifts recorded in Figure 3 are not predicated on any knowledge of the tetramer structures studied here.
Figure 3.
Comparison between the IM distributions and centroids recorded for intact tetramer ions generated under control (200mM NH4Ac, upper panel) and disruption conditions (lower panel). Three replicates are measured and overlaid for each dataset shown. Blue dashed lines indicate those datasets where no significant shift in tetramer drift time is observed (<1%). Red dashed lines indicate those cases where a drift time shift (≥1%) is observed under disrupting solution conditions when compared to control.
While the criteria we use to construct Figure 3 requires similar protein disruption from all tetramers studied, significantly different solution conditions are represented in each of the lower panels in Figure 3 (see Table S1 and Figure S2, Supporting Information), chosen to represent equivalent amounts of protein complex disruption. Despite these differences, we note that the six protein complexes that produce high-fidelity R values when compared to X-ray data do not, under any solution composition probed in this report, generate ions with shifted drift times. For example, TTR tetramer ions retain their centroid drift time when generated from solutions comprised of 50% DMSO and 4M NH4Ac, while CAT displays the shifted drift time profile shown in Figure 3 at 33% DMSO and 2M NH4Ac. Previously published results for the CON tetramer support these findings, where methanol-based disruption of the tetramer results in both dramatically enlarged intact complexes and distorted monomeric subunits upon disruption.51 Thus, taken as a complete dataset, the results shown in Figure 3 constitute a general analytical framework for detecting those conditions likely to produce deformed protein subcomplexes through disruption, as the swelling of intact protein complex CCS appears to accurately predict the formation of such deformed sub-assemblies.
A detailed analysis of the X-ray data available for the eight protein tetramers studied here provides a structural explanation for the data shown in Figures 2 and 3. First, the X-ray structures for the monomeric units within the CAT and AVD tetramers are the least spherical of the complexes studied here (Figure S3A, Supporting Information). Furthermore, AVD and CAT exhibit both the largest number of inter-protein contacts per unit mass (Figure S3B, Supporting Information) and the most closely-packed monomers (Figure S3C, Supporting Information) in our field of tetramers. In summary, therefore, AVD and CAT are comprised of relatively non-spherical subunits, possessed of larger than average protein-protein interfaces, and exhibit comparatively tight inter subunit packing. Given such a description, it is relatively facile to rationalize the data shown in Figures 2 and 3. AVD and CAT subunits are likely more stabilized by inter protein contacts than the subunits that make up the other complexes studied here, thus making them more prone to deformation upon release from their respective tetramers.
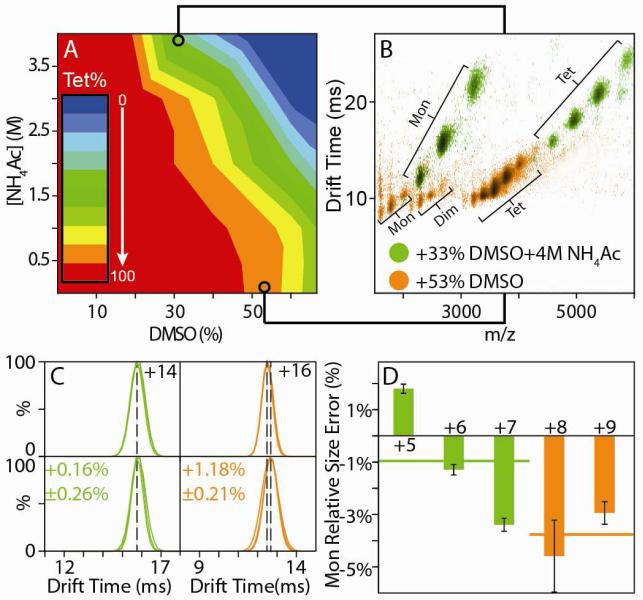
