Abstract
Background
The HBV-X (HBX) protein is believed to contribute to the development of HCC. However, the molecular mechanisms involved in HBX- mediated hepatocarcinogenesis remain obscure. In this study, the effect of hepatitis B virus X gene and its protein product HBxAg on expression of p53 gene in Hep G2 cell line was investigated.
Methods
Viral DNA extracted from HBV-positive serum and HBX gene region was amplified using polymerase chain reaction (PCR). Then, PCR product was cloned into the pcDNA3 vector. After confirmation of cloning, the recombinant plasmid pcDNA3-X was transfected into HepG2 cell line using lipid-mediated DNA-transfection procedure. SDS-PAGE and western blotting methods were used to identify expression of HBX protein. Relative quantification was used to analyze the p53gene expression using the 2-ΔΔ Ct method.
Results
Recombinant plasmid pcDNA3–HBX was confirmed by restriction endonucleases digestion and colony-PCR. The results of SDS-PAGE and western blot assays showed that HBX gene could be expressed in Hep G2 cell line. There was no significant difference between the expression levels of p53 compared with GAPDH gene as housekeeping gene (p < 0.05).
Conclusion
There was no significant difference in the protein levels between the transfected cells with X gene containing HBX130 and HBX131 double mu-tations and p53 gene. It is necessary to do more studies on Hepatitis B virus to understand the role of HBX on the development of liver cancer and its function on p53 tumor suppressor protein.
Keywords: Hepatitis B virus, Hep G2 cell line, p53 gene, X gene
Introduction
Hepatocellular Carcinoma (HCC) is a common malignancy and a leading cause of death worldwide. Recent epidemiological data have demonstrated that liver cancer incidence has been continuously rising for more than a decade, not only in Asia and Africa but also in North America and Europe (1). The risk factor of HCC has been identified based on epidemi ological studies i.e. chronic HBV and HCV infection and prolonged exposure to alfatoxin (2).
Chronic Hepatitis B Virus (HBV) infection is the dominant global cause of HCC, accounting for 55% of cases worldwide and 80% or more in eastern Pacific region and sub-Saharan Africa, the regions with highest rates for incidence of the tumor (3). The mechanisms whereby HBV causes malignant transformation remain uncertain. However, much of the evidence available supports a pathogenetic role for the product of HBVX gene, the HBX protein (4). HBX gene is the smallest of the four partially overlapping open reading frames of HBV. It comprises 456 nucleotides that encode a 154-amino acid regulatory protein with a molecular mass of 17 kDa (5).
In this study, an established human hepatocarcinoma cell line (Hep G2) was used for the expression of HBX protein. Several studies have reported that HBX expression in Hep G2 cell line has an important role in regulation of expression of many genes, which have a variety of cellular functions including oncogenesis, apoptosis and cell cycle regulation (6). Some studies indicate the correlation between mutation/inactivation of p53 and HBV protein X (HBX) in hepatocarcinogenesis. In that process, HBX will suppress p53 function which leads to unlimited liver cell division and results in HCC (2). p53 is a protein which is responsible for protecting, maintaining and repairing cells. HBX may cause inactivation of p53 function, therefore if HBxAg-p53 complex is formed, p53 will lose its function, and it is the beginning of cancer development (2).
Mutations of HBV are known as an escape mechanism for escaping from the host immune system and they may affect the carcinogenesis of chronic HBV diseases. A study showed that double mutations of HBV at nucleotide 1762 and 1764 in the Basal Core Promoter (BCP) region were the most common mutation types in HCC patients (7). Because HBV core promoter overlaps partially with HBX coding sequence, these double mutations in BCP region convert K to M at position 130 and V to I at position 131 in the hepatitis B virus X gene product. HBX130 and HBX131 double mutations were the most common HBX mutations occurring in HCC patients with chronic HBV infection. On the other hand, based on a study in our country, it was reported that prevalence rates of mutations in the precore and basal core regions were 46 and 30%, respectively (8).
In this study, the effect of hepatitis B virus X gene containing HBX130 and HBX131 double mutations and its protein product (HBxAg) on expression of p53 gene in cell line Hep G2 was investigated.
