Abstract
Exploring the ability of organisms to locally adapt is critical for determining the outcome of rapid climate changes, yet few studies have addressed this question in microorganisms. We investigated the role of a heterogeneous climate on adaptation of North American populations of the wild yeast Saccharomyces paradoxus. We found abundant among-strain variation for fitness components across a range of temperatures, but this variation was only partially explained by climatic variation in the distribution area. Most of fitness variation was explained by the divergence of genetically distinct groups, distributed along a north–south cline, suggesting that these groups have adapted to distinct climatic conditions. Within-group fitness components were correlated with climatic conditions, illustrating that even ubiquitous microorganisms locally adapt and harbour standing genetic variation for climate-related traits. Our results suggest that global climatic changes could lead to adaptation to new conditions within groups, or changes in their geographical distributions.
Keywords: Saccharomyces paradoxus, climate adaptation, global warming, temperature-dependent fitness, freeze–thaw survival
1. Introduction
It is broadly accepted that global climatic changes are affecting most living systems. Among these changes, global warming is expected to be a major source of biome perturbation in the next 100 years. During this period, warming could exceed the range of temperatures that many living organisms can tolerate [1,2], especially in taxa that exhibit low adaptability to temperature change. For instance, Deutsch et al. [3] showed that terrestrial ectotherms, which have distributions determined by a narrow thermal variation tolerance, would be highly sensitive to temperature changes. Similarly, Quintero & Wiens [2] found that most locally adapted vertebrate species probably evolve too slowly to overcome the global warming expected in the next century. Finally, many thermal-dependent fitness components varying among populations are correlated with local climatic conditions in diverse species (e.g. [4,5]).
Despite considerable interest in the local adaptation to temperature in animals and plants [1,2,6], climatic adaptation in the cryptic biodiversity, especially microorganisms, has received much less attention [7]. This lack of knowledge has long been justified by the Baas Becking hypothesis: ‘everything is everywhere but the environment selects’ [8]. The first part of this postulate, which has long been debated, implies that if microorganisms form worldwide populations they should not be considered as part of the threatened biodiversity. However, it is now well established that not all microorganisms are ubiquitous [9,10]. The second part of the Baas Becking's hypothesis implies that microbes could freely adapt in response to environmental changes. Microbes could form extremely large populations and are thus expected to have access to a large pool of beneficial mutations when the environment changes. This last assumption is supported by the rapid adaptation of experimental yeast and bacterial populations to changing environments in the laboratory [11,12]. In addition, evidence suggests that the adaptation of microbial populations to changing environments continually increases with artificially introduced migration [13], challenging theoretical predictions that high rate of migration could limit adaptability to new environments (reviewed in [14]).
Microbial local adaptation in the wild has been reported in a limited number of studies. For instance, local adaptation was investigated in several bacterial species by Belotte et al. [15] and Whitaker [16] in different populations associated with contrasting environments. Distances separating these populations were small (from a few metres to 40 km), suggesting that they were not likely to be isolated by geographical barriers. Nonetheless, the authors found evidence of local adaptation in these specialized populations, suggesting that selection was strong enough to overcome migration in patchy environments [15,16]. Studies looking for evidence of local adaptation in diverse populations of specialized microorganisms, such as bacteria and ciliates with more ubiquitous distribution (50 to several thousands of kilometres), showed that limited migration and local adaptation could shape population structure at this scale [16,17]. In these cases, however, it was difficult to distinguish between the respective contributions of environmental conditions and restricted migration, because variation in environmental factors was often correlated with geographical distance among sampling sites. Thus, we still do not know whether natural populations of ubiquitous microbes are limited in space owing to local adaptation or whether they harbour genetic variation that could allow adaptation to changes in climatic conditions.
Although Baas Becking's model has been investigated in natural populations of prokaryotes (reviewed in [18]), studies of eukaryotic microorganisms are more sparse (see, for instance, fungi reviewed in [10], ciliates in [17], diatoms in [19]). There are several differences between bacteria and eukaryotic microbes that prevent the extension of what has been observed in the former to the latter. One of the most important differences is the fact that eukaryote microbial lifestyles, for example those of fungi, are not strictly clonal and involve phases of sexual reproduction, which allows recombination, and may thus affect the spread of advantageous mutations.
The budding yeast Saccharomyces paradoxus is a widespread free-living fungus associated with deciduous trees (see electronic supplementary material, figure S1 and table S1). In North America, S. paradoxus is sympatric with its sister species Saccharomyces cerevisiae, although S. cerevisiae seems to be less common or absent at higher latitudes [20,21], where temperatures are cooler (see electronic supplementary material, figure S2a,b). This distribution is in agreement with previous observations that S. paradoxus is able to grow at lower temperatures than its sister species [22–26]. In North America, days with oscillations below and above 0°C (freeze–thaw cycles) are more frequent in the south than in the north (see electronic supplementary material, figure S2c–e), which is also in agreement with the lower ability of S. paradoxus to tolerate freezing [27]. These observations suggest that the ecological niche of S. paradoxus is defined at least partly by climatic conditions. Given that S. paradoxus is distributed up to the northern boundary of deciduous tree distribution (see electronic supplementary material, figure S1) and that global changes are expected to disturb the northern boundary of tree species distributions [28], one would expect that S. paradoxus populations will also be affected by global changes. This distribution makes it a prime model to study the role of climate change in adaptation. In addition, S. paradoxus shows important genetic and phenotypic variation within population and differentiation at the continental [23,29] and on more local scales (for instance, of the order of 2500 km in North America [30]), consistent with population dynamics and biogeography that are largely independent of human activities and subjected to limited migration. Thus, S. paradoxus provides an ideal model to study the effect of climate warming at the continental scale on a naturally occurring microbe.
