Abstract
Helicobacter hepaticus was discovered in 1992 as a cause of liver cancer in the A/JCr mouse model. In susceptible mice, infection by H. hepaticus causes chronic gastrointestinal inflammation leading to neoplasia. It can also cause morphological changes in breast-glands leading to neoplasm and adenocarcinoma in mouse models. Studies performed on humans have revealed that H. hepaticus may also be a human pathogen since infection by H. hepaticus can be associated with cholecystitis, cholelithiasis and gallbladder cancer. H. hepaticus is a close relative of H. pylori, but it lacks the major virulence factors of H. pylori including vacoulating cytotoxin A (VacA) and cytotoxin associated gene (cagA). Moreover, SabA, AlpA, and BabA, three important adhesin proteins of H. pylori, are absent in its genome. In contrast, the genome of H. hepaticus contains genes encoding some orthologus virulence factors of Campylobacter jejuni such as cytolethal distending toxin (CDT), and PebI adhesin factor. Other genes including 16S rRNA, 18 KDa immunogenic protein, and urease structural subunits are related to H. pylori. Its genome contains a small island consisting of 71 Kbp named HHGI1, which probably encodes a secretion system type IV (T4SS), and some other virulence factors. As far as the immunogenic antigens are concerned, the lipopolysaccharide (LPS) and flagellin of H. hepaticus are weak stimulants of the immune system, while pro-inflammatory responses are mainly induced by its lipoproteins and most likely by the peptidoglycan. Concerning the multidrug efflux pumps, a homologue of H. pylori TolC, HefA, has been observed in H. hepaticus which contributes to resistance to amoxicillin and bile acids.
Keywords: Helicobacter hepaticus, Identification, Metabolism, Virulence factors, Efflux pumps
INTRODUCTION
In 1992, the pathologists at the Fredrick-Institute of the Cancer Research Center, experiencing the effect of chemicals on long cancer in mouse model found that the mice A/JCr serving as negative controls, exhibited an unexpected rate of cancer. Histopathological evaluation of this control group suggested presence of liver tumor in a large number of them. Their food, water and litter were negative for presence of any agent as potential cause of chronic hepatocellular tumors and liver inflammations. Furthermore, the mice that were kept and fed separately also developed liver Pathology. Liver inflammation was more severe in male mice than in females and could be observed in numerous cases of mouse strains except C57BL/6NCr. Moreover, infection could be transmitted to uninfected mice by homogenized liver suspensions. Silver staining of the specimens obtained from the mice livers demonstrated presence of the spiral bacteria in bile and its canaliculi which could grow in microaerophilic conditions (1–2). Later studies have revealed that naturally infected mice can develop a local unpurulent necrosing liver inflammation which progress to active chronic liver inflammation. Development of active chronic inflammation in liver, the mediator of hepatocellular neoplasm in H. hepaticus infected mice, has led to the conclusion that this organism is a mice pathogen (3–4). Further analysis has shown that infection by H. hepaticus increase nitrogen and oxygen active substances, which lead to an oxidative stress in liver, a process that plays an important role in the liver cancer (2). The investigators have also noted that Tumor Necrosis Factor-α (TNF-α), induced through infection by H. hepaticus, have been involved in the development of cancer in abdominal cavity, liver and other organs (5). In addition, several studies performed in mouse models have suggested that H. hepaticus can trigger mammary carcinoma. Mechanism, which have been proposed for development of the mammary carcinoma, would be related to the fact that dysregulation of the host immune responses due to infection by enteric bacteria, may induce development of extraintestinal cancers (5).
H. hepaticus has received the most attention since it was the first Helicobacter sp. that was recognized as a cofactor for the hepatic carcinogenesis. Induction of malignancies in mice after exposure to H. hepaticus provides a useful model to study the pathogenesis of infection and generation of the liver cancer in humans. The pivotal roles of the innate immunity cells in colorectal cancer have also been revealed after infection of immunodeficient mice by H. hepaticus (6–9).
In human, presence of H. hepaticus in the bile samples of patients with cholelithiasis, cholecystitis and gallbladder polyps, has been traced by nested PCR and in situ analysis of the bile samples. Further studies on pathogenesis of H. hepaticus have supported the hypothesis that H. hepaticus could be a human pathogen and associated with diseases of liver and biliary tract (10–12). Investigaters have also reported that H. hepaticus may be a risk factor for the progression of liver disease to cirrhosis and hepatocellular carcinoma, especially among the patients chronically infected with hepatitis C virus (13–14). In addition, higher titers of specific anti-H. hepaticus antibodies in patients with gallbladder cancer, compared to the control group, has suggested that H. hepaticus infection may be associated with gallbladder cancers in human (15–16). The researchers have also observed that patients with cholelithiasis and cholecystitis associated with gastric cancer had significantly higher prevalence of H. hepaticus infection than patients with other diseases (12, 15). Although H. hepaticus infection have been observed in 82% of gallbladders and 87.5% of related malignancies, it is not clear whether this organism is causative of gallstone, conducting to malignancy, or contributes as a risk factor (16).
H. hepaticus may enter the human body through the contaminated food and water. It can invade the tissues and produce chemical carcinogens that potentially damage DNA. Production of multiple gene mutations may then transform the normal cells into the cancer cells. H. hepaticus has also been detected on the intestinal epithelium surface and depth of crypts particularly in cecum. In fact, H. hepaticas may be present more frequently in intestine than in liver. Therefore, it is suggested that intestine is the primary site of H. hepaticus colonization in human (5, 12, 16).
