Abstract
The Philippines is a strategic point in the Asia-Pacific region for the study of human diversity, history and origins, as it is a cross-road for human migrations and consequently exhibits enormous ethnolinguistic diversity. Following on a previous in-depth study of Y-chromosome variation, here we provide new insights into the maternal genetic history of Filipino ethnolinguistic groups by surveying complete mitochondrial DNA (mtDNA) genomes from a total of 14 groups (11 groups in this study and 3 groups previously published) including previously published mtDNA hypervariable segment (HVS) data from Filipino regional center groups. Comparison of HVS data indicate genetic differences between ethnolinguistic and regional center groups. The complete mtDNA genomes of 14 ethnolinguistic groups reveal genetic aspects consistent with the Y-chromosome, namely: diversity and heterogeneity of groups, no support for a simple dichotomy between Negrito and non-Negrito groups, and different genetic affinities with Asia-Pacific groups that are both ancient and recent. Although some mtDNA haplogroups can be associated with the Austronesian expansion, there are others that associate with South Asia, Near Oceania and Australia that are consistent with a southern migration route for ethnolinguistic group ancestors into the Asia-Pacific, with a timeline that overlaps with the initial colonization of the Asia-Pacific region, the initial colonization of the Philippines and a possible separate post-colonization migration into the Philippine archipelago.
Keywords: complete mitochondrial genome, Filipino ethnolinguistic, genetic affinity
Introduction
The Philippines is a strategic point in the Asia-Pacific region for the study of human diversity, history and origins because of several factors. First, the Philippine archipelago was connected to the Sunda land mass,1 which may have facilitated migrations to and/or through the Philippines. Second, Philippine human fossil remains date back to 47–66.7 thousand years ago (kya),2, 3 indicating an ancient occupation. Third, the Filipino population is composed of over 170 Filipino ethnolinguistic (FE) groups with different cultural and demographic backgrounds.4 There are over 30 different FE groups who possess the pygmy phenotype5 and have been historically and collectively called Filipino ‘Negrito' (FEN) groups, in contrast to groups who do not have the pygmy phenotype (FE non-Negrito (FEnN) groups). Fourth, all Philippine languages are Austronesian languages. The expansion of this language family, proposed to have originated from Taiwan about 5 to 6 kya and spread first through the Philippines,6 makes the Philippines an arena to study human cultural and/or demic expansion. Finally, the Philippine archipelago is composed of about 7000 islands, clustered into three main island groups (Luzon, Visayas and Mindanao), and further subdivided into 17 regions, 80 provinces, 138 cities, 1496 municipalities and 42 025 districts.7 The 17 regional divisions were part of the Integrated Reorganization Plan of President Ferdinand Marcos in the 1970s, for political and administrative purposes. Regional centers were established within each regional division, where government offices, services, commerce and livelihood were concentrated. Regional centers immediately became hubs for migrating Filipinos from across the country for livelihood and economic reasons. The current distribution of Filipino populations can thus be divided in two: (1) FE groups settled in their respective communities (within specific provincial cities and/or municipalities, possibly their ancestral lands) and (2) Filipinos in regional centers (RC groups) who may have specific ethnolinguistic ancestry and/or are migrating Filipinos from other Philippine regions. All these factors make the Philippines an interesting region for the study of ancient and recent human migrations, origins and demographic history.
Several population genetic studies have revealed various aspects of Filipino groups and their history. A survey of genome-wide single-nucleotide polymorphisms (SNPs) of Asian groups show that six FE groups (five FEN and one FEnN) and three RC groups do not differ from each other or from other Austronesian speaking groups, and that there was a single migration wave into Asia from which all Asian groups are descended.8 Genome-wide SNPs show that gene flow occurred into the common ancestors of New Guineans, Australians and the Mamanwa FEN group from Denisovans.9 Genome-wide SNPs also show an ancient association between Australia, New Guinea and the Mamanwa FEN group, which supports the view that these populations represent the descendants of an early ‘southern route' migration out of Africa.10 Fifteen autosomal short tandem repeats (aSTRs) from 12 RC groups demonstrate limited population stratification, which supports the pooling of RC aSTR data into a single population genetic database for forensic applications.11 A preliminary comparison of aSTR data from 12 RC groups11 and 12 FE groups (F Delfin, MCA DeUngria and M Stoneking, unpublished data) show that the Filipino population is composed of a homogeneous set of different RC groups with separate, distinct FE groups. Mitochondrial DNA (mtDNA) diversity based on the hypervariable segment (HVS) and complete mtDNA genomes of Filipinos from three regional centers, each within the main Philippine islands of Luzon, Visayas and Mindanao show support for the ‘Out of Taiwan' theory for the expansion of the Austronesian language family.12 Next-generation sequencing methods allowed an unbiased, population-based survey of complete mtDNA genomes revealing novel and ancient mtDNA lineages as well as different demographic histories (effective population size (Ne) changes) among three FE groups.13 A survey of the non-recombining region of the Y-chromosome (NRY) reveal extensive heterogeneity with different ancient and recent genetic affinities for 16 FE groups.14 A subsequent NRY and mtDNA study of the Batak FEN group15 demonstrates that this group is not as distinct and isolated as other FEN groups studied in Delfin et al.14
It can thus be seen that with the exception of our previous study of NRY variation,14 other studies of Philippine genetic diversity have included very few FE groups. We therefore analyzed an unbiased, population-based data set of complete mtDNA genomes from 14 FE groups, consisting of 11 FE groups (4 FEN and 7 FEnN) generated in this study, and 3 FE groups (1 FEN and 2 FEnN) published previously.13 We compare the FE group data set with mtDNA HVS data from Filipino RC groups and with other complete mtDNA genomes retrieved from the data archive of PhyloTree.org (http://www.phylotree.org/mtDNA_seqs.htm) that represent Asian and Pacific groups. This study provides new insights into the maternal genetic history and genetic structure of Filipino groups.
Methods
FE group samples
A total of 260 complete mtDNA genomes representing 11 FE groups were sequenced in this study. This data set was combined with a data set of 97 complete mtDNA genomes from 3 FE groups,13 bringing the FE group data set to a total of 357 complete mtDNA genomes representing 14 FE groups (Supplementary Figure 1, Supplementary Tables 1 and 2, Supplementary Text). Human population sampling procedures followed the guidelines outlined by the Philippine National Bioethics Committee on Scientific Research, the National Commission on Indigenous Peoples (NCIP) and the Max Planck Institute for Evolutionary Anthropology (MPI-EVA). This research was approved by the ethics committee of the University of Leipzig Medical Faculty. Free, prior and informed consent for sample collection was obtained at the level of the community and the individual.
DNA sequencing and sequence assembly
DNA libraries were prepared using a multiplex method developed for the Illumina Genome Analyzer (GA) platform,16 coupled with a target-enrichment method specific for human mtDNA.17 DNA libraries were sequenced on an Illumina GA IIx machine (Illumina Inc., San Diego, CA, USA) with post processing using Illumina software followed by the Improved Base Identification System.18 Sequencing reads were mapped to the revised Cambridge Reference Sequence of the human mitochondrial genome (GenBank: NC012920.1)19 using the program MIA,20 implemented in an MPI-EVA sequence assembly-analyses pipeline for detecting mtDNA heteroplasmy21 and low-level mutations22 (Supplementary Text). Sequence coverage ranged from 10x to >2000x (Supplementary Figure 2, Supplementary Table 2, Supplementary Text). To ensure quality data analyses, all sequences used in the study had <1% missing data (ambiguous bases (N) plus gaps<165) (Supplementary Table 2, Supplementary Text). Complete mtDNA sequences (n=260) have been deposited in GenBank (GenBank accession numbers: KC993902–KC994161).