In an effort to find those disruption conditions capable of generating subcomplex CCS values for AVD and CAT that are in close agreement with X-ray data, we initiated a thorough analysis of all IM-MS data collected in our screens. An example of such an analysis is shown in Figure 4, where we use a contour plot to represent the relationship between AVD tetramer signal intensity, DMSO solution fraction, and NH4Ac concentration (Figure 4A). Within this dataset, each of the 49 data points used for its construction represent a complete IM-MS spectrum (Figure 4B), and comparison of multiple points within this plot reveals stark differences in the dissociation patterns of the AVD tetramer. Two drift time versus m/z plots are shown, overlaid, in Figure 4B, one acquired from a solution containing 53% DMSO with minimal (10 mM) NH4Ac and the other containing 33% DMSO and 4 M NH4Ac. Preparing AVD in higher salt-concentration solutions prior to nESI engenders lower than average charge states for both intact complexes and disruption products as well as an absence of any dimer signal when compared to IM-MS data acquired from solutions containing low salt and higher amounts of organic co-solvent. Charge states for the AVD tetramer under control conditions range from 14+ to 17+, while charge states from 11+ to 15+ are observed under conditions of high salt (green data, Figure 4B) and 15+ to 20+ when low salt is coupled with large amounts of DMSO in solution (orange data, Figure 4B). When the IM drift time profiles from charge states that sufficiently overlap with control data are compared, it is clear that the AVD tetramer shifts its size when large amounts of DMSO are used for protein disruption in the absence of high salt concentrations (Figure 4C, orange), as observed in Figure 3. However, when a similar comparison is made for disruption conditions that add 4M NH4Ac, no shift is detected under conditions that lead to complex disruption (Figure 4C, green). Analysis of the monomer CCS values from both of these datasets reveals an agreement with X-ray data to within acceptable errors when disruption is performed under conditions of high salt (Figure 4D, green), but not when solutions with added organic solvent are used (Figure 4D, orange). A few caveats surround the result presented in Figure 4. Most notably, the lack of a dimeric subcomplex disruption product for the AVD tetramer under the high salt conditions presented means that such a result, on its own, does not provide sufficient information to generate the CG structure and R values analyzed in Figure 2. In addition, similar conditions that lessen the deformation experienced by disrupted monomers have yet to be discovered for the CAT tetramer. Regardless, the results presented in Figures 2, 3, 4, and S3 provide a general picture that illustrates the overall agreement between IM-MS and X-ray datasets, methodologies that enable the detection of distorted subcomplex formation in unknowns, and approaches that enable the recovery of partial subcomplex CCS data from proteins previously distorted in a more general screen.
Figure 4.
AVD topology models from IM-MS data are brought into close agreement with X-ray when disruption conditions incorporating high ionic strength are used. (A) 2D titration results for AVD using DMSO and NH4Ac as disrupting agents. (B) Overlay of two IM-MS contour plots extracted from the dataset shown in A (green and orange). (C) A comparison of the drift time distributions recorded for the remaining intact tetramer ions formed from the two disruption conditions (lower panels) with those measured under control conditions (200 mM NH4Ac, upper panels). (D) The relative errors in monomer size measured by IM when compared to X-ray data for different AVD monomer charge states derived from the disruption conditions defined in C. Solid lines indicate the average monomer size for each solution condition.
