Abstract
Atlantic cod (Gadus morhua) vertebrae from archaeological sites were used to study the history of the Icelandic Atlantic cod population in the time period of 1500–1990. Specifically, we used coalescence modelling to estimate population size and fluctuations from the sequence diversity at the cytochrome b (cytb) and Pantophysin I (PanI) loci. The models are consistent with an expanding population during the warm medieval period, large historical effective population size (NE), a marked bottleneck event at 1400–1500 and a decrease in NE in early modern times. The model results are corroborated by the reduction of haplotype and nucleotide variation over time and pairwise population distance as a significant portion of nucleotide variation partitioned across the 1550 time mark. The mean age of the historical fished stock is high in medieval times with a truncation in age in early modern times. The population size crash coincides with a period of known cooling in the North Atlantic, and we conclude that the collapse may be related to climate or climate-induced ecosystem change.
Keywords: Atlantic cod, coalescence modelling, ancient DNA, effective population size
1. Introduction
The population structure and demographic history of Atlantic cod have been subjects of numerous research projects in the past decades. The first studies on allozyme variation indicated complex population structuring at small geographical scales [1]. Whether such patterns resulted from population divergence or contemporary effects of natural selection, facilitated by high variation in individual reproductive success was later questioned [2], but research on rapidly evolving and functionally important genetic markers has again revealed significant population sub-structuring [3–6]. The differentiation of inshore and offshore stocks in Norway [7,8], Iceland [6,9] and Newfoundland [3] is an example of such sub-structuring. Recently found to have a broad genetic basis [10], genetic divergence by depth was first associated with allele frequencies at the PanI locus [8]. In contrast to this genetic structuring, mitochondrial DNA relationships in Atlantic cod are characterized by shallow gene genealogies, indicating low effective population sizes [11–13]. Disparity between effective (NE) and census (NC) population size is common in marine animals that are often highly fecund broadcast spawners with extreme mortality of sub-adults [14,15]. This mode of reproduction results in high variance in offspring number, i.e. reproductive sweepstakes [14] that may lead to low genetic diversity within populations and stochastic population structuring [14,16].
Archived material from fisheries surveys has revealed the history of many commercial fish populations, although the samples commonly include only few generations [17–19]. Atlantic cod vertebrae have accumulated at historical fishing sites in the North Atlantic since the beginning of commercial fishing [20,21] and in Iceland since the early settlement [22,23]. The quantity and temporal resolution of this material allow for both empirical estimates of population history and structure and improved model estimation of demographic variables. Estimation of population size based on genetic modelling has improved rapidly in recent years [16,24] and the serial coalescent framework [25] has been successfully applied to study historical fluctuations [26–28].
Since the commencement of fisheries surveys in the twentieth century, the Icelandic cod stock has gone through a severe decline [29]. Historical records cite periods of low Atlantic cod fishing as far back as the fourteenth and fifteenth centuries in Iceland [30]. Intense fishing of Atlantic cod in Icelandic waters started in the thirteenth century as the European market for dried fish boomed, taking English and Dutch fishermen to new fishing grounds, particularly in Iceland and Newfoundland [31–33]. Historical fisheries may have impacted the Atlantic cod populations [34], including the Icelandic stock [35]. Commercial cod fishing in Icelandic waters collapsed in the seventeenth century, a period of both political [36] and climatic [37] upheaval. A cold period, ‘the little ice age’, followed ‘the medieval warm period’ in the sixteenth century and continued until the mid-nineteenth century. Recent evidence suggests an earlier drop of 2–3°C in summer temperatures in the thirteenth century and subsequent decrease in sea temperatures in the fourteenth and fifteenth centuries [38–40]. Climate change and the concomitant ecosystem effects may have determined Atlantic cod catches both in historical [21] and modern times [41]; for example, temperatures warmer than at present are expected to be favourable for cod recruitment around Iceland [42].
Inference of historical population dynamics requires adequate sample sizes from known populations and time periods [43]. In this study, we used Atlantic cod vertebrae from historical fishing sites in Western Iceland, dating from 1500 to 1910, to examine genetic variation and demography in the Atlantic cod population and compare it to contemporary variation. Our first objective was to examine the effects of industrial fisheries on the Icelandic cod stock by comparing NE, genetic variation and age composition in historical times and after the onset of modern fisheries. The effects of intense fisheries on fish stocks may include truncation in the mean age of fished stock, changes in population structure and the distribution of genetic variation [44,45]. Our second objective was to evaluate whether historical reports of abundant cod catches in the medieval warm period coincided with high estimates of NE and, similarly, if the early modern collapse of commercial fishing in Iceland and documented historical temperature fluctuations corresponded to changes in NE, diversity or demography. First, we examined empirical variation at two genetic markers that have been extensively studied in modern Atlantic cod, the mitochondrial marker cytochrome b (cytb) and the PanI locus. Second, we modelled effective population size and fluctuation based on cytb variation. Finally, we estimated the age distribution of the fished stock within the study period.