Population data comparisons
FE group mtDNA data were compared with a Filipino data set of unknown FE group affiliation from12 (Supplementary Text). FE group data also were compared with a reference data set composed of 1759 complete mtDNA genomes representing 43 Asian and Pacific groups retrieved from the data archive of PhyloTree.org (http://www.phylotree.org/mtDNA_seqs.htm; Supplementary Table 3, Supplementary Text).
Multiple sequence alignment (MSA) and haplogroup assignment
MAFFT v6.833b23 was used in the MSA of all available data: FE group data; Filipino, non-FE group data;12 the reference data set and the Reconstructed Sapiens Reference Sequence (RSRS)24 (http://www.mtdnacommunity.org). Aligned, complete mtDNA sequences were assigned haplogroups using a custom Perl script (available from the DREEP website: http://dmcrop.sourceforge.net) that combines the MUSCLE v3.825, 26 with mtDNAble v1.1.0.028 (http://www.mtdnacommunity.org). Haplogrep27 (haplogrep.uibk.ac.at) with PhyloTree mtDNA tree build 1328 (http://www.phylotree.org), also was used to check haplogroup assignment (Supplementary Text). Haplogroup assignment served as another gauge for data quality (Supplementary Table 4, Supplementary Text). Before all subsequent analyses, BioEdit29 was used to remove all nucleotide positions with missing data, across all available data (FE group, RC group and reference data sets).
Population genetic statistics
Arlequin 3.5.1.330 was used for the following analyses: genetic diversity indices (Table 1); ΦST genetic distance; Mantel tests and analysis of molecular variance (AMOVA). Diversity indices also were checked using DnaSP v5.31 Statistica v832 was used for the following analyses: multidimensional scaling (MDS) using ΦST genetic distance estimates; correspondence analysis (CA) using haplogroup counts and Mann–Whitney U-tests. Matrix correlation analyses via Mantel tests for genetics–geography and genetics–language association made use of the following components: ΦST genetic distance matrix generated by Arlequin 3.5.1.330; geographic distance matrix generated by Geographic Distance Matrix Generator v1.2.333 (http://biodiversityinformatics.amnh.org/open_source/gdmg) and a language distance matrix kindly provided by Russell Gray.6
Table 1. Genetic diversity indices for FE groups.
| Filipino group | n |
Haplotype (Ht) informationa |
Haplogroup (Hg) informationa |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of haplotypes | Number of polymorphic sites | HtD | SE | MPD | SE | Nucleotide diversity | SE | Number of haplogroups | HgD | SE | ||
| Philippines | 357 | 172 | 512 | 0.98 | 0.005 | 15.38 | 6.88 | 0.0010 | 0.00050 | 54 | 0.94 | 0.006 |
| Ivatan | 29 | 20 | 102 | 0.98 | 0.013 | 16.62 | 7.62 | 0.0011 | 0.00055 | 12 | 0.93 | 0.023 |
| Ifugao | 29 | 21 | 87 | 0.97 | 0.018 | 15.06 | 6.93 | 0.0010 | 0.00050 | 9 | 0.86 | 0.036 |
| Kankanaey | 30 | 20 | 80 | 0.96 | 0.020 | 16.20 | 7.43 | 0.0011 | 0.00054 | 8 | 0.88 | 0.024 |
| Ibaloi | 26 | 17 | 70 | 0.95 | 0.027 | 11.64 | 5.45 | 0.0008 | 0.00039 | 9 | 0.85 | 0.051 |
| Kalangoya | 26 | 17 | 55 | 0.96 | 0.022 | 12.31 | 5.74 | 0.0008 | 0.00042 | 5 | 0.70 | 0.059 |
| Bugkalot | 30 | 14 | 88 | 0.90 | 0.034 | 13.66 | 6.31 | 0.0009 | 0.00046 | 8 | 0.81 | 0.048 |
| AetaZa | 10 | 1a | 0 | 0.00 | 0.000 | 0.00 | 0.00 | 0.0000 | 0.00000 | 3a | 0.69 | 0.104 |
| AetaB | 23 | 10 | 58 | 0.87 | 0.048 | 11.43 | 5.38 | 0.0007 | 0.00039 | 5 | 0.70 | 0.058 |
| AgtaI | 9 | 4 | 82 | 0.58 | 0.183 | 18.22 | 8.94 | 0.0012 | 0.00066 | 6 | 0.83 | 0.127 |
| Abaknon | 30 | 20 | 116 | 0.96 | 0.021 | 15.12 | 6.95 | 0.0010 | 0.00050 | 13 | 0.92 | 0.025 |
| Surigaonon | 24 | 19 | 88 | 0.97 | 0.021 | 12.78 | 5.97 | 0.0008 | 0.00043 | 13 | 0.93 | 0.026 |
| Mamanwa | 33 | 17 | 101 | 0.90 | 0.037 | 16.66 | 7.61 | 0.0011 | 0.00055 | 14 | 0.86 | 0.046 |
| Manobo | 40 | 22 | 140 | 0.95 | 0.017 | 13.09 | 6.02 | 0.0009 | 0.00043 | 17 | 0.92 | 0.021 |
| Maranao | 18 | 17 | 143 | 0.99 | 0.021 | 21.37 | 9.89 | 0.0014 | 0.00072 | 14 | 0.95 | 0.039 |
Abbreviations: HgD, haplogroup diversity; HtD, haplotype diversity; MPD, mean pairwise difference; n, sample size.
FE groups are ordered from top to bottom, according to a north to south geographical location (see Figure 1). FEN groups are underlined.
For haplotype information, nucleotide positions that have missing data (ambiguous bases (Ns) and gaps) across the entire data set were not considered. For haplogroup information, haplogroup assignment was based on full sequence data for each sample in the data set. This explains why diversity indices for the AetaZ FEN group show three haplogroups, but only one haplotype.
Bayesian evolutionary analyses
The program package BEAST v1.7.234 (http://beast.bio.ed.ac.uk/) was used for all Bayesian Markov Chain Monte Carlo (MCMC) analyses. Bayesian MCMC was used to estimate coalescent times and to generate Bayesian Skyline Plots (BSPs; Supplementary Text). The mtDNA coding region (nt577–nt16023) was used for these analyses including that of the RSRS24 to root mtDNA trees. Network version 4.6.1.0 and Network Publisher version 1.3.0.0 (http://fluxus-engineering.com) were used to check for consistency in the clustering of sequences on the Bayesian maximum clade credibility trees (MCC tree; Supplementary Text).