Materials and Methods
Biological and chemical materials
DNA extraction kit, PCR product purification kit, plasmid isolation kit, FuGene®6 transfection kit and qRT PCR (SYBR Green I Light Cycler-based real-time PCR assay) were supplied by Roche Company (Germany). Cloning kit (Inset/Aclone PCR product cloning kit), DNA extraction kit from Agarose gel kit, RT PCR kit and restriction endonuclease were obtained from Fermentase (Switzerland). Hepatoma cell line Hep G2 was obtained from the Cell Bank of Pasteur Institute. RPMI-1640, Fetal Bovine Serum (FBS) and penicillin/streptomycin were from Gibco Company. PCR primers were synthesized by Cinnagen Company. The expression vector pcDNA3 was provided from Invitrogen (USA).
Construction of HBX expression vector
Viral DNA was extracted from HBV positive serum and a 465 bp HBX gene fragment was amplified using PCR. HBX gene was amplified using forward 5′ATGCAAGCTTA TGGCTGCTAGGCTGTACTG-3′and reverse 5′TGCGAATTC T T AGGCAGAGGTGA A A AAGTTG-3′ primers. PCR amplification was performed at 95°C for 5 min followed by 35 cycles at 94°C for 1 min, 61°C for 45 s and 72°C for 1 min. Final extension was performed at 72°C for 10 min. Then, PCR product was purified and directionally cloned into PTZ57 vector and transformed into TG1 strain of Escherichia coli (E. coli) (results not shown).
The positive clones were selected to extract plasmid, and the plasmid was then digested with EcoRI and Hind Ш and finally ligated into digested pcDNA3 to construct the recombinant plasmid pcDNA3-HBX. For confirmation of cloning, colony-PCR, restriction endonucleases digestion and sequencing methods were employed.
Cell culture and DNA transfection
Hep G2 cell line was cultured in RPMI-1640 medium supplemented with 10% heat inactivated Fetal Bovine Serum (FBS), penicillin (100 U/ml), and streptomycin (100 µg/ml). Cells were maintained at 37°C in a saturating humidity atmosphere containing 5% CO2.
About 2.5 ml of trypsin-EDTA was used for passaging the cells every 2-3 days. Hep G2 cells were cultured in flat-bottom culture plates. When the confluence reached 70-80%, pcDNA3-X vector that was established and sequenced, was transfected to Hep G2 cell line by lipid-mediated transfection using FuGene®6 transfection Reagent Kit. According to the protocol of this kit, ratios of 3:1 and 6:1 of FuGene®6 Transfection Reagent and the vector were used, respectively for transfection of pcDNA3vector and pcDNA3-HBX vector to Hep G2 cells. After 72 hr, the cell monolayer in 6-well culture plates was washed with PBS and then collected.
RNA extraction and RT-PCR
Total RNA from 3 kinds of cells (Hep G2 cells, cells that were transfected with Hep G2 + pcDNA3 and Hep G2 + pcDNA3 + HBX) was isolated using Trizol reagent and stored at -70°C. HBx cDNA was synthesized from total RNA (10 ng) using Moloney Murine Leukemia Virus (MMLV), Reverse Transcriptas (Promega) and HBX specific primers at 25°C for 10 min, 42°C for 60 min, 70°C for 10 min by RT-PCR kit and stored at -20°C.
SDS/PAGE and western blot assays
SDS/PAGE and Western blot methods were performed to detect HBX protein expression in Hep G2 cells. Briefly, the cells were collected, washed, and lysed in lysis buffer and heated in denaturing lysis buffer for 10 min. The extraction from each sample was separated on 12% SDS polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Then the membranes were blotted for 60 min at room temperature with nonfat dry milk (5%). The primary antibody dilution was 1:500, followed by 1:3000 dilution of goat anti-rabbit HRP-labeled antibody (Bio- Rad).
Quantification Real-Time PCR (q RT-PCR)
To analyze the relative changes in gene expression, real-time quantitative PCR was used. Primers sequences used for real-time PCR were as follows: p53 sense: 5-TGCGTGTGG AGTATTTGGATG-3’, antisense: 5’-TGGTAC AGTCAGAGCCAACCAG-3’;GAPDH sense: 5’-ACGCATTTGGTCGTATTG GG-3’, antisense:5’-TGATTTTGGAGGGAT CTCGC-3’.
Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) gene was used as the internal control gene for normalization of relative quantification and also, Hep G cell line and Hep G2-pcDNA3 were used as the control samples. Real-time PCR was performed with SYBR Green I Light Cycler-based Real-Time PCR assay. Twenty µl reaction mixtures for any gene were prepared using 4 µg Light Cycler Fast Start DNA master mixtures for SYBR Green I, 0.5 mM of each primer and 5 µl of cDNA.
The amplification reaction was carried out under the following conditions: first 95°C for 10 min, followed by 40 cycles at 95°C for 5 s and 60°C for 10 s and 72°C for 20 s. After PCR amplification, a melting curve was generated by holding the reaction at 95°C for 60 s and the gradient from 72 to 95°C and raising by 1°C for each step. The efficiency of amplification of the target gene (p53) and internal control (GAPDH) was examined using realtime PCR. Serial dilutions of cDNA were amplified by real-time PCR using specific primers for each gene. All reverse transcription-PCRs were performed in triplicate. Then, Δ CT (CT, p53CT, GAPDH) was calculated for each cDNA dilution. The data were analyzed using least-squares linear regression. Relative quantification of p53 expression was performed using 2-ΔΔ Ct method.
Statistical analysis
All data were analyzed with non-parametric Wilcoxon test by using SPSS software package. A p-value less than 0.05 was considered to be significant.
Results
PcDNA3-HBX vector construction
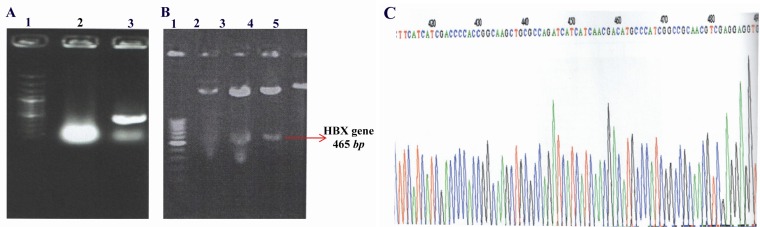
A 465 bp fragment of HBX gene was amplified using HBV- DNA and subcloned into the expression vector pcDNA3, with which the pcDNA3-HBX was constructed. The presence of 465 bp fragment of HBX gene in pcDNA3 was verified by restriction endonucleases digestion, colony PCR and DNA direct sequencing (Figure 1). The result of sequencing showed that there were substitutions in amino acids 130 and 131 of HBV X protein.
Figure 1.
Confirmation of the presence of 465 bp hbx fragment in pcDNA3 vector by 3 methods; A) Colony PCR. Lane 1: 100 bp DNA molecular size marker, Lane 2: PCR product of pcDNA3 plasmid without HBX gene (negative control), Lane 3: PCR product of pcDNA3-HBX vector; B) Restriction endonucleases digestion. Lane 1: 100 bp DNA molecular size marker, Lane 2: pcDNA3- HBX without digestion, Lane 3 and 4: Digestion product of pcDNA3-HBX with EcoRI and Hind III, Lane 5: Digestion product of pcDNA3 (control); C) The direct sequencing result of pcDNA3-HBX
Expression of HBX gene in Hep G2 cells
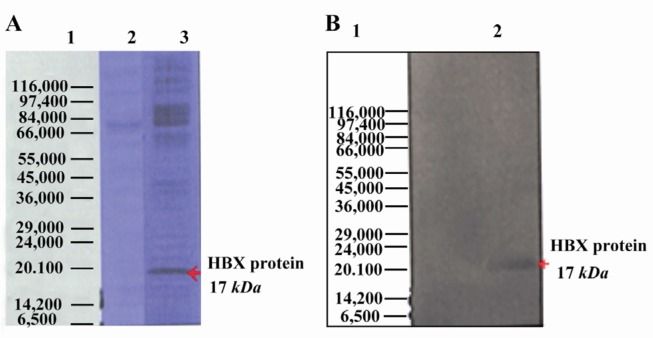
After transfection of pcDNA3-HBX in Hep G2 cell line, expression of HBX gene was confirmed by using RT-PCR (data not shown), SDS-PAGE and Western blot assays. HBX is 154 amino acids in size, with a molecular mass of approximately 17 kDa. In this study, the presence of 17 kDa hepatitis B virus gene product, HBX, was investigated using SDS- PAGE and Western blot assays (Figure 2). The expected band was seen in HBX expressing cells while there was no detectable band in the control group (Hep G2-pcDNA3). These results indicated that HBX was successfully transfixed and expressed in Hep G2 cells.
