Abstract
Cellular senescence is an irreversible proliferation arrest, thought to contribute to tumour suppression, proper wound healing and, perhaps, tissue and organismal aging. Two classical tumor suppressors, p53 and pRB, control cell cycle arrest associated with senescence. Profound molecular changes occur in cells undergoing senescence. At the level of chromatin, for example, senescence associated heterochromatic foci (SAHF) form in some cell types. Chromatin is inherently dynamic and likely needs to be actively maintained to achieve a stable cell phenotype. In proliferating cells chromatin is maintained in conjunction with DNA replication, but how non-proliferating cells maintain chromatin structure is poorly understood. Some histone variants, such as H3.3 and macroH2A increase as cells undergo senescence, suggesting histone variants and their associated chaperones could be important in chromatin structure maintenance in senescent cells. Here, we discuss options available for senescent cells to maintain chromatin structure and the relative contribution of histone variants and chaperones in this process.
Introduction
Cellular senescence is an irreversible proliferation arrest, thought to mediate tumour suppression, wound healing and, perhaps, contribute to aging [1]. This proliferative arrest is accompanied by profound changes in chromatin structure. Activated oncogenes, shortened telomeres that result from repeated rounds of cell division, oxidative stress, inadequate in vitro growth conditions and other cellular stresses trigger cell senescence [1-5].
Maintenance of chromatin structure and its proper regulation are a fundamental cellular requirement. Chromatin consists of DNA, histones and many accessory proteins and RNAs that contribute to its structure and regulation [6]. Proliferating cells maintain chromatin in conjunction with DNA replication during S phase of the cell cycle [7]. However, non-proliferating senescent cells can persist in the body for decades; for example, senescent melanocytes in benign human nevi [8]. In the absence of DNA replication, these cells maintain their senescent phenotype and, presumably, their chromatin structure. This raises the question as to how chromatin dynamics are regulated and chromatin structure is maintained in senescent cells. In this review we discuss how histone chaperones and their substrates might contribute to chromatin structure maintenance in senescent cells. While we focus on non-proliferating senescent cells, the ideas discussed are also relevant to other non-proliferating cells, such as terminally differentiated cells.
Overview of senescence
Triggers of senescence
Since its original description by Leonard Hayflick in early 1960s [9, 10], much has been learnt about senescence. As outlined above, senescence is an irreversible proliferation arrest triggered by a multitude of factors. These triggers can be subdivided into four broad categories: 1, DNA damage and short telomeres which are also recognized as DNA damage and caused by excess rounds of replication of linear chromosomes (the so-called “end replication” problem). Senescence caused by short telomeres is called replicative senescence; 2, oxidative stress; 3, activated oncogenes; 4, other stresses such as inadequate in vitro growth conditions. Irrespective of the specific trigger, these triggers share a key physiological output, which is cell proliferation arrest.
Effectors of senescence
pRB and p5
Senescence associated proliferation arrest can be enforced and reinforced by a network of effector pathways. At the heart of this network are the pRB and p53 pathways, which are activated by most triggers of senescence. One of the effectors of the p53 response is p21CIP1 (p21). p21 is a cyclin dependent kinase inhibitor (CDKI), and therefore, inhibitor of cell proliferation. p21 is a direct transcriptional target of p53 and mediates p53-dependent cell cycle arrest [11]. Another CDKI p16INK4a (p16) governs pRB and the senescence-associated growth arrest. Ultimately, through inhibition of cyclin/cdk kinases, p21 and p16 both maintain pRB in a hypophosphorylated [11]. This, in turn, stops cellular proliferation, in part, by inhibiting expression of E2F target genes that are required for progression through the cell cycle. To activate senescence, numerous upstream effector pathways converge on the pRB and p53 tumor suppressors. For example, DNA damage signaling activated by telomere shortening and/or oncogene activation results in activation of various effector proteins, e.g. kinases such as ataxia telangiectasia mutated (ATM) and CHK2, and subsequent stabilization of p53 and hypophosphorylation of pRB [12-16]. Hence two classical tumor suppressors, p53 and pRB, are the major components of effector pathways required for establishment of senescence.
The senescence secretome
Senescent cells secrete a cocktail of growth factors, proteases, and inflammatory cytokines, termed the senescence-associated secretory phenotype (SASP) [2, 17], or senescence-messaging secretome (SMS) [18], or more simply the senescence secretome. The senescence secretome is complex and includes increased IGFBP, PAI-1, TGFβ1 [19-21], increased expression of immune regulators such as IL6, IL6R, IL8, CXCL1, 5 and 7 [22-24] and increased expression of extracellular remodeling proteins such as matrix metalloproteinase (MMP) 1 and 3 [25]. This altered secretory program contributes to and reinforces the senescence-associated proliferation arrest. For example, cytokines such as IL6 and its receptor IL6R are required in a cell-autonomous manner to implement cell-cycle arrest, and depletion of IL6 results in bypass of oncogene induced senescence (OIS) [22]. Similarly, increased expression of the chemokine receptor, CXCR2, and some of its ligands, such as, IL8, help to reinforce the senescence program [23]. Thus senescence-associated secretion helps senescent cells to enforce the senescent phenotype.
Autophagy
Recent studies have implicated autophagy as important effector of the senescence program [26-28]. Autophagy is a conserved lysosomal degradation process, which recycles cytoplasmic proteins and small organelles. Autophagy genes such as ATG1, ATG6/Beclin1 and ATG8/LC3 drive autophagy through sequestration of cytosolic contents and organelles in double membrane vesicles called autophagosomes, which fuse with lysosomes to form autolysosomes. Autophagy has been found to increase in senescence, by various autophagic markers such as lipidation and cleavage of LC3, upregulation of ATG1-related genes such as ULK1 and 3, and detection of autolysosomes [26, 27]. Senescent cells also express a senescence-associated p-galactosidase (SA β-gal) [29], which partly reflects the increase in lysosomal mass [26, 30]. Other lysosomal proteins such as Cathepsin D are also upregulated in senescent cells [28, 31], suggesting active senescence-associated autophagic processing. Autophagy helps senescent cells to generate raw materials for protein synthesis. Recently it was shown that the mammalian target of rapamycin (mTOR) protein and autolysosomes colocalize in senescent cells, suggesting coupling of protein degradation and biosynthetic pathways. This was shown to be important for generation of components of the secretome, such as IL6 and IL8 [26]. In line with this, previous reports have shown that mTOR inhibition by rapamycin treatment suppresses senescence [32, 33]. The demonstration that autophagy potentiates oncogene-induced senescence in vitro [27], and yet rapamycin, a potent activator of autophagy, can suppress replicative senescence [32, 33] underscores the fine control of the level of autophagy required to enact senescence. Together, these results indicate that senescence-associated autophagy coupled to senescence-associated protein synthesis is involved in production of essential metabolites and secreted components, indicative of collaboration of various effector pathways in imposing senescent associated proliferative arrest.
Functions of senescence
Tumor suppression
Senescence in vivo is thought to be an important tumor suppression process (Figure 1). Senescence triggered by activated oncogenes, such as RAS and BRAF, is thought to block progression to a transformed cell phenotype. For example, in primary human fibroblasts, primary lymphocytes and melanocytes, an oncogene mutation, such as RASG12V or BRAFV600E, results in senescence associated growth arrest [8, 34, 35]. In vivo, senescent cells are known to exist in premalignant neoplasms, including nevi, neurofibromas, prostatic intraepithelial neoplasia (PIN), pancreatic intraepithelial neoplasia (PanIN) and mouse lung adenomas [8, 36-39]. Inactivation of senescence pathways allows progression to cancer in multiple tumors, notably in melanoma, lymphoma, prostate and pancreas [35, 36, 40, 41]. Further highlighting the role of senescence as a tumor suppression mechanism and its therapeutic potential, reactivation of p53 in murine tumors causes cell senescence and associated tumor regression [24, 42].