Results
mtDNA diversity, genetic relationships and demographic history of FE groups
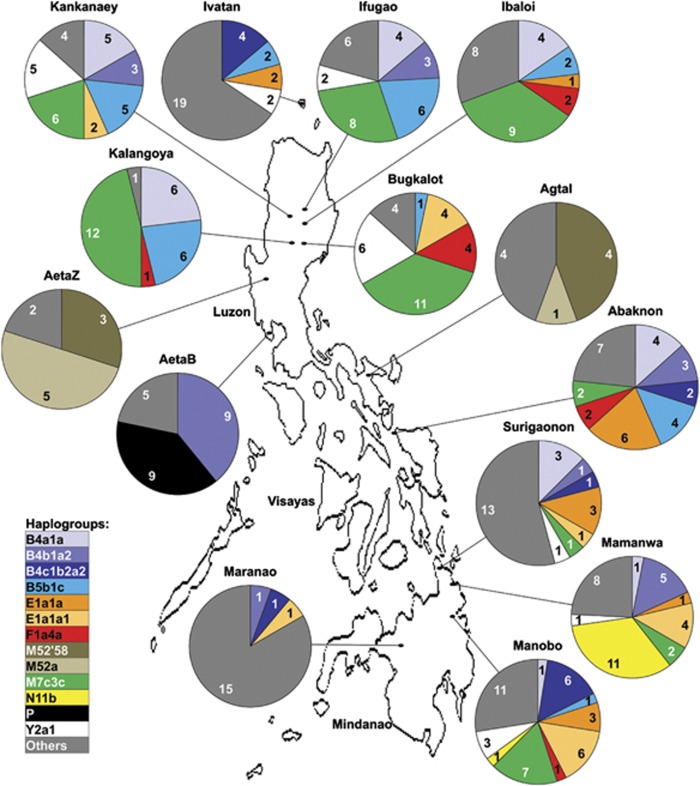
Genetic diversity indices (Table 1) show there are 172 haplotypes and 54 haplogroups (Table 2) among the 14 FE groups in this study. HtD values are higher for FEnN than for FEN groups, whereas the other diversity indices do not show a discrete FEnN and/or FEN grouping pattern, in that some FEN group indices are comparable to those of FEnN groups. Based on Mann–Whitney U-tests, the diversity indices of FEN groups did not significantly differ from FEnN groups (polymorphic sites: Z=1.42, P=0.16; MPD: Z=0.28, P=0.78 and nucleotide diversity: Z=0.28, P=0.78) except for HtD (Z=2.83, P=0.005) and HgD (Z=2.12, P=0.03), which could be influenced by the smaller sample size of some FEN groups. AMOVA (Table 3) also does not support a discrete FEN vs FEnN grouping. The 54 haplogroups have a patchy distribution across the different FE groups (Figure 1, Table 2). These patterns of mtDNA diversity and haplogroup distribution are consistent with those of the NRY,14 which are also probably due to the effects of genetic drift on isolated groups with small population sizes.
Table 2. Haplogroup frequencies for FE groups.
| Filipino group | Philippines |
Northern groups |
Central groups |
Southern groups |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample size | 360 | Ivatan | Ifugao | Kankanaey | Ibaloi | Kalangoya | Bugkalot | AetaZ | AetaB | AgtaI | Abaknon | Surigaonon | Mamanwa | Manobo | Maranao |
| 29 | 29 | 30 | 26 | 26 | 30 | 10 | 23 | 9 | 30 | 24 | 33 | 40 | 18 | ||
| Haplogroup | Frequency (%) | ||||||||||||||
| B4a1 | 0.28 | 5.56 | |||||||||||||
| B4a1a | 7.78 | 13.79 | 16.67 | 15.38 | 23.08 | 13.33 | 12.50 | 3.03 | 2.50 | ||||||
| B4a1a1a | 0.28 | 5.56 | |||||||||||||
| B4a1a3 | 0.83 | 6.90 | 2.50 | ||||||||||||
| B4a1a4 | 0.56 | 6.90 | |||||||||||||
| B4a1a5 | 1.11 | 3.45 | 3.45 | 3.85 | 3.85 | ||||||||||
| B4b1a | 1.39 | 13.04 | 11.11 | 3.03 | |||||||||||
| B4b1a2 | 6.94 | 10.34 | 10.00 | 39.13 | 10.00 | 4.17 | 15.15 | 5.56 | |||||||
| B4b1a2b | 1.39 | 12.50 | 2.50 | 5.56 | |||||||||||
| B4b1a2d | 1.11 | 16.67 | |||||||||||||
| B4c | 0.28 | 2.50 | |||||||||||||
| B4c1b2a2 | 3.89 | 13.79 | 6.67 | 4.17 | 15.00 | 5.56 | |||||||||
| B5 | 0.28 | 11.11 | |||||||||||||
| B5a1d | 0.28 | 3.33 | |||||||||||||
| B5a2a | 0.28 | 3.45 | |||||||||||||
| B5b1c | 7.50 | 6.90 | 20.69 | 16.67 | 7.69 | 23.08 | 3.33 | 13.33 | 2.50 | ||||||
| D5b1c | 1.39 | 3.45 | 3.33 | 11.54 | |||||||||||
| D6a | 0.83 | 3.33 | 3.03 | 2.50 | |||||||||||
| D6c | 1.11 | 22.22 | |||||||||||||
| E1a1a | 4.44 | 6.90 | 3.85 | 20.00 | 12.50 | 3.03 | 7.50 | ||||||||
| E1a1a1 | 5.00 | 6.67 | 13.33 | 4.17 | 12.12 | 15.00 | 5.56 | ||||||||
| E2a | 0.83 | 12.50 | |||||||||||||
| E2a1 | 0.28 | 3.03 | |||||||||||||
| E2b1 | 1.11 | 10.00 | |||||||||||||
| F1a3a | 1.94 | 17.24 | 4.35 | 3.33 | |||||||||||
| F1a4a | 2.78 | 7.69 | 3.85 | 13.33 | 6.67 | 2.50 | |||||||||
| F3b | 2.22 | 6.90 | 10.00 | 11.54 | |||||||||||
| F3b1 | 0.56 | 3.33 | 5.56 | ||||||||||||
| M | 0.83 | 20.00 | 11.11 | ||||||||||||
| M17c | 0.56 | 6.67 | |||||||||||||
| M21a | 0.28 | 4.17 | |||||||||||||
| M50 | 0.28 | 5.56 | |||||||||||||
| M52′58 | 1.94 | 30.00 | 44.44 | ||||||||||||
| M52a | 1.67 | 50.00 | 11.11 | ||||||||||||
| M74b1 | 0.83 | 4.17 | 6.06 | ||||||||||||
| M7b1′2′4′5′6′7′8-(C16192T) | 0.28 | 2.50 | |||||||||||||
| M7b3a | 1.11 | 4.35 | 3.33 | 11.11 | |||||||||||
| M7c3c | 16.11 | 27.59 | 20.00 | 34.62 | 46.15 | 36.67 | 6.67 | 4.17 | 6.06 | 17.50 | |||||
| M7c3c1 | 0.28 | 2.50 | |||||||||||||
| M7c3-T16295C! | 1.39 | 13.79 | 11.11 | ||||||||||||
| M13′46′61-T16362C | 0.28 | 3.85 | |||||||||||||
| N11 | 0.28 | 3.03 | |||||||||||||
| N11b | 3.33 | 33.33 | 2.50 | ||||||||||||
| P | 2.50 | 39.13 | |||||||||||||
| P1d1 | 0.28 | 5.56 | |||||||||||||
| P2′10 | 0.83 | 6.67 | 5.56 | ||||||||||||
| Q1′2 | 0.28 | 4.17 | |||||||||||||
| R | 0.28 | 3.45 | |||||||||||||
| R9b1a1 | 0.28 | 3.03 | |||||||||||||
| R9c1 | 0.83 | 6.90 | 3.33 | ||||||||||||
| R9c1a | 1.39 | 10.34 | 2.50 | 5.56 | |||||||||||
| R-T16189C! | 0.56 | 3.33 | 3.03 | ||||||||||||
| Y2a | 0.28 | 5.56 | |||||||||||||
| Y2a1 | 5.56 | 6.90 | 6.90 | 16.67 | 20.00 | 4.17 | 3.03 | 7.50 | |||||||
Table 3. Analysis of molecular variance (AMOVA) results.
| Grouping (number of groups) |
Haplotype |
Haplogroup |
||||
|---|---|---|---|---|---|---|
| Among groups | Among populations within groups | Within population | Among groups | Among populations within groups | Within populations | |
| Philippines (14) | – | 7.9 | 92.1 | – | 9.8 | 90.2 |
| FEN (4) | – | 16.4 | 83.6 | – | 18.9 | 81.1 |
| FEnN (10) | – | 4.2 | 95.8 | – | 6.0 | 94.0 |
| FEN (4) vs FEnN (10) | 4.5 | 6.0 | 89.5 | 4.6 | 7.9 | 87.6 |
| Geography: | ||||||
| North (8) vs Central (2) vs South (4) | 1.2a | 7.1 | 91.7 | 0.2a | 9.7 | 90.1 |
| NL (3) vs CL (2) vs SL (2) vs Central (2) vs South (3) | 2.0a | 7.0 | 91.0 | 3.8 | 6.8 | 89.5 |
| Language: North (9) vs Central (2) vs South (2) | 3.1a | 6.7 | 90.2 | 0.0a | 10.5 | 89.6 |
All P-values are significant (P<0.05) except for the estimates marked with superscript 'a'.