In this paper we reviewed the literature for the potential pathogenicity of this microaerophilic bacterium and its differences with the human specific pathogen, H. pyori.
General Features of H. hepaticus
H. hepaticus is a spiral bacterium with 1.5-5 µm length and 0.2-0.3 µm width that is smaller than H. pylori. This species can grow in both anaerobic and microaerophilic conditions. Its bipolar flagella are sheathed but contrary to H. pylori, lack the periplasmic fibers (Fig. 1).
Fig. 1.
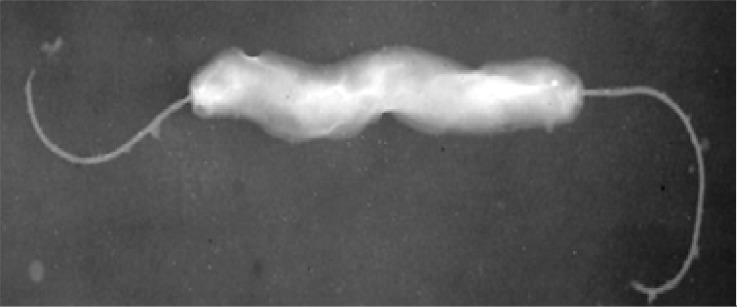
Helicobacter hepaticus with bipolar flagella (2).
H. hepaticus 51449 ATCC strain contains a circle chromosome with 1799146 bp and its G + C content is 35.9% that is between G+ C% of H. pylori and C. jejuni. The size of its genome is a little larger than those of H. pylori and C. jejuni, containing 1875 open reading frame (ORF) that expresses 1875 proteins. Most of H. pylori virulence factors, including almost of cag pathogenicity islands, are absent in H. hepaticus. Interestingly, the homologues of C. jejuni cdt (cytotoxin) and pebI (adhesin) genes have been found in its genome (17). There are 9, and 4 chemosensory proteins in H. hepaticus, and H. pylori, respectively suggesting that H. hepaticus interacts with more chemical agents than H. pylori for its spatial orientation. H. hepaticus lacks the secretion system (comB locus) which is used for natural competition in H. pylori (17–18). Evaluation of 16S rRNA gene sequences has shown that H. hepaticus is the closest relative of Helicobacter muridarum (19).
Similar to H. pylori, hepaticus is catalase and oxidase positive and rapidly hydrolyses urea. However, it can hydrolyse nitrate to nitrite and can also produces H S. It is resistant to cephalotin and nalidixic acid but susceptible to metronidazole (2). Metabolic capabilities of H. hepaticus, and H. pylori are likely to be similar however, there are sufficiently differences between their metabolic potentials to provide interesting view of their basic physiology. There is a possibility for the expression of a NADH-1 and NADH-2 dehydrogenase, cytochrome bd and cytochrome cbb3 terminal oxidase in H. hepaticus. So, with high diversity in respiratory system, H. hepaticus can adapts with harsh conditions of intestinal tract, liver and gallbladder. Respiratory chain of H. pylori with only one NADH-1 dehydrogenase and cytochrome cbb3 terminal oxidase has the lower diversity than H. hepaticus (17). Furthermore, there are important differences between these two species concerning the genes encoding for tricarboxylic acid cycle components. For example, three and four out of the genes that encode five oxidizing metabolite enzymes from α-keto glutarate to oxaloacetate are absent in H. hepaticus and H. pylori, respectively. Among these enzymes, succinyl-CoA-acetoacetyl-CoA transferase is absent in both H. pylori and H. hepaticus but the gene encoding malate dehydrogenase is present in H. hepaticus, only. This suggests the role of a tricarboxylic acid branch in H. hepaticus metabolism, which acts in reductive pathway, a characteristic observed in many anaerobic bacteria (17).
Identification of H. hepaticus
Culture and Isolation
The samples (feces, biopsy and tissue) can be stored in Brain Heart Infusion broth or Brucella broth containing 30% glycerol at -70°C. To isolate H. hepaticus, homogenization of fresh samples in phosphate buffered saline (PBS) and filtration by 0.45 µm filter before cultivation on blood agar containing trimetoprim, vancomycin and polymixin B (11, 20) is recommended. Incubation under microaerophilic condition for a minimum period of 3-7 is also required. Furthermore, it was noted that inoculation of bacteria in Brucella broth with 5% bovine fetal serum and incubation with shaking for 24-48 h would increase its growth speed (11). On culture plates, H. hepaticus has been observed as mucoid film or under spreading form without development of the isolated colonies. The experiments have shown that the dilution methods cannot be used for quantitative identification of H. hepaticus since it cannot produce isolated colonies on solid medium (11). H. pylori produce the isolated small colonies on culture plates however, some strains of H. pylori could also generate the mucoid or spreading colonies, as demonstrated in Fig. 2. Unlike other enterohepatic species of Helicobacter such as H. bilis, H. fennelliae and H. pullorum, H. hepaticus have not yet been cultured from human although its role in inflammatory diseases of human has been demonstrated (11, 20).
Fig. 2.
A H. pylori strain isolated from biopsy specimen producing the spreading colonies on Bleu-horizont agar plate (21).