Solution Disruption followed by IM-MS Reveals the Details of Protein-Protein Interface Chemistry
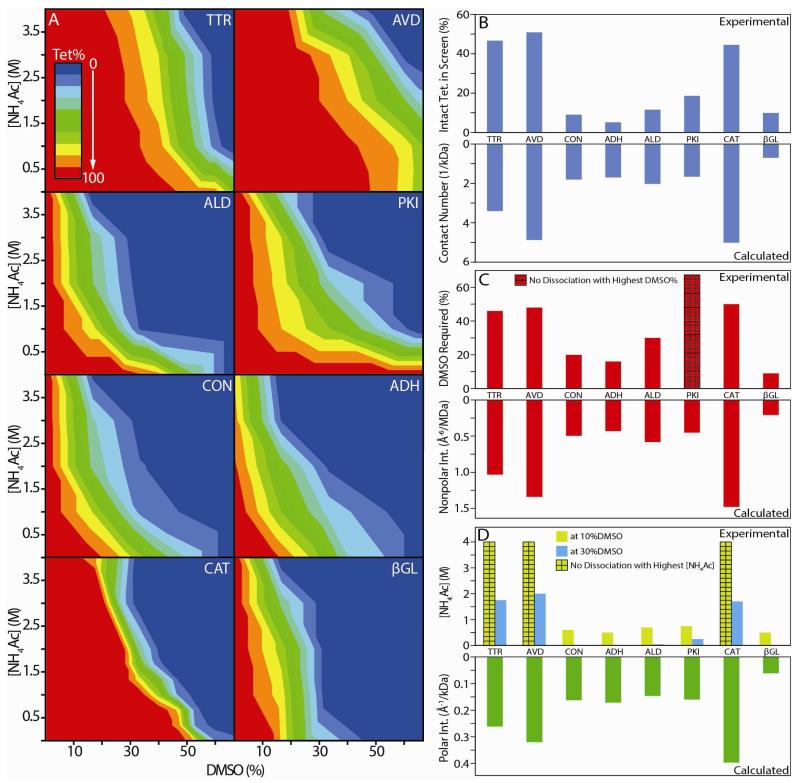
Figure 4A indicates that the AVD tetramer responds more strongly to DMSO as a disruption agent than NH4Ac. For example, by following a vertical line fixed at 10% DMSO concentration in Figure 4A, it is apparent that no AVD tetramer disruption occurs during such an NH4Ac-only titration. This observation could indicate a general correlation between the chemical nature of the protein-protein interfaces within the complex and the disruption of the intact assembly under specific solvent conditions as recorded by IM-MS. In order to pursue this basic premise, we analyzed the multidimensional DMSO/NH4Ac titration data for all eight protein tetramers studied in this report (Figure 5A), and evaluated them for general correlations with X-ray data. Each dataset shown is individually normalized to the tetramer intensity observed for a given complex under control conditions (200 mM NH4Ac, 0% DMSO), and each contour transition shown represents a 10% relative decrease in tetramer signal intensity observed. Every protein complex represented in Figure 5A displays a unique disruption pattern as a function of DMSO and NH4Ac solution content, and qualitative comparisons between the plots illustrate how such data can be used to differentiate assemblies based on the general strength of their non-covalent protein interactions. For example, TTR, AVD, and CAT are amongst the most disruption resistant of the complexes studied here relative to DMSO/NH4Ac titration, as evidenced by the relatively large number of total solution conditions probed in our screen where IM-MS data records only intact tetramer for these three cases. Of these three, CAT exhibits the steepest descent from 100% intact tetramer to complete disruption of any of the complexes studied here, once a sufficient threshold amount of disrupting agent is reached. PKI also represents a unique disruption profile, as the tetramer does not generate subcomplex ion current as a function of DMSO added in solution unless a certain threshold of NH4Ac is present as well. Previous studies have shown that key salt bridges within the PKI protein-protein interfaces exist and stabilize the quaternary structure of the complex.52 Our IM-MS data agrees well with these findings qualitatively.
Figure 5.
(A) 2D titration results obtained for the eight homo-tetrameric protein complexes studied here, where NH4Ac concentration (M) is plotted against DMSO solution content (%). The colors shown indicate the normalized intensity of the tetramer ion signal recorded under each solution condition, from red (100%) to blue (0%). Correlations between our IM-MS titration experiments (top) and X-ray structure data (bottom) are presented in three histogram plots (B), (C) and (D). (B) Normalized intact tetramer intensity integrated over all disruption conditions vs. the number of contacts per unit kDa calculated from X-ray data. (C) The DMSO % in solution, without added NH4Ac, required to initiate tetramer disruption (signal drops by 10%) vs. the estimated average strength of hydrophobic contacts within the protein-protein contacts from X-ray (Å−6/MDa). (D) NH4Ac concentration required for tetramer disruption at fixed 10% (yellow) and 30% DMSO (blue) vs. the estimated average strength of the polar interactions (Å−1/kDa) within the protein-protein contacts from X-ray. For all datasets, bars marked with a grid pattern indicate that the complex did not undergo disruption using any amount of the selected agent under the conditions of the screen. Linear correlation coefficients between IM-MS and X-ray datasets are discussed in the text.
