Abstract
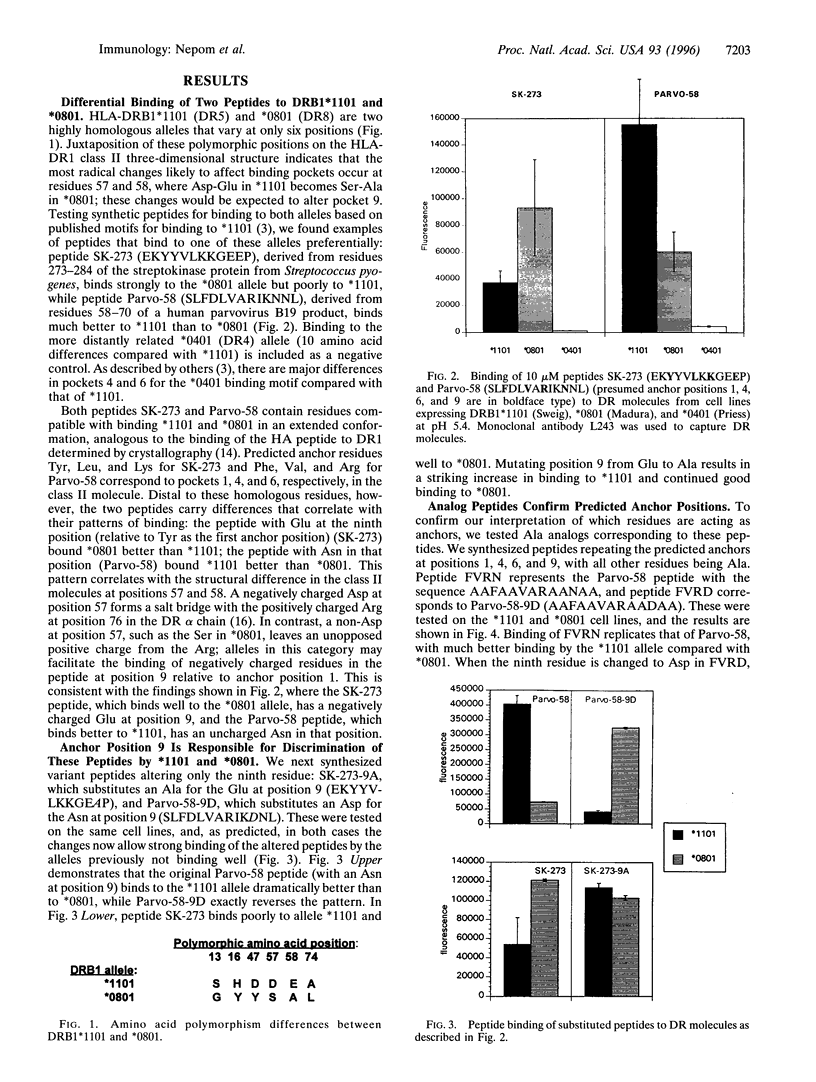
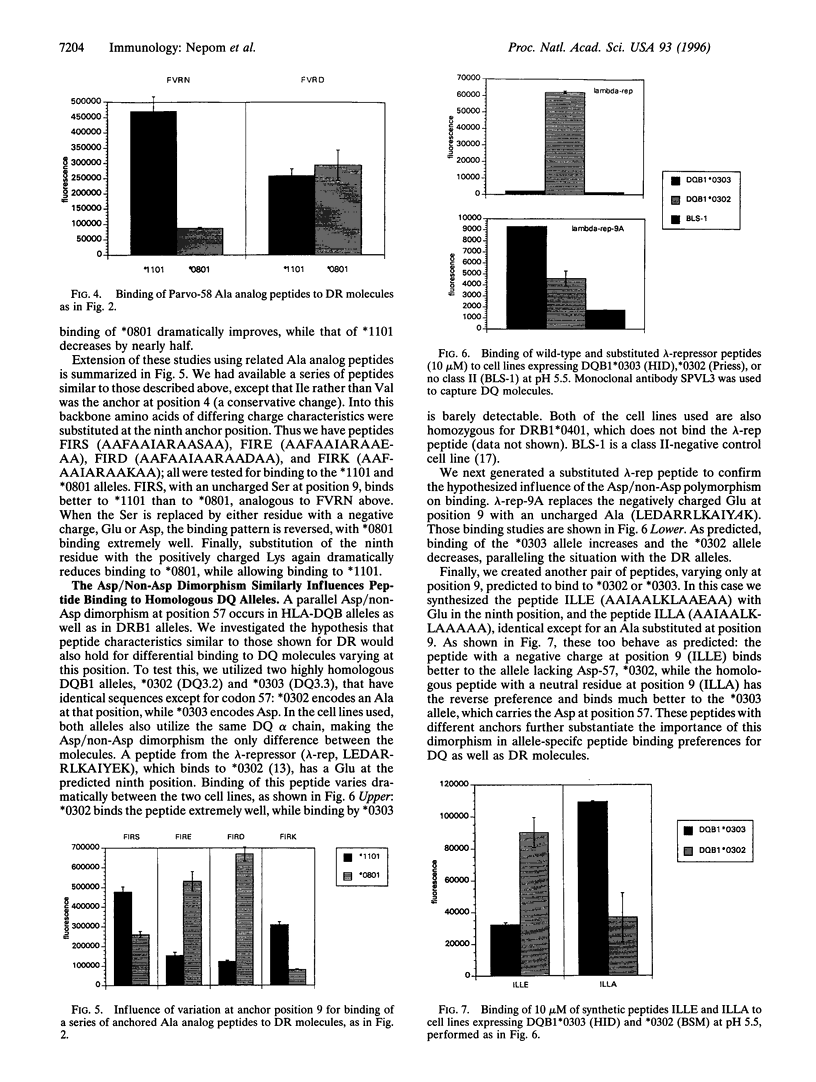
Position 57 in the beta chain of HLA class II molecules maintains an Asp/non-Asp dimorphism that has been conserved through evolution and is implicated in susceptibility to some autoimmune diseases. The latter effect may be due to the influence of this residue on the ability of class II alleles to bind specific pathogenic peptides. We utilized highly homologous pairs of both DR and DQ alleles that varied at residue 57 to investigate the impact of this dimorphism on binding of model peptides. Using a direct binding assay of biotinylated peptides on whole cells expressing the desired alleles, we report several peptides that bind differentially to the allele pairs depending on the presence or absence of Asp at position 57. Peptides with negatively charged residues at anchor position 9 bind well to alleles not containing Asp at position 57 in the beta chain but cannot bind well to homologous Asp-positive alleles. By changing the peptides at the single residue predicted to interact with this position 57, we demonstrate a drastically altered or reversed pattern of binding. Ala analog peptides confirm these interactions and identify a limited set of interaction sites between the bound peptides and the class II molecules. Clarification of the impact of specific class II polymorphisms on generating unique allele-specific peptide binding "repertoires" will aid in our understanding of the development of specific immune responses and HLA-associated diseases.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown J. H., Jardetzky T. S., Gorga J. C., Stern L. J., Urban R. G., Strominger J. L., Wiley D. C. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993 Jul 1;364(6432):33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- Chicz R. M., Lane W. S., Robinson R. A., Trucco M., Strominger J. L., Gorga J. C. Self-peptides bound to the type I diabetes associated class II MHC molecules HLA-DQ1 and HLA-DQ8. Int Immunol. 1994 Nov;6(11):1639–1649. doi: 10.1093/intimm/6.11.1639. [DOI] [PubMed] [Google Scholar]
- Chicz R. M., Urban R. G., Gorga J. C., Vignali D. A., Lane W. S., Strominger J. L. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J Exp Med. 1993 Jul 1;178(1):27–47. doi: 10.1084/jem.178.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicz R. M., Urban R. G., Lane W. S., Gorga J. C., Stern L. J., Vignali D. A., Strominger J. L. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature. 1992 Aug 27;358(6389):764–768. doi: 10.1038/358764a0. [DOI] [PubMed] [Google Scholar]
- Erlich H. A., Bugawan T. L., Scharf S., Nepom G. T., Tait B., Griffith R. L. HLA-DQ beta sequence polymorphism and genetic susceptibility to IDDM. Diabetes. 1990 Jan;39(1):96–103. doi: 10.2337/diacare.39.1.96. [DOI] [PubMed] [Google Scholar]
