Abstract
Background and Objective
Acinetobacter baumannii is an aerobic non-motile Gram-negative bacterial pathogen that is resistant to most antibiotics. Carbapenems are the most common antibiotics for the treatment of infections caused by this pathogen. Mechanisms of antibiotic-resistance in A. baumannii are mainly mediated by efflux pumps-lactamases. The aim of this study was to determine antibiotic susceptibility, the possibility of existence of OXAs genes and fingerprinting by Pulsed-Field Gel Electrophoresis (PFGE) among clinical isolates of Acinetobacter collected from Kermanshah hospitals.
Materials and Methods
One hundred and four isolates were collected from patients attending Imam Reza, Taleghani and Imam Khomeini hospitals of Kermanshah (Iran). Isolates were identified by biochemical tests and API 20NE kit. The susceptibility to different antibiotics was assessed with Kirby-Bauer disk diffusion method. PCR was performed for detection of bla OXA-23, bla OXA-24, bla OXA-51 and bla OXA-58 beta-lactamase genes. Clonal relatedness was estimated by PFGE (with the restriction enzyme Apa I) and DNA patterns were analyzed by Gel compare II 6.5 software.
Results
All isolates showed high-level of resistance to imipenem, meropenem as well as to other antimicrobial agents, while no resistance to polymyxin B, colistin, tigecylcine and minocycline was observed. The bla OXA-23like and bla OXA-24 like were found among 77.9% and 19.2% of the isolates, respectively. All isolates were positive for bla OXA-51, but none produced any amplicon for bla OXA-58. PFGE genotype analysis suggested the existence of eight clones among the 104 strains [A (n = 35), B (n = 29), C (n = 19), D (n = 10), E (n = 4), F (n = 3), G (n = 3), H (n = 1)]. Clone A was the dominant clone in hospital settings particularly infection wards so that the isolates in this group, compared to the other clones, showed higher levels of resistance to antibiotics.
Conclusion
The bla OXA-51-like and bla OXA-23like were the predominant mechanisms of resistance to imipenem in A. baumannii. A high prevalence of clone A, B and C in different parts of the healthcare system showed that hospitalized patients should be safeguarded to prevent the spread of these clones. Early recognition of the presence of carbapenem-resistant A. baumannii clones is useful for preventing their spread within the hospital environment.
Keywords: Acinetobacter, beta-lactamase, carbapenemase, Pulsed-Field Gel Electrophoresis, Kermanshah
INTRODUCTION
Acinetobacter baumannii is an aerobic non-motile Gram-negative coccobacillus and polymorphic bacterial pathogen that is easily spread from one patient to others, persisting in the environment for many days (1). It is the third most common pathogenic bacteria isolated from hospitalized patients with pneumonia that plays a significant role in nosocomial infections (2–3). Acinetobacter is known to be an important nosocomial pathogen, isolated predominantly in intensive care units (ICUs), and responsible for severe infections. A. baumannii are usually multidrug resistant (MDR), showing resistance to the third generation cephalosporins, aminoglycosides and fluoroquinolone (4). Carbapenem resistance among Acinetobacter spp. has been increasing during the last decade so that carbapenem resistant A. baumannii has become a worldwide problem (5, 6). The most common mechanism of imipenem resistance reportedly involves the association between carbapenem-hydrolyzing-β-lactamases belonging to the metallo-β-lactamases (Ambler class B) and oxacillinases (Ambler class D). A. baumannii usually hydrolyze oxacillin more efficiently than benzyl penicillin. (6) Class B carbapenemase including 2 important enzymes IMP and VIM that metallo beta lactamases. (7) Extensive use of antimicrobial chemotherapy, particularly carbapenems, has contributed to the appearance of carbapenem-hydrolyzing class D β-lactamases (CHDLs). These enzymes are frequently identified in A. baumannii. Identification of a CHDL-encoding gene was first reported in 1995 (8). Four major subgroups of acquired CHDLs have been identified in A. baumannii, including OXA-23, OXA-40, OXA-58, and OXA-143 β-lactamase groups in addition to the naturally-occurring OXA-51 β-lactamase (9). The bla OXA-58 like has been identified worldwide, but mostly from France, England, Argentina, Spain, Turkey, Romania, Austria, Greece, Scotland, and Kuwait (10). The significant contribution of these enzymes to carbapenem resistance has been emphasized, particularly when they are accompanied by ISAba1 and ISAba3 in the naturally occurring plasmid (6). Therefore, the present study was aimed to determine the drug susceptibility patterns of A. baumannii. We report an evaluation of CHDL-producing A. baumannii isolates collected from three Hospitals of Kermanshah (Iran). The isolates were assessed for the presence of genes bla OXA-23 like, bla OXA-24 like, bla OXA-51 like and bla OXA-58 like with PCR. Furthermore, in this study, we report an evaluation of CHDL-producing A. baumannii isolates collected from various Hospitals of Kermanshah and the association between their susceptibility and genetic profile as well as PFGE typing for total isolates. Pulsed-Field Gel Electrophoresis (PFGE) was performed to investigate the genetic relation among the isolates and determined wide spread clone (11).