2. Material and methods
(a). Historical sample selection, excavation and sample dating
Atlantic cod vertebrae were sampled from four historical fishing sites in Western Iceland; Breiðavík northwest Iceland (24°24′45.98″ W, 65°32′38.13″ N), Finnbogastaðir northwest Iceland (21°19′37.73″ W, 66° 0′17.39″ N), Gufuskálar western Iceland (23°55′44.92″ W, 64°54′36.93″ N) and Gufuskálar in southwest Iceland (22°36′41.57″ W, 64°3′18.56″ N) (figure 1). Stratigraphical layers were initially aged by context and finds, and ultimately by 14C dating (Scottish Universities Environment Research Centre). Two stratigraphical layers contained no material suitable for 14C dating, and dates were thus based on tephra deposits and context (see electronic supplementary material, table S1). All vertebrae from the excavations were aged by counting the growth rings with a dissection microscope. Temporal changes in age were tested with linear regression. Before calculation of descriptive statistics and haplotype networks, the samples were grouped into five temporal groups to minimize temporal overlap and to increase sample size for each time interval (see electronic supplementary material, table S1).
Figure 1.
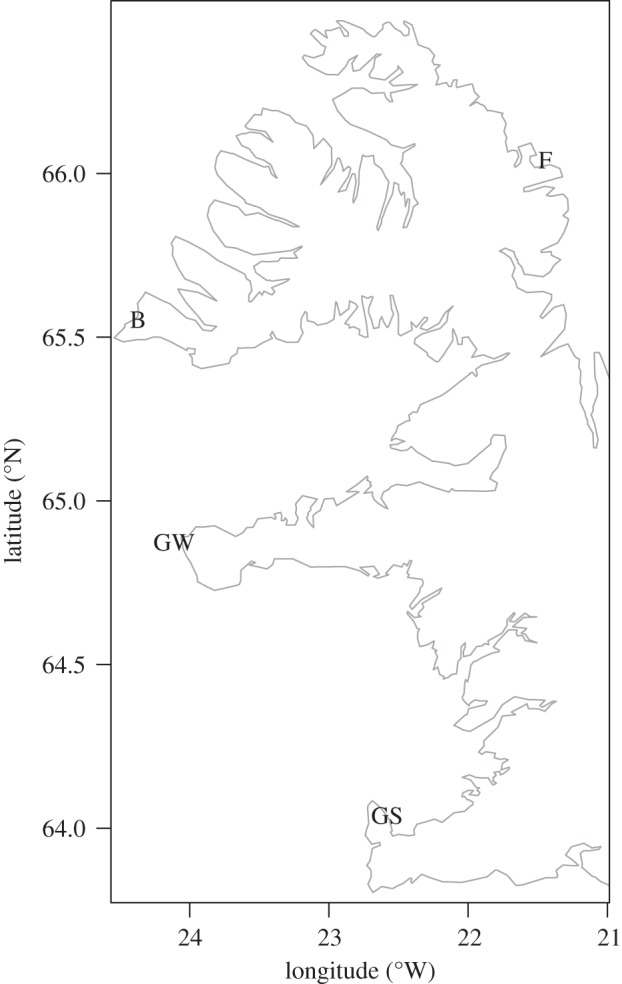
Map of Western Iceland showing the excavation sites. Letters denote abbreviations for site names: GS, Gufuskálar in southwest Iceland; GW, Gufuskálar in Western Iceland; B, Breiðavík; F, Finnbogastaðir. Further information on sites and samples are found in electronic supplementary material, table S1.
(b). DNA extraction, amplification and sequencing
DNA was extracted at Lakehead University Paleo-DNA Laboratory (Thunder Bay, Canada). A 340 base pair fragment of the mitochondrial cytb gene, with known variable sites in Atlantic cod [11], was targeted for PCR amplification with primers L14406 (5′-TTGGCTCTCTTCTAGGCCTTT-3′) and H14746 (5′-GCTCCTCAGAATGATATTTGTCC-3′). A 210 base pair fragment of the Pantophysin I gene was amplified with primers (fwd 5′-GATAGTGAAGGCACTTGACAGC-3′ and rev 5′-AAAAGTCACAGCACAGATTAAGC-3′). PCR of 2–20 ng per reaction was conducted including 2 mM MgCl2, 3 mg bovine serum albumin (BSA), 1.5 µM each primer, 1.5 mM dNTP, 1 U Taq DNA polymerase and a thermostable buffer. Cycling conditions for both genes were: 92°C, 4′; (92°C, 20″; 60°C/55°C ↓Δ0.25°C, 45″; 70°C, 45″) × 20; (92°C, 20″; 55°C, 45″; 70°C, 45″) × 25; 12°C, ∞. PCR products were cleaned using ExoSAP-IT and sequenced using L14406 (cytb) and rev (PanI) on an ABI3500xL (Applied BioSystems). All PCR reactions were performed in a room where no previous work had been done on Atlantic cod DNA. A subset of individuals (n = 16) were re-sequenced for the cytb, using both PCR primers (see electronic supplementary material, table S2). The cytb sequences were easily aligned to known modern sequences covering positions 14494–14768 [46]. Previously published information on the modern Icelandic cod stock was used in all analyses (see electronic supplementary material, table S3).