Histopathology and antibody based tests
It is possible to localize H. hepaticus in samples, using Warthin-Starry or Steiner silver staining. Moreover, the application of immunoflourcent rabbit antisera containing polyclonal anti-H. hepaticus in liver parenchyma and gallbladder is possible (1, 4).
Commercial serologic tests for diagnosis of H. hepaticus infections are not available. However, using cellular particles, membrane digested products or recombinant antigens, the anti H. hepaticus specific IgGs have been evaluated by ELISA test (22). Because of cross-reaction between H. hepaticus and H. pylori, antiserums against H. pylori can be used for detection of H. hepaticus in mice liver tissue by immunohistochemistry with biotin avidin (1). Commercial kits using polyclonal antibodies against H. pylori have been evaluated for specific detection of H. pylori in human biological samples such as stool (23). However, for the reason of cross-reaction between H. pylori and hepaticus, these commercial antibodies cannot be used for the detection of H. hepaticus infection in humans. However, a new monoclonal antibody obtained from hybridoma clone (HRII-51) has shown a high specificity for H. hepaticus without any cross-reaction with other gastrointestinal bacteria. Using ELISA, the sensitivity and specificity of this antibody directed against a 15 KDa molecular weight immune reactive antigen have been reported 87% and 97.6%, respectively (22).
Polymerase chain reaction
Using the commercial kits conceived for DNA extraction from tissue, H. hepaticus have been identified by PCR in scratched samples from cecum (24). Nucleotide sequences of ureAB genes have been employed in RFLP and PCR test for identification of H. hepaticus. Real time PCR has also been developed for identifying H. hepaticus in cecum and fecal samples by detection of cdtB gene (25).
Virulence Factors
Cytolethal distending toxin (CDT)
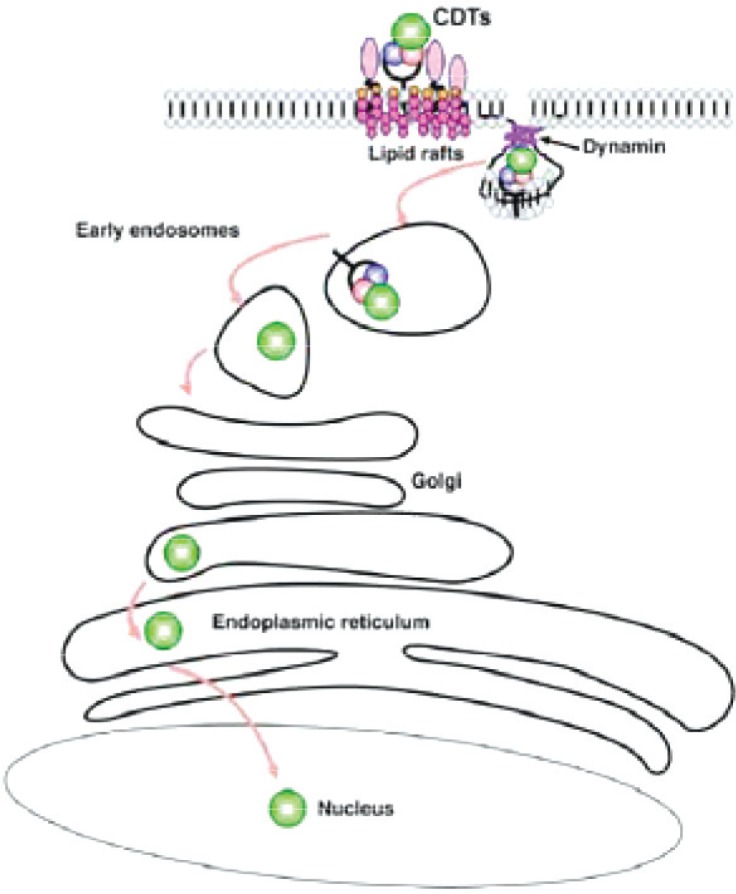
CDT is a heterodimeric A-B2 toxin protein encoded by three neighboring genes namely cdtA, cdtB and cdtC, which are essential for cellular cytotoxicity (20). CdtA and CdtB subunits create non-globular amino acid extensions and these extensions interact with CdtC subunit (26). CdtB is a conserved component of holotoxin in CDT producing bacteria and CdtA is responsible for attaching to the cell membrane, while CdtC helps to transmit CdtB into the nucleus (27). The nature of surface receptor for this toxin is not fully understood but contact of CDT with healthy lipid rafts is needed for its entry via dynamin dependent endocytosis. Fig. 3, schematize the retrograde transmission of toxin via Golgi complex to endoplasmic reticulum and then to the nucleus where its toxic effects may be manifested (27). CdtB is an Mg+2 and Ca+2 dependent neutral nuclease, containing DNA hydrolyzing and cation binding domains (28). It hydrolyzes the double strands DNA via phosphodiester bounds and creates the mono and oligo deoxyribonucleotides. After entry to target cells, CDT can progressively cause cytoplasm and nuclear extension, and stop cell growth in G2/M phase of eukaryotic cell cycle (28). Thus, by preventing the growth of the infected host cells, CDT helps in the persistence of infection. By its direct and /or indirect effects on T cell and antigen presenting cells, CDT is also able to interrupt the immune response. Therefore, CTD play a principal role in colonization of intestinal tract and increases the severity of mucosal inflammation in the liver diseases of sensitive mice strains (29).
