Summary
Mammalian pain-related sensory neurons are derived from TrkA lineage neurons located in the dorsal root ganglion. These neurons project to peripheral targets throughout the body, which can be divided into superficial and deep tissues. Here we find that the transcription factor Runx1 is required for the development of many epidermis-projecting TrkA lineage neurons. Accordingly, knockout of Runx1 leads to the selective loss of sensory innervation to the epidermis, whereas deep tissue innervation and two types of deep tissue pain are unaffected. Within these cutaneous neurons, Runx1 concurrently suppresses a large molecular program normally associated with sensory neurons that innervate deep tissues, such as muscle and visceral organs. Ectopic expression of Runx1 in these deep sensory neurons causes a loss of this molecular program and marked deficits in deep tissue pain. Thus this study provides new insight into a genetic program controlling the segregation of cutaneous versus deep tissue pain pathways.
Introduction
The dorsal root ganglia (DRGs) are composed of a heterogeneous population of primary sensory neurons, including nociceptors, mechanoreceptors, thermoceptors, and pruriceptors (Basbaum et al., 2009; Delmas et al., 2011; Marmigere and Ernfors, 2007). The peripheral targets of DRG neurons can be divided into two main zones based on a radial topographical body plan: (1) superficial tissues, the largest being the epidermis of the skin and (2) deep tissues, including the dermis, muscle, bone, and visceral organs. The rationale for this division is twofold. First, the superficial tissues and the deep tissues stem from distinct germ layers, with the epidermis derived from the ectoderm, and the deep tissues derived from the mesoderm (dermis, muscle, bone, muscular layers of visceral organs) and endoderm (mucosal layers of visceral organs) (Gilbert, 2000). Second, with respect to pain, a focus of this study, there are well-documented clinical differences between cutaneous pain and deep tissue pain, such as cutaneous pain generating more reflexive or protective responses, and deep tissue pain being associated with more emotional and autonomic responses (Bove et al., 2009; McMahon et al., 1995; Ness and Gebhart, 1990; Robinson and Gebhart, 2008).
In mammals, all pain neurons are derived from TrkA lineage neurons, since a mutation in TrkA (the receptor for the nerve growth factor) leads to congenital pain insensitivity (Indo, 2012). In the past decade, progress has been made in defining TrkA lineage neurons that selectively innervate cutaneous structures. For example, polymodal nociceptors marked by the expression of the G-protein coupled receptors Mrgprd, Mrgprb4, and Mrgpra3 innervate mainly the skin epidermis (Cavanaugh et al., 2009; Han et al., 2012; Rau et al., 2009; Zylka et al., 2005). In contrast, no markers have been identified that are exclusively associated with deep tissue TrkA lineage neurons. Deep tissue DRG neurons do express genes such as CGRP, TRPV1, and ASIC3, but these genes are also expressed in cutaneous DRG neurons (Bennett et al., 1996; Castañeda-Corral et al., 2011; Christianson et al., 2006; Jimenez-Andrade et al., 2010; Malin et al., 2011; McCoy et al., 2012; McMahon et al., 1994; Molliver et al., 2005). The dearth of information on deep tissue pain-related sensory neurons belies their clinical relevance, as most medical cases of pain relate to muscle, bone, and visceral pain, as opposed to cutaneous pain (Ness and Gebhart, 1990).
The genetic program that controls the segregation of cutaneous versus deep tissue pain-related sensory neurons is still poorly understood. The runt domain transcription factor Runx1 is initially expressed in ~93% of embryonic TrkA-expressing neurons (Abdel Samad et al., 2010), but the expression of Runx1 and TrkA is reciprocally extinguished in distinct subsets of these neurons during perinatal and postnatal development (Lallemend and Ernfors, 2012; Liu and Ma, 2011). Functionally, Runx1 is necessary for the specification of a large cohort of sensory neurons involved with pain, itch, and temperature sensation (Liu and Ma, 2011). In this study, we will present a series of evidence supporting a pivotal role for Runx1 in controlling the segregation of cutaneous versus deep tissue pain sensory neurons.
Results
Identification of two molecular programs associated with cutaneous versus deep tissue sensory neurons
We first used microarray analysis to determine the extent to which Runx1 regulates gene expression on a genome-wide scale. Adult Runx1 conditional knockouts (Runx1F/F;Wnt1Cre, referred to here as Runx1 CKO) were used, in which the deletion of Runx1 is driven by Wnt-1-cre expression in pre-migratory neural crest cells, including the progenitors of DRG neurons (Chen et al., 2006). By using the dCHIP software (Li and Wong, 2001) to compare the gene expression profiles of DRGs from Runx1 CKO mice and their wild-type littermates, we identified 1) over 200 genes that are markedly downregulated in the Runx1 CKO (Table S1), referred to here as “Runx1-dependent” genes, and 2) over 100 genes whose expression is increased in the Runx1 CKO, referred to here as “Runx1-suppressed” genes (Table S2). The quality of this microarray data is validated by the fact that the genes listed in Table S1 include most of the known Runx1-dependent genes.
Previous genetic marking studies showed that three Runx1-dependent genes, Mrgprd, Mrgprb4, and Mrgpra3, are expressed in DRG neurons that sense mechanical pain, itch and/or pleasant touch, and these neurons innervate exclusively the epidermal tissues, with the majority of the glabrous epidermal fibers being marked by Mrgprd (Cavanaugh et al., 2009,Han, 2012 #2571; Liu et al., 2007; Vrontou et al., 2013; Zylka et al., 2005). Double staining showed that a large set of newly identified Runx1-dependent genes were mainly expressed in Mrpgrd+ polymodal nociceptors (Figure S1A). The Runx1-dependent gene set also includes those encoding vesicular glutamate transporter VGLUT3, which is expressed in unmyelinated low threshold mechanoreceptors (Li et al., 2011; Lou et al., 2013; Seal et al., 2009). Genetic marking analysis showed that VGLUT3-expressing DRG neurons innervate the hair follicles and the skin epidermis (Li et al., 2011; Lou et al., 2013), tissues fully or partly derived from the ectoderm (Gilbert, 2000). Here we further found that VGLUT3 lineage neurons, marked by the expression of the Tomato reporter protein, do not innervate the deep tissues, such as the muscle and the bladder (Figure S1B). Consistently, the expression of Runx1 itself is excluded from adult sensory neurons innervating deep tissues (see below). These data suggest that Runx1-dependent DRG neurons preferentially innervate the superficial ectodermal tissues.
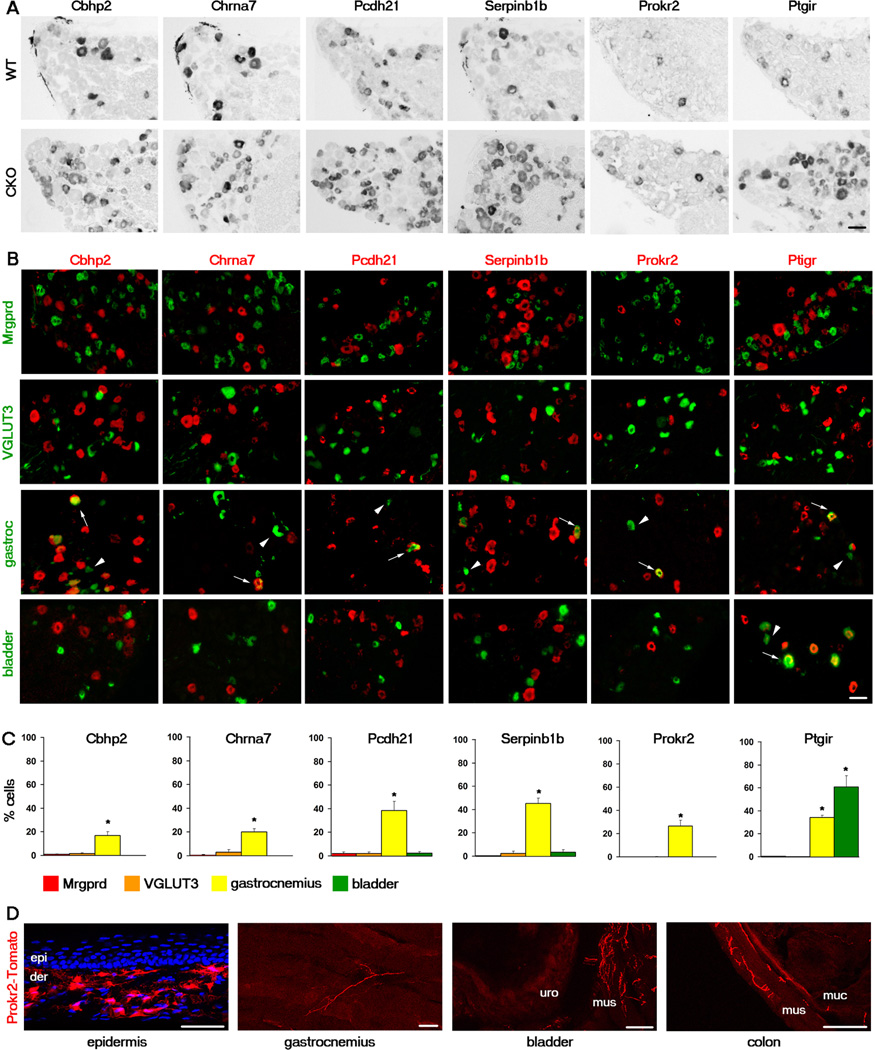
The microarray analysis also revealed over 100 Runx1-suppressed genes, whose expression is markedly upregulated in the Runx1 CKO (Table S2). We validated 25 of these genes by in situ hybridization, and the derepression of these genes in the Runx1 CKO was clearly indicated by their dramatic expansion into neurons labeled by the isolectin B4 (IB4) (Figures 1A and S1; data not shown). Many Runx1-suppressed genes are expressed in a small subset of DRG neurons, as exemplified by Cbhp2, Chrna7, Pcdh21, Serpinb1b, Prokr2, and Ptgir (Figure 1A). Previous genetic fate mapping showed that ~80% of DRG neurons express persistently or transiently the sodium channel Nav1.8, including the majority of TrkA lineage neurons (Agarwal et al., 2004; Liu et al., 2010). We found that expression of Cbhp2, Chrna7, Pcdh21, Serpinb1b, Prokr2, and Ptgir is nearly entirely confined to Nav1.8 lineage neurons (Figure S1D). A double staining with CGRP, a peptidergic neuron marker, showed that while some of these genes are mostly peptidergic (for example, Pcdh21 and Ptgir), others (for example, Serpinb1b) are expressed largely in CGRP– neurons (Figure S1D). A double staining with TrkA showed that ~85% and ~45% of Ptgir+ and Serpinb1b+ neurons coexpressed TrkA, respectively, suggesting that at least a subset of neurons expressing Runx1-suppressed genes represent TrkA lineage neurons (Figure S1E).