MATERIALS METHODS
Bacterial identification and antimicrobial susceptibility testing
A total of 104 Acinetobacter spp. were cultured from sputum (n = 69), blood (n = 32) and urine (n = 3) clinical specimens in different hospitals of Kermanshah (Iran) during 2010–11. The strains were identified as A. baumannii by conventional biochemical tests and API 20NE kit (version 6.0, bio-Mérieux, Marcy L'Etoile, France) (12). All isolates were tested by the Kirby-Bauer method of disk diffusion according to the CLSI guidelines to check their susceptibilities to amikacine (30 μg), ceftriaxone (30 μg), ciprofloxacin (5 μg), trimethoprim/ sulfamethoxazole (30 μg), gatifloxacine (5 μg), colistin (10 μg), gentamicine (10 μg), imipenem (10 μg), meropenem (10 μg), piperacillin (100 μg), polymyxin B (300 unit), levofloxacin (5 μg), minocycline (30 μg), mezlocilline (75 μg), tetracycline (30 μg), tobramycine (10 μg), cefepime (30 μg), cephpodoxime (10 μg), cefotaxime (30 μg), ceftazidime (30 μg), rifampicine (5 μg) (MAST, Merseyside, UK) (13).
MBL and ESBL screening
The isolates were identified for a phenotypic MBL screening with the E-test MBL (bio-Mérieux, Marcy L'Etoile, France) as per the manufacturer's instructions (14). Indistinguishable MBL producing isolates were detected and confirmed by Cica-Beta-Test Strip (Kanto Chemical, Tokyo) as per the manufacturer's instructions.
Screening for ESBL-producing organisms was carried out using Double Disk Synergy Test (DDST). Those strains that showed resistance to imipenem by disk diffusion test were re-checked in T-test (15).
PCR amplification of OXA genes
PCR screening was performed for the carbapenemase-encoding genes (bla OXA-23-like, bla OXA-24-like, bla OXA-51-like and bla OXA-58-like). PCR analysis was performed using the primers described by Kuo et al. (16).
Pulsed-field gel electrophoresis (PFGE)
All A. baumannii isolates were analyzed by CHEF Mapper PFGE according to the protocols previously described by Durmaz et al. with little modifications (17). We used A. baumannii ATCC 19606 for normalization of the gel (17, 18). The solution absorption coefficient was set at 600 nm. The cell lysis solution I consisted of Tris-HCl 50 mM (pH 8), EDTA 50 mM (pH 8), 1 mg/ml proteinase K and 1.5 mg/ml lysozyme (Roche Applied Sciences) that was kept shaking in water bath at 37°C for 45 min. Lysis solution II consisted of EDTA 0.5 M (pH 8), 0.4 mg/ml proteinase K and 1% sarcosyl was transferred to tubes containing plugs while kept shaking in water bath at 50°C for 2 h. Digestion of all organisms were performed with ApaI (New England Biolabs) restriction enzyme. One half of each plug was transferred to 2 ml tubes that contains 100 μl buffer 1X consisting of ApaI enzyme, and then was incubated at 25°C for 2 h.