Here, we used S. paradoxus northeastern North American populations as a model to examine whether populations of microbial fungi could be locally adapted to climatic conditions. We assembled a set of 27 strains from locations that were representative of temperature and freeze–thaw cycle variation across northeastern North America. We assayed these strains for two fitness components: growth rates at temperatures covering the range in which budding yeasts are known to grow (10–42°C) and survival through a freeze–thaw cycle. We observed significant correlations between these fitness components and the climatic conditions at locations where strains were sampled. These correlations could only be detected when considering strong phenotypic and genetic divergences between genetic groups of S. paradoxus in North America. These results support the existence of local adaptation, both for growth at high temperature and for survival to freeze–thaw cycles, which suggests that S. paradoxus populations could be affected by global climate changes.
2. Material and methods
(a). Collection of Saccharomyces paradoxus
We used a collection of 41 S. paradoxus strains that we previously collected in a sampling area covering the Saint Lawrence River Valley and Estuary (Quebec) and British Columbia (see electronic supplementary material, figures S1 and S2, and table S2) [20]. We completed our collection with 42 S. paradoxus strains that we sampled on oak trees from different elevations (140–413 m) on Mont St Hilaire (Quebec) and eight strains we sampled in Wisconsin (see electronic supplementary material, figure S1 and table S2). We also used 25 S. paradoxus strains from different collections, isolated from Mont St Hilaire [31], Michigan, Pennsylvania, New Jersey [29], Saskatchewan, Ontario and Missouri (kindly provided by J. B. Anderson, M. A. Lachance and P. Sniegowski; electronic supplementary material, figures S1 and S2, and table S2).
(b). Climatic data
We collected monthly climatic data for 69 geographical locations uniformly distributed across our sampling area (see electronic supplementary material, figure S3a). We used the mean annual temperature (Tm) at each site as a climatic factor to explain variation in strain growth rate at different temperatures (see electronic supplementary material, figure S2b). We estimated the frequency of freeze–thaw cycles to estimate Do, the number of days per year with alternation of positive and negative temperatures (see electronic supplementary material, figure S2c). We used Do as climatic factor to explain variation in strain survival to a freeze–thaw cycle (see electronic supplementary material, figure S2c–e). Additionally, we defined three main and five subclimatic divisions in our sampling area using clustering analysis of Tm values (see electronic supplementary material, figure S3b) to simplify result descriptions, as many of the climatic variables are correlated with each other.
(c). Fitness component assays
We examined two fitness components potentially related to climatic adaptation for 27 strains representative of our strain collection and for two control reference strains (S. cerevisiae strain BY4743 and S. paradoxus strain CBS432). Growth rates (six replicates per strain) were estimated as follows: 150 µl of fresh liquid culture (yeast extract, peptone, dextrose, YPD) at OD600 = 0.03 (approx. 50 000 cells) were grown for 4 (20, 30, 37 and 42°C) or 7 days (10°C), and OD600 was measured every 15 min. Growth rate at each temperature (GT) was measured as the maximum slope of the growth curves (see the electronic supplementary material). Freeze–thaw survival (two replicates per strain) was estimated as described by Will et al. [27] using colony counting with some modifications. For each strain and each replicate, a fresh liquid culture in YPD at OD600 = 0.3 was frozen for 2 h at −80°C and a replicate culture was stored for 2 h on ice (approx. 4°C) as a control. For each replicate and treatment, cultures were sampled twice, and 150 and 300 cells were plated on solid YPD medium, resulting in four replicates per strain and per treatment. Freeze–thaw survival (FS) was measured as the ratio of colony count on treatment plates (−80°C) over the colony count on control plates (+4°C).
(d). Genetic origin of Saccharomyces paradoxus strains
PCR products of partial CDS sequences of unlinked genes POP2 (1044 pb, Chromosome XIV) and RPB2 (414 pb, Chromosome XV) for 102 strains (see NCBI accession numbers in electronic supplementary material, table S2) were amplified as described in electronic supplementary material (table S3 for PCR primers) and were sequenced by Sanger sequencing. We completed these data with sequences available for 19 reference S. paradoxus strains that are representative of other parts of the world and of all known genetic groups (see electronic supplementary material, table S4) [23]. We used these sequences in a phylogenetic approach to assign S. paradoxus strains to genetic groups previously defined [29] (see the electronic supplementary material). Additionally, we developed an RFLP method for the quick identification of S. paradoxus genetic groups to identify 15 strains from Mont St Hilaire that we did not include in the sequence analysis (see the electronic supplementary material, table S2 and figure S6).
(e). Statistical analyses of fitness component variation
For each strain (n = 27, control strains removed), we first removed outlier measurements and calculated the mean GT (six replicates) and total FS (four replicates) values (see the electronic supplementary material, figure S4 and supplementary method in File S1, and File S2). We used a generalized linear model (GLM) to identify the factors that contribute to variation in individual strain fitness components GT and FS (see the electronic supplementary material). First, we used Tm as an additive climatic factor to explain GT (formula GT1: GT ∼ Tm) and Do as an additive climatic factor to explain FS (formula FS1: FS ∼ Do). We then performed a χ2-test to estimate whether the climatic factor significantly improved the GLM model (assessed by the part of residual deviance explained by the factor). We repeated the analysis by including the information of genetic groups (Gg), first by considering it as both an additive and interacting factor (formula GT2: GT ∼ Tm + Gg + Tm:Gg and FS2: FS ∼ Do + Gg + Do:Gg), and second by repeating the first analysis within each genetic group separately (formula GT3: GT ∼ Tm and FS3: FS ∼ Do). All analyses were performed in R [32].