(c). Population analyses
A haplotype network was generated using Pegas [47] in R v. 2.9.2 [48]. DnaSP v. 5.10 [49] was used to generate mismatch distributions and estimate haplotype (Hd) and nucleotide (π) diversities, Tajima's D and DnaSP and pairwise FST values with 95% confidence intervals from 10 000 bootstrap replicates. Haplotypic richness (hr) [50] was calculated using the hierfstat package in R [51]. The linear relationship of haplotype richness with time was tested by comparing the slope of the observed data with slopes of regression lines obtained by permuting haplotypes across the temporal intervals. The permutation was conducted in R [48], using 1000 permutations.
(d). Coalescent simulations and demographic analyses
The Bayesian Skyride Plot (BSP) [52], implemented in BEAST v. 1.6.1 [53], was used to evaluate effective population size changes using time-stamped cytb sequences. The molecular substitution model (HKY) was selected using the program jModelTest v. 0.1.1 based on the Akaike Information Criterion (AIC) [54]. A range of mutation rates (1.8–2.2% Myr) was employed with a generation time of 5 years, based on previous research [11], while allowing for a level of uncertainty. Sample dates were rounded to the nearest 50 years BP. We used two replicate runs with 50 × 106 iterations after a burn-in of 5 × 106 iterations. Genealogical likelihood and model parameters were sampled every 5000th iteration. Mixing and convergence were assessed in Tracer v. 1.3., using a minimum effective sample size of 100 from combined runs for parameter estimates. We also modelled a constant size coalescent tree and calculated Bayes factor to compare the support for the BSP with the constant size model [55].
The genetic effect of three effective population size scenarios was modelled in a coalescence framework using approximate Bayesian computation (ABC) [56]: (i) constant population size, (ii) a single bottleneck between 0 and 600 years BP and (iii) two bottlenecks, 0–300 years BP and 300–600 years BP, as implemented in diy-ABC v. 1.0.4.37 [57]. We estimated NE prior to any bottleneck (NS), after the first bottleneck (N1) and after the second bottleneck (N2), as well as the times of the first (t1) and second (t2) bottlenecks. One million simulations were obtained for each scenario and the number of haplotypes, pairwise number of haplotypes, number of segregating sites and pairwise FST values were recorded for each simulation. The different scenarios were compared by estimating the fit of 500 simulated datasets closest to the observed data (direct approach) and by logistic regression on the posterior probability of each scenario as predicted by the deviations between simulated and observed summary statistics, using 1% of the best-fitting datasets. Confidence in scenario choice was evaluated using the method of Cornuet et al. [58], using 500 simulated datasets under each scenario. Type I error was estimated as the proportion of instances when the supported true scenario had lower posterior probability than the alternative and type II error as the proportion of instances where the supported scenario was incorrectly chosen. The posterior distributions of population size and the timing of bottlenecks were estimated under the supported model using a local logistic regression on the 1% closest of 1.5 × 106 simulated datasets. To evaluate the performance of the estimation, we generated pseudo-observed datasets with known parameter values drawn from the posterior distribution of the supported scenario. The mean relative bias (MRB) for each parameter was estimated and averaged over the 500 datasets.
3. Results
(a). Age composition and genetic diversity
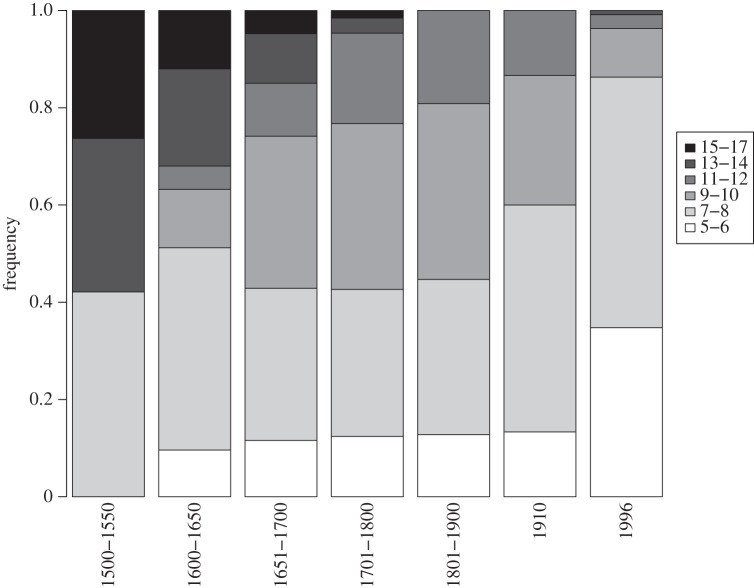
The mean age of the 1500–1550 sample was 13.14 (s.d. = 1.75) and 10.61 (s.d. = 2.11) years in the sample dated from 1910 (table 1 and figure 2). There was a significant decline of mean age over time (b = −0.458 (per 100 years), p < 0.001).