Electrophoresis
1% (w/v) of pulsed-field Mega-base Agaros (Bio-Rad laboratories) added into 100 ml of TBE 0.5X (1 liter of 5X stock solution with pH 8). DNA separation was performed in TBE 0.5X (pH 8) buffer in a pulsed-field electrophoresis system (CHEF Mapper; Bio-Rad Laboratories, Hercules, CA, USA) by program two state with the following conditions: temperature 14°C; voltage 6 V/cm; switch angle, 120°; switch ramp 2.2–35 s for 19 h. We used Lambda Ladder PFGE Marker (NEB: N0340) as molecular size marker. The Dice coefficient was used to calculate similarities, and the unweighted paired group method based on average linkages (UPGMA) was used for cluster analysis with Gel compare II version 6.5 (Applied Maths, St Martens-Latem, Belgium) and with a high similar pattern (similarity > 87%) were considered to be derived from a cluster (closely related strains) (17–19).
Statistical analysis
Data were recorded and entered into a database. Statistical analyses were performed using Stata (Version 11.0). Continuous variables were compared using one-way analysis of variance. Variables were analyzed by chi-square test. A P-value of <0.05 was set as the level of statistical significance for all analyses in the study.
RESULTS
A total of 104 clinical isolates of A. baumannii were collected from 3 hospitals in Kermanshah (Iran). The results showed high-level of resistance to imipenem (79.8%) and meropenem (75%) as well as resistance to other antibiotics is shown in (Table 1). Resistance was observed against polymyxin B (13.5%), minocycline (16.3%), colistin (10.6%). Resistance rates for tigecylcine was low resistance (2.9%). 84 isolates (80.8%) produced MBLs. In our study, 85.6% (n = 89) of isolates were able to produce carbapenemase that was significantly associated with imipenem and meropenem resistance (p value <0.001). Furthermore, 32.7% (n = 34) of isolates showed multidrug resistance phenotype, (MDR, resistant to third generation cephalosporins, amino glycosides and fluoroquinolone) and 8.7% (n = 9) pandrug resistant (PDR, resistant to all available antibacterial agents except polymyxin B and colistin) phenotype, whereas none of the isolates was extensively drug resistant phenotype (XDR). XDR has been defined for the strains showing resistance to some of the most effective anti-bacterial drugs (20). Using PCR assay 77.9% and 19.2% of the isolates were positive for bla OXA-23like and bla OXA-24 like genes, respectively. While all isolates were positive for bla OXA-51like, none gave any amplicon for the bla OXA-58 like. Co-existence of bla OXA-23 and bla OXA-24 like was observed among 16.4% of the isolates. The co-relations between carbapenemase and bla OXA-23 like was statistically significant (P value 0.002).