3. Results
(a). Fitness estimates at different temperatures and freeze–thaw survival
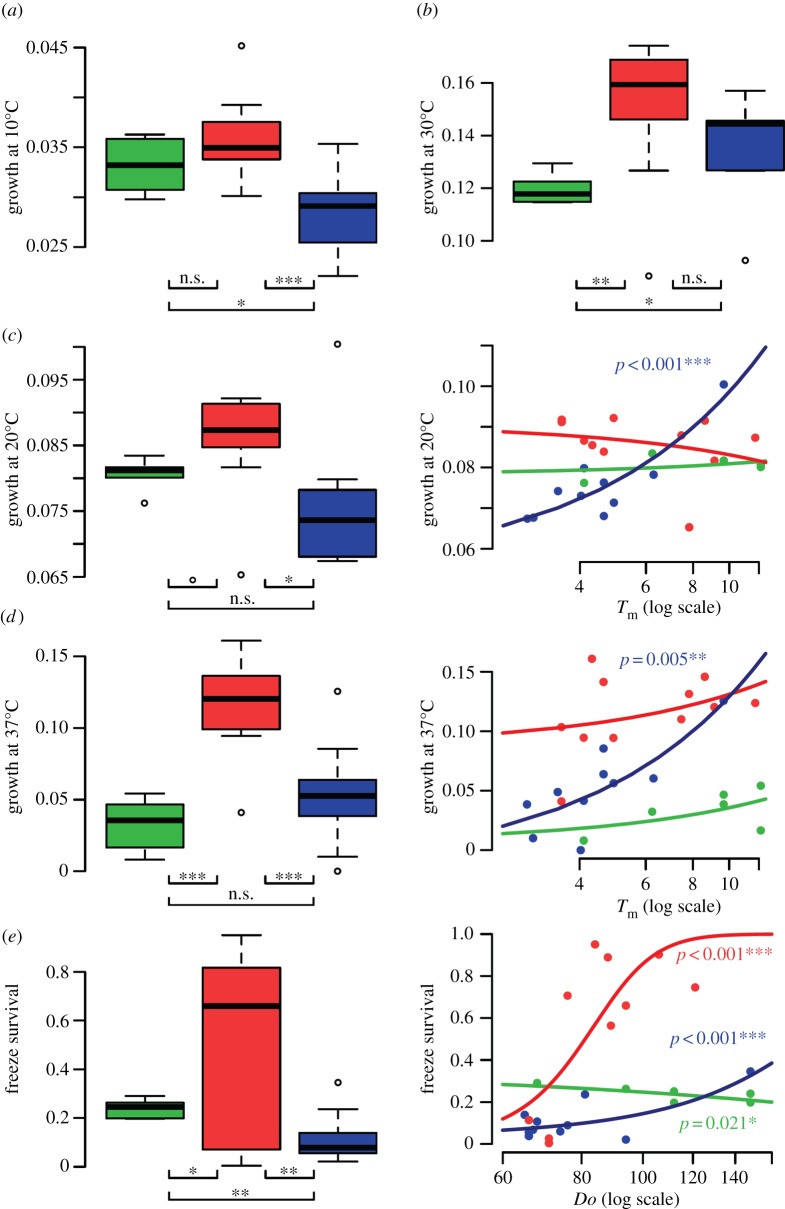
We examined the effect of climatic parameters on fitness components of diverse strains of S. paradoxus collected from across northeastern North America (see electronic supplementary material, figure S2a). To ensure a representative temperature gradient and a broad sample of S. paradoxus diversity, we measured the growth rates (GT) of 28 S. paradoxus strains (including the European-type strain CBS432), and the diploid S. cerevisiae reference strain BY4743 as a control (see the electronic supplementary material, table S1 and figure S4), at 10°C, 20°C, 30°C, 37°C and 42°C, and freeze–thaw survival (FS). Strains came from a representative subsample of our collection with respect to geography and climate (see electronic supplementary material, figure S2a). None of the tested strains showed significant growth at 42°C (data not shown). Saccharomyces paradoxus strain CBS432 had a maximal growth rate at 20°C and showed no growth at 37°C, whereas S. cerevisiae BY4743 had the maximal growth rate at 30°C and showed significant growth at 37°C (see electronic supplementary material, figures S4 and S5). FS was higher for CBS432 (0.20) than for BY4743 (0.00; electronic supplementary material, figures S4 and S5e). We observed high variability among North American S. paradoxus strains for all traits measured, with the exception of growth rate at 10°C, which remained low for all strains (see electronic supplementary material, figures S4 and S5a). Unlike the European strain CBS432, most North American S. paradoxus strains could grow at 37°C and seven of them showed faster growth rate than S. cerevisiae at this temperature (see electronic supplementary material, figure S5d). Similarly, 13 strains were more tolerant to a freeze–thaw cycle than the S. paradoxus-type strain CBS432, and seven of them were almost unaffected by this stress (mean survival > 0.50; electronic supplementary material, figures S4 and S5e).
We tested whether among-strain variation for fitness components was explained by the climatic factors at the sampling locations. We found that the climatic factor Tm had no significant effect on GT at any temperature (p > 0.5; electronic supplementary material, figure S5a–d and table S6). We observed a negative correlation between the number of days per year with alternation of positive and negative temperatures (Do) and latitude (r = 0.83, p < 0.001; electronic supplementary material, figure S2d). FS was positively and significantly correlated with Do (estimate: 0.010 ± 0.001, p < 0.001; electronic supplementary material, figure S5e and table S6) and the proportion of residual deviance explained by Do was weak but significant (0.03; p < 0.001). We tested for the correlation of strain growth rates (GT) among four different temperatures in a two-way crossed ANOVA. The main effect of strains was significant (F26,78 = 2.25, p < 0.01, random-effects model) and the strain × temperature interaction was modest but significant (F78,540 = 1.90, 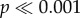 ). Two-thirds of the strain × temperature interaction variance were attributable to genetic correlations among strains at different temperatures (responsiveness; electronic supplementary material, table S5). All pairwise genetic correlations between temperatures were positive, with a Pearson correlation of +0.4 or more (see electronic supplementary material, table S5).
). Two-thirds of the strain × temperature interaction variance were attributable to genetic correlations among strains at different temperatures (responsiveness; electronic supplementary material, table S5). All pairwise genetic correlations between temperatures were positive, with a Pearson correlation of +0.4 or more (see electronic supplementary material, table S5).