Table 1.
The age composition of Atlantic cod vertebrae across the five temporal groups, from 1500 to 1910. Mean age and standard deviation (s.d.) are in years and n presents sample size.
| n | mean | s.d. | |
|---|---|---|---|
| 1910 | 53 | 10.62 | 2.11 |
| 1700–1800 | 126 | 10.31 | 1.95 |
| 1650–1700 | 57 | 10.10 | 1.98 |
| 1600–1650 | 50 | 12.84 | 1.58 |
| 1500–1550 | 29 | 13.14 | 1.75 |
Figure 2.
Frequency of age cohorts estimated by growth ring counts of Atlantic cod vertebrae from each temporal group and previously published age distribution of catch from Icelandic cod fisheries in 1996 [59]. The counts were grouped to facilitate visual presentation. Individuals aged over 17 years were found only in the samples from 1500 to 1650.
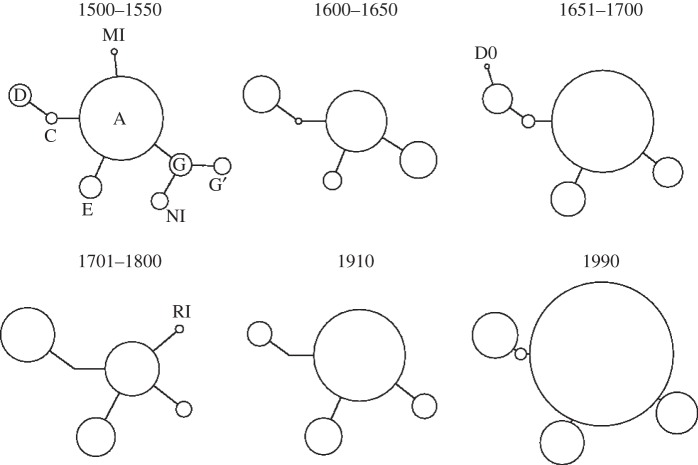
We successfully sequenced 258 bp of the 340 bp cytb target sequence of 156 historical samples (table 2). Ten haplotypes were observed from the historical samples. Two haplotypes had not been described in Atlantic cod from Iceland; a single individual with haplotype DO in the eighteenth century sample and a novel haplotype identified in three individuals from the 1500 to 1550 sample (GB: KF834568). The novel haplotype differed by a single base pair transition at site 14547 [46] from haplotype G. The G-clade [11] was more common in the sixteenth and seventeenth century samples than in more recent samples (figure 3). Haplotypic richness was highest in the earliest sample and decreased over time (p < 0.001, table 2). Pairwise FST values ranged from 0 to 0.053 and were significant among samples dated from before and after the sixteenth century (table 3). Population contraction/expansion was indicated by the mismatch distributions, except within the eighteenth century samples (see electronic supplementary material, figure S1). Tajima's D values ranged from −0.8 to 0.6 and were not significant. Allele frequency at the PanI locus was determined for 55 historical specimens. The frequency of the Pan I B allele was low in all samples (table 2).
Table 2.
Historical diversity in cytb and the PanI loci and data on cytb diversity from modern studies [11]. Sample size is indicated by n. Abbreviations denote the number of segregating sites (SS), number of haplotypes (H), haplotype diversity (HD), haplotypic richness based on 22 sequences (h22), nucleotide diversity (π), the frequency of the Pan IB allele (fB), observed (HO) and expected (HE) PanI heterozygosity.
|
cytb |
PanI |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | SS | H | HD | h22 | π | n | fB | HO | HE | |
| 1990 | 503 | 7 | 10 | 0.7 | 4.53 | 0.006 | — | — | — | — |
| 1910 | 23 | 4 | 4 | 0.64 | 4.00 | 0.005 | 14 | 0 | 0 | 0 |
| 1700–1800 | 22 | 4 | 5 | 0.74 | 5.00 | 0.007 | 12 | 0.042 | 0.080 | 0.083 |
| 1650–1700 | 50 | 4 | 6 | 0.71 | 5.24 | 0.006 | 16 | 0.031 | 0.025 | 0.061 |
| 1600–1650 | 26 | 4 | 5 | 0.74 | 4.84 | 0.007 | 10 | 0.050 | 0.100 | 0.095 |
| 1500–1550 | 36 | 7 | 9 | 0.80 | 7.13 | 0.008 | 3 | 0.167 | 0.333 | 0.277 |
Figure 3.