Table 1.
Antimicrobial-susceptibility for Acinetobacter baumannii isolates.
| Antimicrobial | Susceptibility; no. (%) of isolates: | ||
|---|---|---|---|
|
| |||
| Susceptible | Intermediate | Resistant | |
| Amikacine | 37 (36.6) | 10 (9.6) | 56 (53) |
| Ceftriaxone | 3 (2.9) | 6 (5.8) | 95 (91.3) |
| Ciprofloxacin | 32 (30.8) | 0(0) | 72 (69.2) |
| Sulfamethoxazole | 45 (43.3) | 1 (1) | 58 (55.8) |
| Gatifloxacine | 51 (49) | 8 (7.7) | 45 (43.3) |
| Colistin | 93 (89.4) | 0 (0) | 11 (10.6) |
| Gentamicine | 31 (29.8) | 2 (1.9) | 71 (68.3) |
| Imipenem | 17 (16.3) | 4 (3.8) | 83 (79.8) |
| Meropenem | 20 (19.2) | 6 (5.8) | 78 (75) |
| Piperacillin | 21 (20.2) | 3 (2.9) | 80 (76.9) |
| Polymyxin B | 90 (86.5) | 0 (0) | 14 (13.5) |
| Ceftazidime | 30 (28.8) | 1 (0.96) | 73 (70.2) |
| Levofloxacin | 33 (31.7) | 6 (5.8) | 65 (62.5) |
| Minocycline | 79 (76) | 8 (7.7) | 17 (16.3) |
| Mezlocilline | 15 (14.4) | 4 (3.8) | 85 (81.7) |
| Tetracycline | 30 (28.8) | 1 (0.96) | 73 (70.2) |
| Tobramycine | 57 (54.8) | 6 (5.8) | 41 (39.4) |
| Tigecylcine | 100 (96.2) | 1 (0.96) | 3 (2.9) |
| Cefepime | 26 (25) | 2 (1.9) | 76 (73.1) |
| Cephpodoxime | 3 (2.9) | 0 (0) | 100 (96.2) |
| Cefotaxime | 5 (4.8) | 1 (0.96) | 97 (93.3) |
| Rifampicine | 11 (10.6) | 6 (5.8) | 87 (83.7) |
| AMP-Sulbactam | 66 (63.5) | 3 (2.9) | 35 (33.7) |
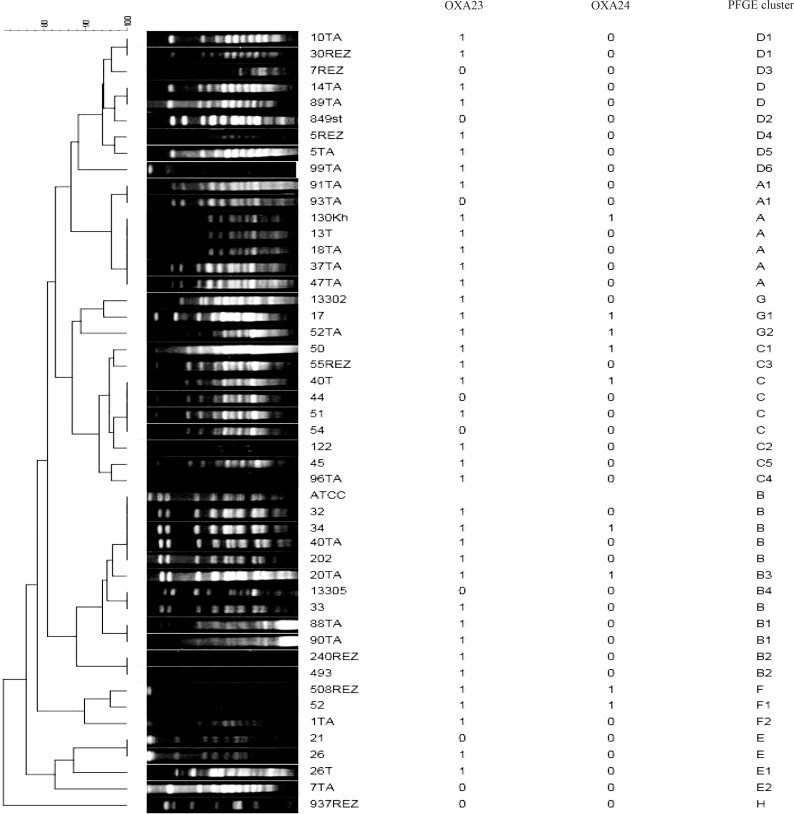
According to Tenover's criteria(18), a major PFGE type (clone A) containing 35 isolates was identified in 3 studied hospitals in this study. A subtyped designated as A1 was found for type A that differed in migration of one to three bands (Fig. 1). Other PFGE patterns include types B (n = 29), C (n = 19), D (n = 10), E (n = 4), F (n = 3), G (n = 3) and H (n = 1)]. The types A, B, C and D were the dominant types found in the hospitals. Type H consisted of single isolate. The PFGE analysis revealed that 18 isolates were closely related. All iso-lates from infection wards belong to clone A (Table 2). All isolates within type F (100%) showed high-level of resistance to imipenem followed by type A (91.4%) (P value 0.03). In addition, 97.1% of the isolates with type A (n = 89) were able to produce carbapenemase (P value 0.023) (Table 2).
Fig. 1.
Pulsed-field gel electrophoresis (PFGE) dendrogram and polymerase chain reaction (PCR) of Acinetobacter baumannii.
Isolates (1= positive, 0= negative for gene)
Table 2.