Figure 1. Cellular characterization of the Runx1-suppressed genes Cbhp2, Chrna7, Pcdh21, Serpinb1b, Prokr2, and Ptgir.
(A) In situ hybridization (ISH) with indicated probes on adult lumbar DRG sections of wild-type (“WT”) or Runx1 conditional knockout (CKO) mice.
(B) mRNAs of Runx1-suppressed genes (red) detected by ISH were excluded from Mrpgrd+ neurons marked by GFP expression (green, from MrpgrdGFP/+ mice) and from VGLUT3+ neurons marked by Tomato expression (pseudo green, from VGLUT3Cre;RosaTomato fate-mapping mice), but were detected in subsets of neurons retrogradely labeled from the gastrocnemius muscle and/or from the bladder (green). Arrows and arrowheads indicate examples of retrogradely labeled neurons expressing or not expressing the indicated gene, respectively.
(C) The percentage of Mrgprd-GFP+, VGLUT3-Tomato+, or neurons retrogradely labeled from the gastrocnemius or bladder that expressed the indicated Runx1-suppressed gene. n ≥ 3 animals for each analysis. Asterisks indicate a significant enrichment of expression in muscle and/or bladder DRG neurons, in comparison with that in Mrgprd-GFP+ or VGLUT3-Tomato+ neurons. Error bars, SEM; * p < 0.05. 924–1124 Mrpgrd-GFP+ cells, 674–1129 VGLUT3-Tomato+ cells, 121–208 muscle retrogradely labeled cells, and 156–364 bladder retrogradely labeled cells were counted for each gene.
(D) Genetic marking of the innervation of Prokr2 lineage neurons by using Prokr2Cre;RosaTomato fate-mapping mice (see also Figure S2). Tomato+ nerve terminals of Prokr2 lineage neurons (red) are virtually excluded from the epidermis (“epi”, thin glabrous skin), with cell nuclei revealed by DAPI staining (blue). Note an uncharacterized group of Prokr2-expressing cells in the dermis (“der”). Tomato+ nerve terminals were detected in the gastrocnemius, the bladder, and the colon. For bladder and colon, “mus” for muscular layer, “uro” for urothelial layer, and “muc” for mucosal layer.
All scale bars represent 50 µm. See also Figures S1 and S2.
As mentioned above, Mrgprd+ and VGLUT3+ neurons, which compose 30% and 10–19% of DRG neurons, respectively, innervate exclusively the ectodermal tissues (Li et al., 2011; Liu et al., 2008; Lou et al., 2013; Seal et al., 2009; Zylka et al., 2005). Expression of Cbhp2, Chrna7, Pcdh21, Serpinb1b, Prokr2, and Ptgir was not detected in either Mrgprd+ or VGLUT3 lineage neurons, as genetically marked by GFP and Tomato reporters, respectively (Figure 1B). Expression of these Runx1-suppressed genes is therefore excluded from two major classes of cutaneous DRG neurons.
We next asked whether Runx1-suppressed genes might be expressed in deep tissue DRG neurons. To determine this, we tested the expression of Runx1-suppressed genes in DRG neurons retrogradely labeled from various deep tissues, including the gastrocnemius muscle, bladder, and colon. To ensure that there was no leakage of the fluorescent tracer into the skin, particularly with the injections into the gastrocnemius, only those DRGs whose retrogradely labeled neurons showed no or minimal overlap with the cutaneous neuron marker Mrgprd were used for analysis (Figure S1F). The results revealed that one Runx1-suppressed gene—Ptgir—was expressed in 63% and 36% of neurons retrogradely labeled from the bladder and the muscle, respectively (Figures 1B–C). Expression of five other Runx1-suppressed genes analyzed—Cbhp2, Chrna7, Pcdh21, Serpinb1b and Prokr2—was detected in 10–50% of neurons retrogradely labeled from the muscle. It should be noted that Nav1.8 lineage neurons do not include proprioceptors marked by the expression of parvalbumin (Agarwal et al., 2004; Liu et al., 2010). Since Runx1-suppressed genes are confined to Nav1.8 lineage neurons (Figure S1), we can conclude that muscle afferents expressing Runx1-suppressed genes do not represent proprioceptors. Statistic analyses show that expression of these six genes is significantly enriched in neurons innervating muscle and/or bladder compared to the virtual lack of their expression in epidermis-projecting, Mrgprd+ and VGLUT3+ neurons (Figure 1C).
To unambiguously determine if there are Runx1-suppressed genes expressed exclusively in deep tissue DRG neurons, we used genetic tracing to directly visualize the peripheral innervation of neurons expressing one of the Runx1-suppressed genes Prokr2, encoding a receptor for the prokineticin peptide (Negri and Lattanzi, 2012). We crossed Prokr2Cre transgenic mice (GENSAT, http://www.gensat.org/Cre.jsp), in which the Cre recombinase is driven from the Prokr2 promoter, with the Cre-dependent Rosa26LSL-tdTomato reporter mice to generate mice referred to here as Prokr2Cre;RosaTomato (Madisen et al., 2010). In these mice, neurons expressing Prokr2 were permanently marked by Tomato expression, irrespective of persistent or transient Prokr2 expression. Most Prokr2+ neurons, as detected by in situ hybridization, are within Tomato+ DRG neurons in the Prokr2Cre;RosaTomato mice (Figure S2). However, there are a greater number of Tomato+ neurons than neurons exhibiting Prokr2 mRNA signal, which could either reflect transient developmental expression of Prokr2 which is captured by the Prokr2Cre;RosaTomato line or higher detection sensitivity of the Tomato reporter. Regardless, like Prokr2 mRNA, Tomato signal was observed in primarily CGRP+ neurons and largely excluded from IB4+ neurons (Figure S2).
Interestingly, Tomato+ fibers were rarely detected in the skin epidermis of glabrous skin (thin or thick) or in the epidermis of hairy skin in Prokr2Cre;RosaTomato mice, although a mesh of Tomato+ local cells were observed in the dermis (Figure 1D and data not shown). In contrast, Tomato+ fibers were visible in the gastrocnemius and the marrow of the femur (Figure 1D and data not shown). In the bladder, there are Tomato+ fibers mostly in the muscular layer with occasional fibers in the urothelial layer (Figure 1D). Similarly, Tomato+ fibers are detectable in the colon, but primarily in the muscular layer (Figure 1D). Among the mice tested, none showed neuronal labeling in the sympathetic ganglia (Figure S1G). These Tomato+ fibers in muscle and bladder are thus derived from the DRG. It should be noted that adult Prokr2 expression is detected in DRG neurons retrogradely labeled from the muscle, but not from the bladder or the colon (see Figure 1B). Collectively, these data suggest that Prokr2-Tomato fibers observed in these visceral tissues may represent neurons expressing Prokr2 transiently, whereas those fibers observed in muscle should at least partly be derived from neurons with persistent Prokr2 expression. In some rare instances, perhaps due to the transgenic nature of the Prokr2Cre line, a Prokr2Cre;RosaTomato progeny exhibited a significantly larger number of labeled DRG neurons with some innervation to the epidermis; however, this “leaky” expression represented only a small minority of the animals examined (data not shown). Taken together, these data suggest that Prokr2 lineage neurons innervate preferentially the deep tissues, and do not innervate the skin epidermis.