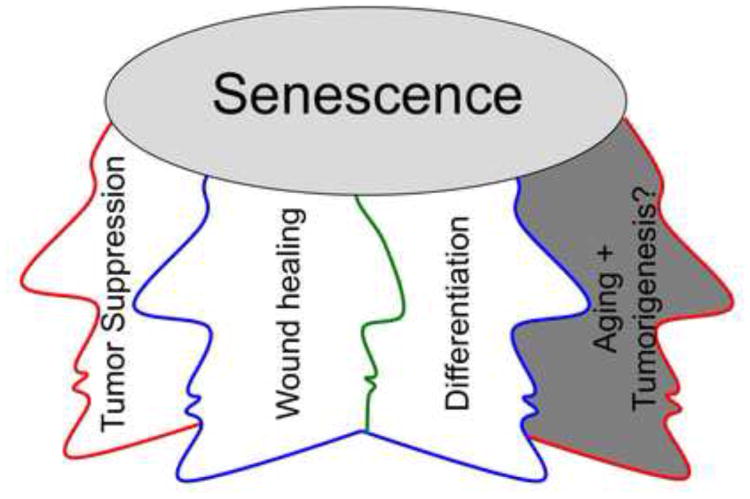
Figure 1. The multifaceted effects of cell senescence.
Senescence is a well- established tumor suppression process. Senescence is likely also important for wound healing, and perhaps for generation of terminally differentiated cells in the body, highlighting the importance of senescence in normal physiology. On the darker side, senescence may also contribute to tissue aging. The secretome and senescence-associated inflammation have been proposed to contribute to tumorigenesis.
Programmed cell differentiation
Recently senescence was also shown to have another normal physiological function in maturation of megakaryocytes [43]. As part of the normal differentiation process, mature megakaryocytes enter a senescent state in which cell division stops. Megakaryocytes express increased SA β-gal, p21 and Cathepsin D, indicative of cell senescence. By studying megakaryocytes in culture, the authors were able to determine that thrombopoetin induces a differentiation program that leads to gene activation associated with senescence. Thus, it appears that senescence can be a normal programmed physiological endpoint in differentiated cell lineages (Figure 1).
Wound healin
Another proposed normal physiological function of the senescence program is wound healing, a process which is accompanied by inflammation, new tissue formation and tissue remodelling (Figure 1). The role of senescence in wound healing was first described by Lowe and coworkers [25] in a mouse model of chemical-induced liver damage. Upon liver damage, stellate cells differentiate into myofibroblasts and secrete a similar cocktail of inflammatory mediators as cells undergoing replicative senescence and oncogene induced senescence, including MMPs. MMPs are thought to help in the wound healing response through restraining and reversing the fibrotic component of the wound, and the senescence secretome also triggers clearance of senescent stellate cells by NK cells. In mice deficient in master senescence regulators, senescence of liver stellate cells is impaired, resulting in excessive liver fibrosis. This implicates senescence in maintenance and repair of normal tissue integrity.
Pathological impact of senescence
On the one hand the senescence secretome is beneficial for the cell and organism. But, senescence can also be harmful to neighboring cells through its effects on the microenvironment (Figure 1). For example, senescent stromal cells can promote malignant transformation of epithelial cells, in part by altering epithelial cell differentiation [44]. Similarly, senescent human fibroblasts can stimulate premalignant and malignant epithelial cells to proliferate in culture and form tumors in mice [45]. Further underscoring the harmful effects of senescence, two recent papers provide direct genetic evidence that inactivation of proteins included in the senescence secretome, such as IL6 and MMP7, results in decreased tumorigenesis [46, 47]. Both groups utilized the KRASG12D mouse model, which develop benign pancreatic intraepithelial neoplasia (PanINs), due to oncogene-induced senescence [37, 39, 48]. Some of these PanIN lesions acquire additional mutations that bypass senescence program and progress to pancreatic ductal adenocarcinoma (PDAC). Specifically, Lesina et al. (2011) found that genetic deletion of IL6 in these mice significantly decreased development of PDACs, highlighting the importance of interleukin signaling in promoting cancerous transformation of PanIN lesions into PDACs. Similarly, Fukuda et al. (2011) found that deletion of MMP7 in the same KRASG12D mice limits tumor size, progression and metastasis, without affecting PanIN formation. This suggests that onset of senescence is not affected by loss of MMP7, but MMP7 can drive tumor progression and metastasis. This argues that senescence signaling can also promote tumorigenesis under certain contexts.
Overview of chromatin
A chromatin language
Chromatin consists of DNA and a large number of proteins that contribute toits proper folding and regulation in the cell nucleus [6]. The basic repeatingunit of chromatin is the nucleosome, approximately 147 base pairs of DNAfolded around a histone octamer comprised of two copies each of histones H2A, H2B, H3 and H4. The DNA between each nucleosome is called linkerDNA and can be bound by H1 histone. The N and C terminal tails of thehistones in nucleosomes are unstructured and protrude from the globularnucleosome itself [49]. These tails are decorated with various posttranslational modifications (PTMs), such as acetylation, phosphorylation, ubiquitination and methylation. These modifications impact function of the underlying DNA. For example, histone acetylation is generally associated with active transcription [6]. In contrast, histone lysine methylation can be associated with either active or repressed chromatin [6]. For example H3K4 methylation is generally associated with active transcription [50, 51]. H3K9 momomethylation has been reported in association with promoters of active genes [52], and di and trimethylation of H3K9 are typically associated with gene repression, constitutive and facultative heterochromatin [52-57]. Similarly, H3K27 trimethylation is associated with gene repression and silenced chromatin [58-61]. Therefore, the sites of lysine methylation and mono-, di- or tri- status govern the activity status of genes.
Histone PTMs frequently mediate interactions between nucleosomes and other regulatory proteins. For example, acetylated lysines serve as a recognition platform for proteins with bromodomains such as, P/CAF (p300/CBP-associated factor) and SWI/SNF chromatin remodeling complexes [62-66]. Such chromatin remodelers remodel chromatin structure by utilizing energy generated from ATP hydrolysis to restructure the nucleosomes, and consequently are called ATP-dependent chromatin remodelers. Similar to recognition of acetylated lysines by bromodomains, methylated lysines can serve as a recognition platform for proteins with chromodomains such as HP1 and CHD chromatin remodeler complexes [67, 68]. These molecular switches behave in a context dependent way but generally, the acetylation-bromo-remodeler interactions result in gene activation [62, 66, 69, 70], and the methylation-chromo-remodeler interactions result in gene repression [67, 68, 71].
Histone post-translational modifications also dictate access and release of various chromatin-binding proteins, which establish a repressive or permissive state of chromatin. These modifications interact in complex ways to regulate protein binding and the functional output of chromatin. For example, Heterochromatin Protein 1 (HP1) is a transcriptional repressor that directly binds to di or tri methylated lysine 9 residue of histone H3 (H3K9me2/3), a modification that is often associated with transcriptionally silenced heterochromatin [71-73]. However, phosphorylation of neighboring serine 10 results in displacement of HP1 from chromatin, and this helps facilitate chromatin condensation and proper chromosome segregation in mitosis [74, 75]. Similarly H3K27 methylation recruits the repressive polycomb PRC1 complex of proteins [76-78]. Phosphorylation of neighboring S28 results in the displacement of polycomb proteins, leading to gene activation [79]. Such phospho-switches help to change the chromatin status without the requirement for active histone demethylation.