Median joining haplotype network of all haplotypes in each temporal group and the common haplotypes in the modern sample (1990). The haplotypes are represented by circles, the area being proportional to their frequencies. Each trace corresponds to a single mutation. Letters identify different haplotypes (electronic supplementary material, table S3).
Table 3.
Differentiation among temporal samples estimated by pairwise FST values based on haplotypic variation in cytb sequences. Asterisks denote a significant p-value, based on 10 000 permutations.
| 1990 | 1910 | 1700–1800 | 1650–1700 | 1600–1650 | |
|---|---|---|---|---|---|
| 1990 | |||||
| 1910 | 0.01 | ||||
| 1700–1800 | 0.00 | 0.01 | |||
| 1650–1700 | 0.01 | 0.00 | 0.00 | ||
| 1600–1650 | 0.01 | 0.00 | 0.00 | 0.017 | |
| 1500–1550 | 0.021* | 0.01 | 0.053* | 0.024* | 0.015 |
*p < 0.05
(b). Estimation of historical population size
The BSP suggested large changes in effective population size in historical times, most notably a rapidly growing medieval population with a severe bottleneck at around 1440 (figure 4). The NE estimate before the bottleneck was high at 440 000 individuals (95% CI 191 000–1 037 000). Subsequent population size fluctuations were smaller and NE estimates in modern times were low at 1955 individuals (95% CI 402–7550). A Bayes factor of −71.7 supported the BSP model over a model of constant size. The estimated substitution rate was 5.12 × 10−7 (95% highest posterior density interval: 1.05 × 10−8 − 8.33 × 10−6) substitutions per site per year.
Figure 4.
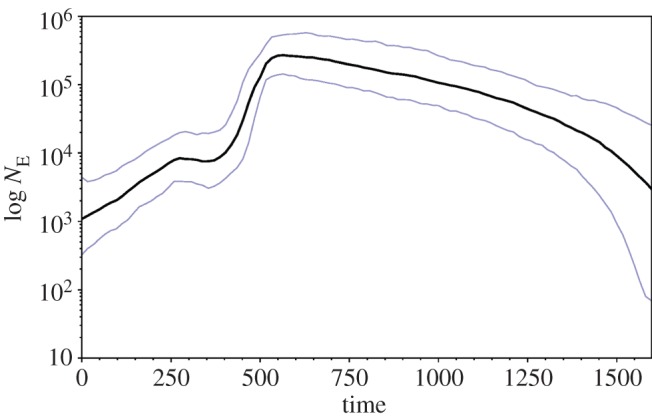
Effective female population size (NE) through time estimated with BSP on cytb variation. The skyride plots shows mean effective female population sizes (black line) and 95% highest posterior density intervals (blue lines). The time unit is calendar years before 1990. The population size axis is on a logarithmic scale.
The ABC estimation supported the scenario of two bottlenecks over the scenarios of either a single bottleneck or a constant population size with both the direct estimate and logistic regression (see electronic supplementary material, figure S2). The scenarios with bottleneck effects had high statistical support compared with constant population size, but the model with two bottlenecks was weakly supported. Estimated type I and type II error rates for the choice of a two bottleneck scenario compared with the constant size scenario were 44% and 46%, respectively, but much lower for single bottleneck scenario or 0.05% for both type I and type II errors. The median estimate of effective population size before the first bottleneck was 330 000 (95% quantile: 89 000–852 000) decreasing to 75 000 (95% quantile: 44 800–98 000) at the earlier bottleneck and to 21 300 (95% quantile: 3290–40 900) at the modern-day bottleneck. The time of the earlier bottleneck was estimated at 96.2 (95% quantile: 72.5–118.3) generations before the present (BP) (or between 1400 and 1630) and the modern bottleneck at 23.8 (95% quantile: 1.9–40.0) generations BP (or in the past 200 years), both dating from the samples collected at 1990. The bias (MRB) was low for all estimates of population size, Ns (1.002), N1 (0.411) and N2 (1.005), and for the bottleneck time estimates, t1 (0.002) and t2 (0.988).
4. Discussion
Our results indicate a large, growing Atlantic cod population during the medieval warm period and a marked population size bottleneck in the fifteenth or early sixteenth century with concomitant loss of haplotype variation. Fish age declines over the study period, with a marked drop in the mid-seventeenth century.