Fig. 3.
Pathway of CDT entry to the host cell (27).
It was also revealed that CDT of H. hepaticus plays a principal role in the activation of pre-inflammatory NF-kB pathway during the progress of infectious hepatitis to dysplasia injuries and increases hepatocytes development. Pre-inflammatory NF-kB pathway is activated during the progress of infectious hepatitis to dysplasia injuries; therefore, CDT may have the carcinogenic potential in vivo conditions (30). Induction of apoptosis throughout H. hepaticus infection was also observed and apoptotic bodies are formed in the last apoptotic stage, which can induce anti-inflammatory cytokines and reduce the pre-inflammatory cytokines for facilitating immune inhibitory effects. Apoptosis could be induced via two major pathways: Extrinsic pathway that starts with activation of death receptors while intrinsic pathway is activated with a change in mitochondrial membrane potential. Experiments have revealed that contact of the INT407 cells with CDT activates caspase 3, 7, 9, suggesting involvement of mitochondrial apoptosis in the case of H. hepaticus infection (31).
Other in vitro cytotoxic activity of CDT is granulating activity that is different from the activity of VacA in H. pylori (29). There is no cdt gene in H. pylori but, regarding its mechanism of action, CDT may be an equivalent gene of VacA as immune regulatory toxin in H. hepaticus.
Urease
Like as H. pylori, hepaticus expresses a multimeric urease enzyme that uses nickel as a cofactor. In H. pylori, active urease hydrolyses the urea into ammonium and bicarbonate, which protects this bacterium from the gastric acidic microenvironment and acts as a nitrogen source. H. hepaticus is sensitive to acid and addition of urea into the cultured medium, cannot protect the bacteria from acidic pH 3. Therefore, H. hepaticus urease is not involved in resistance to acid. Urease gene cluster of H. pylori and H. hepaticus contains structural genes (ureAB) and accessory genes (ure IEFGH) however, the distance between ureI and ureB in H. hepaticus is 9 bp smaller than that of H. pylori, suggesting that there is no promoter in upstream of ureI in H. hepaticus. This fact limits the probability of regulation at transcription level or afterwards (32–33).
In H. pylori, urease system is induced via nickel at transcription level; it is dependent on nickel and regulatory protein NikR that regulates the nickel adsorption via another gene, namely nixA (32).
Presence of nikR in H. hepaticus genome has been demonstrated, but its role in the regulation of urease was not quite determined. It was proposed that NikR affects independently urease activity, via regulation of nickel adsorption. In addition, the urease system of H. hepaticus lacks a homologue of nixA although; an ABC transporter gene was detected near its urease operon that may be involved in nickel uptake. Therefore, unlike H. pylori, regulation of urease via nickel in H. hepaticus may not be at transcriptional level. It is proposed that this regulation may at translational level, presumably via activation of urease apoenzyme, like the case of Streptococcus salivarius (33). It should be noted that enteropathogenic Helicobacter species of rodent's intestinal tract does not require the high level of urease activity but there is a high level of urease activity in gastric Helicobacter sp. Therefore, Enteropathogenic Helicobacter sp. use urease system for nitrogen metabolism and ammonium store, only.
Table 1.
General features of H. hepaticus genome (17).
| Total size | 1, 799,146 |
|---|---|
| GC content,% | 35.9 |
| Coding sequences | 1,875 |
| Average gene length, bp | 1,082 |
| Coding density,% | 93,04 |
| Predicted secreted proteins | 347 |
| Predicted membrane proteins | 358 |
| Predicted proteins with assigned function | 1,022 |
| Ribosomal RNA | 1×16s-23s-5s |
| tRNA | 37 (7clusters, 15 single genes) |
DNA binding protein from starved cells (DPS)
DPS is a member of ferritin like proteins, identified in H. hepaticus. The mutant of H. hepaticus lacking DPS cannot grow in conditions with 3% oxygen. It is more sensitive to oxidative reagent such as H2O2, cumene, hydroperoxide and t-butyl hydroperoxide and has more damaged DNA, which can lead to lysis or change to coccoid form (34). Pure DPS protein from H. hepaticus is able to bind to both iron and DNA, and compared to the natural DPS or DPS without iron, the DPS-iron form has higher ability to attach to DNA. Phylogenetically, H. hepaticus DPS protein is relative to H. pylori NapA protein. DPS and NapA act by scavenging of irons and protect the bacterial DNA under oxidative conditions. Therefore, in absence of iron, DPS proteins are oligomerized (34). N-terminal of DPS, at its first N-terminal α-helix, is rich in lysine and plays an important role in attachment of DPS to DNA (35).
Catalase
Chronic infection of mice with H. hepaticus has been distinguished by infiltration of neutrophil and macrophages, which lead to produc-tion of reactive oxygen intermediates in cecum or liver. The free radicals of oxygen, secreted during infection may increase the damage of DNA in intestinal hepatocytes or epithelial cells that leads to colitis or hepatitis. To circumvent these substances, H. hepaticus produces a catalase that may be cytoplasmic or periplasmic. Its periplasmic location is similar to that of other Gram-negative bacteria such as Pseudomonas syringae, Brucella abortus and Vibrio fischeri (36). Periplasmic location of catalase in H. hepaticus and other Gram-negative bacteria facilitate the antigenic presentation to mammalian immune system and mediate the immune responses. Therefore, the catalase of H. hepaticus acts as immunogenic target since host can differentiate this catalase from the endogen one (36).