- Falk K., Rötzschke O., Stevanović S., Jung G., Rammensee H. G. Pool sequencing of natural HLA-DR, DQ, and DP ligands reveals detailed peptide motifs, constraints of processing, and general rules. Immunogenetics. 1994;39(4):230–242. doi: 10.1007/BF00188785. [DOI] [PubMed] [Google Scholar]
- Fu X. T., Bono C. P., Woulfe S. L., Swearingen C., Summers N. L., Sinigaglia F., Sette A., Schwartz B. D., Karr R. W. Pocket 4 of the HLA-DR(alpha,beta 1*0401) molecule is a major determinant of T cells recognition of peptide. J Exp Med. 1995 Mar 1;181(3):915–926. doi: 10.1084/jem.181.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer J., Belunis C., Bolin D., Papadopoulos J., Walsky R., Higelin J., Danho W., Sinigaglia F., Nagy Z. A. High-affinity binding of short peptides to major histocompatibility complex class II molecules by anchor combinations. Proc Natl Acad Sci U S A. 1994 May 10;91(10):4456–4460. doi: 10.1073/pnas.91.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer J., Takacs B., Sinigaglia F. Identification of a motif for HLA-DR1 binding peptides using M13 display libraries. J Exp Med. 1992 Oct 1;176(4):1007–1013. doi: 10.1084/jem.176.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer J., Valsasnini P., Tolba K., Bolin D., Higelin J., Takacs B., Sinigaglia F. Promiscuous and allele-specific anchors in HLA-DR-binding peptides. Cell. 1993 Jul 16;74(1):197–203. doi: 10.1016/0092-8674(93)90306-b. [DOI] [PubMed] [Google Scholar]
- Hill C. M., Liu A., Marshall K. W., Mayer J., Jorgensen B., Yuan B., Cubbon R. M., Nichols E. A., Wicker L. S., Rothbard J. B. Exploration of requirements for peptide binding to HLA DRB1*0101 and DRB1*0401. J Immunol. 1994 Mar 15;152(6):2890–2898. [PubMed] [Google Scholar]
- Hume C. R., Shookster L. A., Collins N., O'Reilly R., Lee J. S. Bare lymphocyte syndrome: altered HLA class II expression in B cell lines derived from two patients. Hum Immunol. 1989 May;25(1):1–11. doi: 10.1016/0198-8859(89)90065-7. [DOI] [PubMed] [Google Scholar]
- Jardetzky T. S., Gorga J. C., Busch R., Rothbard J., Strominger J. L., Wiley D. C. Peptide binding to HLA-DR1: a peptide with most residues substituted to alanine retains MHC binding. EMBO J. 1990 Jun;9(6):1797–1803. doi: 10.1002/j.1460-2075.1990.tb08304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen B. H., Buus S., Vartdal F., Viken H., Eriksen J. A., Thorsby E., Sollid L. M. Binding of peptides to HLA-DQ molecules: peptide binding properties of the disease-associated HLA-DQ(alpha 1*0501, beta 1*0201) molecule. Int Immunol. 1994 Mar;6(3):453–461. doi: 10.1093/intimm/6.3.453. [DOI] [PubMed] [Google Scholar]