Comparison of PFGE pattern with antimicrobial susceptibility, OXA genes and source (wards) of isolates (%).
| CloneAntibiotics, genes and wards | Clone A | Clone B | Clone C | Clone D | Clone E | Clone F | Clone G | Clone H | P-value |
|---|---|---|---|---|---|---|---|---|---|
| IPM | 32(91) | 20(69) | 17(89) | 7(70) | 3(75) | 3(100) | 1(33) | 0 | 0.030 |
| MEM | 31(88) | 19(65) | 13(68) | 8(80) | 2(50) | 3(100) | 2(66) | 0 | 0.146 |
| CRO | 32(91) | 27(93) | 18(94) | 10(100) | 3(75) | 3(100) | 1(33) | 1(100) | 0.027 |
| CPD | 35(100) | 29(100) | 19(100) | 9(90) | 2(50) | 3(100) | 2(66) | 1(100) | <0.001 |
| AMP | 35(100) | 29(100) | 18(94) | 9(90) | 2(50) | 3(100) | 3(100) | 1(100) | <0.001 |
| SXT | 20(57) | 15(51) | 14(73) | 2(20) | 3(75) | 1(33) | 3(100) | 0 | 0.079 |
| TGC | 1(2) | 0 | 0 | 2(20) | 0 | 0 | 0 | 0 | 0.094 |
| Carbapenemase | 34(98) | 22(76) | 17(89) | 7(70) | 4(100) | 3(100) | 2(66) | 0 | 0.02 |
| MBL | 30(85) | 22(75) | 16(84) | 7(70) | 3(75) | 3(100) | 2(66) | 1(100) | 0.854 |
| ESBL | 19(54) | 12(42) | 12(63) | 5(50) | 2(50) | 1(33) | 1(33) | 0 | 0.768 |
| MDR | 12(34) | 10(34) | 7(36) | 1(10) | 1(25) | 2(66) | 1(33) | 0 | 0.705 |
| PDR | 4(11) | 2(7) | 2(10) | 1(10) | 0 | 0 | 0 | 0 | 0.823 |
| Oxa23 | 28(80) | 23(79) | 16(84) | 7(70) | 1(25) | 3(100) | 3(100) | 0 | 0.081 |
| Oxa24 | 7(20) | 5(17) | 3(15) | 0 | 1(25) | 2(66) | 2(66) | 0 | 0.113 |
| Urgency | 6(17) | 2(7) | 3(15) | 1(10) | 1(25) | 1(33) | 1(33) | 1(100) | 0.198 |
| ICU | 21(60) | 24(82) | 15(79) | 8(80) | 3(75) | 2(66) | 2(66) | 0 | 0.198 |
| Children | 1(2.8) | 3(10) | 1(5.2) | 1(10) | 0 | 0 | 0 | 0 | 0.198 |
| Infection | 7(20) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.198 |
IPM: Imipenem, MEM: Meropenem, CRO: Ceftriaxone, CPD: Cephpodoxime, AMP: Ampicillin, SXT: trimethoprim/sulfamethoxazole, TGC: Tigecylcine, MBL:, ESBL: extended-spectrum-beta-lactamase, MDR: multidrug resistant, PDR: pandrug resistant, ICU: intensive care units, P marked in bold if <0.05.
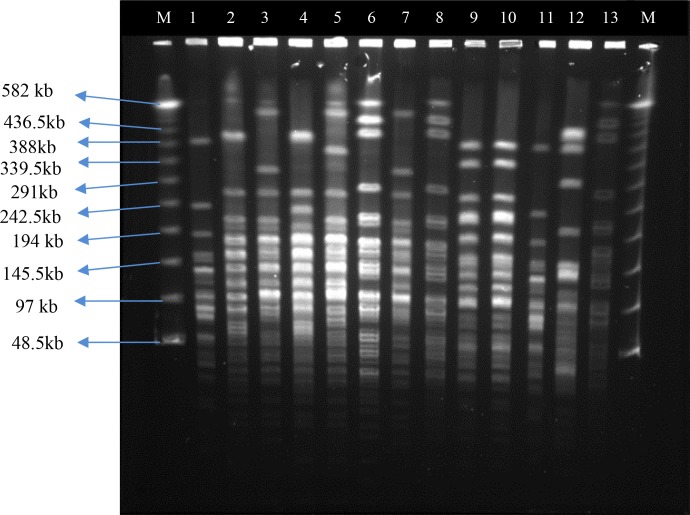
Fig. 2.
CHEF Profiles of A. baumannii strains isolated from different patients. Lateral lanes contain Lambda Ladder PFG Marker, (8 and 13) A. baumannii ATCC 19606. (1, 2 and 4) clone D, D3 and D1, (9 and 10) clone A, (3, 7) clone C5 and C, (5) G1, (6) B, (11) clone F, (12) clone H.