(b). Identification of Saccharomyces paradoxus genetic groups
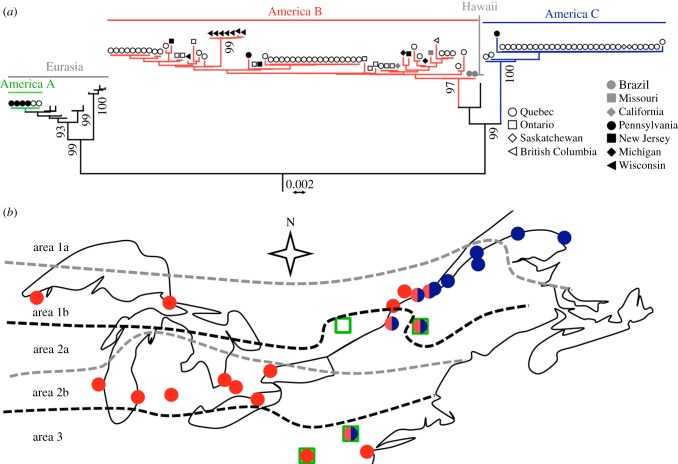
We tested whether the large unexplained fraction of fitness component variation could be explained by heterogeneity in the lineages of S. paradoxus strains, because North American S. paradoxus strains belong to three genetic groups, as previously described by Kuehne et al. [29]. From the phylogenetic tree based on the nucleotide sequences of two unlinked loci, we assigned a representative sample of 102 strains from North America to these groups, including the 27 strains we used for fitness component assays (figure 1). The two markers were sufficient to discriminate the three groups (namely A, B and C) previously described with a larger number of loci (see electronic supplementary material, figure S7). Grouping of strains within groups A, B and C was strongly supported by bootstrap analyses (figure 1). Two strains from southern Quebec clustered with four strains from Pennsylvania previously associated with the non-indigenous West Eurasian genetic group (group A; figure 1a) [29]. Fifty-seven strains from across our sampling area (except Saskatchewan and Eastern Quebec) clustered with 12 isolates belonging to the main previously described indigenous American S. paradoxus group (group B; figure 1a) [23,29,33,34]. The 32 remaining strains clustered in the third group, which is genetically distinct from all other S. paradoxus strains (group C; figure 1a). This group was previously described as a single diverging isolate from Pennsylvania (YPS667) (see electronic supplementary material, figure S7) [29], but our collection expands its range further north to Quebec (30) and Saskatchewan (2). Group A showed 2.63 ± 0.53% of nucleotide divergence with group B, in agreement with previous studies [23,29,33]. Group C showed 3.01 ± 0.57% and 2.06 ± 0.43% of divergence with groups A and B, respectively (see electronic supplementary material, figure S7). We found significant genetic variation within groups B (0.33 ± 0.09%) and C (0.04 ± 0.01%), and none in group A (see electronic supplementary material, table S8 and figure S7).
Figure 1.
Two main genetic groups of S. paradoxus are found in northeastern North America. (a) Assignment of S. paradoxus strains to the main genetic groups found in North America. The tree was drawn from 1458 nucleotide positions in 118 strains of S. paradoxus and four relatives as out-groups (not shown). Symbols indicate sample locations of American strains (see legend). The three American groups A, B and C are coloured in green, blue and red, respectively. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test is shown next to the branches (10 000 replicates, values lower than 90% are not shown). (b) Distribution of S. paradoxus groups in sampling area. The occurrence of genetic groups A, B and C is indicated by green squares, red circles and blue circles, respectively. The dotted lines indicate the limit between climatic areas.
Figure 2.
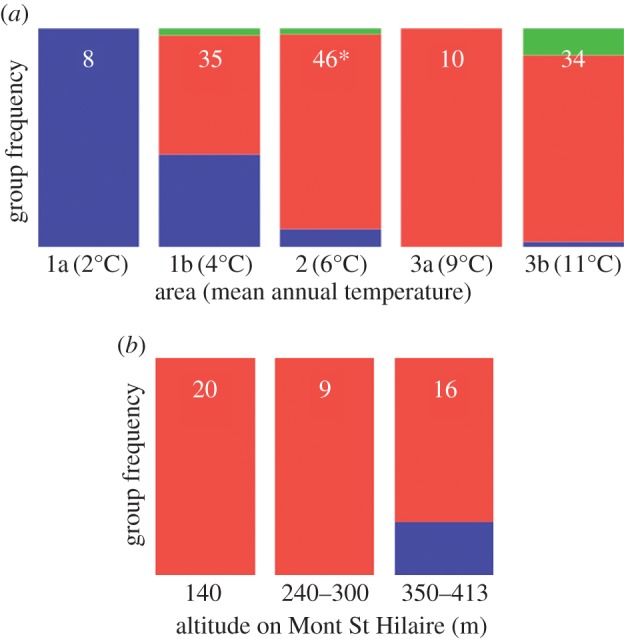
Saccharomyces paradoxus groups are distributed along a north–south cline. (a) Frequency of three North American S. paradoxus genetic groups (A, green; B, red; C, blue) in five climatic areas (figure 1). The total number of strains in each area is indicated by white. All strains identified by Kuehne et al. [29] were also included in these calculations. The asterisk (*) indicates 45 strains of area 2 collected in Mont St Hilaire. (b) Frequency of genetic groups B and C at different altitudes on Mont St Hilaire. Group C is only found at higher than 350 m (see electronic supplementary material, table S2).
Our genetic screen of 102 S. paradoxus strains in North America revealed that, with the exception of the location where strain YPS667 was isolated (Buck Hill Falls, PA [29]), groups B and C were largely spatially segregated, and locations where groups B and C were found together were all located close to the 5°C isocline separating climatic areas 1 and 2 (figure 1b). Strains from group A were only found close to the 5°C isocline or further south, often in the same locations as other groups (figure 1b). We found that groups B and C were distributed following a north–south gradient, with B strains more frequent in the south (areas 2 and 3) and C strains more frequent in the north (area 1; figure 2a). We observed the same pattern with altitude at a very local scale in Mont St Hilaire (Quebec), where group B was present at all altitudes, and group C was only found at sites near the mountain top (figure 2b).