Pathogenicity Islands (HHGI1)
The genome of H. hepaticus contains a large and some small regions with a different G + C content suggesting the horizontal transfer of latter region. The largest region encodes the proteins including three proteins with homology to structural components of type IV secretion system. Unlike many other pathogenicity islands, HHGI1 of H. hepaticus, is not interspersed by the direct repeats and does not have any gene for tRNA. Furthermore, there is an integrase gene like the integrates gene of P4 (HH269) that is present in pathogenicity islands. Existence of secretion systems and other secretary proteins suggest that HHGI1 would be the true pathogenicity islands however, these genomic or pathogenicity islands are not present in all strains of this species. Different studies have suggested that the strains containing HHGI1 are more virulent than the strains lacking these regions (17, 37).
Flagella
H. hepaticus is a spiral bacterium with a bipolar-sheathed flagellum. This filamentous structure is composed of two flagellin subunits, FlaA and FlaB. In relative bacteria, flagellin genes are mainly regulated by sigma factor FliA (σ28). H. hepaticus has two similar copies of the flaA (flaA1 and flaA2) genes that encode major subunits of flagellin FlaA. Inactivation of each copy of these flaA genes has small effect on flagellum morphology and expression of flaA, however; inactivation of flaA-1 has a more prominent effect on the motility of bacterium (18). Mutations in two genes of flaA or in fliA cease FlaA synthesis or produce small flagella; these mutants cannot colonize the mice (38). Genetic documents suggest that the components and regulatory genes of H. hepaticus flagellum are completely relative to H. pylori. In addition, like those of H. pylori, the flagellar genes of H. hepaticus, are distributed throughout the bacterial chromosome. In H. hepaticus, flagella are the important antigenic targets for innate and adaptive immune systems. Moreover, its hook protein, FlgE is a T-cell dependent dominant antigen (38–39).
Adhesins and outer membrane proteins
The factors involved in colonization and virulence of H. hepaticus are not similar to those of H. pylori. Most of H. pylori adhesin proteins including sabA, alpA, and babA are absent in H. hepaticus. H. hepaticus carries 11 genes that encode the proteins homologue to a large family of outer membrane proteins (17). This family consists of 33 analogous genes classified into two subfamilies; Hop and Hor. The adhesins of H. pylori, BabA, SabA, AlpA, AlpB and HopA-E may be considered as prototypes for Hop family. This subfamily has a sectile amino terminal motif while Hor proteins with unknown function lack this motif. Slight comparison of H. hepaticus outer membrane proteins with other outer membrane proteins, provides no clear result about their functions (17). None of Hop proteins in H. hepaticus contains the typical Hop protein amino terminal motif. Phylogenetically comparison of outer membrane, either Hor, or Hop proteins of H. hepaticus with porins of E. coli (HH0525, HH1713, HH0661, HH1453, HH0812), suggests that these proteins may be the porins. A few OMPs of H. hepaticus are related to Hor proteins such as Hor G of H. pylori. In general, there is no significant similarity between outer membrane proteins of H. hepaticus and those of H. pylori. H. hepaticus does not colonize the human gastric epithelium and among its proteins involved in the attachment to epithelial cells, one protein (HH1481) demonstrates 72% homology with PebI of C. jejuni, a protein not found in H. pylori (17, 20).
Lipopolysaccharide (LPS)
LPS and its lipid A, is not well studied in H. hepaticus however, the lipid content (small chains of fatty acids) of H. hepaticus is different from that of H. pylori. Among Helicobacter sp., LPS of H. hepaticus and enterohepatic Helicobacter sp. display lower activity in limulus amebocyte assay (39). In susceptible mice, H. hepaticus can escapes or inhibit innate immune response of gastric epithelium (18). It is revealed that bacterial lysate and especially soluble components of LPS disturb innate immune responses via TLR4, and TLR5. Inhibition of innate immune responses by H. hepaticus LPS, can affect the responses to resident microbial flora, epithelial homeostasis and inflammatory conditions of intestine (39).
Antibiotic-resistance and efflux pumps
Resistance to commonly used antibiotics is frequent in the case of H. pylori infection (40–41). H. hepaticus is able to resist to antimicrobial agents but contrary to H. pylori, it is also resistant to bile acids. Multiple studies on the mechanisms of resistance to antibiotics have shown that multidrug efflux pumps are involved in the resistance of H. pylori to structurally unrelated antibiotics (42–44). A homologue of H. pylori TolC, named HefA, have been observed in H. hepaticus, which is involved in the resistance to amoxicillin and bile acids (45). An outer membrane protein that is a component of resistance-nodulation-cell division (RND) family is involved in multidrug efflux of antibiotics in H. pylori. A difference between H. pylori TolC and its homologue in H. hepaticus is that, in H. pylori, TolC is not involved in resistance to amoxicillin (45). Two other genes, HH0174, and HH0175, are identified in the genomic sequence of H. hepaticus that are homologous to inner and periplasmic proteins of CmeABC in H. pylori, which may be involved in resistance to bile, macrolide and tetracycline. So, resistance to bile in H. hepaticus may be regulated by both hefA and CmeAB orthologus may be regulated by both hefA and CmeAB orthologus (45, 46).