Philippine groupings are shown in Supplementary Figure 1 and enumerated in Supplementary Table 1.
Geography: Luzon groups also were tested as northern Luzon (NL), central Luzon (CL) and southern Luzon (SL).
Figure 1.
Distribution of haplogroups of FE groups. This figure shows the haplogroups that are present in ≥10 individuals across the entire FE group data set and those of major interest (haplogroups M52'58 and M52a). The figure legend on the lower left section of the figure shows the list of haplogroups whose color code correspond to those on the pie charts. Haplogroups represented by <10 individuals across the entire FE group data set are pooled into the dark gray pie chart sections and labeled as ‘Others' in the figure legend. Numbers in each pie chart section indicate the number of individuals that possess the haplogroup and total to each FE groups' sample size (Table 2).
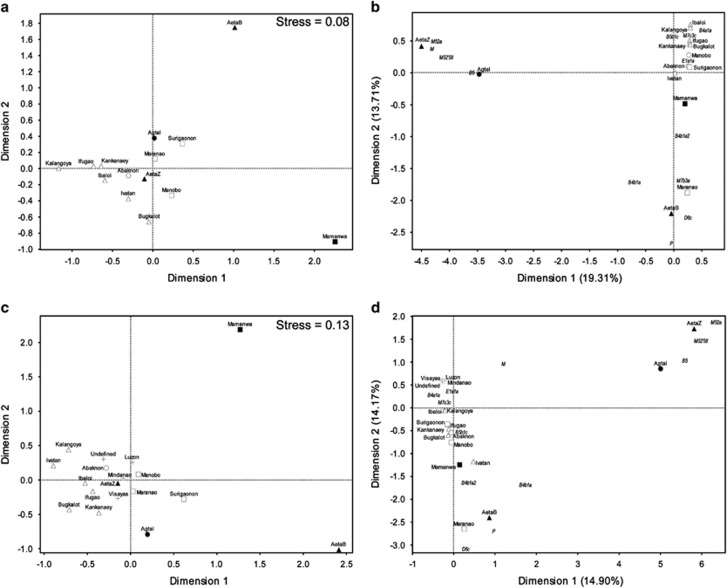
MDS analysis (Figure 2a) shows no tight, group-specific clustering among FE groups, not even for FEnN and/or FEN groups. However, there are some subtle patterns such as northern FEnN groups in one plot section and central FE groups in the plot center. CA (Figure 2b) shows that the cluster of all FEnN groups and the Mamanwa FEN group is associated with haplogroups B4a1a, B5b1c, E1a1a and M7c3c. Three FEN groups and the Maranao are distinct from the others because of haplogroups found only in these groups, namely: haplogroups M52'58 and M52a in AetaZ and Agta; haplogroup B5 in Agta; haplogroup P in AetaB and haplogroups D6c and M7b3a in the Maranao. Haplotype sharing analyses (Supplementary Figure 3) show haplotypes shared exclusively among northern FE groups (Ht# 1–9); haplotypes shared only among southern groups (Ht# 10–13) and several shared haplotypes that also have a patchy distribution across FE groups (Ht# 14–20). The AetaZ group shares its one haplotype (Ht# 21) with all FE groups except the AetaB, Surigaonon and Maranao and the AetaB group shares one haplotype (Ht# 22) only with the Agta and Abaknon. Multivariate analyses (MDS and CA) and haplotype sharing analyses do not show any discrete grouping among the FE groups. Matrix correlation analyses show a weak and nonsignificant positive correlation between genetics and geography (r=0.10, P=0.24) as well as a nonsignificant negative correlation between genetic and linguistic distances (r= −0.20, P=0.85). AMOVA (Table 3) also does not support any particular grouping of FE groups. Instead AMOVA results highlight the higher heterogeneity for FEN groups than for FEnN groups and the Philippines as a whole. Overall, FE groups appear to be heterogeneous, and more so among FEN groups despite having in common the ‘pygmy' phenotype.
Figure 2.
Multivariate analyses of Filipino mtDNA data. (a) MDS plot of ΦST genetic distances based on complete mtDNA data of FE groups. (b) CA plot of haplogroup counts in FE groups. (c) MDS plot of ΦST genetic distances based on HVS-1 sequence data of FE groups and RC groups (Luzon, Visayas, Mindanao, undefined) from Tabbada et al..12 (d) CA plot of haplogroup counts in FE and RC groups. FE group names are indicated and are adjacent to shapes. Shapes indicate northern (triangles), central (circles) and southern (squares) geographical locations of FE groups. Open shapes indicate FEnN groups, while filled shapes indicate FEN groups. Plus signs (+) indicate RC groups from Tabbada et al..12 In CA plots (b, d), haplogroup names are in narrow italicized font. Other relevant details are in Supplementary Text.
Bayesian MCMC estimates (BE) and credible intervals (CI) of haplogroup coalescent times are within the range of previously published estimates and standard deviations (SD; Table 4).12, 24 Overall, BEs of haplogroup coalescent times for FE groups extend from about 2800 years ago (2.8 kya), corresponding to the Holocene and the Neolithic period in the Philippines, to >10 kya in the Late Pleistocene and the time of early human occupation in the Philippines. BSPs (Supplementary Figures 4 and 5, Supplementary Text) show that from >20 kya to about 10 kya, FE groups experienced different trends in Ne change with generally, a decline in Ne between 10 and 5 kya and a drop in Ne around 2 kya to the present time. Estimates of current Ne (Supplementary Table 5) show that the female Ne are but a fraction of the actual group sizes with groups such as the AetaZ and the Agta having very low Ne. Ne estimates (Supplementary Table 5) also coincide with mtDNA diversity (Table 1) in that groups with the smallest Ne have the lowest diversity because of the effects of genetic drift. The differences in the trends of Ne change and the current Ne estimates suggests different demographic histories for the FE groups included in this study.