In addition to the qualitative analysis above, a quantitative comparison between IM-MS and X-ray datasets are necessary to extend the application of the data shown in Figure 5A to unknown protein complexes. We begin by noting the strong correlation between the integrated areas found by IM-MS multi-dimensional titration corresponding to those conditions where tetramers remain completely un-disrupted (red shaded areas within Figure 5A) and the normalized number of inter-protein contacts contained within the X-ray structures for the complexes studied here (Figure 5B). These two parameters appear to be linearly correlated in our dataset, having an R2 value of 0.826, despite the fact that our analysis clearly over-simplifies the complex nature of the underlying disruption mechanism. We extended this analysis to include narrower definitions for the inter-protein contacts and discovered additional strong correlations between IM-MS and X-ray structure data. By keeping the NH4Ac concentration of protein complex containing solutions at fixed minimum values, and titrating DMSO content, we observe a strong correlation between the amount of tetramer disruption observed by IM-MS and the calculated strength of the inter-chain hydrophobic contacts revealed by X-ray (Figure 5C), generating an R2 value of 0.931 to a linear relationship. We find a somewhat less strong linear correlation coefficient of 0.797 between the strength of polar inter protein interactions and tetramer disruption efficiency as a function of NH4Ac concentration in solutions with fixed DMSO content (yellow data, Figure 5D). The weaker correlation we observe for polar interactions in our IM-MS data can be attributed, in part, to the lack of orthogonality between the types of molecular interactions likely disrupted by DMSO and NH4Ac. While polar and non-polar interactions can be accessed exclusively by DMSO and NH4Ac respectively in our screens, both solvents likely affect H bonds within protein interfaces.50 In addition, the long range nature of polar and electrostatic interactions, in contrast to the forces involved in non-polar interactions, may elicit non-native contacts within complexes during disruption, thus degrading the observed correlation.53,54 There are a number of caveats associated with these correlations, however. For example, NH4Ac content alone does not dissociate any of the complexes studied here; therefore, a baseline level of DMSO is required to generate the correlations associated with polar interaction strength. Specifically, when DMSO content of the solutions is locked at 10%, and NH4Ac concentration is ramped, TTR, CAT and AVD do not generate any measurable disruption products over the range of salt probed in this report (10mM to 4M). Similarly, by increasing the fixed DMSO value to 30%, CON, ADH and βGL all undergo disruption such that the intact tetramer is 50% depleted. The linear correlation coefficient recorded under this condition is similar to that generated by our data when minimal DMSO is used during the titration (blue data, Figure 5D, R2 = 0.775). Also, PKI is not observed to undergo disruption as a function of DMSO content when the NH4Ac concentration is kept at 10mM (the maximum DMSO solution content in our screen is 67%). We attribute this observation to the critical salt bridges known to stabilize the protein-protein contacts in the PKI tetramer, as discussed above. Despite these caveats, we find that the correlations between IM-MS titration data and X-ray protein-protein contact data are surprisingly strong, and likely pave the way for future use of such quantitative values for different types of inter chain interactions to constrain models of unknown protein complexes.