Selective reduction of epidermal innervation in Runx1 CKO mice
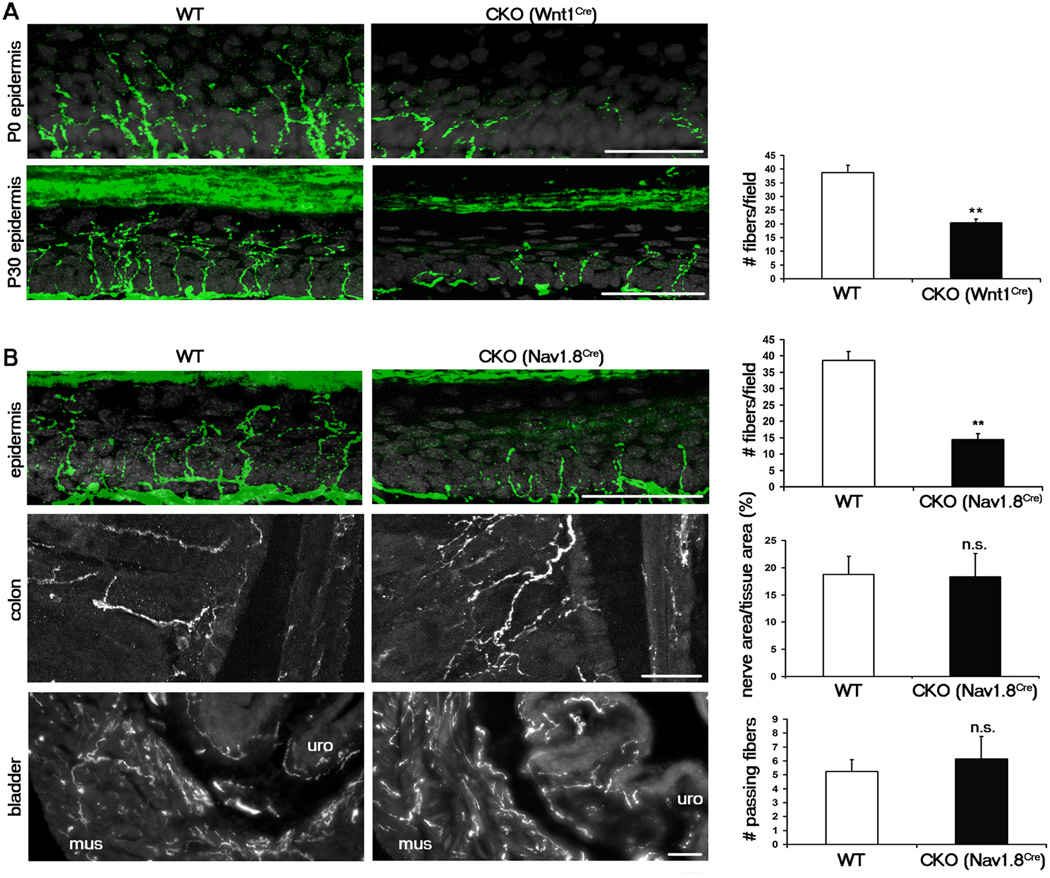
The finding that Runx1-dependent genes are preferentially associated with cutaneous sensory neurons led us to ask whether Runx1 plays a role in controlling the innervation of these neurons to their peripheral targets. To answer this question, we first examined the innervation to the epidermis of the glabrous hindpaw in Runx1 CKO mice, using immunostaining against the pan-neuronal marker PGP9.5 to visualize sensory fibers. In the Runx1 CKO, there is a 50% loss of fibers entering the epidermis (Figure 2A). Notably, those fibers that terminate in the most superficial layer (stratum granulosum) and exhibit wavy and branching morphologies are essentially eliminated; such fibers are derived from Mrgprd+ polymodal nociceptors (Zylka et al., 2005), which represent the largest group of Runx1-dependent DRG neurons. This loss could be observed at every developmental stage examined, starting from P0, the stage at which fibers appear to first cross the basal layer of the epidermis (Figure 2A and data not shown). Thus Runx1 is indeed essential for DRG neurons to innervate cutaneous targets, specifically the most superficial lamina within the epidermis.
Figure 2. Runx1 is required selectively for sensory innervation to the epidermis.
(A) Sensory innervation to the thin glabrous epidermis of the hindpaw at P0 and P30 in the WT (Runx1F/F) and early CKO (Runx1F/F;Wnt1Cre), represented by PGP9.5 staining (green). Keratinocytes are represented by DAPI staining (grey). The number of PGP9.5+ fibers/field of 100 DAPI+ basal keratinocytes at P30 was quantitated.
(B) Sensory innervation to indicated regions of WT and late CKO mice at P30, by PGP9.5 staining for epidermis and by GFP immunostaining for colon and bladder. For the epidermis, “WT” is Runx1F/F, and “CKO” is Runx1F/F;Nav1.8Cre. The number of PGP9.5+ fibers/field of 100 DAPI+ basal keratinocytes at P30 was quantitated. For the colon and the bladder, “WT” is Nav1.8Cre;TauLSL-,EGFP/+, and “CKO” is Runx1F/F;Nav1.8Cre;TauLSL-,EGFP/+. The percentages of colon tissue area that is innervated and the number of fibers passing through lines drawn through the muscular layer of the bladder were quantitated. Two methods were used to quantitate bladder innervation, both producing similar results (see Supplementary Methods and Figure S5). n = 3 animals for each genotype. Error bars, SEM; ** p < 0.01; n.s. (not significant) p ≥ 0.05. Methods of quantitation are detailed in Supplementary Experimental Procedures and in Figure S5.
All scale bars represent 50 µm.
To visualize deep tissue innervation, we needed to mark sensory fibers derived from DRG neurons, but not neurons in sympathetic or enteric ganglia. To do this, we crossed Runx1F/F mice with Nav1.8Cre mice and the Cre-dependent TauLSL-mEGFP reporter line (Hippenmeyer et al., 2005). In the resulting Runx1F/F;Nav1.8Cre;TauLSL-mEGFP/+ mice, Runx1 is removed in Nav1.8Cre-expressing neurons whose nerve terminals are marked by the expression of the membrane-associated enhanced green fluorescent protein or mEGFP. Nav1.8Cre was chosen because it is expressed in 80% of DRG neurons, including most TrkA lineage neurons, but not in sympathetic or enteric neurons (Agarwal et al., 2004; Liu et al., 2010). Wnt1Cre, used above to eliminate Runx1 in neural crest cell precursors, is not suitable for this purpose because Wnt1Cre is also expressed in sympathetic and enteric neurons. As in Runx1F/F;WntCre CKO mice, Runx1F/F;Nav1.8Cre;TauLSL-mEGFP/+ CKO mice showed a similar reduction in epidermal fiber density (Figure 2B). In contrast, no reduction in fiber density in the colon or the bladder was observed in Runx1F/F;Nav1.8Cre;TauLSL-mEGFP/+ mice compared to control mice (Nav1.8Cre;TauLSL-mEGFP/+) (Figure 2B). We also did not detect qualitative differences in innervation to the periosteum or bone marrow (data not shown). Thus while Runx1 is necessary for sensory innervation to the epidermis, it is dispensable for sensory innervation to the deep tissues.