Treatment
Different treatment regimens for eradication of H. hepaticus infection in mice are described but in many cases, treatment has not been successful although H. hepaticus may be sensitive to many antibiotics. Two weeks treatment with amoxicillin via drinking water has not been effective in elimination of infections (41). Prescription from drinking water is less effective than that of gavages since H. hepaticus was isolated from the mice that were treated via drinking water. Treatment regiments composed of three drug including combination of amoxicillin, metronidazole and bismuth administrated three times by day for two weeks was more effective via gavages in non-immune mice (6-8 weeks-old). However, this difficult method may limit the success of therapy. In general, contradictory results were obtained concerning the effectiveness of the different regimens. For example, in one study, treatment with four drug regimens including amoxicillin, metronidazole, clarithromycin and omperazole was not able to eradicate H. hepaticus and the role of amoxicillin was not definite in this treatment regiment (47–49). Also, treatment of naturally infected mice (8-10 weeks-old) suggested that single dose of amoxicillin, metronidazole and tetracycline was not able to eradicate infection of intestinal tract but amoxicillin or tetracycline in combination with metronidazole and bismuth for two weeks was effective in eradication of liver, cecum and colon infection (47). Today, three drug regimens (inhibitor of proton pumps, amoxicillin and clarithromycin or metronidazole) that are frequently recommended for eradication of human Helicobacter infection are also used for treatment of H. hepaticus infection in mice. This regiment includes three drug administration including metronidazole, amoxicillin/tetracycline and bismuth for two weeks in mice A/JCr (47–49).
CONCLUSION
H. hepaticus is a Gram-negative bacterium with pathogenicity in mice and humans. Although it has some similarities with H. pylori, but shares some characteristics with C. jejuni, suggesting that some of its genes may have been acquired from C. jejuni. This would be consistent with the suggestion that the intestine is the primary site of H. hepaticus colonization in humans. Since this bacterium is implicated in human associated diseases such as gallbladder cancer, cholecystitis, cholelithiasis and other yet unidentified diseases, research pertaining to this field will be of utmost importance in this region as well as in other parts of the world.
REFERENCES
- 1.Ward JM, Anver MR, Haines DC, Benvenistet RE. Chronic active hepatitis in mice caused by Helicobacter hepaticus . A J Pathol. 1994;145:959–968. [PMC free article] [PubMed] [Google Scholar]
- 2.Fox JG, Dewhirst FE, Tully JG, Paster BJ, Yan L, Taylor NS, et al. Helicobacter hepaticus sp. nov: a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol. 1994;32:1238–1245. doi: 10.1128/jcm.32.5.1238-1245.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fox JG, Li X, Yan L, Cahill RJ, Hurley R, Lewis R, Murphy JC. Chronic proliferative hepatitis in A/ JCr mice associated with persistent Helicobacter hepaticus infection: a model of Helicobacter-induced carcinogenesis. Infect Immun. 1996;64:1548–1558. doi: 10.1128/iai.64.5.1548-1558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward JM, Fox JG, Anver MR, Haines DC, George CV, Collins MJ, et al. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J Natl Cancer Inst. 1994;86:1222–1227. doi: 10.1093/jnci/86.16.1222. [DOI] [PubMed] [Google Scholar]
- 5.Rao VP, Pootahidis T, Fox JG, Fridman SE. Breast cancer: should gastrointestinal bacteria be on our Radar screen. Cancer Res. 2007;67:847–850. doi: 10.1158/0008-5472.CAN-06-3468. [DOI] [PubMed] [Google Scholar]
- 6.Erdman SE, Poutahidis T, Tomczak M, et al. CD4 + CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2 deficient mice. Am J Pathol. 2003;162:691–702. doi: 10.1016/S0002-9440(10)63863-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powrie F, Maloy KJ. Immunology: Regulating the regulators. Science. 2003;299:1030–1031. doi: 10.1126/science.1082031. [DOI] [PubMed] [Google Scholar]
- 8.Corpet DE, Pierre F. Point: from animal models to prevention of colon cancer. Systematic review of chemoprevention in min mice and choice of the model system. Cancer Epidemiol Biomarkers Prev. 2003;12:391–400. [PMC free article] [PubMed] [Google Scholar]
- 9.Sterzenbach T, Lee SK, Brenneke B, von Goetz F, Schauer DB, Fox JG. Inhibitory Effect of Enterohepatic Helicobacter hepaticus on Innate Immune Responses of Mouse Intestinal Epithelial Cells. Infect Immun. 2007;75:2717–2728. doi: 10.1128/IAI.01935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nilson I, Lindgren S, Eriksson S, Wadström T. Serum antibodies to Helicobacter hepaticus and Helicobacter pylori in patients with chronic liver diseases. Gut. 2000;46:410–414. doi: 10.1136/gut.46.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whary MT, Fox JG. Natural and Experimental Helicobacter Infection . American Association for Laboratory Animal Science. 2004;54:128–158. [PubMed] [Google Scholar]