An additional layer of complexity is overlaid by the presence of histone variants and their associated marks in chromatin [80]. In mammals, except for histone H4, all histones have variants. Variant histones differ in amino acid sequence from the canonical histones, and, unlike the canonical histones, are typically encoded as single copy genes in the genome, are expressed throughout the cell cycle and in non-proliferating cells, and give rise to polyadenylated mRNAs [80]. Variant histones are implicated in diverse chromatin regulation events such as gene activation, differentiation, DNA repair and gene silencing. Histones H2A and H3 have the most documented variants [81]. H2A variants include macroH2A, H2AX, H2AZ and H2ABbd. MacroH2A has roles in gene silencing, for example X-chromosome inactivation [82]. H2AX has roles in DNA repair [83]. H2AZ has roles linked to active transcription [84] Variants of canonical H3.1 include H3.2, H3.3, CENP-A and a testis specific variant H3.1t. Histone H3.3 often marks sites of active transcription and is enriched for histone modifications associated with active transcription in flies, plants and mammals [85-88]. More specifically, histone H3.3 is enriched at transcription start sites (TSS) and gene bodies of actively transcribed genes, TSS of genes that are likely poised for activation and some other transcription regulatory sites, likely including promoters and enhancers [89, 90]. At the TSS, nucleosomes containing both H3.3 and histone H2A variant H2A.Z are more labile, hence facilitating access to transcription factors [91].
Regulation of chromatin by histone modifications and variants is invariably context dependent, and, illustrating this, histone H3.3 is also deposited at telomeres and pericentromeres, regions considered to be transcriptionally silenced [89, 92-94]. Histone H3.3 is also incorporated into the × and Y-chromosomes during formation of the transcriptionally silent sex body by meiotic sex chromosome inactivation [95]. Similarly H3.3 is also required for the establishment of heterochromatin in the mouse embryo [96]. This context-dependent combinatorial effect of histone modifications, histone variants and binding proteins culminates in a so-called “chromatin language” [97], which orchestrates a vast array of physiological processes from gene transcription to epigenetic inheritance.
Histone chaperones
One class of molecules important in setting out and maintaining this language are histone chaperones [98]. Histone chaperones facilitate the assembly of nucleosomes from DNA and histones. During S-phase of proliferating cells, histone chaperones, such as Chromatin Assembly Factor 1 (CAF1), contribute to assembly of nucleosomes on newly replicated DNA. While non-proliferating senescent cells do not undergo S-phase and S-phase coupled chromatin assembly, other processes such as transcription and DNA repair are active in senescent cells and are associated with nucleosome disassembly and reassembly. Presumably, therefore, senescent cells have active DNA replication-independent chromatin assembly pathways.
Histone H3.3 is one histone variant that is incorporated into chromatin predominantly via the replication-independent pathway [99]. Several histone chaperone complexes facilitate deposition of histone H3.3 into chromatin, including the mammalian HIRA/UBN1/CABIN1/ASF1a (HUCA) complex, DAXX/ATRX complex and the DEK complex. Like H3.3 itself (see above), these chaperones are implicated in diverse processes from gene activation to gene repression. For example, HUCA (and orthologous complexes in other species) is implicated in both gene silencing and gene activation [19, 95, 100-105]. DAXX/ATRX deposits H3.3 at pericentromeres and telomeres, regions of chromatin that are generally considered to be heterochromatic [89, 93, 94]. Drosophila DEK (dDEK) facilitates H3.3 assembly during transcription puff formation [106]. Subsequently, it was also shown that DEK is also crucial to global heterochromatin integrity [107]. In vivo, loss of DEK in Drosophila leads to a Suppressor of Variegation Su(var) phenotype and global reduction in heterochromatin formation. In sum, several different histone chaperones are known to be involved in deposition of H3.3, and the full extent to which these deposition pathways are functionally distinct or overlapping likely remains to be determined. H3.3 and its chaperones have functions linked to both gene activation and repression. Understanding regulation and function of histone variant H3.3 is likely to be instructive regarding chromatin regulation in non-proliferating senescent cells, because H3.3 is deposited into chromatin in a DNA replication independent manner.
Chromatin and Senescence
Structure and formation of SAHF
Chromatin reorganization plays an important role in the senescence program. In many senescent human cells, this reorganization is visible at the global level when senescent cells are stained with 4′-6-Diamidino-2-phenylindole (DAPI). Proliferating cells exhibit a diffuse distribution of DNA throughout the cell nucleus. However in DAPI-stained senescent cells, punctate DNA foci become visible (Figure 2). These foci have been described as heterochromatic (so-called senescent associated heterochromatin foci (SAHF)), based on the presence of heterochromatic proteins, such as HP1, repressive histone modifications such as H3K9 methylation and generally hypoacetylated histones [108]. Each SAHF focus in a senescent cell is thought to represent an individual chromosome [109, 110]. Significantly, SAHF do not contain pericentromeres and telomeres, pointing to massive heterochromatization of euchromatin in senescent cells [109-112]. Formation of SAHF has been reported by some to be wholly or partly dependent on major effectors of senescence, such as pRB and p53 [111-113] (but see also below). pRB colocalizes with SAHF and its inactivation blocks formation of SAHF [111, 112]. Formation of SAHF is also dependent on the histone chaperone activity of the HUCA histone chaperone complex and so is likely to involve histone variant H3.3 [114]. Further, SAHF-positive senescent cells lose linker histone H1 and exhibit increased levels of chromatin-bound high mobility group A proteins (HMGA) [108, 109], which cooperate with p16 to promote SAHF formation. Therefore, there is large-scale reorganization and heterochromatization during formation of SAHF in senescent cells
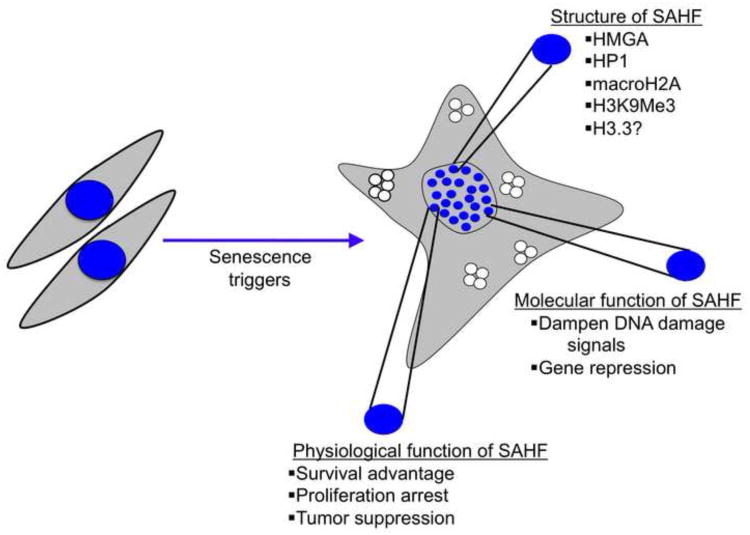
Figure 2. Structure function of SAHF.
Normal proliferating cells have diffuse DAPI staining associated with euchromatin. Senescent cells form punctate foci termed SAHF, which are heterochromatic based on markers such as HP1 proteins, H3K9Me3 and hypoacetylated histones. On the one hand, SAHF might contribute to tumor suppression via repression of proliferating promoting genes such as cyclin A. On the other hand, SAHF may dampen the DNA damage response, suppress apoptosis and promote viability of senescent cells.