DISCUSSION
Carbapenem resistance has been increasingly common issue among A. baumannii isolates in Iranian hospitals in recent years. This study was aimed to evaluate the molecular epidemiology of carbapenem-resistant A. baumannii in Kermanshah hospitals, with the aim of identifying predominant clonal circulating. Our study showed low susceptibility rates to most of the clinically available antimicrobial agents for the treatment of A. baumannii-induced infections. There was a resistance to polymyxin B, colistin, minocycline, whereas low resistance to tigecylcine (Table 1). This antibiotic can be helpful in treating A. baumannii-related infections in hospital settings. This study reports one of the first large outbreaks of MDR 32.7% A. baumannii in west of Iran. The global incidence of meropenem resistance in A. baumannii was approximately 6% in 1998 but it has dramatically increased to approximately 29% in 2005 (10). This rapid increase has also been happened in the Kermanshah hospitals, where resistance rates to imipenem and meropenem were 79.8% and 75%, respectively. Findings of the present study showed that susceptibility to tigecylcine in 82 out of 83 (98.7%) isolates was significant. A similar increase in the resistance of A. baumannii isolates to imipenem and meropenem was found in the previous studies conducted in Iran during 2006–2010 (5–21). In addition, A. baumannii isolates (obtained in 2006–2007) from Singaporean hospitals were also highly resistant to carbapenems (22). All the clinical A. baumannii isolates obtained in 2006–2007 from Malaysia, exhibited high resistance to all the examined antimicrobial agents except for polymyxin B (Different mechanisms are involved in the A. baumannii resistance to imipenem. β-Lactamase is important factor to carbapenem-resistance (23). The acquisition of carbapenem resistance in A. baumannii is mainly because of the production of two types of β-lactamases: Metallo- β-lactamases (MBLs) and carbapenem-hydrolysing class D β-lactamases (CHDLs). The CHDLs such as bla OXA-23, bla OXA-24, bla OXA-51 appear to be more prevalent and important to carbapenem resistance in this bacterium in some countries including Bahrain, UAE and Kuwait (10). However, the bla OXA-23-like and bla OXA-24-like genes were responsible for the majority of carbapenem resistance in the isolates studied in this research.
Carbapenemase were found in 89 strains (85.6%), including 74 (83.2%) bla OXA-23like that the bla OXA-23like was associated with carbapenemase (P = 0.002) because the bla OXA-23like gene is associated with Tn2006 that is capable of transposing from bacteria to bacteria could be located on the plasmid or a chromosome (24). bla OXA-23like was associated with imipenem (P 0.002) and also with meropenem (P < 0.001). In conclusion, bla OXA-51-like and bla OXA-23like were the predominant mechanisms of resistance to imipenem in A. baumannii. However, for a global epidemiologic analysis, further studies in large scale and different places should be conducted.
Various molecular typing systems have been developed so far to facilitate better understanding of epidemiology of infection with A. baumannii (11). This is the first published study of PFGE typing among the clinical strains of A. baumannii conducted in west of Iran. We obtained 8 clones and 20 sub clone. Clone A was involved in the majority of outbreaks in Kermanshah. It occurred at different hospitals wards and was the predominant pattern 100% cultured from Infectious Disease wards. Clone B was the second most common pattern involved in outbreaks. Isolates within this clone were mainly positive for bla OXA-23like where clone A was dominant for the presence of this gene. The clone (A, A1) were associated with cefotaxime (P value 0.004) indicating a close relationship with spreading dominant clone. Most of MDR and PDR phenotypes were presented in the clones A, B and C. Clonal outbreaks of A. baumannii-induced infections containing different OXA-type carbapenemases were reported in Brazil, Taiwan, Iran, Spain, Malaysia, Italy, Turkey, Korea and Argentina (5, 16, 20, 21–25–26). It is conjectured that these genes are closely associated with outbreaks in some countries. Similarly, the results of this study support the viewpoint indicating clonal spread as the main reason for the increasing trend of imipenem, meropenem resistance as well as other antibiotics studied in the hospitals of this study. It is possible that the transfer of these clones to other wards by staff or hospital equipments. A high prevalence of the clone A, B and C in different parts of the healthcare system showed that hospitalized patients should be highly careful to prevent the spread of these clones. Finally, all isolates collected from the hospital 3 in Kermanshah contained bla OXA-23like, and all isolates belonged to the clone A; therefore, this clone was responsible for the outbreak in this hospital. Early recognition of the presence of carbapenem-resistant A. baumannii clones is useful for preventing their spread within the hospital environment.