Table 4. Coalescent time estimates (years) for FE group haplogroups.
|
This study |
Behar et al.24 |
Tabbada et al.12 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Haplogroup | n | HPD lower | BE | HPD upper | Haplogroup | MLE | SD | Haplogroup | Estimate | SD |
| B5b1c | 27 | 4263 | 9915 | 16 220 | B5b1c | 9486 | 4352 | B5b1 | 4700 | 2400 |
| E1a1a | 16 | 2438 | 6557 | 11 638 | E1a1a | 6884 | 2429 | E1a1a | 7300 | 4500 |
| E1a1a1 | 17 | 1427 | 4965 | 9139 | E1a1a1 | 5153 | 2947 | no | no | no |
| E1a1a+E1a1a1 | 33 | 2346 | 6145 | 10 729 | ||||||
| N11b | 12 | 2068 | 7605 | 13 894 | N11b | 6748 | 4591 | no | no | no |
| B4c1b2a2 | 14 | 5528 | 11 739 | 18 826 | B4c1b2a | 9139 | 4337 | B4c1b | 16 300 | 14 500 |
| Y2a1 | 20 | 2524 | 7139 | 12 477 | Y2a1 | 2488 | 2658 | Y2 | 5000 | 2300 |
| B4a1a | 28 | 3531 | 8050 | 13 656 | B4a1a | 9541 | 2290 | B4a1a | 7900 | 2400 |
| F1a4a | 10 | 987 | 4393 | 8437 | F1a4 | 5660 | 3297 | F1a4 | 10 100 | 6000 |
| M7c3c | 58 | 3729 | 8582 | 14 416 | M7c3c | 10 538 | 4608 | M7c3c | 11 400 | 3600 |
| B4b1a2 | 25 | 3066 | 7329 | 12 268 | B4b1a2 | 15 237 | 3838 | B4b1 | 8800 | 3200 |
| M52′58 | 7 | 324 | 2853 | 6212 | M52′58 | 42 448 | 8131 | no | no | no |
| M52a | 6 | 311 | 2844 | 6301 | M52a | 24 424 | 7766 | no | no | no |
| M52′58+M52a | 13 | 426 | 2808 | 5905 | ||||||
| P | 9 | 14 281 | 27 291 | 41 722 | P | 54 801 | 2501 | no | no | no |
This study: n (sample size); BE (Bayesian estimate); HPD (95% highest posterior density or credible intervals (CI)).
Behar et al.24 and Tabbada et al.12: in these studies, some haplogroups in ′This study′ were not observed or time was not estimated, hence the relevant upper haplogroup lineages (underlined) were included in the table for comparison.
Tabbada et al.12: no (not observed).
FE group mtDNA and Filipino RC group mtDNA
The Luzon, Visayas and Mindanao groups, who were previously surveyed using mtDNA HVS data,12 can be considered RC groups because individuals were sampled in regional centers. More haplogroups were detected among FE groups than were previously reported for RC groups12 (Supplementary Table 6). Although, the same East Asian and Southeast Asian haplogroups previously observed for RC groups (B4, B5, D, E, F, R9, M7 and Y)12 also were observed among FE groups, but with differences in the array of haplogroup sublineages (Supplementary Table 6). MDS analysis shows that HVS-1 sequences of RC groups appear to be in the middle of FE groups in this study (Figure 2c). The Filipino group of ‘Undefined' geographical origins and/or FE affiliation appears in the same general central MDS plot position as the other RC groups. A separate MDS analysis (data not shown) of complete mtDNA genomes of FE groups and 30 complete mtDNA genomes of undefined geographical origins and/or FE group affiliation,12 showed the same MDS plot position for the ‘Undefined' Filipino group as in Figure 2c. CA (Figure 2d) shows that RC groups (including the ‘Undefined' group) are close to most FE groups in the first dimension of the plot, with the association driven by common haplogroups (B4a1a, E1a1a and M7c3c). In the second CA plot dimension, RC groups are closer to the AetaZ and Agta FE groups because of M haplogroup sublineages. In the same dimension, differences in the array of haplogroup sublineages (Supplementary Table 6) drive the distinction between RC and FE groups. These results further characterize Filipino population structure in that apart from RC groups there are distinct FE groups.
FE groups and the Asia-Pacific region
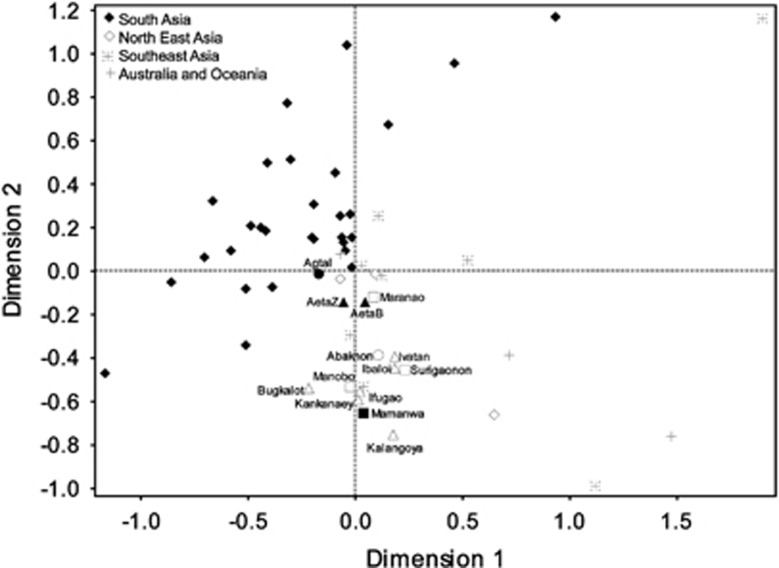
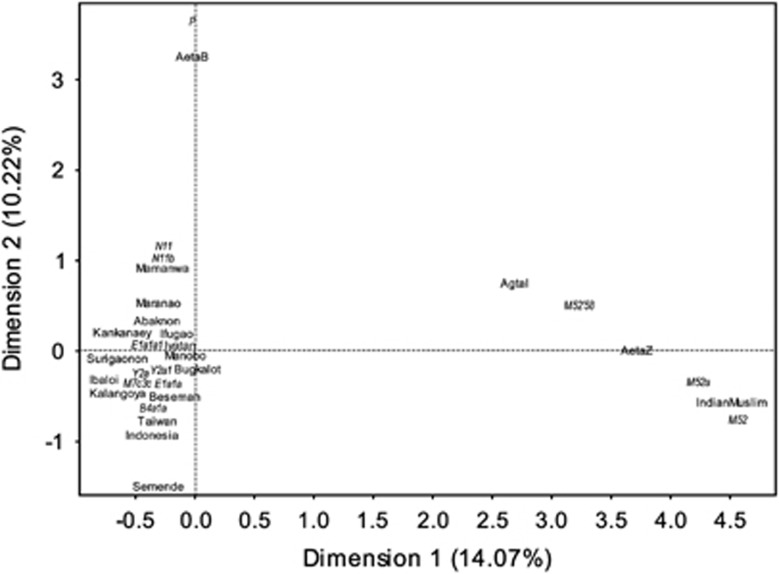
Multivariate analyses (MDS: Figure 3, Supplementary Figure 6; CA: Figure 4, Supplementary Figure 7, Supplementary Tables 7 and 8; Supplementary Text), show an overview of the relation of FE groups with Asian and Pacific groups (Supplementary Table 3, Supplementary Text). All MDS biplots (Figure 3, Supplementary Figure 6) showed the same overall pattern in that FE groups are most associated with northern East Asian, Southeast Asian and Pacific groups. However, there also appears to be an associations between FE groups and Indian groups. Haplotype sharing analyses showed that FE groups share only five haplotypes with only Taiwan, Indonesia (including Besemah and Semende) and Near Oceania (Supplementary Table 9). Like MDS (Figure 3), CA (Figure 4) shows Taiwan and Indonesia (including Besemah and Semende) to be closest to FE groups. There also appears to be an association between the Agta and AetaZ groups and Indian Muslim groups (Suni and Shia)35 driven by the sharing of haplogroups M52'58 and M52a.
Figure 3.
MDS plot of FE groups and the reference data set. MDS plot of ΦST genetic distances based on complete mtDNA data of FE groups and the reference data set. Shown here is the biplot of dimensions 1 and 2. The biplots of dimensions 2 to 5 are shown in Supplementary Figure 6. Stress value is 0.12. FE group names are indicated and are adjacent to shapes. These shapes indicate northern (triangles), central (circles) and southern (squares) geographical locations. Open shapes indicate FEnN groups, while filled shapes indicate FEN groups. Asian and Pacific groups of the reference data set are represented by the following symbols: filled diamonds (South Asia: 28 Indian groups, including Andaman Islanders); asterisks (Southeast Asia: Vietnam, the Besemah and Semende groups of Sumatra, Indonesia and the Bidayuh, Jehai, Seletar and Temuan groups of Malaysia); open diamonds (North East Asia: Han Chinese, Japan and Taiwan); plus signs (Australia, Near Oceania and Remote Oceania). Other relevant details are in Supplementary Text and Supplementary Table 3.