Figure 2.
Expression of HBX in Hep G2 cell line by SDS PAGE and Western blot. The purified bands had a molecular size of ~17 kDa, corresponding with the size of HBX protein; A) Hepatitis B virus X protein was separated on 12% SDS-polyacrylamide gel electrophoresis; Lane 1: Protein marker (Sigma marker, M 3913), Lane 2: Non-transfected cell (negative control), Lane 3: transfected cell; B) Expression of HBX 16.7 kDa was analyzed using Western blot by serum samples from patients with Liver Cirrhosis. Lane 1: Protein molecular weight marker (Sigma marker, M 3913), Lane 2: transfected cell with HBX
Expression of p53 gene with real-time PCR
Expression of p53 gene in transfected and non-transfected cells with HBX gene compared to GAPDH gene was evaluated by relative quantification and melting curve analysis. The results showed a 0.7 fold decrease in the expression of p53 compared to GAPDH as the housekeeping gene. However, there was no significant difference in expression level of p53 gene in transfected and non-transfected cells with HBX gene containing HBX130 and HBX131 double mutations.
Discussion
The small, 3.2 kb DNA genome of HBV contains four known open reading frames, called S, C, P, and X. The hepatitis B virus X (HBX) gene is the smallest, with a length of 465 nucleotides. HBX is 154 amino acids in size, with a molecular mass of approximately 17 kDa. Comparative analyses of HBV X gene sequences from mammalian hepadenoviruses of different species revealed areas of high conservation, including presumptive helical domains located in the amino and carboxy-terminal regions, and a potential coiledcoil motif (9).
The highest rate of HBX gene expression has been seen in hepatoma cells and Qu ZL et al showed that the transfection efficiency was 46.4% in Hep G2 cells and 29.6% in QBC939 cells for both HBX gene expression vector and empty vector (10). HBX gene expression in HL-7702 and CCL13 cell lines by Hong-Ying Chen (11) and Di-Peng Ou (12) and also, frequently in Hep G2 cells (13) was employed. Theretofore, Hep G2 cell line was selected for investigation of HBX gene expression and its effect on p53 was evaluated. As expected, presence of the approximately 17 kDa band confirmed expression of HBX gene using SDS-PAGE and Western blot assays.
HBX protein has been shown to be associated with liver diseases such as Chronic Hepatitis B (CHB), Liver Cirrhosis (LC) and HCC. Antigen x of hepatitis B virus (HBxAg) induces viral replication and gene expression, which are important for survival and chronic state of this carrier and the progression of HCC. Several studies indicated that the antibodies to HBxAg (antiHBX) may serve as a pre-neoplastic marker for diagnosis of HCC (14–16). In this research, expression of recombinant HBX protein in Hep G2 line was confirmed using Western blot and serum samples from patients with LC whereby the presence of anti-HBX in sera of these patients can be used as a marker for diagnosis of HCC in the future.
HBX is a multifunctional transactivator for various viral and cellular genes. It is assumed that HBX is a protein kinase with autophosphorylation activity and the phosphorylized HBX in human hepatoma cell indicates that phosphorylation has a contribution in carcinogenesis (2). Transactivation function of HBX involves both activation of signal transduction cascades and direct protein-protein interactions. HBX can transactivate several signaling transduction pathways including protein kinase C, ras/raf/ MAP, Jak1 kinase and src kinase. In addition, HBX directly interacts with transcription machinery such as RNA polymerase, TATA- binding protein and TF IIB and other cellular proteins such as p53, a tu-mor suppressor protein. Thus, the pleiotropic activity of HBX may contribute to the modulation of gene expression, which finally leads to the formation of liver cancer (17, 18).
One of the most important molecules relating to cancer is p53. p53 protein is a common target site in molecular pathogenesis of DNA tumor virus. Many viral oncoproteins disrupt p53 activity through direct protein-protein interaction (19). These include large T antigen of simian virus 40, E6 protein of human papillomavirus (20), E1B 55 kDa of adenovirus (21) and HBX protein of HBV (22). Direct association between HBX and p53 proteins is observed with HBX-GST fusion proteins in vitro or with in vitro translated proteins (17, 23).