Conclusions
Here, we utilize robotically assisted multidimensional titration of disrupting agents in solution to probe the ability of IM-MS to produce topology models in agreement with existing X-ray data for a series of eight tetrameric proteins. Our final dataset contains 8 2D screens of protein complex disruption, using both ionic strength and organic content axes, containing ca. 400 individual protein complex IM-MS datasets, each rich with size and mass data and many individual signals. As such, the acquisition of this data was greatly enabled by our automated approach. In addition, the ability to robotically-manipulate samples with great precision facilitates the cross-dataset comparisons discussed throughout this report. We demonstrate that R values computed from IM-MS data agree well with the majority of those extracted from X-ray. Clear disagreements are found in the case of the AVD and CAT tetramers, and we rationalize these observations based on the large number of inter subunit contacts found within these two complexes that likely stabilize their relatively aspherical subunits. Furthermore, a careful analysis of IM-MS data allows deformed protein subcomplexes to be detected, even in the absence of X-ray comparisons, as tetramer CCS increases and broadened R value ranges are recorded in concert with such subcomplex distortions. Also, careful analysis of multi-dimensional titration data enables the collection of IM-MS information under conditions that better match X-ray structures, as we demonstrate in the case of the AVD tetramer. In addition to providing foundational data for future protein topology discovery efforts using IM-MS, the data presented here provides evidence of solution phase memory effects for proteins and their associated complexes, adding substantially to previous reports.22,51,55,57 The data shown here is the first to indicate that there are broad sets of solution conditions that are cable of reorganizing protein structure in such a way to influence the resulting gas-phase conformation of intact protein ions, and it is clear that in future IM-MS efforts to constrain protein topology models data similar to those shown in Figures 3 and 4 should be acquired to direct and evaluate the use of subcomplex size information in protein complex model generation.
Our IM-MS titration data are also used to generate quantitative values associated with the number and type of stabilizing interactions that populate protein-protein interfaces. By determining the ratios of intact tetramers to disrupted subcomplexes over a controlled range of solution conditions, we find strong correlations between IM-MS and X-ray datasets. We estimate the average relative errors for this type of measurement at +/− 27% for quantifying polar interactions, 14% for hydrophobic interactions, and 33% for total numbers of interactions per unit mass (see Supporting Information for the details of this error calculation). Such data is potentially highly-complimentary to CXL12 and HDX13 MS information. While providing a lower resolution estimate of protein-protein interface area than these other MS approaches, such IM-MS based multi-dimensional titration results may provide quantitative values for different types of inter-chain contacts within protein assemblies not accessible to protease digestion or present at low levels, following appropriate calibration with proteins of known structure.
Supplementary Material
Acknowledgments
The authors thank Charles Brooks (University of Michigan) for aiding in the computational analysis presented here. BTR acknowledges support from the National Institutes of Health (1-R01-GM-095832-01).
References
- (1).Alberts B. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- (2).Pierce K, Premont R, Lefkowitz R. Nature Rev. Mol. Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- (3).Wu C, Murray M, Bernstein S, Condron M, Bitan G, Shea J-E, Bowers M. J. Mol. Biol. 2009;387:492–501. doi: 10.1016/j.jmb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Steven A, Baumeister W. J. Struct. Biol. 2008;163:186–195. doi: 10.1016/j.jsb.2008.06.002. [DOI] [PubMed] [Google Scholar]