- 12.Hamada T, Yokota K, Ayada K, Hirai K, Kamada T, Haruma K, et al. Detection of Helicobacter hepaticus in human bile samples of patients with biliary disease. Helicobacter. 2009;14:545–551. doi: 10.1111/j.1523-5378.2009.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishra RR, Tewari M, Shukla HS. Helicobacter species and pathogenesis of gallbladder cancer. Hepatobiliary Pancreat Dis Int. 2010;9:129–134. [PubMed] [Google Scholar]
- 14.Pellicano R, Ménard A, Rizzetto M, Mégraud F. Helicobacter species and liver diseases: association or causation? Lancet Infect Dis. 2008;8:254–260. doi: 10.1016/S1473-3099(08)70066-5. [DOI] [PubMed] [Google Scholar]
- 15.Shimoyama T, Takahashi R, Abc D, Mizuki I, Endo T, Fukuda S. Serological analysis of Helicobacter hepaticus infection in patients with biliary and pancreatic diseases. J Gastroenterol Hepatol. 2010;25:S86–89. doi: 10.1111/j.1440-1746.2010.06224.x. [DOI] [PubMed] [Google Scholar]
- 16.Pradhan SB, Dali S. Relation between gallbladder neoplasm and Helicobacter hepaticus infection. Kathmands University Medical Journal. 2004;2:331–335. [PubMed] [Google Scholar]
- 17.Suerbaum S, Josenhans C, Sterzenbach T, Drescher B, Brandt P, Bell M, et al. The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus . Proc Natl Acad. Sci USA. 2003;100:7901–7906. doi: 10.1073/pnas.1332093100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sterzenbach T, Bartonickova L, Behrens W, Brenneke B, Schulze J, Kops F, et al. Role of the Helicobacter hepaticus flagellar sigma factor fliA in gene regulation and murine colonization. J Bacteriol. 2008;190:6398–6408. doi: 10.1128/JB.00626-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Battles JK, Williamson JC, Pike KM, Gorelick PL, Ward JM, Gonda MA. Diagnostic assay for Helicobacter hepaticus based on nucleotide sequence of its 16S rRNA gene. J Clin Microbiol. 1995;33:1344–1347. doi: 10.1128/jcm.33.5.1344-1347.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen LP. New Helicobacter species in Humans. Digest Dis. 2001;19:112–115. doi: 10.1159/000050664. [DOI] [PubMed] [Google Scholar]
- 21.Falsafi T, Valizadeh N, Najafi M, Ehsani A, Khani A, Landarani Z, Falahi Z. Culture of Helicobacter pylori from Stool Sample in Children. Can J Microbiol. 2007;53:411–416. doi: 10.1139/W06-144. [DOI] [PubMed] [Google Scholar]
- 22.Fukuda Y, Shimoyama T, Ohmura T, Sano Y, Nakabayashi N, Takahashi R, et al. Characterization and application of new monoclonal antibody with high specificity for Helicobacter hepaticus . Helicobacter. 2009;14:66–71. doi: 10.1111/j.1523-5378.2009.00652.x. [DOI] [PubMed] [Google Scholar]
- 23.Falsafi T, Valizadeh N, Sepehr S, Najafi M. Application of a stool antigen test to evaluate the incidence of Helicobacter pylori infection in children and adolescents from Tehran, Iran. Clin Diagn Lab Immunol. 2005;12:1094–1097. doi: 10.1128/CDLI.12.9.1094-1097.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge Z, White DA, Whary MT, Fox JG. Fluorogenic PCR-based quantitative detection of a murine pathogen, Helicobacter hepaticus . J Clin Microbiol. 2001;39:2598–2602. doi: 10.1128/JCM.39.7.2598-2602.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shames B, Fox JG, Dewhirst F, Yan L, Shen Z, Taylor NS. Identification of widespread Helicobacter hepaticus infection in feces of commercial mouse clones by culture and PCR assay. J Clin Microbiol. 1995;33:2968–2972. doi: 10.1128/jcm.33.11.2968-2972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arenavd P, Castroviejo M, Claret S, Rosenbaum Q, Megraud F. Expression and activity of the cytolethal distending toxin of Helicobacter hepaticus . Biochem Biophys Res Commun. 2004;318:739–745. doi: 10.1016/j.bbrc.2004.04.089. [DOI] [PubMed] [Google Scholar]
- 27.Gurra L, Cortes-Bratti X, Guidi R, Frisan T. The biology of the cytolethal Distending toxins. Toxins. 2011;3:172–190. doi: 10.3390/toxins3030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dassanayake RP, Griep MA, Duhamel G. The cytolethal distending toxin B subunit of Helicobacter hepaticus is a Ca2+ and Mg2+ depndent neutral nuclease. FEMS Microbiol Let. 2005;25:219–225. doi: 10.1016/j.femsle.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Pratt QS, Sachen KI, Mwood HD, Eaton KA, Young VB. Modulation of host immune responses by the cytolethal distending toxin of Helicobacter hepaticus . Infect Immun. 2006;74:4496–4504. doi: 10.1128/IAI.00503-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young VB, Knox KA, Scaver DB. Cytolethal distending toxin sequence and activity in the enterohepatic pathogen Helicobacter hepaticus . Infect Immun. 2000;68:184–191. doi: 10.1128/iai.68.1.184-191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lianage NPM, Manthe KC, Dassanayake RP, Kuszynski CA, Oakley GC, Doamel GE. Helicobacter hepaticus cytolethal distending toxin causes cell death in intestinal epithelial cell via mitochondrial apoptotic pathway. Helicobacter. 2010;15:98–107. doi: 10.1111/j.1523-5378.2010.00749.x. [DOI] [PubMed] [Google Scholar]