Impaired deep tissue neuron development in Runx1 Gain-of-function or GOF mutants
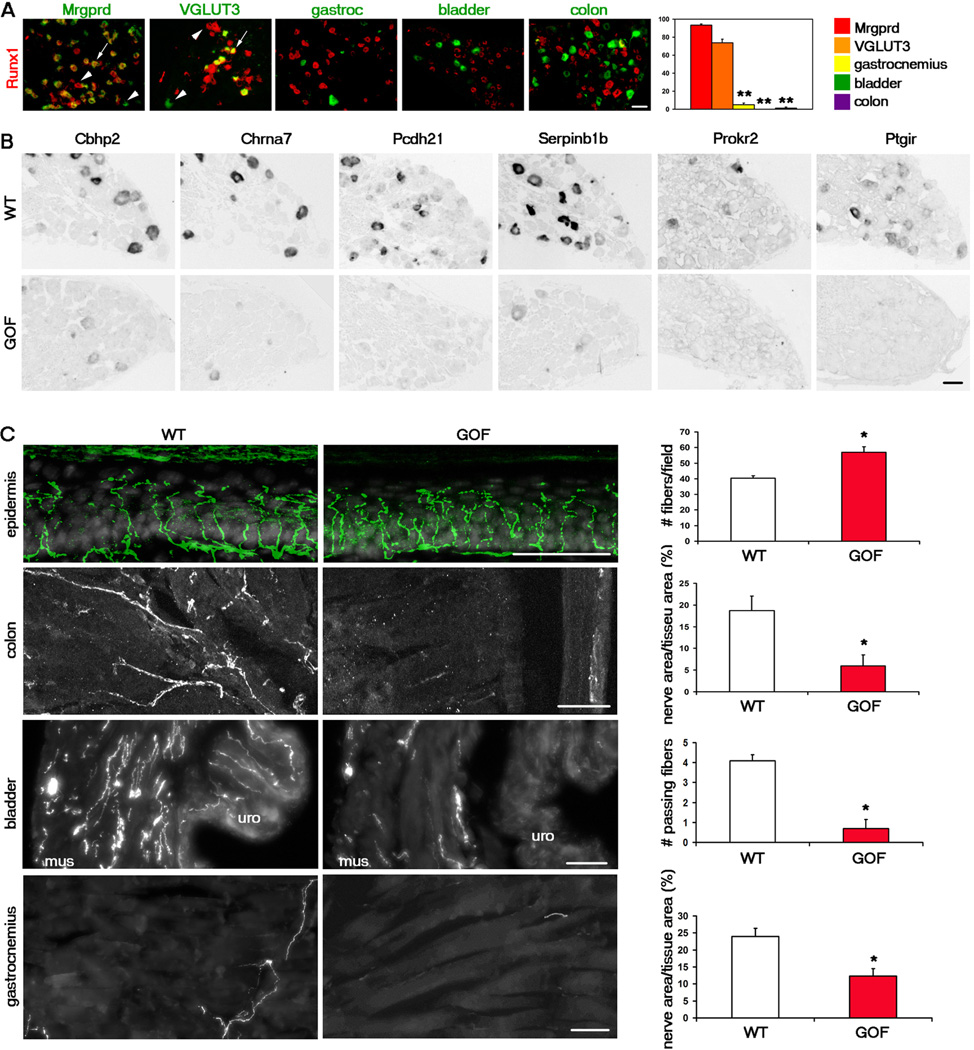
Runx1 is initially expressed in most, if not all, TrkA lineage neurons, at early embryonic stages, but is downregulated in ~50% of these neurons during perinatal and postnatal development (Liu and Ma, 2011). The association that we found between Runx1-suppressed genes and deep tissue neurons naturally led to the hypothesis that Runx1 itself should be either downregulated or excluded in these neurons. This is indeed the case as Runx1 expression is largely absent in adult DRG neurons retrogradely labeled from the muscle, bladder, or the colon in marked contrast with persistent Runx1 expression in most Mrgprd+ and VGLUT3+ cutaneous sensory neurons (Figure 3A).
Figure 3. Downregulation of Runx1 is necessary for the development of deep tissue sensory neurons.
(A) Runx1 mRNA (red) was detected in cutaneous neurons marked by Mrgprd-GFP (962 neurons counted) and VGLUT3-Tomato (844 neurons counted), but was largely excluded from neurons retrogradely labeled from the gastrocnemius (129 neurons counted), bladder (329 neurons counted), and colon (338 neurons counted) as indicated by the percentages of these labeled neurons expressing Runx1. Asterisks indicate a significant difference in expression between Mrgprd-GFP+/VGLUT3-Tomato+ neurons and muscle, bladder, or colon neurons. Error bars, SEM; ** p < 0.01.
(B) In situ hybridization with indicated probes on adult lumbar DRG sections of wild-type (“WT”, TauLSL-Runx1/+) or Runx1 gain-of-function (“GOF”, Nav1.8Cre;TauLSL-Runx1/+) mice.
(C) Sensory innervation to indicated regions of adult WT and GOF mice. For the epidermis, “WT” is TauLSL-Runx1/+, and “GOF” is Nav1.8Cre;TauLSL-Runx1/+. The number of PGP9.5+ fibers/field of 100 DAPI+ basal keratinocytes was quantitated. For the colon, “WT” is Nav1.8Cre;TauLSL-mEGFP/+, and “GOF” is Nav1.8Cre;TauLSL-Runx1/LSL-mEGFP. The percentage of tissue area that is innervated was quantitated. For the bladder and the gastrocnemius, “WT” is Nav1.8Cre;RosaLSL-Tomato/+, and “GOF” is Nav1.8Cre;TauLSL-Runx1/+;RosaLSL-Tomato/+. The number of fibers passing through lines drawn through the muscular layer of the bladder and the percentage of gastrocnemius tissue area that is innervated were quantitated. Two methods were used to quantitate bladder innervation, both producing similar results (see Supplementary Methods and Figure S5). n = 3 animals for each genotype. Error bars, SEM; *p < 0.05. Methods of quantitation are detailed in Supplementary Experimental Procedures and in Figure S5.
All scale bars represent 50 µm. See also Figure S3.
We next examined whether Runx1 downregulation or exclusion is necessary for the development of deep tissue neurons. For this line of study, we used Nav1.8Cre;TauLSL-Runx1/+ mice as a conditional Runx1 gain-of-function (Runx1 GOF) animal model (Abdel Samad et al., 2010). In these mice, upon Cre-mediated removal of a STOP cassette, Runx1 expression is driven from the pan-neuronal Tau promoter; as a result, Runx1 expression is no longer downregulated and becomes constitutive in all Nav1.8Cre-positive neurons. In these mice the number of DRG neurons expressing Runx1 nearly doubles and the numbers of CGRP-, TrkA-,and TRPV1-expressing neurons are significantly reduced (Abdel Samad et al., 2010)
We first found that the expression of the Runx1-suppressed genes tested was either completely lost or dramatically reduced in Runx1 GOF DRGs (Figure 3B). Thus Runx1 downregulation or exclusion is indeed required for deep tissue neurons to acquire proper molecular identities. We next examined the peripheral innervation of these mice. Using PGP9.5 as a pan-neuronal marker, we found that there is no loss and in fact, even a slight increase, in epidermal innervation to the glabrous skin of Runx1 GOF mice (Figure 3C).
To examine innervation to the deep tissues, we crossed Runx1 GOF mice to TauLSL-mEGFP reporter mice to create Nav1.8Cre;TauLSL-Runx1/LSL-mEGFP GOF mice, in which Nav1.8Cre-positive DRG neurons express Runx1 constitutively while being labeled with mEGFP expression. Sensory innervation to the colon, bladder, and gastrocnemius muscle is significantly reduced in these GOF mice (Figure 3C). In the bladder, at least, this loss of innervation was apparent as early as P0 (data not shown). It should be noted that the gain-of-function allele of Runx1 is also a knockin of Runx1 at the Tau locus (Kramer et al., 2006). As a result, the Nav1.8Cre;TauLSL-Runx1/LSL-mEGFP GOF mutant mice lose both wild-type alleles of the microtubule-associated protein Tau. To exclude the possibility that deep tissue innervation defects are due to the loss of Tau, we generated and analyzed Nav1.8Cre;TauLSL-mEGFP/LSL-mEGFP homozygous mice that also lose two copies of Tau. These mice have normal innervation to the visceral organs (Figure S3). This, in addition to the fact that there is no loss of innervation to the epidermis in the Runx1 GOF mice, suggests that the loss of Tau does not affect peripheral sensory innervation. Taken together, these results suggest that downregulation or a lack of Runx1 expression is a prerequisite for proper development of deep tissue DRG neurons.
Impaired deep tissue pain behavior in Runx1 GOF mutants
We have so far shown that molecularly and anatomically, the development of cutaneous and deep tissue sensory neurons is reciprocally compromised in Runx1 CKO mice and Runx1 GOF mice, respectively. We next asked if behaviorally, cutaneous versus deep tissue pain will be preferentially impaired in these mutants.