- (5).Sali A, Glaeser R, Earnest T, Baumeister W. Nature. 2003;422:216–225. doi: 10.1038/nature01513. [DOI] [PubMed] [Google Scholar]
- (6).Robinson C, Sali A, Baumeister W. Nature. 2007;450:973–982. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- (7).Dutta S, Berman H. Structure. 2005;13:381–388. doi: 10.1016/j.str.2005.01.008. [DOI] [PubMed] [Google Scholar]
- (8).Benesch J, Robinson C. Curr. Opin Struct. Biol. 2006;16:245–251. doi: 10.1016/j.sbi.2006.03.009. [DOI] [PubMed] [Google Scholar]
- (9).Hernandez H, Robinson CV. Nature Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- (10).Parrish J, Gulyas K, Finley R. Curr. Opin. Biotech. 2006;17:387–393. doi: 10.1016/j.copbio.2006.06.006. [DOI] [PubMed] [Google Scholar]
- (11).Gingras A-C, Gstaiger M, Raught B, Aebersold R. Nature Rev. Mol. Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- (12).Sinz A. J. Mass Spectrom. 2003;38:1225–1237. doi: 10.1002/jms.559. [DOI] [PubMed] [Google Scholar]
- (13).Wales T, Engen J. Mass Spectrom. Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- (14).Konermann L, Tong X, Pan Y. J. Mass Spectrom. 2008;43:1021–1036. doi: 10.1002/jms.1435. [DOI] [PubMed] [Google Scholar]
- (15).Konermann L, Stocks B, Pan Y, Tong X. Mass Spectrom. Rev. 2010;29:651–667. doi: 10.1002/mas.20256. [DOI] [PubMed] [Google Scholar]
- (16).Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- (17).Heck A. Nature Meth. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- (18).Hernandez H, Dziembowski A, Taverner T, Seraphin B, Robinson CV. EMBO Rep. 2006;7:605–610. doi: 10.1038/sj.embor.7400702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Taverner T, Hernandez H, Sharon M, Ruotolo BT, Matak-Vinkovic D, Devos D, Russell RB, Robinson CV. Accounts Chem. Res. 2008;41:617–627. doi: 10.1021/ar700218q. [DOI] [PubMed] [Google Scholar]
- (20).Sharon M, Robinson C. Annual Rev. Biochem. 2007;76:167–193. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- (21).Leary JA, Schenauer MR, Stefanescu R, Andaya A, Ruotolo BT, Robinson CV, Thalassinos K, Scrivens JH, Sokabe M, Hershey JWB. J. Am. Soc. Mass Spectrom. 2009;20:1699–1706. doi: 10.1016/j.jasms.2009.05.003. [DOI] [PubMed] [Google Scholar]
- (22).Pukala TL, Ruotolo BT, Zhou M, Politis A, Stefanescu R, Leary JA, Robinson CV. Structure. 2009;17:1235–1243. doi: 10.1016/j.str.2009.07.013. [DOI] [PubMed] [Google Scholar]
- (23).Levy ED, Boeri Erba E, Robinson CV, Teichmann SA. Nature. 2008;453:1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zhou M, Robinson CV. Trends Biochem.Sci. 2010;35:522–529. doi: 10.1016/j.tibs.2010.04.007. [DOI] [PubMed] [Google Scholar]
- (25).Benesch JLP. J .Am .Soc. Mass Spectrom. 2009;20:341–348. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- (26).Zhou M, Dagan S, Wysocki V. Angew.Chemie Int. Ed. 2012;51:4336–4339. doi: 10.1002/anie.201108700. [DOI] [PubMed] [Google Scholar]
- (27).Deng L, Broom A, Kitova E, Richards M, Zheng R, Shoemaker G, Meiering E, Klassen J. J. Am.Chem. Soc. 2012;134:16586–16596. doi: 10.1021/ja305213z. [DOI] [PubMed] [Google Scholar]
- (28).Deng L, Kitova E, Klassen J. J. Am. Soc. Mass Spectrom. 2013;24:988–996. doi: 10.1007/s13361-013-0651-8. [DOI] [PubMed] [Google Scholar]
- (29).Benesch J, Aquilina J, Ruotolo B, Sobott F, Robinson C. Chem. Biol. 2006;13:597–605. doi: 10.1016/j.chembiol.2006.04.006. [DOI] [PubMed] [Google Scholar]
- (30).Joanna F, Matthew FB, Carol VR, Brandon TR. Chem. Phys. Lett. 2012;524 [Google Scholar]
- (31).Pagel K, Hyung SJ, Ruotolo BT, Robinson CV. Anal. Chem. 2010;82:5363–5372. doi: 10.1021/ac101121r. [DOI] [PubMed] [Google Scholar]
- (32).Zhong Y, Hyung S-J, Ruotolo B. Expert Rev. Proteomics. 2012;9:47–58. doi: 10.1586/epr.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Hall Z, Politis A, Robinson C. Structure. 2012;20:1596–1609. doi: 10.1016/j.str.2012.07.001. [DOI] [PubMed] [Google Scholar]