(c). Fitness component variation in the light of genetic groups
We revisited the analysis of growth and freeze–thaw survival, this time considering genetic groups. In our set of strains, six belonged to group A, 11 to group B and 10 to group C (see electronic supplementary material, table S2). A substantial and significant part of among-strain fitness component residual deviance (0.23–0.66) was explained by differences among groups (electronic supplementary material, table S7). At 10°C and 20°C, group B grew significantly faster than group C, and group A was intermediate (figure 3a,c, respectively). At 30°C, groups B and C were indistinguishable, and grew significantly faster than strains of group A (figure 3b). We observed the strongest effect of genetic groups for growth at 37°C and for freeze–thaw survival (p < 0.001; electronic supplementary material, table S7). At 37°C, group B grew significantly better than both groups A and C, which showed similar fitness (figure 3d). Freeze–thaw survival was significantly higher in group B than in group A, and significantly lower in group C than in both other groups (figure 3e). For growth at 20°C and freeze–thaw survival, the interaction between climatic indicators and genetic groups explained an additional and significant part of residual deviance (0.32 and 0.18, respectively, p < 0.001; electronic supplementary material, table S7). We thus repeated the analysis within each genetic group independently for each fitness component and found that strains responded differently to climatic factors at 20°C and 37°C, and for freeze–thaw survival (see electronic supplementary material, table S7), but not at 10°C and 30°C (data not shown). In group C, most of residual deviance at 20°C and 37°C (0.80 and 0.65, respectively) was explained by mean annual temperature Tm (p < 0.001 and p < 0.01, respectively; electronic supplementary material, table S7). We observed no such tendency in groups A and B. Higher among-strain variation in freeze–thaw survival was observed in group B (figure 3e). A main part of the residual deviance in freeze–thaw survival in group B was explained by the number of freeze–thaw days per year Do at sampling sites of the strains (0.41, p < 0.0001; electronic supplementary material, table S7). Although the variation was about 100-fold lower in group A and 15-fold lower in group C, most of freeze–thaw survival in groups A and C was also explained by Do (0.47, p < 0.001 and 0.49, p < 0.05, respectively; electronic supplementary material, table S7). Overall, growth at 20°C and 37°C in group C (estimates: 0.004 ± 0.001, p < 0.001 and 0.015 ± 0.004, p < 0.001, respectively) and freeze–thaw survival in groups B and C (estimates: 0.095 ± 0.005, p < 0.001 and 0.022 ± 0.002, p < 0.001, respectively) were significantly positively correlated with climatic factors (figure 3c–e). In group A, freeze–thaw survival was significantly negatively correlated with Do (estimates: −0.005 ± 0.002, p < 0.05).
Figure 3.
Among-strain variation in fitness components (growth at different temperatures and freeze–thaw survival) is explained by differences among groups and by variation within groups. The following conditions were tested for 27 S. paradoxus strains: growth in liquid-rich medium at (a) 10°C, (b) 30°C, (c) 20°C, (d) 37°C and (e) survival to a freeze–thaw cycle. (a–e) T-test comparison between fitness of three genetic groups represented by boxplots (A, green; B, red; C, blue); (c–e) dots indicate the measurements of fitness components; curves indicate fitness component values expected under GLM models within each group separately (formula 3) as a function of climatic factors (c,d) Tm or (e) Do. Significance of the climatic estimate is shown next to the curve only for p ≤ 0.05. Within-group analyses are not shown for growth at 10°C and 30°C (where differences were not significant). p-value ranges are indicated by the following symbols: ***p < 0.001; **p < 0.01; *p < 0.05; °p < 0.1.
4. Discussion
Here, we used S. paradoxus to investigate whether populations of budding yeasts were adapted to local climatic conditions. We found evidence that these populations are locally adapted and that the global patterns of adaptation are also affected by historical factors that are reflected in the presence of distinct evolutionary lineages in our sampling area. Our results support previous observations suggesting that climatic factors play a major role in the ecological differentiation of budding yeasts [22–27,35,36], but this time within species. For instance, S. paradoxus and its sister species S. cerevisiae naturally occur in very similar ecological niches in terms of available substrates, and they are sympatric in many places in the world [24,37], including North America (see electronic supplementary material, figure S1). Interestingly, the natural distribution of S. paradoxus seems to be slightly shifted further north in America (see electronic supplementary material, figure S1) and Europe [38] when compared with S. cerevisiae. These differences between S. cerevisiae and S. paradoxus distributions are surprising, considering that their worldwide distribution suggests large dispersion capabilities. This contrast provides evidence for their adaptive divergence, rather than limited migration, at least at local geographical scales [30]. Phenotypic divergence between S. cerevisiae and S. paradoxus is limited to only a few traits, but their ability to grow at different temperatures [22–26] is the most consistent with their geographical distribution [20]. Another consequence of temperature gradients in North America is that soils and surfaces of trees may remain frozen during longer periods in the north than in the south, where temperature oscillates around freezing temperature for longer periods (see electronic supplementary material, figure S2d). This phenomenon could be another explanation for the shifted geographical distributions of S. cerevisiae and S. paradoxus [20], as it was previously shown that they have contrasted tolerance to freeze–thaw cycles [27]. Additional evidence for the prominent role of temperature on budding yeast distribution was found in Saccharomyces eubayanus, a cold-adapted yeast that has only been reported in Patagonian forests, where climate is comparable with that of northeastern North America [35]. The importance of adaptation to temperature in yeast ecology has also been suggested for Saccharomyces kudriavzevii and Saccharomyces uvarum, which have maximum growth temperatures of 32–35°C, and naturally coexist with S. cerevisiae and S. paradoxus in some parts of the world [24]. Hence, temperature appears to be one of the only traits that had been systematically associated with the divergence of ecological niches among budding yeasts [26]. Our study system therefore establishes a new paradigm for the study of ecological genetics in a model organism, and potentially its link with speciation. In addition, we found that temperature plays a major role in the divergence and local adaptation of S. paradoxus, suggesting that climatic conditions may affect population divergence within species.