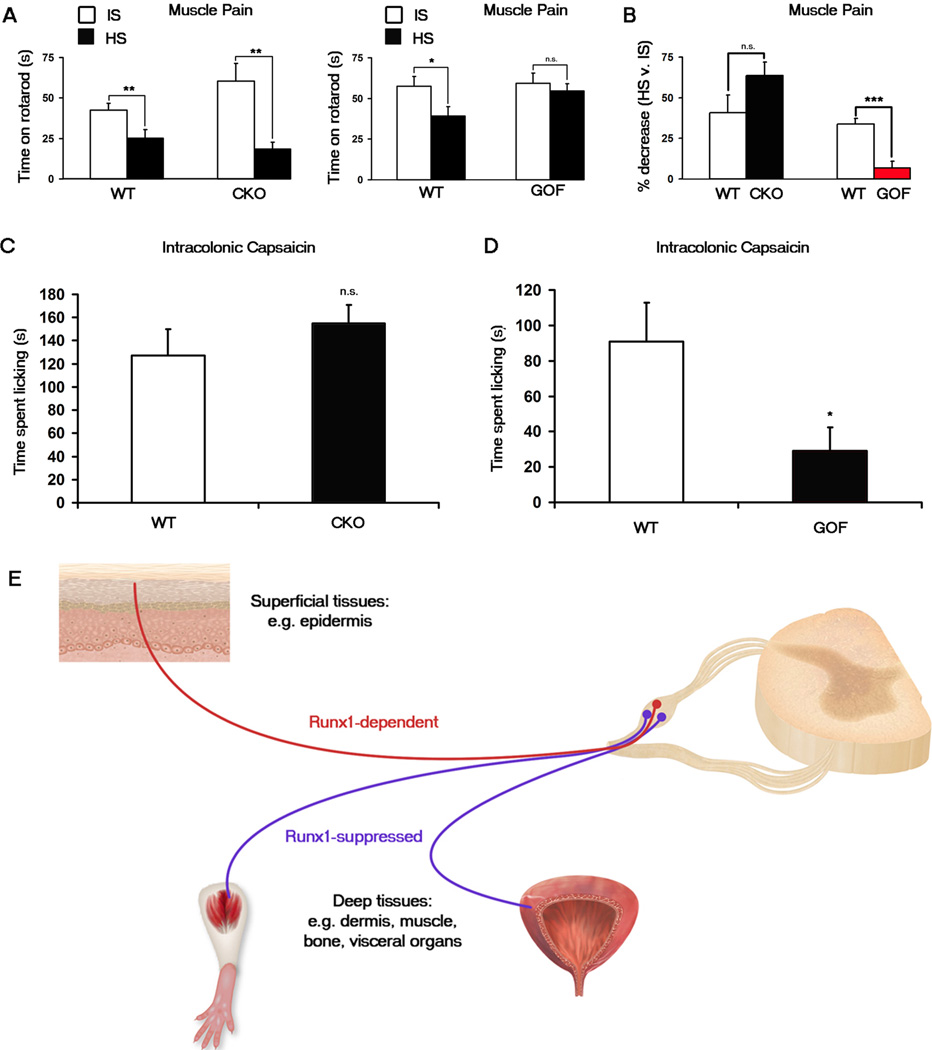
An acute muscle pain behavior was used as one measure of deep tissue pain. To do this, a modified hypertonic saline assay was developed. An injection of hypertonic saline (HS) is known to cause acute leg muscle pain in humans (Graven-Nielsen and Mense, 2001) and more recently in rats, as reflected by paw withdrawal and flexing (Sánchez et al., 2010). We found that mice displayed these behaviors more variably when allowed to roam freely within a chamber. Thus we adapted the protocol by injecting HS into both gastrocnemius muscles and putting them through a forced ambulation assay, namely an accelerating rotarod. In pilot experiments, C57/BL6 mice showed a 69% drop in time spent on the rotarod after HS injection compared to baseline. A modest 12% drop after the control injection of isotonic saline (IS) was also observed, possibly caused by pain generated by the needle itself (Figure S4B). The HS-induced effect is acute, as 30 minutes later, the duration spent on the rotarod returns to baseline levels. For all proceeding experiments, times following IS versus HS injections were compared.
To determine whether Runx1 CKO or GOF mice exhibit any baseline locomotor defects, we first measured their baseline rotarod times compared to those of their wild-type littermates. All genotypes tested showed similar baseline times, suggesting that proprioceptors and neurons necessary for muscle function are unaffected in these mutant mice (Figure S4A). After HS injection, we found that Runx1 CKO mice exhibited a similar drop in time spent on the rotarod to their wild-type littermates, suggesting that acute muscle pain is unaffected in CKO mice (Figure 4A). In contrast, HS injection in Runx1 GOF mice does not cause a statistically significant reduction in time spent on the rotarod (Figure 4A). We also calculated the percentage decrease in rotarod times each animal exhibited following HS injection compared to IS injection. While the CKO mice and their WT littermates showed no statistical difference in this value, the GOF mice displayed a much smaller decrease in performance times compared to their WT littermates (Figure 4B). Finally, whereas their wild-type littermates displayed leg extensions after HS injections (which seemed to contribute to the mice falling prematurely off the rotarod), the GOF mice did not display such behaviors (data not shown). Thus this type of muscle pain is essentially abolished in Nav1.8Cre;TauLSL-Runx1/+ GOF mice. That the Nav1.8Cre;TauLSL-Runx1/+ GOF mice fail to respond to HS also validates that the HS-induced fall from the rotarod is mediated by Nav1.8-expressing neurons, rather than by an adverse effect on proprioceptors, which, as marked by the expression of parvalbumin, do not belong to Nav1.8 lineage neurons (Agarwal et al., 2004; Liu et al., 2010; Shields et al., 2012; Stirling et al., 2005).
Figure 4. Muscle and visceral pain are impaired in Runx1 GOF mice but not in Runx1 CKO mice.
(A, B) Muscle pain assay. Time spent on an accelerating rotarod after the injection of hypertonic saline (“HS”) was compared with that after the injection of isotonic saline (“IS”). (A) Runx1 CKO (Runx1F/F;Wnt1Cre) mice (n = 9) displayed a significant decrease in rotatod performance after HS injection as did their WT (Runx1F/F) littermates (n = 9). Runx1 GOF (Nav1.8Cre;TauLSL-Runx1/+) mice (n = 8) displayed no significant decrease after HS injection in contrast to a significant decrease by their WT (TauLSL-Runx1/+) littermates (n = 8). (B) The percentage decrease after HS was calculated for each animal. Shown are the comparisons of the percentage decrease for the CKO and the GOF with their wild-type littermates. Error bars, SEM; * p < 0.05; ** p < 0.01; n.s., p ≥ 0.05.
(C, D) Intracolonic capsaicin injection visceral pain assay. (C) No difference in time spent licking between Runx1 CKO (Runx1F/F;Wnt1Cre) mice (n = 7) compared to their WT (Runx1F/F) littermates (n = 6). (D) Runx1 GOF (Nav1.8Cre;TauLSL-Runx1/+) mice (n = 7) displayed a significant decrease in time spent licking compared to their WT (TauLSLRunx1/+) littermates (n = 7). Error bars, SEM; * p < 0.05; n.s., p ≥ 0.05.
(E) Dynamic Runx1 expression and activity control the segregation of DRG neurons innervating the superficial ectodermal versus deep mesodermal/endodermal tissues. Neurons expressing Runx1-dependent genes innervate the skin epidermis and hair follicles, tissues fully or partly derived from the ectoderm; these neurons are required to sense cutaneous pain. Neurons expressing Runx1-suppressed genes innervate the mesodermal/endodermal tissues; these neurons are required to sense deep tissue pain. See also Figure S4.
We next tested a visceral pain behavior as another measure of deep tissue pain, by using the intracolonic capsaicin assay (Laird et al., 2001). In this assay, capsaicin is injected into the distal colon via the anus, and pain is measured by the time spent licking the posterior part of the body within a twenty-minute period. In our wild-type strains (C57BL/6 and mixed C57/S129), we observed increased licking not only in the abdominal area, but also the back near the tail, the tail itself, and the feet. This licking of a broad area of the posterial body is consistent with projection of visceral sensory neurons to multiple spinal segments and the central convergence upon the same spinal neurons that receive inputs from cutaneous sensory afferents, thereby leading to referred cutaneous pain (Brumovsky and Gebhart, 2010; Sugiura et al., 1989). In some cases, regardless of the genotype, there were moments of “freezing”; however, as this behavior was more variable, only licking responses were recorded. We found that the Runx1 CKO mice displayed licking responses indistinguishable from their wild-type littermates (Figure 4C), suggesting that Runx1 is dispensable for this type of visceral pain. In contrast, the duration of licking response was greatly reduced in the Runx1 GOF mice compared to their wild-type littermates, suggesting a marked impairment of visceral pain (Figure 4D). We previously reported a reduction in TRPV1-expressing DRG neurons in the Runx1 GOF mice (Abdel Samad et al., 2010) and now show here a marked loss of general colon innervation in these mutants (see Figure 3). Thus, a possible of loss of TRPV1+ fibers in the GOF colon might account for the impairment of capsaicin-evoked pain compared to the wild-type mice, although our data could not rule out that the pain defect is caused by a loss of expression of TrkA or other Runx1-suppressed genes in GOF mice.