Molecular function of chromatin changes in senescence
Inclusion of proliferation-promoting genes, such as cyclin A, into SAHF correlates with silencing of expression of these genes and senescence-associated cell cycle arrest [110, 111]. So, it has been suggested that the heterochromatinization of proliferation genes through SAHF could directly contribute to senescence-associated silencing of these genes and proliferation arrest (Figure 2).
Recently, an alternate viewpoint emerged for the function of SAHF from Fabrizio d'Adda di Fagagna's lab [115]. These authors concluded that SAHF is formed as a result of persistent DNA damage and helps to dampen the cellular responses to DNA damage. In fact, d'Adda di Fagagna's recent work showed the existence of two types of heterochromatin in senescent cells; one which represses proliferation-promoting genes and another, SAHF, which suppresses the DNA damage response [115]. Inactivation of ATM or p53 was sufficient to release proliferation-promoting genes from heterochromatin-mediated repression and reactivate cell proliferation. Remarkably, these proliferating cells retained SAHF, showing that SAHF are not sufficient to repress proliferation genes and drive proliferation arrest. Previous reports have similarly shown that inactivation of p53 does not completely bypass SAHF formation [111, 112]. Also some studies have shown an shown increase in abundance of heterochromatin proteins and marks in senescence, without formation of SAHF [116]. Hence, local heterochromatinization of proliferation-promoting genes, but not large scale SAHF, appears to mediate silencing of proliferation genes and senescence-associated proliferation arrest. Instead, d'Adda di Fagagna and coworkers went on to show that SAHF serve to restrict senescence-associated DNA damage signaling to sublethal levels, thereby contributing to the prolonged viability of senescent cells (Figure 2).
Another recent report suggests that, rather than suppressing DNA damage signaling, chromatin changes in senescence amplify DNA damage signaling. Karlseder and coworkers showed that levels of all core histones and the linker histone H1 decrease during senescence [117]. This coincides with reduced expression of the stem loop–binding protein SLBP, which is important for stability and expression of histone mRNAs [117, 118]. Karlseder and coworkers proposed that DNA damage signals at short telomeres destabilize SLBP, leading, in turn, to reduced production of H3 and H4. This leads to genome-wide chromatin changes, which amplify the damage signal that triggers senescence. Moreover, chromatin changes at telomeres allow DNA damage and repair proteins to associate with chromosome ends, further elevating the DNA damage signal. Eventually, the DNA damage signal exceeds a threshold and triggers cell cycle arrest. Taken together with the work of d'Adda di Fagagna this work suggest that chromatin finely balances the level of DNA damage signaling in a senescent cell. On the one hand, chromatin changes, particularly at telomere ends, upregulate DNA damage signaling to promote cell cycle arrest. On the other hand, chromatin changes in the form of SAHF suppress DNA damage signaling to prevent cell death. This dual role is consistent with our previous observation that telomeres in senescent cells are excluded from SAHF [110].
Physiological function of chromatin changes in senescence
Since cell senescence is a tumor suppression process, it may be that SAHF and other chromatin changes contribute to tumor suppression (Figure 2). Defining the molecular function of SAHF will shed light on whether or not these structures are likely to contribute to tumor suppression. If SAHF suppress cell proliferation, they might be tumor suppressive. On the other hand, if they suppress DNA damage-induced cell killing in mutated cells, they might actually be oncogenic (Figure 2). Significantly, inactivation of effectors involved in SAHF formation, such as HMGA2, contributes to bypass of senescence [108]. This implies that SAHF are tumor suppressive and inactivation of gene products that are required for SAHF formation, such as HUCA and HMGA2, might contribute to development of human tumors. Interestingly, a member of the HUCA complex, CABIN1, is hypermethylated in ovarian carcinoma [119], and missense mutations have been reported in schwannomatosis, a rare tumor of the tissue covering nerves [120]. Similarly, HIRA was reported to be deleted in 22.7% of meningiomas, which are brain tumors that develop in the meninges, the tissue that surrounds and protects the brain and spinal cord [121]. Further, other chaperones that deposit H3.3, such as DAXX and ATRX, while not yet implicated in SAHF formation, have been reported to be mutated in 43% of pancreatic neuroendocrine tumors (PanNETs) [122]. Other chromatin remodelling factors, such as BRG1 and SNF5, also mediate tumor suppression, in part by upregulation of p16 and activation of senescence [123-133]. Taken together, it is clear that some chromatin regulators act as tumor suppressors, but whether any do so through formation of SAHF remains to be established.
Chromatin maintenance in non-proliferating senescent cells
Chromatin is a highly dynamic entity in which nucleosomes and associated proteins actively turnover. By controlling accessibility of DNA to chromatin binding proteins, regulated nucleosome turnover controls gene expression states and facilitates processes, such as replication and DNA repair [134]. The dynamic nature of chromatin is highlighted by the fact that nucleosomes at active genes and at epigenetic regulatory elements are replaced multiple times during a cell cycle time of approximately 20 hours [135]. Similarly, regulatory proteins associated with chromatin also show dynamic binding and dissociation. For example even apparently stable heterochromatin domains show transient binding and dynamic exchange of HP1 on chromatin [136]. Moreover, heterochromatin seems to be a dynamic structure which can spread to neighboring sites [137]. Together, these results imply that there is active maintenance of euchromatin and heterochromatin in eukaryotic cells.
In proliferating cells, chromatin is maintained in conjunction with DNA replication. Nucleosomes are transiently disrupted ahead of the replication fork and are rapidly deposited and positioned behind the replication fork. One factor, which actively participates in DNA replication-coupled nucleosomal deposition is CAF1 that is recruited to the DNA by interacting with proliferating cell nuclear antigen (PCNA) [138-141]. Consistent with this, human CAF-1 is found in a complex containing two of the major histones expressed in S phase, H3.1 and H4 [99, 142]. Loss of CAF-1 leads to defects in DNA replication and S-phase progression, suggesting that in human cells, CAF-1 couples chromatin maintenance and assembly to DNA replication [143, 144]. PCNA, in addition to recruiting CAF1, also serves as platform for recruitment of DNMT1 (a maintenance DNA methyltransferase) [145], various chromatin remodelling complexes and histone modifying enzymes such as HDACs [146, 147]. DNMT1 preferentially methylates hemimethylated DNA, suggesting the parental strand serves as template for copying methylation patterns to the daughter strand [145]. Likewise DNMT1 also interacts with EZH2 and helps in establishment of H3K27Me3 repressive chromatin coupled to DNA replication [148]. In sum, the activities of DNMT1, CAF1, PCNA, HDACs and other chromatin remodelers recruited to the replication fork can achieve faithful propagation of DNA methylation and histone PTMs and “replication coupled” maintenance of chromatin.
However, since we have already noted that chromatin is inherently dynamic, even outside of S phase, this raises the question of how chromatin structure is maintained in a non-proliferating cell. For example, in a senescent nevus melanocyte chromatin structure should be maintained for decades, and a failure to do so could result in malignant transformation of such a cell, which already harbors an activated oncogene.