Figure 4.
CA plot of haplogroup counts in FE groups and reference data set. The CA result here involved five reference data set groups (Supplementary Table 8) and is the last of a series of CA that started with 18 reference data set groups (Supplementary Table 7; Supplementary Figure 7). FE group and reference data set group names are indicated in bold print. Haplogroup names are in narrow italicized font. Other relevant details are in Supplementary Text and Supplementary Table 3.
Analyses of specific mtDNA lineages
The B4a1a MCC tree (Supplementary Figure 8) showed two major clades. One clade contains all FE group (except Mara3683) B4a1a, associated with Taiwan and Indonesian sequences and coalesces at 9298 years (yrs) (CI: 5879–13 159 yrs), approximating a previous B4a1a estimate24 (Table 4). The only B4a1a1a FE group sample, Mara3683 (Maranao), coalesces with a Remote Oceanian sequence in the other major clade composed of only B4a1a1 and its sublineages and coalesces at 7152 yrs (CI: 4725–9858 yrs), again corresponding to a previous estimate.24 The E1a1a MCC tree (Supplementary Figure 9), also bifurcates into a clade of 6936 yrs (CI: 3182–10 817 yrs) and a younger clade of 3943 yrs (CI: 886–7985). The older clade, which approximates previous time estimates,24 contains all FE group E1a1a sequences along with sequences from Indonesia and Taiwan. With only one Taiwan and two Malaysian M7c3c complete mtDNA genomes available in the reference data set, all sequences in the M7c3c MCC tree (Supplementary Figure 10) coalesced at 12 510 yrs (CI: 6289–20 162 yrs). Reanalysis without the Taiwan and Malaysia sequences yielded a coalescent time of 10 138 yrs (CI: 5286–16 154 yrs), which then approximates previous estimates24 (Table 4). The M7c3c MCC tree also bifurcates into an older (8682 yrs (CI: 6355–12 648 yrs)) and younger (5758 yrs (2417–10 243 yrs)) clade. Sequences from FE groups, Taiwan, Indonesia and Malaysia fall within the older clade while only FE groups are within the younger clade. The general trend here is that majority of FE group B4a1a, E1a1a and M7c3c sequences are within clades of older coalescent ages and are associated with Taiwan and Indonesian sequences. Although with limited data from Taiwan, phylogeographic analysis for the three haplogroups (Supplementary Table 10) show decreasing variation from Taiwan to the Philippines and Indonesia. Although variation appears higher in Indonesia than in FE groups, the SEs overlap. In addition, while the coalescent time CIs overlap, the coalescent mean estimates (BE) are slightly higher for FE groups than for Indonesia. The analyses here would have certainly benefited from more representative and population-based sampling in Taiwan and Indonesia, as was done here for the Philippines, rather than biased sampling of complete mtDNA genomes, as discussed elsewhere.13 Nevertheless, overall the results are consistent with the prevailing view of a southward dispersal of B4a1a, E1a1a and M7c3c that originated from Taiwan.
Haplogroups M52'58 and M52a were observed among the AetaZ and Agta FE groups (Figure 1, Table 2). The M52'58 MCC tree coalesces at 18 482 yrs (CI: 10 638–27 148 yrs; Supplementary Figure 11). In general, the coalescent estimates made here are more recent than those previously published.24, 35, 36 The older ages in previous estimates can be attributed to stochastic variance, most likely to be caused by smaller sample sizes in these previous estimations. The MCC tree (Supplementary Figure 11) bifurcates into two clades. One clade is composed of South Asian sequences that coalesce at 13 871 yrs (CI: 7457–20 540 yrs). The other clade is composed of AetaZ and Agta sequences and one South Asian sequence that coalesce at 12 557 yrs (CI: 4791–20 587 yrs). Although there may be Ns at some haplogroup diagnostic nts, all FE group sequences were consistently classified by mtDNAble24 (Supplementary Table 4) and Haplogrep.27 Furthermore, mtDNAble results suggest M52a1 as partial descendant for all M52a samples (Supplementary Table 4). All FE group sequences coalesce at a recent age of 4996 yrs (CI: 1181–9786 yrs), overlapping with the M52a1 age previously estimated.24 Results show an association of mtDNA lineages between specific FE groups, in particular FEN groups and South Asian groups. However, the estimated timelines are more recent than the initial colonization of the Philippines.2 The estimates range from the Late Pleistocene to the Holocene suggesting a post-colonization link between FEN and South Asian groups.
Previously classified as an unresolved N haplogroup in the Mamanwa and the Manobo,13 the survey of 11 other FE groups still finds haplogroup N11b unique to the Mamanwa and the Manobo (Figure 1, Table 2). A MCC tree (Supplementary Figure 12) of haplogroup N and its sublineages shows FE group coding region sequences coalesce at 8415 yrs (CI: 3616–14 325 yrs). FE group sequences further coalesce with two sequences from India at 42 994 yrs (CI: 32 494–53 711 yrs). This particular result suggests another possible mtDNA link between a FEN group, the Mamanwa and South Asia. However; compared with M52'58 and M52a, the observed timeline for the Mamanwa-South Asia mtDNA link overlaps with the initial settlement of the Philippines2 and the Asia-Pacific region.37
Haplogroup P was observed in the AetaB FEN group and P sublineages in the Maranao and the Bugkalot (Figure 1, Table 2). Results of mtDNAble (Behar et al.24) (Supplementary Table 4) suggest AetaB, P haplogroup sequences may be further classified as P9. The haplogroup P, MCC tree (Supplementary Figure 13) shows Bugkalot and Maranao sequences coalesce with Near Oceania (46 078 yrs (CI: 33 532–59 717 yrs)) and Australia (44 107 yrs (CI: 30 343–57 854 yrs)), while all AetaB sequences are in a subclade that coalesces at 40 353 yrs (CI: 26 970–53 525 yrs). These results show an association of FE groups with Australian and Near Oceanian mtDNA lineages that extend well into the Late Pleistocene.
Other haplogroups
Haplogroups B4b1a and B5b1c are of appreciable frequency (> 5%) in FE groups (Table 3). However, population comparison was limited to FE groups and Japan because Japan was the only population-based and geographically defined group in the reference data set that possessed B4b1 and B5b1 lineages. FE group and Japanese B4b1a and B5b1 coding sequences fall into distinct clades that diverged some 15–20 kya (Supplementary Figures 14 and 15), suggesting an ancient link. But as macrohaplogroups B4 and B5 and their sublineages are generally associated with mainland East and Southeast Asia,38, 39, 40 more population-based samples of complete mtDNA genomes from these regions are required in order to verify the observed Filipino–Japanese association. Haplogroups B4c1b2a2, F1a4a and Y2a1 are present in >10 FE group individuals (Tables 2 and 4) and also have patchy distributions across the entire FE group data set (Figure 1). The average coalescent time estimates for these haplogroups reflect a timeline within the Holocene (for F1a4a and Y2a1) and extends into the Late Pleistocene (for B4c1b2a2; Table 4). These three haplogroups help drive the association between FE groups and Indonesia (Figure 4) as they are present only in the Semende and Besemah reference data set groups, albeit at lower frequencies than FE groups (Supplementary Tables 7 and 8). Unfortunately, the limited distribution of these three haplogroups in the reference data set does not allow phylogeographic and/or phylogenetic inference. The same can be said of the remaining haplogroups in the FE group data set that have sporadic distributions in the reference data set (Supplementary Table 7).