Some studies reported that HBX protein binds to the C-terminus of p53 and induces its sequestration from the nucleus to the cytoplasm, thereby leading to inhibition of its effects on cell cycle arrest and DNA repair (9). Chung et al (24) studied the expression of p53 in transfected cells compared with non transfected cells by eukaryotic recombinant vector pEGFP-C2-X and Chang cell line and confirmed that HBX had no effect on expression of p53. Alvin et al demonstrated that cells expressing HBX increased sensitivity to UV damage but in the absence of UV damage, cells expressing HBX were found to be similar to control cells in their morphology, cell cycle, and apoptotic profiles and untreated HBX-expressing cells were also able to repair damaged DNA as efficiently as the control cells. The ability of HBX to alter the capability of the cell to repair damaged DNA may predispose a chronically HBV infected individual to cancer. HBV and its HBX protein contribute to carcinogenesis by destabilizing the cellular genome, where random genetic alterations due to integration of HBV genome into cellular DNA can cause chromosomal rearrangements and affect key genes involved in the cell growth (25).
Conclusion
Our results were compatible with these findings that HBX does not change the level of p53 expression. HBX130 and HBX131 double mutations did not increase the expression levels of p53. However, the molecular mechanisms responsible for HBX proteininduced HCC remain uncertain. In the future, regarding the essential role of HBX in the replication of HBV and its effect on tumor suppressor elements such as p53, we propose that HBX can be used in diagnostic kits for diagnosis of hepatic carcinoma.
Acknowledgement
This study has been fulfilled with the support of the High Institute for Research and Education in Transfusion Medicine, Blood Transfusion Research Center and Shahid Beheshti University. We gratefully acknowledge the advice and technical support of the staff of Virology laboratory of the Iranian Blood Transfusion Organization. The authors declare that they have no conflict of interests.
References
- 1.Neuveut C, Wei Y, Buendia MA. Mechanisms of HBV-related hepatocarcinogenesis. J Hepatol. 2010;52(4):594–604. doi: 10.1016/j.jhep.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 2.Dewantoro O, Gani RA, Akbar N. Hepatocarcinogenesis in viral hepatitis B infection: The role of HBx and p53. Acta Med Indones. 2006;38(3):154–159. [PubMed] [Google Scholar]
- 3.Kew MC. Epidemiology of hepatitis B virus infection, hepatocellular carcinoma, and hepatitis B virus-induced hepatocellular carcinoma. Pathol Biol. 2010;58(4):273–277. doi: 10.1016/j.patbio.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Kew MC. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J Gastroenterol Hepatol. 2011;26(Suppl 1):144–152. doi: 10.1111/j.1440-1746.2010.06546.x. [DOI] [PubMed] [Google Scholar]
- 5.Miller RH, Robinson WS. Common evolutionary origin of hepatitis x virus and retroviruses. Proc Natl Acad Sci USA. 1986;83(8):2531–2535. doi: 10.1073/pnas.83.8.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lian Z, Liu J, Li L, Li X, Clayton M, Wu MC, et al. Enhanced cell survival of Hep3B cells by the hepatitis B x antigen effector, URG11, is associated with upregulation of beta-catenin. Hepatology. 2006;43(3):415–424. doi: 10.1002/hep.21053. [DOI] [PubMed] [Google Scholar]
- 7.Lee JH, Han KH, Lee JM, Park HJ, Kim HS. Impact of hepatitis B virus (HBV) x gene mutations on hepatocellular carcinoma development in chronic HBV infection. Clin Vaccine Immunol. 2011;18(6):914–921. doi: 10.1128/CVI.00474-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghabeshi S, Sharifi Z, Hosseini SM, Mahmoodian Shooshtari M. Correlation between viral load of HBV in chronic hepatitis B patients and precore and basal core promoter mutations. Hepat Mon. 2013;13(2):e7415. doi: 10.5812/hepatmon.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdel-Hafiz HA. Role of hepatitis B virus x protein in DNA repair during hepatocellular carcinoma development. J Carcinogene Mutagene. 2011;S3:001. [Google Scholar]