Chromatin maintenance linked to transcription
In non-proliferating cells, processes other than DNA replication disrupt chromatin and provide an opportunity for maintenance and/or remodelling. For example, during transcription, passage of RNA polymerase II results in displacement of histones followed by reassembly. An important factor in facilitating transcription through chromatin is FACT, which removes one copy of the H2A–H2B dimer in a transcription-coupled manner [149, 150]. FACT acts by destabilizing nucleosomes as RNA polymerase II passes along chromatin and restores nucleosomal structure after the DNA has been transcribed [150]. Although senescent cells are replication deficient, they are transcription competent, metabolically active and secrete lots of inflammatory proteins. Thus, senescent cells have active ongoing transcription, which could provide a window of opportunity for chromatin maintenance (Figure 3).
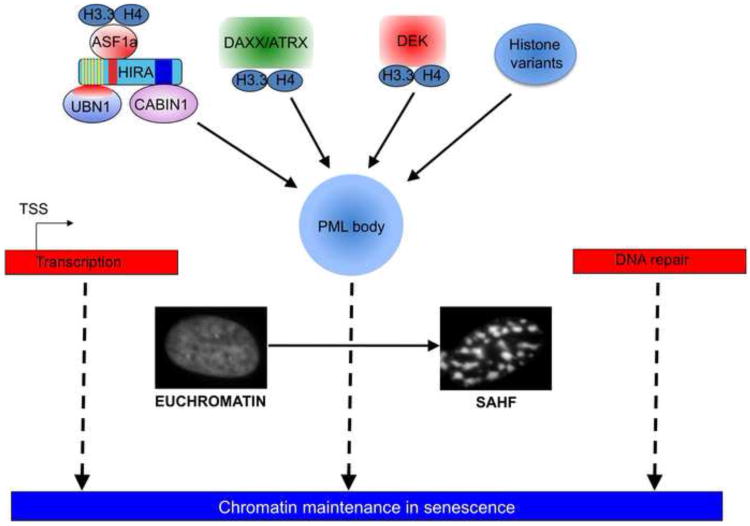
Figure 3. Some options available for chromatin regulation and maintenance in senescence.
Multiple histone chaperones and some histone variants, e.g. H3.3, converge on PML bodies and help in formation of SAHF and chromatin maintenance. Other histone variants such as macroH2A also get incorporated into SAHF and may contribute to maintenance of chromatin structure in senescenc Finally e chromatin can also be maintained in senescent cells in conjunction with ongoing transcription and DNA repair, processes that depend on turnover of chromatin.
Chromatin maintenance linked to DNA repair
Another example of chromatin metabolism independent of replication is DNA repair, which involves remodeling of the chromatin via nucleosome exchange and eviction. DNA damage results in rapid recruitment of histone acetyltransferases at the damage loci, which, via acetylation of histones, open up the chromatin for repair factors [151]. Damage loci also show rapid phosphorylation of the histone H2AX variant [83]. After completion of the repair, the chromatin must be restored to its original format. In flies, the histone acetyltransferase dTip60 and the ATPase Domino/p400 catalyze the replacement of phospho-H2Av by unmodified H2Av (the fly ortholog of H2Ax) [152]. Another PTM that needs to be removed is acetylation. The acetylated histones present at the DNA damage site could result in inappropriate gene activation and so should be either removed or deactylated. Indeed, there is decrease in histone acetylation post DNA repair, via recruitment of HDACs at the repaired loci [153]. Thus DNA damage repair provides an opportunity for chromatin restructuring and maintenance independent of DNA replication. Indeed, the DNA damage-signaling pathway is a major effector of the cell senescence program [12-14, 115]. Therefore, replication-independent chromatin assembly coupled to DNA damage repair may be important for chromatin maintenance in senescent cells (Figure 3).
Chromatin maintenance and histone variants
Transcription is known to be coupled to deposition of the histone variant, H3.3 which marks actively transcribed genes [85]. Given H3.3′s diverse role in gene activation and silencing it could be involved in both active transcription and heterochromatin maintenance in senescent and/or non-proliferating cells. Although differentiation is different from senescence in many aspects, both processes share absence of cell division and, perhaps, utilization of histone variants. Many lines of evidence points to H3.3's role in senescent and/or non proliferating differentiated cells. Firstly, mouse erythroleukemia (MEL) cells undergoing differentiation induce expression of both H3.3 genes, H3.3B and H3.3A [154]. This was shown at the mRNA level, but presumably precedes H3.3 deposition. Secondly, histone H3 variant, H3.3 accumulates in fibroblasts approaching senescence and in non-dividing differentiated cells [154-158]. Thirdly, H3.3 also accumulates in quiescent T lymphocytes and the proportion of H3.3 in the chromatin of a cell is related to how long that cell has been quiescent [159]. Fourthly, differentiating and mature rat brain cortical neurons accumulate H3.3 [160]. Consistent with an increase of H3.3 in senescent and differentiated cells, the H3.3 chaperone HUCA pathway is apparently activated in primary senescent cells and in vivo in the skin of aging primates [114, 161, 162]. In addition to H3.3, other histone variants are also associated with chromatin of senescent cells. Specifically, histone H2A variant, macroH2A, is enriched in SAHF of senescent human fibroblasts [114].
Consistent with this, levels of macroH2A increase in human diploid fibroblasts (TIG3) undergoing replicative and oncogene induced senescence [163, 164]. Further, differentiating and mature rat brain cortical neurons accumulate histone variants such as H2AX in chromatin, whereas, H3.1, and H3.2 decrease [160]. Taken together this suggests that histone variants and their chaperones, such as HUCA, could provide a molecular framework for chromatin maintenance in non-proliferating and senescent cells (Figure 3). Therefore, incorporation of histone variants, such as H3.3, into chromatin of senescent cells might be important for maintenance of chromatin structure and function and cell phenotype.
A role for PML bodies? Interestingly, HIRA, a member of the HUCA H3.3 chaperone complex, is recruited to PML nuclear bodies in senescent cells. PML bodies are 0.2–1.0-micrometer diameter structures that are implicated in diverse biological processes, such as senescence, the antiviral response, apoptosis and tumor suppression [165]. Why HIRA goes to PML bodies in senescence is not known, but disruption of HIRA's translocation to PML bodies impairs the ability of senescent cells to form SAHF [112]. Specifically, dominant negative HIRA mutants that block HIRA's localization to PML bodies prevent formation of SAHF. Similarly, a PML-RARα fusion protein which disrupts PML bodies also prevents formation of SAHF [112]. Interestingly, UBN1 and CABIN1 also localize to PML bodies in senescent cells [166] (TSR and PDA, unpublished). This implies that the HUCA histone chaperone's localization to PML bodies might be important for remodeling or maintenance of chromatin structure in senescent cells. Further, PML bodies contribute to senescence by recruiting pRB/E2F complexes and suppressing E2F target gene expression [167]. In fact, in senescent cells there may be multiple replication independent nucleosome assembly pathways operating via PML bodies. For example, DAXX/ATRX also co-localize to PML bodies [168, 169], perhaps for replication independent chromatin assembly [106, 169]. Since both HUCA and DAXX/ATRX complex interact with PML nuclear bodies, it's tempting to speculate that several replication-independent histone assembly pathways converge at PML nuclear bodies for chromatin maintenance. Therefore, PML bodies might be important regulatory sites of replication-independent chromatin assembly in senescence (Figure 3).
Concluding remarks
Although much as been learnt about histone variants, chaperones and chromatin remodelers, and their respective roles in chromatin structure and function in proliferating cells, little is known in the context of senescence and/or differentiation and quiescent stem cells. Recent identification of multiple replication-independent histone deposition pathways has opened doors for investigation of chromatin structure maintenance and function in the context of senescence, differentiation and quiescence. Future studies will address the relative contributions of these chaperones and their associated histone variants in senescence-associated replication-independent chromatin maintenance, and whether or not alterations in these factors are drivers of cancers.