The historical population size estimates are extreme compared with modern-day values but may be interpreted in the context of historical documents that claim vast numbers of cod in the North Atlantic at the height of the European fisheries expansion to Iceland and Newfoundland [31,32]. The scale of the early bottleneck observed in this study and the concomitant loss of genetic variation may be consistent with large-scale ecological change. Summer temperatures in Icelandic waters decreased by 2°C in the late fourteenth and early fifteenth centuries as minimal summer temperatures reached 4°C [38]. Written sources from the same period describe icebergs [37], unusually cold weather and low fish catches [30]. This period, the onset of ‘the little ice age’, immediately precedes the large-scale reduction in NE indicated by the current data and we conclude that the two may be linked, although without any assumptions of causality. Previous research on Atlantic cod growth using otoliths from archaeological excavations in Norway showed disruptions of growth patterns in the early sixteenth century, best explained by a significant drop in sea temperatures [60]. Although adult Atlantic cod are robust to a range of temperatures [61], early life stages are dependent on sea currents and the favourable timing of algae blooms and zooplankton availability [62,63], where the latter may be affected by even moderate changes in sea temperature [64–66]. The North Atlantic climate oscillations in medieval times provide the best current proxy for historical environmental change [67] and may have prompted unknown ecosystem changes, e.g. climatic events have been linked to severe changes in ocean circulation and currents [68,69].
Extensive fishing of the Icelandic cod stock started as early as the thirteenth century [23], and previous studies have suggested that historical fisheries affected the Atlantic cod stock [21,34,70]. Although the total catches of historical fishing fleets are difficult to estimate, historical documents refer to multiple fleets of up to 500 English and Dutch long liners fishing simultaneously in Icelandic waters in the sixteenth century [71]. Modern-day research finds that early signs of intensive fisheries included changes in the age composition of stock [44]. There was a truncation in mean age in our samples that started in the seventeenth century and continued into modern times (figure 2). Although these correlations may not imply causation, they correspond to modern studies on fisheries effects [44]. However, our samples represent the age of the fished stock rather than the population demography and the lack of younger individuals in our earliest samples does suggest that the early fisheries targeted older, larger fish (figure 2). As the decline in age occurred later than the early bottleneck, our data do not particularly support the hypothesis that historical fishing pressure influenced the population collapse.
Fisheries pose strong predation pressure on fish populations and may cause evolutionary change [72]. Two recent studies indicate fisheries effects on the relative frequencies of inshore (Pan IAA) and offshore (Pan IBB and Pan IAB) genotypes in the Icelandic Atlantic cod population [9,18]. The frequency of the offshore Pan I B allele in the historical samples is low (table 2) in comparison with samples from modern-day inshore spawning stock [9]. These results could be biased by the development of fisheries and fishing gear throughout the period although, based on the geography and expected range of local fishing boats, it is probably that the current samples represent inshore cod. Thus, we tentatively conclude that the B-allele was at low frequency in the inshore stock during historical times, supporting a reduction in inshore allele frequencies in the twentieth century. Higher frequency of offshore alleles in the inshore stock after the onset of industrial fisheries may reflect changes in the target groups of the fishing industry but alternatively reflect a change in the relative proportions of inshore and offshore populations owing to greater fishing pressure on inshore stock. Given the low number of samples currently analysed, further studies are required to reach a firm conclusion on temporal changes in PanI allele frequencies.
Genetic estimates of population size assume an isolated population and are biased by gene flow or population admixture [73], which could effect the current results. The distribution range of Atlantic cod in the North Atlantic is dependent on temperature [13] and the genetic structure can be fluid: for example, some spawning populations in Greenland show genetic continuity over several decades, while others appear to have been replaced by neighbouring stock [74]. The Icelandic cod population is genetically structured by geography and depth [6], although the structure is weak or absent for neutral markers, and not detected by mtDNA markers [11]. Cod feeding migrations [75] are independent of genetic structure but could bias the historical samples. In our earlier samples, we observe a single individual with a haplotype currently found in low frequency in the Faroe Islands. This could be consistent with a historically panmictic population or increased structure at the present day. However, the reduction in haplotype richness, loss of rare haplotypes and concomitant increase in the frequency of intermediate haplotypes after 1550 concur with a population size bottleneck [76]. Distinguishing among these factors can only be speculative based on the current data but highlights the potential of archaeological material for understanding historical ecology.
The current results have both theoretical and applied implications irrespective of the cause of the population size changes. Genetic patterns of low nucleotide diversity and singleton polymorphism, characteristic of the cytb of Atlantic cod in modern times, indicate low NE and low NE/NC ratios [11,77] that may have resulted from the severe bottleneck and loss of haplotypic variation in the sixteenth century. This may result in shallow genealogies with multiple merging coalescent events complicating reconstructions of population evolutionary history [78,79]. Moreover, the large discrepancy between current and historical NE and the possible correlation with sea temperature raise questions about the mechanisms underlying fluctuations of NE/NC ratios in Atlantic cod. The NE/NC ratio affects harvesting tolerance, resilience to environmental change and adaptability [16] and has been found to depend on both environmental factors and population size in several other species [80–82].