ACKNOWLEDGMENTS
This work was performed in partial fulfillment of the requirements for M.Sc thesis in medical microbiology (Abbas Farahani). The authors would like to acknowledge Kermanshah University of Medical Sciences. The study was financially supported by the Kermanshah University of Medical Sciences (project 91013).
The authors also thank members of department of microbiology in Pasteur Institute of Iran, Particularly Dr. Shahcheraghi, Fahimeh Shooraj, Shaghaghi and Vajihe Nikbin for their technical assistance. Furthermore, we are very grateful to Baharak Norozi and Zhila Rostami for their assistance with the PCR assessment in this study.
REFERENCES
- 1.Metan G, Sariguzel F, Sumerkan B, Reijden TV, Dijkshoorn L. Clonal diversity and high prevalence of OXA-58 among Acinetobacter baumannii isolates from blood cultures in a tertiary care centre in Turkey. Infect Genet Evol. 2012 doi: 10.1016/j.meegid.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Lin YC, Sheng WH, Chen YC, Chang SC, Hsia KC, Li SY. Differences in carbapenem resistance genes among Acinetobacter baumannii, Acinetobacter genospecies 3 and Acinetobacter genospecies 13TU in Taiwan. Int J Antimicrob Agents. 2010;35:439–443. doi: 10.1016/j.ijantimicag.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 3.Cefai C, Richards J, Gould FK, McPeake P. An outbreak of Acinetobacter respiratory tract infection resulting from incomplete disinfection of ventilatory equipment. J Hosp Infect. 1990;15:177–182. doi: 10.1016/0195-6701(90)90128-b. [DOI] [PubMed] [Google Scholar]
- 4.Sinha M, Srinivasa H. Mechanism of resistance to crabapenem- resistant Acinetobacter isolates from clinical sample. Ind J Med Microbiol. 2007;25:121–125. doi: 10.4103/0255-0857.32717. [DOI] [PubMed] [Google Scholar]
- 5.Shahcheraghi F, Abbasalipour M, Feizabadi M, Ebrahimipour G, Akbari N. Isolation and genetic characterization of metallo-β-lactamase and carbapenamase producing strains of Acinetobacter baumannii from patients at Tehran hospitals. Iran J Microbiol. 2011;3:68–74. [PMC free article] [PubMed] [Google Scholar]
- 6.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–82. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-β-Lactamases: the Quiet before the Storm? J Clin Microbiol. 2002;18:3798–3801. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scaife W, Young HK, Paton RH, Amyes SG. Transferable imipenem-resistance in Acinetobacter species from a clinical source. J Antimicrob Chemother. 1995;36:585–586. doi: 10.1093/jac/36.3.585. [DOI] [PubMed] [Google Scholar]
- 9.Poirel L, Marque′ S, He′ ritier C, Segonds C, Chabanon G, Nordmann P. OXA-58, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii . Antimicrob Agents Chemother. 2005b;49:202–208. doi: 10.1128/AAC.49.1.202-208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Sweih N.A, Al-Hubail M, Rotimi V.O. Three distinct clones of carbapenem-resistant Acinetobacter baumannii with high diversity of carbapenemases isolated from patients in two hospitals in Kuwait. Journal of Infection and Public Health. 2012;5:102–108. doi: 10.1016/j.jiph.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Kaufmann M. E. Pulsed-field gel electrophoresis. Methods Mol Med. 1998;15:33–50. doi: 10.1385/0-89603-498-4:33. [DOI] [PubMed] [Google Scholar]
- 12.Bosshard PP, Zbinden R, Abels S, Böddinghaus B, Altwegg M, Böttger EC. 16S rRNA gene sequencing versus the API 20 NE system and the VITEK 2 ID-GNB card for identification of nonfermenting Gram-negative bacteria in the clinical laboratory. J Clin Microbiol. 2006;44:1359–1366. doi: 10.1128/JCM.44.4.1359-1366.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. Wayne, PA: CLSI; 2007. M7-A7. Dilution antimicrobial susceptibility tests for bacteria that grow aerobically. [Google Scholar]