Collectively, these results suggest that Runx1-dependent DRG neurons are dispensable for sensing deep tissue pain; instead those DRG neurons whose development requires extinguishment of Runx1 expression are involved with muscle pain and visceral pain.
Discussion
Developmental segregation of cutaneous versus deep tissue pain sensory neurons
The study described here illustrates a genetic program that controls the segregation of two topographically distinct populations of pain-related sensory neurons. Runx1- dependent neurons innervate the most superficial ectodermal tissues and control cutaneous pain, whereas neurons whose development requires downregulation or a lack of Runx1 expression innervate the deep mesodermal/endodermal tissues and control deep tissue pain (summarized in Fig. 4E).
Runx1-dependent neurons (Mrgprd+, Mrgpra3+, Mrgprb4+, VGLUT3+, Trpm8+, and TRPV1high) are composed of a variety of sensory modalities, involved with the detection of pain, itch, temperature, and/or sensory touch (Han et al., 2012; Knowlton et al., 2013; Liu and Ma, 2011; Lou et al., 2013; McKemy, 2013; Pogorzala et al., 2013; Vrontou et al., 2013). Several lines of evidence demonstrated in this study support the model that Runx1-dependent neurons predominantly innervate epidermal tissues. Firstly, genetic marking described here (Figure S1) and previously demonstrates that several Runx1-dependent sensory neurons, including VGLUT3+, Mrgprd+, Mrgpra3+, and Mrgprb4+ innervate the skin epidermis and hair follicles, but not the deep tissues such as the muscle and the bladder (Cavanaugh et al., 2009; Han et al., 2012; Rau et al., 2009; Zylka et al., 2005). Consistently, in Runx1 CKO mice, while innervation to the skin epidermis is greatly reduced, no obvious loss is observed in muscle or visceral organs (Figures 2). Secondly, in contrast to marked deficits in sensing cutaneous pain (Chen et al., 2006), two types of deep tissue pain tested here, muscle pain induced by hypertonic solution injection and visceral pain induced by intracolonic capsaicin injection, are preserved in Runx1 CKO mice (Figure 4).
Peripheral tissues develop from different germ layers, with superficial tissues being derived primarily from the ectoderm, and the deep tissues being derived primarily from the mesoderm and endoderm. Interestingly, the innervation of Runx1-dependent neurons extends not only to the epidermis, but to other non-cutaneous tissues that are also derived from the ectoderm. For example, Mrgprd+ neurons do not innervate mesoderm-derived bone tissues in the leg (Jimenez-Andrade et al., 2010), but do innervate the tooth pulp, which, although considered a bone tissue, is actually derived from the ectoderm (Chung et al., 2012; Gilbert, 2000). TRPM8+ neurons also innervate the tooth pulp, as well as the cornea, another ectoderm-derived tissue (Dhaka et al., 2008; Gilbert, 2000; Parra et al., 2010; Takashima et al., 2007). Only less than 1% of bladder-innervating neurons express Runx1-dependent TRPM8 (Hayashi et al., 2009). Thus Runx1 is required predominantly, but not exclusively, for the development of sensory neurons innervating ectodermal tissues.
Within cutaneous sensory neurons, Runx1 also acts as a genetic repressor to prevent these neurons from acquiring a large molecular program represented by Runx1-suppressed genes. We recognize two categories of Runx1-suppressed genes. One is relatively broadly expressed in DRG neurons, such as CGRP. We reported previously that Runx1-mediated CGRP repression is context-dependent (Abdel Samad et al., 2010), and this “Runx1-suppressed gene” is in fact associated with both Runx1-dependent cutaneous neurons, such as Mrgpra3+ and TRPA1+ neurons (Abdel Samad et al., 2010; Han et al., 2012; Story et al., 2003), and Runx1-independent deep tissue neurons (Fig. S1). The other category of Runx1-suppressed genes, however, is expressed in small subsets of Nav1.8 lineage neurons that innervate the deep tissues, and their expression is excluded from major populations of Runx1-dependent cutaneous sensory neurons (Figure 1 and S1). One of these genes is Prokr2, and our genetic fate mapping demonstrates that the Prokr2-Cre mouse line can for the first time be used to mark DRG neurons that innervate the deep tissues but not the superficial cutaneous tissues. Like Runx1-dependent cutaneous sensory neurons, deep tissue sensory neurons are heterogeneous. Firstly, neurons innervating the bladder show a different molecular profile than neurons innervating the muscle (Figure 1). Secondly, muscle-innervating neurons are both peptidergic and non-peptidergic, as demonstrated by Pcdh21+ and Serpinb1b+ neurons, respectively (Figure S1). This heterogeneity is consistent with the existence of different populations of muscle afferents that respond to different types of stimuli (Jankowski et al., 2013).
The mechanism by which Runx1 prevents cutaneous sensory neurons from acquiring the molecular identities belonging to deep tissue Nav1.8 lineage neurons is still unknown. Besides a transcriptional activation domain, Runx1 also contains a C-terminal peptide capable of binding the Groucho class transcriptional repressor complex, and thus can act as a direct repressor (Nishimura et al., 2004) (Figure S3). We found that removal of this repression domain in Δ446 mice did not cause an expansion of Cbhp2, Chrna7, Ptigr, and Serpinb1b into the IB4+ neurons (Figure S3B), which is contrast to the marked expansion observed in Runx1 CKO mice (Figure S1). Thus, Runx1 does not use its c-terminal repression domain to suppress these Runx1-suppressed genes within cutaneous sensory neurons.
Consistent with the association of Runx1-suppressed genes with deep tissue sensory neurons, Runx1 is not detected in these neurons at adult ages (Figure 3). We reported previously that a vast majority of embryonic TrkA+ neurons (~93%) express Runx1 (Abdel Samad, 2010), and about half of these neurons retain TrkA but switch off Runx1 during perinatal and postnatal development (Chen et al., 2006). A majority of Ptgir+ neurons are TrkA+ (Figure S1), suggesting a transient Runx1 expression in these deep tissue neurons. The GOF studies further suggest that downregulation or exclusion of Runx1 is a prerequisite for the acquisition of the Runx1-suppresed molecular program in deep tissue DRG neurons. However, our data do not rule out the possibility that early transient Runx1 expression might play a role in establishing certain unknown features of deep tissue sensory neurons, as early transient TrkA expression is required to establish molecular identities of Runx1-dependent cutraneous sensory neurons (Luo et al., 2007). Thus, future studies are needed to determine if there is any deep tissue pain not tested here that is impaired in Runx1 knockout mice.
There is a well-documented difference between cutaneous and deep tissue pain, with the former one generating more reflexive or protective responses and the latter one associated with more emotional and autonomic responses (Bove et al., 2009; McMahon et al., 1995; Ness and Gebhart, 1990; Robinson and Gebhart, 2008). It is interesting to note that Runx1 also plays a role in guiding Runx1-dependent neurons to innervate inner lamina II of the dorsal horn of the spinal cord, and a knockout of Runx1 leads to a switch of innervation to more superficial laminae (Chen et al., 2006). Combined with our current finding that Runx1 is required predominantly for the development of the cutaneous sensory neurons, these Runx1 activities may form a developmental basis for allowing cutaneous and deep tissue pain to be processed along distinct central pathways, even though some spinal neurons do receive convergent inputs from these two pathways (Brumovsky and Gebhart, 2010; Willis and Westlund, 2001).
Implications on pain treatment
The existence of two molecularly distinct pain pathways highlights the necessity for the development of distinct strategies for treating pain originating from cutaneous versus deep tissues. Notably, while most clinical cases of pain originate from deep mesodermal and endodermal tissues, the majority of current preclinical studies measure pain from the skin. This discrepancy might contribute to the lack of success in the translation of preclinical animal models into human pain medications (Mogil, 2009).
It should be pointed out that among the six Runx1-suppressed genes presented in this study, five are expressed in neurons innervating the muscle (Figure 1), bone, or joint (data not shown). Their expression is excluded not only from known cutaneous neurons but also from neurons innervating the bladder or the colon (Figure 1 and data not shown), suggesting that a majority of Runx1-suppressed genes may preferentially mark sensory neurons projecting to mesodermal tissues. Accordingly, drugs targeted at DRG neurons expressing this set of Runx1-suppressed genes might be ideal for treating pain originating from the muscle, bone and/or joint because they would be less likely to cause side effects on cutaneous somatic sensations (such as the sense of temperature) or visceral organ functions, both of which are vital for our daily life. Thus, as we enter an era of considering personalized pain medicine (von Hehn et al., 2012), the existence of molecularly distinct DRG neurons innervating specific peripheral targets suggests that we should consider “tissue/organ-based” pain medicines, as well.
Experimental Procedures
Detailed protocols and information on the mouse lines are listed in Supplemental Experimental Procedures.