To conclude, we estimate high historical NE with a marked bottleneck in the fifteenth or early sixteenth century and continuing low NE at modern times. Although causality is difficult to assume, we find support for both climate- and fisheries-induced change. The current results identify several lines of future studies, including: (i) the estimation of historical migration/gene flow and/or admixture to investigate the high historical effective population estimates from cytb by including older samples or samples from other regions, (ii) the changes in PanI frequency over time from inshore/offshore comparisons and (iii) concurrent shifts in population size in other Atlantic regions inferred from mtDNA or other genetic markers.
Acknowledgements
We thank Tom McGovern, Lilja Björk Pálsdóttir and Frank Feeley for providing some of the samples used.
Data accessibility
Genbank accession numbers and haplotype frequencies can be found in electronic supplementary material, table S3.
Funding statement
This study was funded by grants from the Icelandic Ministry of Fisheries Research Fund to G.Á.Ó., R.E. and S.P.
References
- 1.Jamieson A, Jónsson J. 1971. The Greenland component of spawning cod at Iceland. Rapports et Procèsverbaux des Réunions du Conseil International pour l'Exploration de la Mer 161, 65–71 [Google Scholar]
- 2.Williams GC. 1975. Sex and evolution. Princeton, NJ: Princeton University Press [Google Scholar]
- 3.Ruzzante DE, Taggart CT, Cook D, Goddard S. 1996. Genetic differentiation between inshore and offshore Atlantic cod (Gadus morhua) off Newfoundland: microsatellite DNA variation and antifreeze level. Can. J. Fish. Aquat. Sci. 53, 634–645 (doi:10.1139/f95-228) [Google Scholar]
- 4.Nielsen EE, Hansen MM, Meldrup D. 2006. Evidence of microsatellite hitch-hiking selection in Atlantic cod (Gadus morhua L.): implications for inferring population structure in nonmodel organisms. Mol. Ecol. 15, 3219–3229 (doi:10.1111/j.1365-294X.2006.03025.x) [DOI] [PubMed] [Google Scholar]
- 5.Nielsen E, Hemmer-Hansen J, Poulsen N, Loeschcke V, Moen T, Johansen T, Carvalho G. 2009. Genomic signatures of local directional selection in a high gene flow marine organism; the Atlantic cod (Gadus morhua). BMC Evol. Biol. 9, 276 (doi:10.1186/1471-2148-9-276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pampoulie C, Ruzzante DE, Chosson V, Jörundsdóttir TD, Taylor L, Thorsteinsson V, Daníelsdóttir AK, Marteinsdóttir G. 2006. The genetic structure of Atlantic cod Gadus morhua around Iceland: Insights from microsatellites, the Pan I locus, and tagging experiments. Can. J. Fish. Aquat. Sci. 63, 2660–2674 (doi:10.1139/f06-150) [Google Scholar]
- 7.Pogson GH, Mesa KA, Boutilier RG. 1995. Genetic population structure and gene flow in the Atlantic cod Gadus morhua: a comparison of allozyme and nuclear RFLP loci. Genetics 139, 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pogson GH, Fevolden SE. 2003. Natural selection and the genetic differentiation of coastal and Arctic populations of the Atlantic cod in northern Norway: a test involving nucleotide sequence variation at the pantophysin (PanI) locus. Mol. Ecol. 12, 63–74 (doi:10.1046/j.1365-294X.2003.01713.x) [DOI] [PubMed] [Google Scholar]
- 9.Árnason E, Hernandez UB, Kristinsson K. 2009. Intense habitat-specific fisheries-induced selection at the molecular PanI locus predicts imminent collapse of a major cod fishery. PLoS ONE 4, e5529 (doi:10.1371/journal.pone.0005529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hemmer-Hansen J, Nielsen EE, Therkildsen NO, Taylor MI, Ogden R, Geffen AJ, Carvalho GR. 2013. A genomic island linked to ecotype divergence in Atlantic cod. Mol. Ecol. 22, 2653–2667 (doi:10.1111/mec.12284) [DOI] [PubMed] [Google Scholar]
- 11.Árnason E. 2004. Mitochondrial cytochrome b DNA variation in the high-fecundity Atlantic cod: trans-Atlantic clines and shallow gene genealogy. Genetics 166, 1871–1885 (doi:10.1534/genetics.166.4.1871) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carr SM, Marshall HD. 1991. Detection of intraspecific DNA sequence variation in the mitochondrial cytochrome b gene of Atlantic cod (Gadus morhua) by the polymerase chain reaction. Can. J. Fish. Aquat. Sci. 48, 48–52 (doi:10.1139/f91-007) [Google Scholar]