*Highlights.
Overview of cell senescence
Overview of chromatin structure
Implications of senescence for chromatin maintenance
Model for maintenance of chromatin structure in senescence.
Acknowledgments
The lab of PDA is funded by the NIA (P01 AG031862) and CRUK (C10652/A10250). Thanks for all members of the Adams lab for critical discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Adams PD. Healing and hurting: molecular mechanisms, functions, and pathologies of cellular senescence. Mol Cell. 2009;36:2–14. doi: 10.1016/j.molcel.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 2.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Herbig U, Sedivy JM. Regulation of growth arrest in senescence: telomere damage is not the end of the story. Mech Ageing Dev. 2006;127:16–24. doi: 10.1016/j.mad.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Ramirez RD, Morales CP, Herbert BS, Rohde JM, Passons C, Shay JW, Wright WE. Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes Dev. 2001;15:398–403. doi: 10.1101/gad.859201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright WE, Shay JW. Historical claims and current interpretations of replicative aging. Nat Biotechnol. 2002;20:682–688. doi: 10.1038/nbt0702-682. [DOI] [PubMed] [Google Scholar]
- 6.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 8.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 9.Hayflick L. The Limited in Vitro Lifetime of Human Diploid Cell Strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 10.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 11.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 12.d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 13.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d'Adda di Fagagna F. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 14.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 15.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 16.Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- 19.Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF Induces Senescence and Apoptosis through Pathways Mediated by the Secreted Protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kortlever RM, Higgins PJ, Bernards R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat Cell Biol. 2006;8:877–884. doi: 10.1038/ncb1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tremain R, Marko M, Kinnimulki V, Ueno H, Bottinger E, Glick A. Defects in TGF-beta signaling overcome senescence of mouse keratinocytes expressing v-Ha-ras. Oncogene. 2000;19:1698–1709. doi: 10.1038/sj.onc.1203471. [DOI] [PubMed] [Google Scholar]
- 22.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 23.Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, Takatsu Y, Melamed J, d'Adda di Fagagna F, Bernard D, Hernando E, Gil J. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 24.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, Yee H, Zender L, Lowe SW. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narita M, Young AR, Arakawa S, Samarajiwa SA, Nakashima T, Yoshida S, Hong S, Berry LS, Reichelt S, Ferreira M, Tavare S, Inoki K, Shimizu S. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science. 2011;332:966–970. doi: 10.1126/science.1205407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavare S, Arakawa S, Shimizu S, Watt FM. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee BY, Han JA, Im JS, Morrone A, Johung K, Goodwin EC, Kleijer WJ, DiMaio D, Hwang ES. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 2006;5:187–195. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 31.Byun HO, Han NK, Lee HJ, Kim KB, Ko YG, Yoon G, Lee YS, Hong SI, Lee JS. Cathepsin D and eukaryotic translation elongation factor 1 as promising markers of cellular senescence. Cancer Res. 2009;69:4638–4647. doi: 10.1158/0008-5472.CAN-08-4042. [DOI] [PubMed] [Google Scholar]
- 32.Demidenko ZN, Blagosklonny MV. Growth stimulation leads to cellular senescence when the cell cycle is blocked. Cell Cycle. 2008;7:3355–3361. doi: 10.4161/cc.7.21.6919. [DOI] [PubMed] [Google Scholar]
- 33.Demidenko ZN, Zubova SG, Bukreeva EI, Pospelov VA, Pospelova TV, Blagosklonny MV. Rapamycin decelerates cellular senescence. Cell Cycle. 2009;8:1888–1895. doi: 10.4161/cc.8.12.8606. [DOI] [PubMed] [Google Scholar]
- 34.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 35.Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dorken B, Jenuwein T, Schmitt CA. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Pandolfi PP. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, Beach D, Serrano M. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 38.Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, Hollstein PE, MacCollin M, Cichowski K. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459–472. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B, Jamieson NB, Oien KA, Lowy AM, Brunton VG, Frame MC, Evans TR, Sansom OJ. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci U S A. 2010;107:246–251. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cordon-Cardo C, Horner JW, 2nd, DePinho RA. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11:2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bardeesy N, Morgan J, Sinha M, Signoretti S, Srivastava S, Loda M, Merlino G, DePinho RA. Obligate roles for p16(Ink4a) and p19(Arf)-p53 in the suppression of murine pancreatic neoplasia. Mol Cell Biol. 2002;22:635–643. doi: 10.1128/MCB.22.2.635-643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 43.Besancenot R, Chaligne R, Tonetti C, Pasquier F, Marty C, Lecluse Y, Vainchenker W, Constantinescu SN, Giraudier S. A senescencelike cell-cycle arrest occurs during megakaryocytic maturation: implications for physiological and pathological megakaryocytic proliferation. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parrinello S, Coppe JP, Krtolica A, Campisi J. Stromal-epithelial interactions in aging and cancer: senescent fibroblasts alter epithelial cell differentiation. J Cell Sci. 2005;118:485–496. doi: 10.1242/jcs.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci U S A. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fukuda A, Wang SC, Morris JPT, Folias AE, Liou A, Kim GE, Akira S, Boucher KM, Firpo MA, Mulvihill SJ, Hebrok M. Stat3 and MMP7 Contribute to Pancreatic Ductal Adenocarcinoma Initiation and Progression. Cancer Cell. 2011;19:441–455. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Kloppel G, Yoshimura A, Reindl W, Sipos B, Akira S, Schmid RM, Algul H. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19:456–469. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 48.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 49.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 50.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 51.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 53.Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Differentially methylated forms of histone H3 show unique association patterns with inactive human × chromosomes. Nat Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- 54.Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- 55.Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 56.Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 57.Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 58.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 2009;7:e13. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 61.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 62.Matangkasombut O, Buratowski S. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol Cell. 2003;11:353–363. doi: 10.1016/s1097-2765(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 63.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 64.Ornaghi P, Ballario P, Lena AM, Gonzalez A, Filetici P. The bromodomain of Gcn5p interacts in vitro with specific residues in the N. terminus of histone H4. J Mol Biol. 1999;287:1–7. doi: 10.1006/jmbi.1999.2577. [DOI] [PubMed] [Google Scholar]
- 65.Matangkasombut O, Buratowski RM, Swilling NW, Buratowski S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 2000;14:951–962. [PMC free article] [PubMed] [Google Scholar]
- 66.Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 67.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 68.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 69.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 70.Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 72.Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Snowden AW, Gregory PD, Case CC, Pabo CO. Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr Biol. 2002;12:2159–2166. doi: 10.1016/s0960-9822(02)01391-x. [DOI] [PubMed] [Google Scholar]