The fact that isolates of S. paradoxus are adapted to the local conditions at which they were sampled may reflect adaptation at two different time scales. First, based on the assignment of the strains to the three known North American groups [29], we observed a north–south gradient in the distribution of groups B and C (figure 2a), which is consistent with our hypothesis that climatic variables determine strain distributions. The shift in frequencies between the two groups is found around the 5°C isocline. This gradient was also observed at a very local scale on Mont St Hilaire (Quebec), where group C was only found at high altitude near the mountain top, whereas group B was present at all altitudes (figure 2b). Mont St Hilaire is located close to the 5°C isocline and its 400 m of elevation probably captures the transition to a slightly colder climate. We observed strong phenotypic differences between these S. paradoxus genetic groups, suggesting divergent evolution for growth rate at these temperatures and for freeze–thaw survival (figure 3). However, group B strains grew on average systematically faster at all temperatures, except for 30°C, and it survived freeze–thaw cycles better than group C. The geographical distribution of groups B and C is in agreement with their respective abilities to grow at high temperature (37°C) and to survive to a freeze–thaw cycle, which both represent extreme conditions faced in nature. The fact that group B has faster growth rate at 10°C than group C is counterintuitive, because it is group C that is mainly found in cold regions. One possibility is that growth at 10°C and 37°C could be genetically correlated because they share some of the same physiological mechanisms, which we know to be partly true for S. cerevisiae [36]. Shared mechanisms could also be an explanation for the positive correlations we observed between strain individual performances at different temperatures in S. paradoxus, indicating no specialization to a particular temperature (see electronic supplementary material, table S5). In these cases, adaptation to high temperatures could exapt strains to low temperatures at the same time. We must consider other adaptive traits that could have been fixed among groups and might provide an advantage to group C in its habitat, for example the ability to use a larger spectrum of substrates than group B. For instance, the nature of sugar sources greatly varies among oak species, which may allow for the maintenance of stable budding yeast communities composed of species with different optimal growth temperatures [24]. Furthermore, freezing tolerance seems to be linked to other adaptive traits, such as copper resistance [39] and carbon metabolism [27,40]. For example, Will et al. [27] demonstrated that the loss of genes involved in freeze–thaw survival in S. cerevisiae provided a higher fitness in environments with high sugar concentration. A comprehensive survey of genomic variation underlying adaptation to temperature and metabolism in North American S. paradoxus populations, together with additional investigations of fitness components under competition between groups, is needed to map and better understand the functional mechanisms underlying these complex ecological trade-offs.
We found within-group genetic variation in agreement with previous genomic estimations in American, European and East Asian S. paradoxus groups [23,30,41]. Together with the genomic evidence that these groups are not clonal lineages [41], the phenotypic data show that there is ample genetic diversity within groups. For instance, as we observed for nucleotide polymorphism (see electronic supplementary material, figure S7 and table S8), variation in freeze–thaw survival was higher in group B than in groups A and C. Phenotypic variation was correlated with local climatic conditions within groups, suggesting adaptation on smaller geographical and temporal time scales. Also, we observed a significant interaction between climatic and genetic factors, suggesting that strains responded differently to climate, depending on their genetic group. Within group C, growth rates at 20°C and 37°C were positively correlated with climatic conditions. Similarly, freeze–thaw survival was positively correlated with climatic conditions in group B and, to a lesser extent, in group C (figure 3). We also observed a weak and negative but significant correlation between freeze–thaw survival and climatic conditions in group A. Given the low phenotypic variation we observed in this group, the absence of polymorphism owing to its recent introduction [29] and its sparse distribution in North America, this counterintuitive effect should be carefully interpreted and must be validated with a larger sampling effort.
Short-term adaptability is expected to be a major factor in the response of organisms to rapid global changes [1]. The strong phenotypic differences among budding yeast species suggest that, with global warming, their distribution could be shifted to higher latitudes, provided the trees on which they live also expand their distribution (but see [28]). Similarly, one would expect the areas of sympatry between species to move towards high latitudes. Such a scenario would probably have impacts at both the species and group levels in S. paradoxus. In North America, the fact that adaptation to local temperature and nucleotide diversity seems to be more limited for group C than for group B suggests that the former harbour less standing genetic variation and could be less likely to adapt to global warming. Moreover, with the possible shift of S. cerevisiae further north, one can speculate that competition would increase in S. paradoxus populations between groups B and C, possibly enhancing the exclusion of the less adapted group. Hence, local adaptation could potentially have profound impacts on microbial populations subjected to global warming. Just as S. cerevisiae has become a prime model for laboratory studies over the years, our work suggests that its sister species S. paradoxus could be a very powerful model to study the ecological genomics of global warming in the wild.
Acknowledgements
We thank M.-A. Lachance, J. Anderson and P. Sniegowski for providing strains, and N. Aubin-Horth, I. Gagnon-Arsenault, M. Filteau, J.-P. Verta, S. Rochette, G. Diss and two anonymous reviewers for helpful comments on the manuscript. J.-B.L., C.R.L. and G.C. designed research and J.-B.L. and G.C. performed experiments. J.-B.L., G.B. and G.C. analysed the results. J.-B.L. wrote the manuscript with contributions from C.R.L. and G.C. P.S., A.K.D., B.J., K.S., P.A., J.P.S., C.T.H. and G.B. contributed to the systematic sampling efforts across the study area, and C.T.H., J.P.S., P.S. and G.B. provided comments on the manuscript.
Data accessibility
Supplementary method and supplementary tables (including list of strains and sequences accession numbers) and figures are available in electronic supplementary material file S1. Growth and survival raw and mean data are available in electronic supplementary material file S2. Meteorological data with monthly mean temperatures for 69 stations, geographical coordinates and calculation of Do values are available in electronic supplementary material file S3.