- Kropshofer H., Max H., Halder T., Kalbus M., Muller C. A., Kalbacher H. Self-peptides from four HLA-DR alleles share hydrophobic anchor residues near the NH2-terminal including proline as a stop signal for trimming. J Immunol. 1993 Nov 1;151(9):4732–4742. [PubMed] [Google Scholar]
- Kwok W. W., Nepom G. T., Raymond F. C. HLA-DQ polymorphisms are highly selective for peptide binding interactions. J Immunol. 1995 Sep 1;155(5):2468–2476. [PubMed] [Google Scholar]
- Marshall K. W., Liu A. F., Canales J., Perahia B., Jorgensen B., Gantzos R. D., Aguilar B., Devaux B., Rothbard J. B. Role of the polymorphic residues in HLA-DR molecules in allele-specific binding of peptide ligands. J Immunol. 1994 May 15;152(10):4946–4957. [PubMed] [Google Scholar]
- O'Sullivan D., Sidney J., Appella E., Walker L., Phillips L., Colón S. M., Miles C., Chesnut R. W., Sette A. Characterization of the specificity of peptide binding to four DR haplotypes. J Immunol. 1990 Sep 15;145(6):1799–1808. [PubMed] [Google Scholar]
- Rammensee H. G., Friede T., Stevanoviíc S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41(4):178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- Sette A., Sidney J., Oseroff C., del Guercio M. F., Southwood S., Arrhenius T., Powell M. F., Colón S. M., Gaeta F. C., Grey H. M. HLA DR4w4-binding motifs illustrate the biochemical basis of degeneracy and specificity in peptide-DR interactions. J Immunol. 1993 Sep 15;151(6):3163–3170. [PubMed] [Google Scholar]
- Sidney J., Oseroff C., del Guercio M. F., Southwood S., Krieger J. I., Ishioka G. Y., Sakaguchi K., Appella E., Sette A. Definition of a DQ3.1-specific binding motif. J Immunol. 1994 May 1;152(9):4516–4525. [PubMed] [Google Scholar]
- Stern L. J., Brown J. H., Jardetzky T. S., Gorga J. C., Urban R. G., Strominger J. L., Wiley D. C. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994 Mar 17;368(6468):215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- Todd J. A., Bell J. I., McDevitt H. O. HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987 Oct 15;329(6140):599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- Verreck F. A., van de Poel A., Termijtelen A., Amons R., Drijfhout J. W., Koning F. Identification of an HLA-DQ2 peptide binding motif and HLA-DPw3-bound self-peptide by pool sequencing. Eur J Immunol. 1994 Feb;24(2):375–379. doi: 10.1002/eji.1830240216. [DOI] [PubMed] [Google Scholar]
- Wucherpfennig K. W., Strominger J. L. Selective binding of self peptides to disease-associated major histocompatibility complex (MHC) molecules: a mechanism for MHC-linked susceptibility to human autoimmune diseases. J Exp Med. 1995 May 1;181(5):1597–1601. doi: 10.1084/jem.181.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]