- 74.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 75.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 76.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 77.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 78.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gehani SS, Agrawal-Singh S, Dietrich N, Christophersen NS, Helin K, Hansen K. Polycomb group protein displacement and gene activation through MSK-dependent H3K27me3S28 phosphorylation. Mol Cell. 2010;39:886–900. doi: 10.1016/j.molcel.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 80.Sarma K, Reinberg D. Histone variants meet their match. Nat Rev Mol Cell Biol. 2005;6:139–149. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- 81.Polo SE, Almouzni G. Chromatin assembly: a basic recipe with various flavours. Curr Opin Genet Dev. 2006;16:104–111. doi: 10.1016/j.gde.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 82.Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive × chromosome of female mammals. Nature. 3931998:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- 83.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 85.Ahmad K, Henikoff S. The histone variant h3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 86.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci U S A. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chow CM, Georgiou A, Szutorisz H, Maia e Silva A, Pombo A, Barahona I, Dargelos E, Canzonetta C, Dillon N. Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division. EMBO Rep. 2005;6:354–360. doi: 10.1038/sj.embor.7400366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Johnson L, Mollah S, Garcia BA, Muratore TL, Shabanowitz J, Hunt DF, Jacobsen SE. Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res. 2004;32:6511–6518. doi: 10.1093/nar/gkh992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sugimura K, Fukushima Y, Ishida M, Ito S, Nakamura M, Mori Y, Okumura K. Cell cycle-dependent accumulation of histone H3.3 and euchromatic histone modifications in pericentromeric heterochromatin in response to a decrease in DNA methylation levels. Exp Cell Res. 2010;316:2731–2746. doi: 10.1016/j.yexcr.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 93.Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, George AJ, Morgan KA, Mann JR, Choo KH. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–360. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wong LH, Ren H, Williams E, McGhie J, Ahn S, Sim M, Tam A, Earle E, Anderson MA, Mann J, Choo KH. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 2009;19:404–414. doi: 10.1101/gr.084947.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van der Heijden GW, Derijck AA, Posfai E, Giele M, Pelczar P, Ramos L, Wansink DG, van der Vlag J, Peters AH, de Boer P. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat Genet. 2007;39:251–258. doi: 10.1038/ng1949. [DOI] [PubMed] [Google Scholar]
- 96.Santenard A, Ziegler-Birling C, Koch M, Tora L, Bannister AJ, Torres-Padilla ME. Torres-Padilla, Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat Cell Biol. 2010;12:853–862. doi: 10.1038/ncb2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 98.De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 99.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 100.Phelps-Durr TL, Thomas J, Vahab P, Timmermans MC. Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis, Plant. Cell. 2005;17:2886–2898. doi: 10.1105/tpc.105.035477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dimova D, Nackerdien Z, Furgeson S, Eguchi S, Osley MA. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol Cell. 1999;4:75–83. doi: 10.1016/s1097-2765(00)80189-6. [DOI] [PubMed] [Google Scholar]
- 102.Kaufman PD, Cohen JL, Osley MA. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol Cell Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Anderson HE, Kagansky A, Wardle J, Rappsilber J, Allshire RC, Whitehall SK. Silencing mediated by the Schizosaccharomyces pombe HIRA complex is dependent upon the Hpc2-like protein, Hip4. PLoS One. 2011;5:e13488. doi: 10.1371/journal.pone.0013488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dutta D, Ray S, Home P, Saha B, Wang S, Sheibani N, Tawfik O, Cheng N, Paul S. Regulation of angiogenesis by histone chaperone HIRA-mediated Incorporation of lysine 56-acetylated histone H3.3 at chromatin domains of endothelial genes. J Biol Chem. 2010;285:41567–41577. doi: 10.1074/jbc.M110.190025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang JH, Song Y, Seol JH, Park JY, Yang YJ, Han JW, Youn HD, Cho EJ. Myogenic transcriptional activation of MyoD mediated by replication-independent histone deposition. Proc Natl Acad Sci U S A. 2011;108:85–90. doi: 10.1073/pnas.1009830108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sawatsubashi S, Murata T, Lim J, Fujiki R, Ito S, Suzuki E, Tanabe M, Zhao Y, Kimura S, Fujiyama S, Ueda T, Umetsu D, Ito T, Takeyama K, Kato S. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev. 2010;24:159–170. doi: 10.1101/gad.1857410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kappes F, Waldmann T, Mathew V, Yu J, Zhang L, Khodadoust MS, Chinnaiyan AM, Luger K, Erhardt S, Schneider R, Markovitz DM. The DEK oncoprotein is a Su(var) that is essential to heterochromatin integrity. Genes Dev. 2011;25:673–678. doi: 10.1101/gad.2036411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Narita M, Narita M, Krizhanovsky V, Nunez S, Chicas A, Hearn SA, Myers MP, Lowe SW. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell. 2006;126:503–514. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 109.Funayama R, Saito M, Tanobe H, Ishikawa F. Loss of linker histone H1 in cellular senescence. J Cell Biol. 2006;175:869–880. doi: 10.1083/jcb.200604005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang R, Chen W, Adams PD. Molecular Dissection of Formation of Senescent Associated Heterochromatin Foci. Mol Cell Biol. 2007;27:2343–2358. doi: 10.1128/MCB.02019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 112.Ye X, Zerlanko B, Zhang R, Somaiah N, Lipinski M, Salomoni P, Adams PD. Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol Cell Biol. 2007;27:2452–2465. doi: 10.1128/MCB.01592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chan HM, Narita M, Lowe SW, Livingston DM. The p400 E1A-associated protein is a novel component of the p53 --> p21 senescence pathway. Genes Dev. 2005;19:196–201. doi: 10.1101/gad.1280205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, Pehrson JR, Berger JM, Kaufman PD, Adams PD. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell. 2005;8:19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 115.Di Micco R, Sulli G, Dobreva M, Liontos M, Botrugno OA, Gargiulo G, Dal Zuffo R, Matti V, d'Ario G, Montani E, Mercurio C, Hahn WC, Gorgoulis V, Minucci S, d'Adda di Fagagna F. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat Cell Biol. 2011;13:292–302. doi: 10.1038/ncb2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kosar M, Bartkova J, Hubackova S, Hodny Z, Lukas J, Bartek J. Senescence-associated heterochromatin foci are dispensable for cellular senescence, occur in a cell type- and insult-dependent manner and follow expression of p16(ink4a) Cell Cycle. 2011;10:457–468. doi: 10.4161/cc.10.3.14707. [DOI] [PubMed] [Google Scholar]
- 117.O'Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol. 2010;17:1218–1225. doi: 10.1038/nsmb.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA. Gene. 1999;239:1–14. doi: 10.1016/s0378-1119(99)00367-4. [DOI] [PubMed] [Google Scholar]
- 119.Feng Q, Deftereos G, Hawes SE, Stern JE, Willner JB, Swisher EM, Xi L, Drescher C, Urban N, Kiviat N. DNA hypermethylation, Her-2/neu overexpression and p53 mutations in ovarian carcinoma. Gynecol Oncol. 2008;111:320–329. doi: 10.1016/j.ygyno.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Buckley PG, Mantripragada KK, Diaz de Stahl T, Piotrowski A, Hansson CM, Kiss H, Vetrie D, Ernberg IT, Nordenskjold M, Bolund L, Sainio M, Rouleau GA, Niimura M, Wallace AJ, Evans DG, Grigelionis G, Menzel U, Dumanski JP. Identification of genetic aberrations on chromosome 22 outside the NF2 locus in schwannomatosis and neurofibromatosis type 2. Hum Mutat. 2005;26:540–549. doi: 10.1002/humu.20255. [DOI] [PubMed] [Google Scholar]