Funding statement
This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) discovery grant to C.R.L. and partly by a Human Frontier Science Programme (HFSP) grant RGY0073/2010. This material is based upon work supported by the National Science Foundation under grant no. DEB-1253634 to C.T.H. and funded in part by the DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494). J.P.S. was supported by FCT (Portugal) grant nos. SFRH/BD/77390/2011 (P.A.) and PEST/OE/BIA/UI0457/2011, PTDC/BIA-EVF/118618/2010, PTDC/AGR-ALI/118590/2010. J.-B.L. was supported by a fellowship from the Fonds de Recherche en Santé du Québec (FRSQ). G.C. was supported by a PROTEO graduate student scholarship. P.S. was supported by a fellowship from the Fonds de la Recherche sur la Nature et les Technologies du Québec (FQRNT).
References
- 1.Chen IC, Hill JK, Ohlemuller R, Roy DB, Thomas CD. 2011. Rapid range shifts of species associated with high levels of climate warming. Science 333, 1024–1026 (doi:10.1126/Science.1206432) [DOI] [PubMed] [Google Scholar]
- 2.Quintero I, Wiens JJ. 2013. Rates of projected climate change dramatically exceed past rates of climatic niche evolution among vertebrate species. Ecol. Lett. 16, 1095–1103 (doi:10.1111/Ele.12144) [DOI] [PubMed] [Google Scholar]
- 3.Deutsch CA, Tewksbury JJ, Huey RB, Sheldon KS, Ghalambor CK, Haak DC, Martin PR. 2008. Impacts of climate warming on terrestrial ectotherms across latitude. Proc. Natl Acad. Sci. USA 105, 6668–6672 (doi:10.1073/pnas.0709472105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eliason EJ, et al. 2011. Differences in thermal tolerance among sockeye salmon populations. Science 332, 109–112 (doi:10.1126/Science.1199158) [DOI] [PubMed] [Google Scholar]
- 5.Willett CS. 2010. potential fitness trade-offs for thermal tolerance in the intertidal copepod Tigriopus californicus. Evolution 64, 2521–2534 (doi:10.1111/J.1558-5646.2010.01008.X) [DOI] [PubMed] [Google Scholar]
- 6.Maclean IMD, Wilson RJ. 2011. Recent ecological responses to climate change support predictions of high extinction risk. Proc. Natl Acad. Sci. USA 108, 12 337–12 342 (doi:10.1073/Pnas.1017352108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteban GF, Finlay BJ. 2010. Conservation work is incomplete without cryptic biodiversity. Nature 463, 293–293 (doi:10.1038/463293c) [DOI] [PubMed] [Google Scholar]
- 8.Baas Becking LGM. 1934. Geobiologie of Inleiding Tot de Milieukunde. The Hague, The Netherlands: Van Stockum & Zoon [Google Scholar]
- 9.Lachance MA. 2004. Here and there or everywhere? Bioscience 54, 884–884 (doi:10.1641/0006-3568(2004)054[0884:Hatoe]2.0.Co;2) [Google Scholar]
- 10.Taylor JW, Turner E, Townsend JP, Dettman JR, Jacobson D. 2006. Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom Fungi. Phil. Trans. R. Soc. B 361, 1947–1963 (doi:10.1098/Rstb.2006.1923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper VS, Bennett AF, Lenski RE. 2001. Evolution of thermal dependence of growth rate of Escherichia coli populations during 20,000 generations in a constant environment. Evolution 55, 889–896 (doi:10.1554/0014-3820(2001)055[0889:Eotdog]2.0.Co;2) [DOI] [PubMed] [Google Scholar]
- 12.Cowen LE, Nantel A, Whiteway MS, Thomas DY, Tessier DC, Kohn LM, Anderson JB. 2002. Population genomics of drug resistance in Candida albicans. Proc. Natl Acad. Sci. USA 99, 9284–9289 (doi:10.1073/pnas.102291099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perron GG, Gonzalez A, Buckling A. 2008. The rate of environmental change drives adaptation to an antibiotic sink. J. Evol. Biol. 21, 1724–1731 (doi:10.1111/J.1420-9101.2008.01596.X) [DOI] [PubMed] [Google Scholar]
- 14.Lenormand T. 2002. Gene flow and the limits to natural selection. Trends Ecol. Evol. 17, 183–189 (doi:10.1016/S0169-5347(02)02497-7) [Google Scholar]
- 15.Belotte D, Curien JB, Maclean RC, Bell G. 2003. An experimental test of local adaptation in soil bacteria. Evolution 57, 27–36 (doi:10.1554/0014-3820(2003)057[0027:Aetola]2.0.Co;2) [DOI] [PubMed] [Google Scholar]
- 16.Whitaker RJ. 2006. Allopatric origins of microbial species. Phil. Trans. R. Soc. B 361, 1975–1984 (doi:10.1098/Rstb.2006.1927) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gachter E, Weisse T. 2006. Local adaptation among geographically distant clones of the cosmopolitan freshwater ciliate Meseres corlissi. I. Temperature response. Aquat. Microb. Ecol. 45, 291–300 (doi:10.3354/Ame045291) [Google Scholar]
- 18.Soininen J. 2012. Macroecology of unicellular organisms—patterns and processes. Environ. Microb. Rep. 4, 10–22 (doi:10.1111/J.1758-2229.2011.00308.X) [DOI] [PubMed] [Google Scholar]
- 19.Telford RJ, Vandvik V, Birks H.JB 2006. Dispersal limitations matter for microbial morphospecies. Science 312, 1015–1015 (doi:10.1126/Science.1125669) [DOI] [PubMed] [Google Scholar]
- 20.Charron G, Leducq JB, Bertin C, Dube AK, Landry CR. In press. Exploring the northern limit of the distribution of Saccharomyces cerevisiae and Saccharomyces paradoxus in North America. FEMS Yeast Res. (doi:10.1111/1567-1364.12100) [DOI] [PubMed] [Google Scholar]
- 21.Maganti H, Bartfai D, Xu J. 2012. Ecological structuring of yeasts associated with trees around Hamilton, Ontario, Canada. FEMS Yeast Res. 12, 9–19 (doi:10.1111/j.1567-1364.2011.00756.x) [DOI] [PubMed] [Google Scholar]