Discussion
This study describes the mtDNA landscape of the Philippines as represented by 14 FE groups. The results appear to be consistent with those of our previous survey of FE groups using the NRY.14 As with the NRY, complete mtDNA genomes showed that FE groups are quite heterogeneous; albeit, there is greater heterogeneity in FE groups based on the NRY than for mtDNA. Apart from population genetic factors (isolation, genetic drift and migration), anthropological processes also affect NRY and mtDNA differentiation patterns.41 Among these processes, the patrilocal residence rule can be responsible for greater NRY differentiation. However, the differentiation of FE groups based on mtDNA may in turn be explained by matrilocality. The reality for FE groups is that patrilocality and matrilocality, as well as neolocality are practiced.42, 43 Residence starts as matrilocal, until bride services are fulfilled, followed by either patrilocal or neolocal residence. However more recently, there is greater tendency toward neolocality, but in close proximity to either side of the family for security (economic, psychological and/or social). Another reality is that marriage is more common between individuals of the same FE group.42, 43 Apart from the effects of genetic drift on small isolated groups, within-group marriage is likely to be one of the main reasons for the distinction between FE groups. But this is not to say that there has not been any admixture because there is sharing of haplogroups and haplotypes between FE groups. Reflected in both the NRY and mtDNA, FEN groups are heterogeneous despite having a relatively similar phenotype compared with FEnN groups; furthermore, mtDNA diversity, heterogeneity and population structure do not support a simple dichotonomy between FEN and FEnN groups. The mtDNA data also contradict genome-wide SNP data that says FEN and FEnN groups do not differ.8 As previously suggested, the isolation of FEN group ancestors from those of FEnN groups with subsequent recent admixture could support the SNP data.14 Group isolation is well documented in FE group ethnography and is fairly evident in contemporary groups, reinforced by cultural practices such as within-group marriages.42, 43 But it also should be noted that genetic drift has a greater effect on uniparental markers than on nuclear markers, which have a larger Ne. Drift also has greater effect on small, isolated groups. Thus overall, the combined effects of group isolation, genetic drift and anthropological processes could have influenced FE group differentiation. As with genetic differentiation, Ne can also be affected by anthropological processes.41 In the case of FE groups, these processes also include historical, demographic and sociocultural factors (group-specific cultural practices, marginalization, and so on).42, 43 Hence, while the different trends in Ne change and different current Ne can suggest different demographic histories for each FE group, more theoretical and statistical work is needed to assess and if possible, quantify the relationship and effects of anthropological processes on Ne and genetic variation.
The mtDNA control region data of RC groups12 and FE groups showed another perspective of Filipino population structure, namely, a heterogeneous population composed of different FE groups and RC groups. This is consistent with the preliminary results of a comparison of aSTR data from 12 RC groups11 and 12 FE groups (F Delfin, MCA DeUngria and M Stoneking, unpublished data). The fact that regional centers are hubs for migrations (Filipino, Chinese, Spanish, and so on), which result in admixed RC groups, whereas FE groups stay isolated in their respective ancestral territories, exposed to greater effects of drift, has probably influenced RC and FE group differentiation.
As with the NRY, different genetic affinities also were observed for FE group mtDNA. FE group mtDNA lineages are mainly associated with Taiwan and Southeast Asian groups. These affinities are driven by mtDNA haplogroups (B4a1a, E1a1a and M7c3c) associated with the ‘Out of Taiwan' hypothesis for the Austronesian language family, with coalescent time estimates overlapping with the Austronesian expansion. This further aligns with the fact that all languages in the Philippines are Austronesian languages.4 Haplogroups M52'58 and M52a are said to have originated in South Asia.35, 36 The presence of these haplogroups in the AetaZ and Agta FEN groups, demonstrates a South Asian-FEN group mtDNA link. This is consistent with STRUCTURE/frappe results from a genome-wide SNP study where a South Asian component is present in Southeast Asian groups including the AetaZ and Agta FEN groups.8 It could be argued that through the Srivijaya and Majapahit, thalassocratic empires based in Indonesia,44 Indian influence and possibly haplogroups M52'58 and M52a were brought to the Philippines as early as the fifth century AD. However, Indian influence through these trade empires were indirect and mainly commercial; moreover, other Southeast Asian groups served as filters that diluted and/or enriched any Indian influence that reached the Philippines.44, 45 As haplogroups M52'58 and M52a or their sublineages have not been observed in Southeast Asian groups, the presence of these haplogroups only in South Asia and in the AetaZ and Agta FEN groups seems to demonstrate a direct mtDNA link between India and the Philippines. However, the estimated ages of haplogroups M52'58 and M52a (16.7–24.5 kya) in South Asia35 and the coalescent ages (CI: 4791–20 587) of the South Asian–FEN group mtDNA association are more recent than the initial colonization of the Asia-Pacific region37 and the Philippines.2 This link could therefore be the result of a separate migration that took place after the initial colonization of the region, possibly the same as that inferred in a recent study, which showed Holocene gene flow between India and Australia 5.7 kya.10 Haplogroup N11b, which is still unique to the Mamanwa FEN group, presents another possible, but more ancient South Asian link that overlaps with the initial colonization of the region. These South Asian affinities support the hypothesis of a migration path through South Asia for the ancestors of FEN groups (and other Asia-Pacific groups) as they peopled the Asia-Pacific region.8, 9, 10 Ancient, Late Pleistocene affinities with Australia and Near Oceania is evident with haplogroup P. FEN group Y-STR haplotypes under haplogroups C-RPS4Y and K-M9 also were observed to have associations with Australian and Melanesian haplotypes.14 Genome-wide SNPs also showed an ancient association between the Mamanwa FEN group and groups from Australia and New Guinea.9, 10 Overall, FE group mtDNA genetic affinities illustrate a Filipino genetic history that extends from the initial colonization of the Asia-Pacific region,37 to the initial colonization of the Philippines, with possibly a separate post-colonization migration into the Philippine archipelago,2 up to the time of the Austronesian expansion.
In conclusion, this study has demonstrated various features of the mtDNA landscape of the Philippines. First, mtDNA showed that the Filipino population is heterogeneous and composed of diverse FE groups and RC groups, with no simple dichotomy between FEN and FEnN groups. Second, there are different trends in Ne changes that could suggest different demographic histories for the FE groups included in the study. Third, FE groups have genetic affinities primarily with northern East Asia and Southeast Asia, with an estimated timeline in the Holocene period that overlaps with the Austronesian expansion. And fourth, FE groups have genetic affinities with South Asia, Near Oceania and Australia. These associations are consistent with a southern migration route for FE group ancestors into the Asia-Pacific, with an estimated timeline that corresponds to the initial colonization of the Asia-Pacific region, the initial colonization of the Philippines and a possible separate post-colonization migration into the Philippine archipelago. Although the NRY and mtDNA landscapes of the Filipino population are now described; these genetic systems are just two loci and specifically reflect respectively, male and female genetic histories. A more comprehensive view of Filipino diversity and history can still be sought through genome-wide variation.