- 10.Qu ZL, Zou SQ, Cui NQ, Wu XZ, Qin MF, Kong D, et al. Upregulation of human telomerase reverse transcriptase mRNA expression by in vitro transfection of hepatitis B virus X gene into human hepatocarcinoma and cholangiocarcinoma cells. World J Gastroenterol. 2005;2811(36):5627–5632. doi: 10.3748/wjg.v11.i36.5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen HY, Tang NH, Li XJ, Zhang SJ, Chen ZX, Wang XZ. Transfection and expression of hepatitis B virus x gene and its effect on apoptosis in HL- 7702 cells. World J Gastroenterol. 2004;10(7):959–964. doi: 10.3748/wjg.v10.i7.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ou DP, Tao YM, Tang FQ, Yang LY. The hepatitis B virus X protein promotes hepatocellular carcinoma metastasis by upregulation of matrix metalloproteinases. Int J Cancer. 2007;120(6):1208–1214. doi: 10.1002/ijc.22452. [DOI] [PubMed] [Google Scholar]
- 13.Lin N, Chen HY, Li D, Zhang SJ, Cheng ZX, Wang XZ. Apoptosis and its pathway in X genetransfected HepG2 cells. World J Gastroenterol. 2005;11(28):4326–4331. doi: 10.3748/wjg.v11.i28.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vitvitski-Trépo L, Kay A, Pichoud C, Chevallier P, de Dinechin S, Shamoon BM, et al. Early and frequent detection of HBxAg and/or anti-HBx in he-patitis B virus infection. Hepatology. 1990;12(6):1278–1283. doi: 10.1002/hep.1840120605. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Wu LY, Zhang S, Qiu LY, Li N, Zhang X, et al. Anti-hepatitis B virus x protein in sera is one of the markers of development of liver cirrhosis and liver cancer mediated by HBV. J Biomed Biotechnol. 2009;2009:289068. doi: 10.1155/2009/289068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang GY, Lin CY, Huang LM, Wang YH, Wang JC, Hsu CT, et al. Detection of the hepatitis B virus x protein (HBx) antigen and anti-HBx antibodies in cases of human hepatocellular carcinoma. J Clin Microbiol. 2003;41(12):5598–5603. doi: 10.1128/JCM.41.12.5598-5603.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yun C, Lee JH, Park H, Jin YM, Park S, Park K, et al. Chemotherapeutic drug, adriamycin, restores the function of p53 protein in hepatitis B virus X (HBx) protein-expressing liver cells. Oncogene. 2000;19(45):5163–5172. doi: 10.1038/sj.onc.1203896. [DOI] [PubMed] [Google Scholar]
- 18.Neuveut Ch, Wei Y, Buendia MA. Mechanisms of HBV-related hepatocarcinogenesis. J Hepatol. 2010;52(4):594–604. doi: 10.1016/j.jhep.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 19.Teodoro JG, Branton PE. Regulation of apoptosis by viral gene products. J Virol. 1997;71(3):1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas M, Matlashewski G, Pim D, Banks L. Induction of apoptosis by p53 is independent of its oligomeric state and can be abolished by HPV-18 E6 through ubiquitin mediated degradation. Oncogene. 1996;13(2):265–273. [PubMed] [Google Scholar]
- 21.Rubenwolf S, Schutt H, Nevels M, Wolf H, Dobner T. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J Virol. 1997;71(2):1115–1123. doi: 10.1128/jvi.71.2.1115-1123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin Y, Nomura T, Yamashita T, Dorjsuren D, Tang H, Murakami S. The transactivation and p53-interacting functions of hepatitis B virus X protein are mutually interfering but distinct. Cancer Res. 1997;57(22):5137–5142. [PubMed] [Google Scholar]
- 23.Truant R, Antunovic J, Greenblatt J, Prives C, Chromlish JA. Direct interaction of the hepatitis B virus HBx protein with p53 leads to inhibition by HBx of p53 response element-directed transactivation. J Virol. 1995;69(3):1851–1859. doi: 10.1128/jvi.69.3.1851-1859.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung TW, Lee YC, Ko JH, Kim CH. Hepatitis B virus x protein modulates the expression of PTEN by inhibiting the function of p53, a transcriptional activator in liver cells. Cancer Res. 2003;63(13):3453–3458. [PubMed] [Google Scholar]
- 25.Lee AT, Ren J, Wong ET, Ban KH, Lee LA, Lee CG. The hepatitis B virus x protein sensitizes HepG2 cells to UV light-induced DNA damage. J Biol Chem. 2005;280(39):33525–33535. doi: 10.1074/jbc.M506628200. [DOI] [PubMed] [Google Scholar]