Microarray analyses were performed using L4-L5 lumbar DRGs from P60-P90 Runx1F/F control and Wnt1Cre;Runx1F/F conditional knockout animals.
In situ hybridization, immunohistochemistry, and IB4 labeling were performed using established procedures.
For the retrograde labeling studies in the muscle, 2–4 µl of Fluorogold was injected into one site in the gastrocnemius muscle, and lumbar DRGs were dissected out after one week. Retrograde labeling studies in the bladder and the colon were performed as previously described (Christianson et al., 2006).
Muscle pain behavior was done by injecting 100 µl isotonic saline (IS, 0.9% NaCl) or hypertonic saline (HS, 5% NaCl) into both gastrocemius muscles of adult mice. The mice were immediately placed upon an accelerating rotarod, and performance was measured by time spent on the rotatrod before falling off. The intracolonic capsaicin assay was done by injecting 50 µl 0.1% capsaicin into the colon via the anus as previously described (Laird et al., 2001). The number of licking behaviors around the abdomen, back of the tail, the tail itself, and the hindlimbs were measured within a 20-minute period.
Supplementary Material
Highlights.
Runx1-dependent DRG neurons innervate superficial ectodermal tissues.
Runx1-suppressed genes mark DRG neurons innervating mesodermal/endodermal tissues.
Cutaneous and deep pain is differentially impaired in complementary Runx1 mutants.
A genetic program for segregation of topographically distinct DRG neurons.
Acknowledgments
We thank Dr. Nancy Speck and Dr. Gary Gilliland for the floxed Runx1 mice, Dr. Rohini Kuner for the Nav1.8Cre/+ mice, Dr. David Rowitch for the Wnt1Cre/+ mice, Dr. Sylvia Arber for TauLSL-mEGFP mice, Dr. Mark Zylka for the MrgprdGFP/+ mice, Dr. Bradford Lowell for VGLUT3Cre/+ mice, and Dr. Tsukasa Okuda for Δ-446 mice. We also thank the Allen Brain Institute and the Jackson Laboratory for the Rosa26LSL-tdTomato mice, and GENSAT and the MMRRC at University of California, Davis for the Prokr2Cre/+ mice. We thank Dr. Clifford Woolf for helpful discussions and comments. This work was supported by the NIH grants from NIDCR (R01 DE018025 for the Ma lab) and NINDS (P01 NS047572 for the Ma lab and R01 NS050758 for the Davis lab).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdel Samad O, Liu Y, Yang FC, Kramer I, Arber S, Ma Q. Characterization of two Runx1-dependent nociceptor differentiation programs necessary for inflammatory versus neuropathic pain. Mol Pain. 2010;6:45. doi: 10.1186/1744-8069-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal N, Offermanns S, Kuner R. Conditional gene deletion in primary nociceptive neurons of trigeminal ganglia and dorsal root ganglia. Genesis. 2004;38:122–129. doi: 10.1002/gene.20010. [DOI] [PubMed] [Google Scholar]
- Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139:267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DL, Dmietrieva N, Priestley JV, Clary D, McMahon SB. trkA, CGRP and IB4 expression in retrogradely labelled cutaneous and visceral primary sensory neurones in the rat. Neurosci Lett. 1996;206:33–36. doi: 10.1016/0304-3940(96)12418-6. [DOI] [PubMed] [Google Scholar]
- Bove SE, Flatters SJ, Inglis JJ, Mantyh PW. New advances in musculoskeletal pain. Brain Res Rev. 2009;60:187–201. doi: 10.1016/j.brainresrev.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumovsky PR, Gebhart GF. Visceral organ cross-sensitization - an integrated perspective. Auton Neurosci. 2010;153:106–115. doi: 10.1016/j.autneu.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda-Corral G, Jimenez-Andrade JM, Bloom AP, Taylor RN, Mantyh WG, Kaczmarska MJ, Ghilardi JR, Mantyh PW. The majority of myelinated and unmyelinated sensory nerve fibers that innervate bone express the tropomyosin receptor kinase A. Neuroscience. 2011;178:196–207. doi: 10.1016/j.neuroscience.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh DJ, Lee H, Lo L, Shields SD, Zylka MJ, Basbaum AI, Anderson DJ. Distinct subsets of unmyelinated primary sensory fibers mediate behavioral responses to noxious thermal and mechanical stimuli. Proc Natl Acad Sci U S A. 2009;106:9075–9080. doi: 10.1073/pnas.0901507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Broom DC, Liu Y, de Nooij JC, Li Z, Cen C, Samad OA, Jessell TM, Woolf CJ, Ma Q. Runx1 determines nociceptive sensory neuron phenotype and is required for thermal and neuropathic pain. Neuron. 2006;49:365–377. doi: 10.1016/j.neuron.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Christianson JA, McIlwrath SL, Koerber HR, BM D. Transient receptor potential vanilloid 1-immunopositive neurons in the mouse are more prevalent within colon afferents compared to skin and muscle afferents. Neuroscience. 2006;140:247–257. doi: 10.1016/j.neuroscience.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Christianson JA, Traub RJ, Davis BM. Differences in spinal distribution and neurochemical phenotype of colonic afferents in mouse and rat. J Comp Neurol. 2006;494:246–259. doi: 10.1002/cne.20816. [DOI] [PubMed] [Google Scholar]
- Chung MK, Jue SS, Dong X. Projection of non-peptidergic afferents to mouse tooth pulp. J Dent Res. 2012;91:777–782. doi: 10.1177/0022034512450298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas P, Hao J, Rodat-Despoix L. Molecular mechanisms of mechanotransduction in mammalian sensory neurons. Nat Rev Neurosci. 2011;12:139–153. doi: 10.1038/nrn2993. [DOI] [PubMed] [Google Scholar]
- Dhaka A, Earley TJ, Watson J, Patapoutian A. Visualizing cold spots: TRPM8-expressing sensory neurons and their projections. Journal of Neuroscience. 2008;28:566–575. doi: 10.1523/JNEUROSCI.3976-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SF. Developmental Biology. Sixth edn. Sunderland (MA): Sinauer Associates; 2000. [Google Scholar]
- Graven-Nielsen T, Mense S. The peripheral apparatus of muscle pain: evidence from animal and human studies. Clin J Pain. 2001;17:2–10. doi: 10.1097/00002508-200103000-00002. [DOI] [PubMed] [Google Scholar]
- Han L, Ma C, Liu Q, Weng HJ, Cui Y, Tang Z, Kim Y, Nie H, Qu L, Patel KN, et al. A subpopulation of nociceptors specifically linked to itch. Nat Neurosci. 2012;16:174–182. doi: 10.1038/nn.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Kondo T, Ishimatsu M, Yamada S, Nakamura K, Matsuoka K, Akasu T. Expression of the TRPM8-immunoreactivity in dorsal root ganglion neurons innervating the rat urinary bladder. Neurosci Res. 2009;65:245–251. doi: 10.1016/j.neures.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indo Y. Nerve growth factor and the physiology of pain: lessons from congenital insensitivity to pain with anhidrosis. Clin Genet. 2012;82:341–350. doi: 10.1111/j.1399-0004.2012.01943.x. [DOI] [PubMed] [Google Scholar]
- Jankowski MP, Rau KK, Ekmann KM, Anderson CE, Koerber HR. Comprehensive Phenotyping of Group III and IV Muscle Afferents in Mouse. J Neurophysiol. 2013 doi: 10.1152/jn.01067.2012. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Andrade JM, Mantyh WG, Bloom AP, Xu H, Ferng AS, Dussor G, Vanderah TW, Mantyh PW. A phenotypically restricted set of primary afferent nerve fibers innervate the bone versus skin: therapeutic opportunity for treating skeletal pain. Bone. 2010;46:306–313. doi: 10.1016/j.bone.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton WM, Palkar R, Lippoldt EK, McCoy DD, Baluch F, Chen J, McKemy DD. A sensory-labeled line for cold: TRPM8-expressing sensory neurons define the cellular basis for cold, cold pain, and cooling-mediated analgesia. J Neurosci. 2013;33:2837–2848. doi: 10.1523/JNEUROSCI.1943-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer I, Sigrist M, de Nooij JC, Taniuchi I, Jessell TM, Arber S. A role for Runx transcription factor signaling in dorsal root ganglion sensory neuron diversification. Neuron. 2006;49:379–393. doi: 10.1016/j.neuron.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Laird JM, Martinez-Caro L, Garcia-Nicas E, Cervero F. A new model of visceral pain and referred hyperalgesia in the mouse. Pain. 2001;92:335–342. doi: 10.1016/S0304-3959(01)00275-5. [DOI] [PubMed] [Google Scholar]