Acknowledgments
We thank all volunteers who donated their DNA sample. We are grateful to all individuals, groups and organizations who facilitated sample collection (for details, see Delfin et al.14). We acknowledge Maia Wang Zegura for technical assistance and Miriam Ruth Dalet and Jethro Magdaraog for administrative assistance. This study was supported by the L'Oreal-For Women in Science Program grant to MCADU, a University of the Philippines, Natural Sciences Research Institute grant (NSR-12-01-01) to FD, and by the Max Planck Society.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Scott WH. Prehispanic Source Materials For the Study of Philippine History. Quezon City, Philippines: New Day Publishers; 1984. [Google Scholar]
- Détroit F, Dizon E, Falguères C, Hameau S, Ronquillo W, Sémah F. Upper Pleistocene Homo sapiens from the Tabon cave (Palawan, The Philippines): description and dating of new discoveries. C R Palevol. 2004;3:705–712. [Google Scholar]
- Mijares A, Détroit F, Piper P, et al. New evidence for a 67,000-year-old human presence at Callao Cave, Luzon, Philippines. J Hum Evol. 2010;59:123–132. doi: 10.1016/j.jhevol.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Lewis MP.(ed): Ethnologue: Languages of the World, Sixteenth edition. Dallas, TX: SIL InternationalOnline version http://www.ethnologue.com/ , 2009
- Reid LA.Unravelling the linguistic histories of Philippine Negritosin: Dutton T, Tyron DT (eds): Language Contact and Change in the Austronesian World Mouton de Gruyter; 1994443–475. [Google Scholar]
- Gray RD, Drummond AJ, Greenhill SJ. Language phylogenies reveal expansion pulses and pauses in Pacific settlement. Science. 2009;323:479–483. doi: 10.1126/science.1166858. [DOI] [PubMed] [Google Scholar]
- NSCB . "List of Regions". Makati City, Philippines: National Statistical Coordination Board; 2010. [Google Scholar]
- Abdulla MA, Ahmed I, Assawamakin A, et al. Mapping human genetic diversity in Asia. Science. 2009;326:1541–1545. doi: 10.1126/science.1177074. [DOI] [PubMed] [Google Scholar]
- Reich D, Patterson N, Kircher M, et al. Denisova admixture and the first modern human dispersals into southeast Asia and Oceania. Am J Hum Genet. 2011;89:516–528. doi: 10.1016/j.ajhg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugach I, Delfin F, Gunnarsdóttir E, Kayser M, Stoneking M. Genome-wide data substantiate Holocene gene flow from India to Australia. Proc Nat Acad Sci. 2013;110:1803–1808. doi: 10.1073/pnas.1211927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiquilla S, Salvador J, Calacal G, et al. Expansion of the Philippine autosomal short tandem repeat population database for DNA-based paternity testing. Philipp J Sci. 2011;140:1–6. [Google Scholar]
- Tabbada KA, Trejaut J, Loo J-H, et al. Philippine mitochondrial DNA diversity: a populated viaduct between Taiwan and Indonesia. Mol Biol Evol. 2010;27:21–31. doi: 10.1093/molbev/msp215. [DOI] [PubMed] [Google Scholar]
- Gunnarsdóttir ED, Li M, Bauchet M, Finstermeier K, Stoneking M. High-throughput sequencing of complete human mtDNA genomes from the Philippines. Genome Res. 2011a;21:1–11. doi: 10.1101/gr.107615.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfin F, Salvador JM, Calacal GC, et al. The Y-chromosome landscape of the Philippines: extensive heterogeneity and varying genetic affinities of Negrito and non-Negrito groups. Eur J Hum Genet. 2011;19:224–230. doi: 10.1038/ejhg.2010.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes C, Siddle K, Ducourneau A, et al. Genetic diversity and evidence for population admixture in Batak Negritos from Palawan. Am J Phys Anthropol. 2011;146:62–72. doi: 10.1002/ajpa.21544. [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harbor Protocols. 2010;2010:1–10. doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- Maricic T, Whitten M, Pääbo S. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS One. 2010;5:e14004. doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Stenzel U, Kelso J. Improved base calling for the Illumina Genome Analyzer using machine learning strategies. Genome Biol. 2009;10:R83. doi: 10.1186/gb-2009-10-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147–147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Briggs AW, Good JM, Green RE, et al. Targeted retrieval and analysis of five neandertal mtDNA genomes. Science. 2009;325:318–321. doi: 10.1126/science.1174462. [DOI] [PubMed] [Google Scholar]
- Li M, Schönberg A, Schaefer M, Schroeder R, Nasidze I, Stoneking M. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am J Hum Genet. 2010;87:237–249. doi: 10.1016/j.ajhg.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Stoneking M. A new approach for detecting low-level mutations in next-generation sequence data. Genome Biol. 2012;13:R34. doi: 10.1186/gb-2012-13-5-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K-i, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar Doron M, van Oven M, Rosset S, et al. A copernican reassessment of the human mitochondrial DNA tree from its root. Am J Hum Genet. 2012;90:675–684. doi: 10.1016/j.ajhg.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004b;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004a;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss-Brandstätter A, Pacher D, Schönherr S, et al. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Human Mutat. 2010;32:25–32. doi: 10.1002/humu.21382. [DOI] [PubMed] [Google Scholar]
- van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Human Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- StatSoft I. STATISTICA (Data Analysis Software System) 2007.
- Ersts PJ. Geographic Distance Matrix Generator v1.2.3. New York, USA: Center for Biodiversity and Conservation, American Museum of Natural History; 2006. [Google Scholar]
- Drummond A, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaaswarkhanth M, Haque I, Ravesh Z, et al. Traces of sub-Saharan and Middle Eastern lineages in Indian Muslim populations. Eur J Hum Genet. 2009;18:354–363. doi: 10.1038/ejhg.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekar A, Kumar S, Sreenath J, et al. Updating phylogeny of mitochondrial DNA macrohaplogroup M in India: dispersal of modern human in South Asian corridor. PLoS One. 2009;4:e7447. doi: 10.1371/journal.pone.0007447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker G, Barton H, Bird M, et al. The ‘human revolution' in lowland tropical Southeast Asia: the antiquity and behavior of anatomically modern humans at Niah Cave (Sarawak, Borneo) J Hum Evol. 2007;52:243–261. doi: 10.1016/j.jhevol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Soares P, Rito T, Trejaut J, et al. Ancient voyaging and polynesian prigins. Am J Hum Genet. 2011;88:239–247. doi: 10.1016/j.ajhg.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C, Soares P, Mormina M, et al. A mitochondrial stratigraphy for island southeast Asia. Am J Human Genet. 2007;80:29–43. doi: 10.1086/510412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trejaut JA, Kivisild T, Loo JH, et al. Traces of archaic mitochondrial lineages persist in Austronesian-speaking formosan populations. PLoS Biol. 2005;3:e247. doi: 10.1371/journal.pbio.0030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer E, Chaix R, Pavard S, Austerlitz F. Sex-specific demographic behaviours that shape human genomic variation. Mol Ecol. 2012;21:597–612. doi: 10.1111/j.1365-294X.2011.05406.x. [DOI] [PubMed] [Google Scholar]
- Jocano FL. Anthropology of the Filipino People II: Filipino Indigenous Ethnic Communities, Patterns, Variations and Typologies. Diliman, Quezon City, Philippines: PUNLAD Research House, Inc; 1998b. [Google Scholar]
- Jocano FL. Anthropology of the Filipino People III: Filipino Social Organization. Traditional Kinship and Family Organization. Diliman, Quezon City, Philippines: PUNLAD Research House, Inc; 1998c. [Google Scholar]
- Munoz PM. Early Kingdoms of the Indonesian Archipelago and the Malay Peninsula. Singapore: Editions Didier Millet; 2006. [Google Scholar]
- Jocano FL. Anthropology of the Filipino People I: Philippine Prehistory, Rediscovering Precolonial Heritage. Diliman, Quezon City, Philippines: PUNLAD Research House, Inc; 1998a. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.