- 121.Wada K, Maruno M, Suzuki T, Kagawa N, Hashiba T, Fujimoto Y, Hashimoto N, Izumoto S, Yoshimine T. Chromosomal and genetic abnormalities in benign and malignant meningiomas using DNA microarray. Neurol Res. 2005;27:747–754. doi: 10.1179/016164105X35648. [DOI] [PubMed] [Google Scholar]
- 122.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, Velculescu VE, Diaz LA, Jr, Vogelstein B, Kinzler KW, Hruban RH, Papadopoulos N. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kuwahara Y, Charboneau A, Knudsen ES, Weissman BE. Reexpression of hSNF5 in malignant rhabdoid tumor cell lines causes cell cycle arrest through a p21(CIP1/WAF1)-dependent mechanism. Cancer Res. 2010;70:1854–1865. doi: 10.1158/0008-5472.CAN-09-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kang H, Cui K, Zhao K. BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI. Mol Cell Biol. 2004;24:1188–1199. doi: 10.1128/MCB.24.3.1188-1199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hendricks KB, Shanahan F, Lees E. Role for BRG1 in cell cycle control and tumor suppression. Mol Cell Biol. 2004;24:362–376. doi: 10.1128/MCB.24.1.362-376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Oruetxebarria I, Venturini F, Kekarainen T, Houweling A, Zuijderduijn LM, Mohd-Sarip A, Vries RG, Hoeben RC, Verrijzer CP. P16INK4a is required for hSNF5 chromatin remodeler-induced cellular senescence in malignant rhabdoid tumor cells. J Biol Chem. 2004;279:3807–3816. doi: 10.1074/jbc.M309333200. [DOI] [PubMed] [Google Scholar]
- 127.Betz BL, Strobeck MW, Reisman DN, Knudsen ES, Weissman BE. Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated with induction of p16ink4a and activation of RB. Oncogene. 2002;21:5193–5203. doi: 10.1038/sj.onc.1205706. [DOI] [PubMed] [Google Scholar]
- 128.Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, Begemann M, Crabtree GR, Goff SP. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 129.Schneppenheim R, Fruhwald MC, Gesk S, Hasselblatt M, Jeibmann A, Kordes U, Kreuz M, Leuschner I, Martin Subero JI, Obser T, Oyen F, Vater I, Siebert R. Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. Am J Hum Genet. 2010;86:279–284. doi: 10.1016/j.ajhg.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 131.Wong AK, Shanahan F, Chen Y, Lian L, Ha P, Hendricks K, Ghaffari S, Iliev D, Penn B, Woodland AM, Smith R, Salada G, Carillo A, Laity K, Gupte J, Swedlund B, Tavtigian SV, Teng DH, Lees E. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 2000;60:6171–6177. [PubMed] [Google Scholar]
- 132.Yuge M, Nagai H, Uchida T, Murate T, Hayashi Y, Hotta T, Saito H, Kinoshita T. HSNF5/INI1 gene mutations in lymphoid malignancy. Cancer Genet Cytogenet. 2000;122:37–42. doi: 10.1016/s0165-4608(00)00274-0. [DOI] [PubMed] [Google Scholar]
- 133.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 134.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 135.Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 137.Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nat Rev Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 138.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 139.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 140.Smith S, Stillman B. Immunological characterization of chromatin assembly factor I, a human cell factor required for chromatin assembly during DNA replication in vitro. J Biol Chem. 1991;266:12041–12047. [PubMed] [Google Scholar]
- 141.Krude T. Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp Cell Res. 1995;220:304–311. doi: 10.1006/excr.1995.1320. [DOI] [PubMed] [Google Scholar]
- 142.Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 143.Hoek M, Stillman B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc Natl Acad Sci U S A. 2003;100:12183–12188. doi: 10.1073/pnas.1635158100. Epub 12003 Sep 12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, Adams PD. Defective S-phase chromatin assembly causes DNA damage, activation of the S-phase checkpoint and S-phase arrest. Molecular Cell. 2003;11:341–351. doi: 10.1016/s1097-2765(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 145.Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 146.Poot RA, Bozhenok L, van den Berg DL, Steffensen S, Ferreira F, Grimaldi M, Gilbert N, Ferreira J, Varga-Weisz PD. The Williams syndrome transcription factor interacts with PCNA to target chromatin remodelling by ISWI to replication foci. Nat Cell Biol. 2004;6:1236–1244. doi: 10.1038/ncb1196. [DOI] [PubMed] [Google Scholar]
- 147.Milutinovic S, Zhuang Q, Szyf M. Proliferating cell nuclear antigen associates with histone deacetylase activity, integrating DNA replication and chromatin modification. J Biol Chem. 2002;277:20974–20978. doi: 10.1074/jbc.M202504200. [DOI] [PubMed] [Google Scholar]
- 148.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 149.Mason PB, Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol. 2003;23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 151.Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 152.Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 153.Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Krimer DB, Cheng G, Skoultchi AI. Induction of H3.3 replacement histone mRNAs during the precommitment period of murine erythroleukemia cell differentiation. Nucleic Acids Res. 1993;21:2873–2879. doi: 10.1093/nar/21.12.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bosch A, Suau P. Changes in core histone variant composition in differentiating neurons: the roles of differential turnover and synthesis rates. Eur J Cell Biol. 1995;68:220–225. [PubMed] [Google Scholar]
- 156.Brown DT, Wellman SE, Sittman DB. Changes in the levels of three different classes of histone mRNA during murine erythroleukemia cell differentiation. Mol Cell Biol. 1985;5:2879–2886. doi: 10.1128/mcb.5.11.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Grove GW, Zweidler A. Regulation of nucleosomal core histone variant levels in differentiating murine erythroleukemia cells. Biochemistry. 1984;23:4436–4443. doi: 10.1021/bi00314a030. [DOI] [PubMed] [Google Scholar]
- 158.Rogakou EP, Sekeri-Pataryas KE. Histone variants of H2A and H3 families are regulated during in vitro aging in the same manner as during differentiation. Exp Gerontol. 1999;34:741–754. doi: 10.1016/s0531-5565(99)00046-7. [DOI] [PubMed] [Google Scholar]
- 159.Wu RS, Tsai S, Bonner WM. Changes in histone H3 composition and synthesis pattern during lymphocyte activation. Biochemistry. 1983;22:3868–3873. doi: 10.1021/bi00285a023. [DOI] [PubMed] [Google Scholar]
- 160.Pina B, Suau P. Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Dev Biol. 1987;123:51–58. doi: 10.1016/0012-1606(87)90426-x. [DOI] [PubMed] [Google Scholar]
- 161.Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Cellular senescence in aging primates. Science. 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- 162.Jeyapalan JC, Ferreira M, Sedivy JM, Herbig U. Accumulation of senescent cells in mitotic tissue of aging primates. Mech Ageing Dev. 2007;128:36–44. doi: 10.1016/j.mad.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Maehara K, Takahashi K, Saitoh S. CENP-A reduction induces a p53-dependent cellular senescence response to protect cells from executing defective mitoses. Mol Cell Biol. 2010:30. 2090–2104. doi: 10.1128/MCB.01318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sporn JC, Kustatscher G, Hothorn T, Collado M, Serrano M, Muley T, Schnabel P, Ladurner AG. Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene. 2009;28:3423–3428. doi: 10.1038/onc.2009.26. [DOI] [PubMed] [Google Scholar]
- 165.Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell. 2002;108:165–170. doi: 10.1016/s0092-8674(02)00626-8. [DOI] [PubMed] [Google Scholar]
- 166.Banumathy G, Somaiah N, Zhang R, Tang Y, Hoffmann J, Andrake M, Ceulemans H, Schultz D, Marmorstein R, Adams PD. Human UBN1 is an ortholog of yeast Hpc2p and has an essential role in the HIRA/ASF1a chromatin-remodeling pathway in senescent cells. Mol Cell Biol. 2009;29:758–770. doi: 10.1128/MCB.01047-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vernier M, Bourdeau V, Gaumont-Leclerc MF, Moiseeva O, Begin V, Saad F, Mes-Masson AM, Ferbeyre G. Regulation of E2Fs and senescence by PML nuclear bodies. Genes Dev. 2011:25. 41–50. doi: 10.1101/gad.1975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Natl Acad Sci U S A. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]