- 14.Yang CH, Lee S, Su PW, Yang CS, Chuang LY. Genotype and antibiotic susceptibility patterns of drug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii isolated in Taiwan. Microb Drug Resist. 2008;14:281–287. doi: 10.1089/mdr.2008.0861. [DOI] [PubMed] [Google Scholar]
- 15.Sinha M, Srinivasa H, Macaden R. Antibiotic resistance profile and extended spectrum beta-lactamase (ESBL) production in Acinetobacter species . Ind J Med Res. 2007;126:63–67. [PubMed] [Google Scholar]
- 16.Kuo HY, Yang CM, Lin MF, Cheng WL, Tien N, Liou ML. Distribution of blaOXA-carrying imipenem-resistant Acinetobacter spp. in 3 hospitals in Taiwan. Diagn Microbiol Infect Dis. 2010;66:195–199. doi: 10.1016/j.diagmicrobio.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 17.Durmaz R, Otlu B, Koksal F, Hosoglu S, Ozturk R, Ersoy Y, Aktas E, Gursoy NC, Caliskan A, et al. The optimization of a rapid pulsed-field gel electrophoresis protocol for the typing of Acinetobacter baumannii, Escherichia coli and Klebsiella spp. Jpn J Infect Dis. 2009;62:372–377. [PubMed] [Google Scholar]
- 18.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, Heersma H, Dijkshoorn L, et al. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii . J Clin Microbiol. 2005;43:4328–4335. doi: 10.1128/JCM.43.9.4328-4335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong BH, Hanifah YA, Yusof MY, Thong KL. Antimicrobial susceptibility profiling and genomic diversity of multidrug-resistant Acinetobacter baumannii isolates from a teaching hospital in Malaysia. Jpn J Infect Dis. 2011;64:337–340. [PubMed] [Google Scholar]
- 21.Taherikalani M, Fatolahzadeh B, Emaneini M, Soroush S, Feizabadi MM. Distribution of different carbapenem resistant clones of Acinetobacter baumannii in Tehran hospitals. New Microbiol. 2009;32:265–271. [PubMed] [Google Scholar]
- 22.Tan T.Y, Hsu L.Y, Koh T.H, et al. Antibiotic resistance in gram-negative bacilli: a Singapore perspective. Ann Acad Med Singapore. 2008;37:819–825. [PubMed] [Google Scholar]
- 23.Hu WS, Yao SM, Fung CP, Hsieh YP, Liu CP, Lin JF. An OXA-66/OXA-51-like carbapenemase and possibly an efflux pump are associated with resistance to imipenem in Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51:3844–52. doi: 10.1128/AAC.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee HY, Chang RC, Su LH, Liu SY, Wu SR, Chuang CH, Chen CL, Chiu CH, et al. Wide spread of Tn2006 in an AbaR4-type resistance island among carbapenem-resistant Acinetobacter baumannii clinical isolates in Taiwan. Int J Antimicrob Agents. 2012;40:163–167. doi: 10.1016/j.ijantimicag.2012.04.018. [DOI] [PubMed] [Google Scholar]
- 25.Gonçalves CR, Vaz TM, Araujo E, Boni Rd, Leite D, Irino K. Biotyping, serotyping and ribotyping as epidemiological tools in the evaluation of Acinetobacter baumannii dissemination in hospital units, Sorocaba, São Paulo, Brazil. Rev Inst Med Trop Sao Paulo. 2000;42:277–282. doi: 10.1590/s0036-46652000000500007. [DOI] [PubMed] [Google Scholar]
- 26.Yum Jh, Yi K, Lee H, Yong D, Lee K, Kim JM, et al. Molecular characterization of metallo-β-lactamase-producing Acinetobacter baumannii and Acinetobacter genomospeicies 3 from Korea: identification of two new integrons carrying the blaVIM-2 gene cassettes. J Antimicrob Chemother. 2002;49:837–840. doi: 10.1093/jac/dkf043. [DOI] [PubMed] [Google Scholar]