- 22.Goncalves P, Valerio E, Correia C, de Almeida JM, Sampaio JP. 2011. Evidence for divergent evolution of growth temperature preference in sympatric Saccharomyces species. PLoS ONE 6, e20739 (doi:10.1371/journal.pone.0020739) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liti G, et al. 2009. Population genomics of domestic and wild yeasts. Nature 458, 337–341 (doi:10.1038/nature07743) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sampaio JP, Goncalves P. 2008. Natural populations of Saccharomyces kudriavzevii in Portugal are associated with oak bark and are sympatric with S. cerevisiae and S. paradoxus. Appl. Environ. Microbiol. 74, 2144–2152 (doi:10.1128/AEM.02396-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sweeney JY, Kuehne HA, Sniegowski PD. 2004. Sympatric natural Saccharomyces cerevisiae and S. paradoxus populations have different thermal growth profiles. FEMS Yeast Res. 4, 521–525 (doi:10.1016/S1567-1356(03)00171-5) [DOI] [PubMed] [Google Scholar]
- 26.Salvado Z, Arroyo-Lopez FN, Guillamon JM, Salazar G, Querol A, Barrio E. 2011. Temperature adaptation markedly determines evolution within the genus Saccharomyces. Appl. Environ. Microb. 77, 2292–2302 (doi:10.1128/Aem.01861-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Will JL, Kim HS, Clarke J, Painter JC, Fay JC, Gasch AP. 2010. Incipient balancing selection through adaptive loss of aquaporins in natural Saccharomyces cerevisiae populations. PLoS Genet. 6, e1000893 (doi:10.1371/journal.pgen.1000893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ordonez A, Williams JW. 2013. Climatic and biotic velocities for woody taxa distributions over the last 16 000 years in eastern North America. Ecol. Lett. 16, 773–781 (doi:10.1111/ele.12110) [DOI] [PubMed] [Google Scholar]
- 29.Kuehne HA, Murphy HA, Francis CA, Sniegowski PD. 2007. Allopatric divergence, secondary contact, and genetic isolation in wild yeast populations. Curr. Biol. 17, 407–411 (doi:10.1016/j.cub.2006.12.047) [DOI] [PubMed] [Google Scholar]
- 30.Hyma KE, Fay JC. 2013. Mixing of vineyard and oak-tree ecotypes of Saccharomyces cerevisiae in North American vineyards. Mol. Ecol. 22, 2917–2930 (doi:10.1111/Mec.12155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Replansky T, Bell G. 2009. The relationship between environmental complexity, species diversity and productivity in a natural reconstructed yeast community. Oikos 118, 233–239 (doi:10.1111/J.1600-0706.2008.16948.X) [Google Scholar]
- 32.R-Developement-Core-Team 2011. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org [Google Scholar]
- 33.Koufopanou V, Hughes J, Bell G, Burt A. 2006. The spatial scale of genetic differentiation in a model organism: the wild yeast Saccharomyces paradoxus. Phil. Trans. R. Soc. B 361, 1941–1946 (doi:10.1098/Rstb.2006.1922) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sniegowski PD, Dombrowski PG, Fingerman E. 2002. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res. 1, 299–306 [DOI] [PubMed] [Google Scholar]
- 35.Libkind D, Hittinger CT, Valerio E, Goncalves C, Dover J, Johnston M, Goncalves P, Sampaio JP. 2011. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl Acad. Sci. USA 108, 14 539–14 544 (doi:10.1073/pnas.1105430108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schade B, Jansen G, Whiteway M, Entian KD, Thomas DY. 2004. Cold adaptation in budding yeast. Mol. Biol. Cell 15, 5492–5502 (doi:10.1091/Mbc.E04-03-0167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Skelton A, Gardner RC, Goddard MR. 2010. Saccharomyces paradoxus and Saccharomyces cerevisiae reside on oak trees in New Zealand: evidence for migration from Europe and interspecies hybrids. FEMS Yeast Res. 10, 941–947 (doi:10.1111/j.1567-1364.2010.00681.x) [DOI] [PubMed] [Google Scholar]
- 38.Johnson LJ, Koufopanou V, Goddard MR, Hetherington R, Schafer SM, Burt A. 2004. Population genetics of the wild yeast Saccharomyces paradoxus. Genetics 166, 43–52 (doi:10.1534/genetics.166.1.43) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahashi S, Ando A, Takagi H, Shima J. 2009. Insufficiency of copper ion homeostasis causes freeze–thaw injury of yeast cells as revealed by indirect gene expression analysis. Appl. Environ. Microb. 75, 6706–6711 (doi:10.1128/AEM.00905-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tulha J, Lima A, Lucas C, Ferreira C. 2010. Saccharomyces cerevisiae glycerol/H+ symporter Stl1p is essential for cold/near-freeze and freeze stress adaptation: a simple recipe with high biotechnological potential is given. Microb. Cell Factories 9, 82 (doi:10.1186/1475-2859-9-82) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai IJ, Bensasson D, Burt A, Koufopanou V. 2008. Population genomics of the wild yeast Saccharomyces paradoxus: quantifying the life cycle. Proc. Natl Acad. Sci. USA 105, 4957–4962 (doi:10.1073/pnas.0707314105) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplementary method and supplementary tables (including list of strains and sequences accession numbers) and figures are available in electronic supplementary material file S1. Growth and survival raw and mean data are available in electronic supplementary material file S2. Meteorological data with monthly mean temperatures for 69 stations, geographical coordinates and calculation of Do values are available in electronic supplementary material file S3.