- Lallemend F, Ernfors P. Molecular interactions underlying the specification of sensory neurons. Trends Neurosci. 2012 Apr 17; doi: 10.1016/j.tins.2012.03.006. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Rutlin M, Abraira VE, Cassidy C, Kus L, Gong S, Jankowski MP, Luo W, Heintz N, Koerber HR, et al. The functional organization of cutaneous low-threshold mechanosensory neurons. Cell. 2011;147:1615–1627. doi: 10.1016/j.cell.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Vrontou S, Rice FL, Zylka MJ, Dong X, Anderson DJ. Molecular genetic visualization of a rare subset of unmyelinated sensory neurons that may detect gentle touch. Nat Neurosci. 2007;10:946–948. doi: 10.1038/nn1937. [DOI] [PubMed] [Google Scholar]
- Liu Y, Abdel Samad O, Duan B, Zhang L, Tong Q, Ji RR, Lowell B, Ma Q. VGLUT2-dependent glutamate release from peripheral nociceptors is required to sense pain and suppress itch. Neuron. 2010;68:543–556. doi: 10.1016/j.neuron.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ma Q. Generation of somatic sensory neuron diversity and implications on sensory coding. Curr Opin Neurobiol. 2011;21:52–60. doi: 10.1016/j.conb.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yang FC, Okuda T, Dong X, Zylka MJ, Chen CL, Anderson DJ, Kuner R, Ma Q. Mechanisms of compartmentalized expression of Mrg class G-protein-coupled sensory receptors. J Neurosci. 2008;28:125–132. doi: 10.1523/JNEUROSCI.4472-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou S, Duan B, Vong L, Lowell BB, Ma Q. Runx1 controls terminal morphology and mechanosensitivity of VGLUT3-expressing C-mechanoreceptors. J Neurosci. 2013;33:870–882. doi: 10.1523/JNEUROSCI.3942-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Wickramasinghe SR, Savitt JM, Griffin JW, Dawson TM, Ginty DD. A Hierarchical NGF Signaling Cascade Controls Ret-Dependent and Ret-Independent Events during Development of Nonpeptidergic DRG Neurons. Neuron. 2007;54:739–754. doi: 10.1016/j.neuron.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin S, Molliver D, Christianson JA, Schwartz ES, Cornuet P, Albers KM, Davis BM. TRPV1 and TRPA1 function and modulation are target tissue dependent. J Neurosci. 2011;31:31. doi: 10.1523/JNEUROSCI.2992-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmigere F, Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat Rev Neurosci. 2007;8:114–127. doi: 10.1038/nrn2057. [DOI] [PubMed] [Google Scholar]
- McCoy ES, Taylor-Blake B, Zylka MJ. CGRPα-expressing sensory neurons respond to stimuli that evoke sensations of pain and itch. PLoS One. 2012;7:e36355. doi: 10.1371/journal.pone.0036355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKemy DD. The molecular and cellular basis of cold sensation. ACS Chem Neurosci. 2013;4:238–247. doi: 10.1021/cn300193h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB, Armanini MP, Ling LH, Phillips HS. Expression and coexpression of Trk receptors in subpopulations of adult primary sensory neurons projecting to identified peripheral targets. Neuron. 1994;12:1161–1171. doi: 10.1016/0896-6273(94)90323-9. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Dmitrieva N, Koltzenburg M. Visceral pain. Br J Anaesth. 1995;75:132–144. doi: 10.1093/bja/75.2.132. [DOI] [PubMed] [Google Scholar]
- Mogil JS. Animal models of pain: progress and challenges. Nat Rev Neurosci. 2009;10:283–294. doi: 10.1038/nrn2606. [DOI] [PubMed] [Google Scholar]
- Molliver DC, Immke DC, Fierro L, Pare M, Rice FL, McCleskey EW. ASIC3, an acid-sensing ion channel, is expressed in metaboreceptive sensory neurons. Mol Pain. 2005;1:3. doi: 10.1186/1744-8069-1-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri L, Lattanzi R. Bv8/PK2 and prokineticin receptors: a druggable pronociceptive system. Curr Opin Pharmacol. 2012;66:62–66. doi: 10.1016/j.coph.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Ness TJ, Gebhart GF. Visceral pain: a review of experimental studies. Pain. 1990;41:167–234. doi: 10.1016/0304-3959(90)90021-5. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Fukushima-Nakase Y, Fujita Y, Nakao M, Toda S, Kitamura N, Abe T, Okuda T. VWRPY motif-dependent and -independent roles of AML1/Runx1 transcription factor in murine hematopoietic development. Blood. 2004;103:562–570. doi: 10.1182/blood-2003-06-2109. [DOI] [PubMed] [Google Scholar]
- Parra A, Madrid R, Echevarria D, del Olmo S, Morenilla-Palao C, Acosta MC, Gallar J, Dhaka A, Viana F, Belmonte C. Ocular surface wetness is regulated by TRPM8-dependent cold thermoreceptors of the cornea. Nat Med. 2010;16:1396–1399. doi: 10.1038/nm.2264. [DOI] [PubMed] [Google Scholar]
- Pogorzala LA, Mishra SK, Hoon MA. The cellular code for Mammalian thermosensation. J Neurosci. 2013;33:5533–5541. doi: 10.1523/JNEUROSCI.5788-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau KK, McIlwrath SL, Wang H, Lawson JJ, Jankowski MP, Zylka MJ, Anderson DJ, Koerber HR. Mrgprd enhances excitability in specific populations of cutaneous murine polymodal nociceptors. Journal of Neuroscience. 2009;29:8612–8619. doi: 10.1523/JNEUROSCI.1057-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DR, Gebhart GF. Inside information: the unique features of visceral sensation. Mol Interv. 2008;8:242–253. doi: 10.1124/mi.8.5.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez EM, Bagües A, Martín MI. Contributions of peripheral and central opioid receptors to antinociception in rat muscle pain models. Pharmacol Biochem Behav. 2010;96:488–495. doi: 10.1016/j.pbb.2010.07.009. 2010 Oct;96(4):488–95. [DOI] [PubMed] [Google Scholar]
- Seal RP, Wang X, Guan Y, Raja SN, Woodbury CJ, Basbaum AI, Edwards RH. Injury-induced mechanical hypersensitivity requires C-low threshold mechanoreceptors. Nature. 2009;462:651–655. doi: 10.1038/nature08505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields SD, Ahn HS, Yang Y, Han C, Seal RP, Wood JN, Waxman SG, Dib-Hajj SD. Na(v)1.8 expression is not restricted to nociceptors in mouse peripheral nervous system. Pain. 2012 doi: 10.1016/j.pain.2012.04.022. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Stirling LC, Forlani G, Baker MD, Wood JN, Matthews EA, Dickenson AH, Nassar MA. Nociceptor-specific gene deletion using heterozygous NaV1.8-Cre recombinase mice. Pain. 2005;113:27–36. doi: 10.1016/j.pain.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Story GM, Peier AM, Reeve AJ, Eid SR, Mosbacher J, Hricik TR, Earley TJ, Hergarden AC, Andersson DA, Hwang SW, et al. ANKTM1, a TRP-like channel expressed in nociceptive neurons, is activated by cold temperatures. Cell. 2003;112:819–829. doi: 10.1016/s0092-8674(03)00158-2. [DOI] [PubMed] [Google Scholar]
- Sugiura Y, Terui N, Hosoya Y. Difference in distribution of central terminals between visceral and somatic unmyelinated (C) primary afferent fibers. J Neurophysiol. 1989;62:834–840. doi: 10.1152/jn.1989.62.4.834. [DOI] [PubMed] [Google Scholar]
- Takashima Y, Daniels RL, Knowlton W, Teng J, Liman ER, McKemy DD. Diversity in the neural circuitry of cold sensing revealed by genetic axonal labeling of transient receptor potential melastatin 8 neurons. Journal of Neuroscience. 2007;27:14147–14157. doi: 10.1523/JNEUROSCI.4578-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hehn CA, Baron R, Woolf CJ. Deconstructing the neuropathic pain phenotype to reveal neural mechanisms. Neuron. 2012;73:638–652. doi: 10.1016/j.neuron.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrontou S, Wong AM, Rau KK, Koerber HR, Anderson DJ. Genetic identification of C fibres that detect massage-like stroking of hairy skin in vivo. Nature. 2013;493:669–673. doi: 10.1038/nature11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis WDJ, Westlund KN. The role of the dorsal column pathway in visceral nociception. Curr Pain Headache Rep. 2001;5:20–26. doi: 10.1007/s11916-001-0006-1. [DOI] [PubMed] [Google Scholar]
- Zylka MJ, Rice FL, Anderson DJ. Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to Mrgprd. Neuron. 2005;45:17–25. doi: 10.1016/j.neuron.2004.12.015. 2005 Jan 6;45(1):17–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.