- (34).Zhou M, Sandercock A, Fraser C, Ridlova G, Stephens E, Schenauer M, Yokoi-Fong T, Barsky D, Leary J, Hershey J, Doudna J, Robinson C. Proc. Nat.Acad. Sci. U.S.A. 2008;105:18139–18144. doi: 10.1073/pnas.0801313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Mason EA, McDaniel EW. Transport Properties of Ions in Gases. John Wiley & Sons; New York: 1988. [Google Scholar]
- (36).Scarff C, Thalassinos K, Hilton G, Scrivens J. Rapid Comm. Mass Spectrom. 2008;22:3297–3304. doi: 10.1002/rcm.3737. [DOI] [PubMed] [Google Scholar]
- (37).Ruotolo BT, Benesch JLP, Sandercock AM, Hyung SJ, Robinson CV. Nature Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- (38).Zhou M, Morgner N, Barrera N, Politis A, Isaacson S, Matak-Vinković D, Murata T, Bernal R, Stock D, Robinson C. Science. 2011;334:380–385. doi: 10.1126/science.1210148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).van Duijn E, Barbu I, Barendregt A, Jore M, Wiedenheft B, Lundgren M, Westra E, Brouns SJ, Doudna J, van der Oost J, Heck A. Molec. Cell. Proteomics. 2012;11:1430–1441. doi: 10.1074/mcp.M112.020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Baldwin A, Lioe H, Hilton G, Baker L, Rubinstein J, Kay L, Benesch J. Structure. 2011;19:1855–1863. doi: 10.1016/j.str.2011.09.015. [DOI] [PubMed] [Google Scholar]
- (41).Politis A, Park A, Hyung S-J, Barsky D, Ruotolo B, Robinson C. Plos One. 2010;10:e12080. doi: 10.1371/journal.pone.0012080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Bernstein S, Dupuis N, Lazo N, Wyttenbach T, Condron M, Bitan G, Teplow D, Shea J-E, Ruotolo B, Robinson C, Bowers M. Nature Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Shoemaker G, van Duijn E, Crawford S, Uetrecht C, Baclayon M, Roos W, Wuite G, Estes M, Prasad BV, Heck A. Molec. Cell. Proteomics. 2010;9:1742–1751. doi: 10.1074/mcp.M900620-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Smith D, Woods L, Radford S, Ashcroft A. Biophys. J. 2011;101:1238–1247. doi: 10.1016/j.bpj.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Giles K, Williams JP, Campuzano I. Rapid Comm. Mass Spectrom. 2011;25:1559–1566. doi: 10.1002/rcm.5013. [DOI] [PubMed] [Google Scholar]
- (46).Zhong Y, Hyung SJ, Ruotolo BT. Analyst. 2011;136:3534–3541. doi: 10.1039/c0an00987c. [DOI] [PubMed] [Google Scholar]
- (47).Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Anal. Chem. 2010;82:9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- (48).David EC, Martin FJ. J. Mass Spectrom. 1997;32 [Google Scholar]
- (49).Benesch J, Ruotolo B. Curr. Opin. Sruct.Biol. 2011;21:641–649. doi: 10.1016/j.sbi.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Jackson M, Mantsch H. Biochim. Biophys. Acta. 1991;1078:231–235. doi: 10.1016/0167-4838(91)90563-f. [DOI] [PubMed] [Google Scholar]
- (51).Han L, Ruotolo B. Angew. Chem. Int. Ed. 2013;52:8329–8332. doi: 10.1002/anie.201301893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Wooll J, Friesen R, White M, Watowich S, Fox R, Lee J, Czerwinski E. J. Mol. Biol. 2001;312:525–540. doi: 10.1006/jmbi.2001.4978. [DOI] [PubMed] [Google Scholar]
- (53).Paci E, Vendruscolo M, Karplus M. Biophys. J. 2002;83:3032–3038. doi: 10.1016/S0006-3495(02)75308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Zarrine-Afsar A, Wallin S, Neculai A, Neudecker P, Howell P, Davidson A, Chan H. Proc. Nat.Acad. Sci. U.S.A. 2008;105:9999–10004. doi: 10.1073/pnas.0801874105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Wyttenbach T, Bowers M. The journal of physical chemistry. B. 2011;115:12266–12275. doi: 10.1021/jp206867a. [DOI] [PubMed] [Google Scholar]
- (56).Jianwei L, John AT, Anne EC, David EC. Int. J. Mass Spectrom. 1999;185-187 [Google Scholar]
- (57).Pierson N, Chen L, Valentine S, Russell D, Clemmer D. J. Am. Chem. Soc. 2011;133:13810–13813. doi: 10.1021/ja203895j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.