- 32.Belzer C, Stoof Q, Beckwith CS, Kuipes EQ, Kusters QG, Vanuliet AHM. Differential regulation of urease activity in Helicobacter hepaticus and Helicobacter pylori . Microbiol. 2005;151:3989–3995. doi: 10.1099/mic.0.28188-0. [DOI] [PubMed] [Google Scholar]
- 33.Beckwith CS, McGee DQ, Mobley HL, Riley LK. Cloning and expression and catalytic activity of Helicobacter hepaticus urease. Infect Immun. 2001;69:5914–5920. doi: 10.1128/IAI.69.9.5914-5920.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong Y, Wang GF, Maier RQ. Helicobacter hepaticus DpS Protein plays an important role in protecting DNA from oxidative damage. Free Radic Res. 2006;40:597–605. doi: 10.1080/10715760600618882. [DOI] [PubMed] [Google Scholar]
- 35.Cooksley C, Qenks PQ, Green A, Cockayne A, Logan RP, Hardie KR. NapA Protects Helicobacter pylori from oxidative stress damage and its production is influenced by the ferric uptake regulator. J Med Microbiol. 2003;52:461–469. doi: 10.1099/jmm.0.05070-0. [DOI] [PubMed] [Google Scholar]
- 36.Alyamani EG, Brandt P, Pana QA, Major AM, Fox GG, Suerbaum S, Versalovic G. Helicobacter hepaticus catalase shares surface predicted epitopes with mammalian catalases. Microbiol. 2007;153:1006–1016. doi: 10.1099/mic.0.29184-0. [DOI] [PubMed] [Google Scholar]
- 37.Hofreuter D, Odenbreit S, Haas R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol Microbiol. 2001;41:379–391. doi: 10.1046/j.1365-2958.2001.02502.x. [DOI] [PubMed] [Google Scholar]
- 38.Fox JG, Ge Z, Whary MT, Erdman SE, Horwitz BH. Helicobacter hepaticus infection in mice: models for understanding lower bowel inflammation and cancer. Nature. 2011;4:22–30. doi: 10.1038/mi.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hynes SO, Ferris QA, Szponar B, Wadstrom T, Fox JG, Roorke J, et al. Comparative chemical and biological characterization of the lipopolysaccharides of gastric and enterohepatic Helicobacters. Helicobacter. 2004;9:313–323. doi: 10.1111/j.1083-4389.2004.00237.x. [DOI] [PubMed] [Google Scholar]
- 40.Alarcon T, Domingo D, Lopez-Brea Antibiotic resistance problems with Helicobacter pylori . Int J Antimicrob Agents. 1999;12:19–26. doi: 10.1016/s0924-8579(99)00051-5. [DOI] [PubMed] [Google Scholar]
- 41.Bontems P, Devaster JM, Corvaglia L, Dezofi LA, Van Den Borre C, Goutier S. Twelve years observation of primary and secondary antibiotic-resistant Helico-bacter pylori strains in children. Pediatr Infect Dis J. 20:1033–1038. doi: 10.1097/00006454-200111000-00006. [DOI] [PubMed] [Google Scholar]
- 42.Kutschke A, de Jonge BLM. Compound Efflux in Helicobacter pylori . Antimicrob Agents Chemother. 2005;49:3009–10. doi: 10.1128/AAC.49.7.3009-3010.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anoushiravani M, Falsafi T, Vahid Niknam V. Proton motive force-dependent efflux of tetracycline in clinical isolates of Helicobacter pylori . J Medical Microbiol. 2009;58:1309–1313. doi: 10.1099/jmm.0.010876-0. [DOI] [PubMed] [Google Scholar]
- 44.Falsafi T, Ehsani A, Niknam V. The Role of Active Eflux in Antibiotic-Resistance of Clinical Isolates of Helicobacter pylori . Ind J Medical Microbiol. 2009;27:335–340. doi: 10.4103/0255-0857.55452. [DOI] [PubMed] [Google Scholar]
- 45.Belzer C, Stoof G, Breiger S, Kusters GG, Kuipers EG, Vanuliet AHM. The Helicobacter hepaticus hefA gene is involved in resistance to amoxicillin. Helicobacter. 2009;14:72–79. doi: 10.1111/j.1523-5378.2009.00661.x. [DOI] [PubMed] [Google Scholar]
- 46.Johnson JM, Church GM. Alignment and structure prediction of divergent protein families: periplasmic and outer membrane proteins of bacterial efflux pumps. J Mol Biol. 1999;287:695–715. doi: 10.1006/jmbi.1999.2630. [DOI] [PubMed] [Google Scholar]
- 47.Kerton A, Warden P. Review of successful treatment for Helicobacter species in laboratory. Mice Laboratory Animals. 2006;40:115–122. doi: 10.1258/002367706776319033. [DOI] [PubMed] [Google Scholar]
- 48.Foltz CJ, Fox JG, Yan L, Shames B. Evaluation of antibiotic therapies for eradication of Helicobacter hepaticus . Antimicrob Agents Chemother. 1995;39:1292–1294. doi: 10.1128/aac.39.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foltz CJ, Fox JG, Yan L, Shames B. Evaluation of various oral antimicrobial formulations for eradication of Helicobacter hepaticus . Lab Anim Sci. 1996;46:193–197. [PubMed] [Google Scholar]