- 13.Carr SM, Marshall HD. 2008. Intraspecific phylogeographic genomics from multiple complete mtDNA genomes in Atlantic cod (Gadus morhua): origins of the ‘codmother,’ transatlantic vicariance and midglacial population expansion. Genetics 180, 381–389 (doi:10.1534/genetics.108.089730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hedgecock D, Pudovkin AI. 2011. Sweepstakes reproductive success in highly fecund marine fish and shellfish: a review and commentary. Bull. Mar. Sci. 87, 971–1002 (doi:10.5343/bms.2010.1051) [Google Scholar]
- 15.Palstra FP, Fraser DJ. 2012. Effective/census population size ratio estimation: a compendium and appraisal. Ecol. Evol. 2, 2357–2365 (doi:10.1002/ece3.329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedgecock D. 1994. Temporal and spatial genetic structure of marine animal populations in the California Current. Calif. Cooper. Oceanic Fish. Invest. Rep. 35, 73–81 [Google Scholar]
- 17.Nielsen EE, Hansen MM. 2008. Waking the dead: the value of population genetic analyses of historical samples. Fish Fish. 9, 450–461 (doi:10.1111/j.1467-2979.2008.00304.x) [Google Scholar]
- 18.Jakobsdóttir KB, Pardoe H, Magnússon Á, Björnsson H, Pampoulie C, Ruzzante DE, Marteinsdóttir G. 2011. Historical changes in genotypic frequencies at the Pantophysin locus in Atlantic cod (Gadus morhua) in Icelandic waters: evidence of fisheries-induced selection? Evol. Appl. 4, 562–573 (doi:10.1111/j.1752-4571.2010.00176.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwamoto EM, Myers JM, Gustafson RG. 2012. Resurrecting an extinct salmon evolutionarily significant unit: archived scales, historical DNA and implications for restoration. Mol. Ecol. 21, 1567–1582 (doi:10.1111/j.1365-294X.2011.05419.x) [DOI] [PubMed] [Google Scholar]
- 20.Perdikaris S. 1998. The transition to a commercial economy: Lofoten fishing in the Middle ages, a preliminary report. Anthropozoologica 25, 505–510 [Google Scholar]
- 21.Barrett JH, Locker AM, Roberts CM. 2004. The origins of intensive marine fishing in medieval Europe: the English evidence. Proc. R. Soc. Lond. B 271, 2417–2421 (doi:10.1098/rspb.2004.2885) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amundsen C, et al. 2005. Fishing booths and fishing strategies in medieval Iceland: an Archaeofauna from the of Akurvík, North-West Iceland. Environ. Arch. 10, 141–198 [Google Scholar]
- 23.Edvardsson R. 2010. The role of marine resources in the medieval economy of Vestfirðir, Iceland. PhD thesis City University of New York, New York, NY, USA [Google Scholar]
- 24.Luikart G, Ryman N, Tallmon DA, Schwartz MK, Allendorf FW. 2010. Estimation of census and effective population sizes: the increasing usefulness of DNA-based approaches. Conserv. Gen. 11, 355–373 (doi:10.1007/s10592-010-0050-7) [Google Scholar]
- 25.Rodrigo AG, Felsenstein J. 1999. Coalescent approaches to HIV-1 population genetics. In Coalescent approaches to HIV population genetics (ed. Crandall K.), pp. 233–272 Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- 26.Chan YL, Anderson CN, Hadly EA. 2006. Bayesian estimation of the timing and severity of a population bottleneck from ancient DNA. PLoS Genet. 2, e59 (doi:10.1371/journal.pgen.0020059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campos PF, et al. 2010. Ancient DNA sequences point to a large loss of mitochondrial genetic diversity in the saiga antelope (Saiga tatarica) since the Pleistocene. Mol. Ecol. 19, 4863–4875 (doi:10.1111/j.1365-294X.2010.04826.x) [DOI] [PubMed] [Google Scholar]
- 28.Alter SE, Newsome SD, Palumbi SR. 2012. Pre-whaling genetic diversity and population ecology in Eastern pacific gray whales: insights from ancient DNA and stable isotopes. PLoS ONE 7, e35039 (doi:10.1371/journal.pone.0035039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marine Research Institute 2009. State of Marine Stocks in Icelandic Waters 2008/2009. Prospects for the Quota Year 2009/2010. Marine Research Technical Report. Marine Research Institute, Reykjavik, Iceland [Google Scholar]
- 30.Annales Islandici I. 1924 1, pp. 24, 145 & 187. Reykjavík, Iceland: Hið Íslenska bókmenntafélag. http://handrit.is/en/manuscript/view/is/AM04-0420a.
- 31.Cell GT. 1969. English Enterprise in Newfoundland 1577 – 1660. Toronto, Canada: University of Toronto Press [Google Scholar]
- 32.Þorsteinsson B. 1976. Tíu Þorskastríð, 1415 – 1976. Reykjavík, Iceland: Sögufélagið [Google Scholar]
- 33.Þorsteinsson B. 1970. Enska öldin í sögu íslendinga. Reykjavík, Iceland: Mál og Menning [Google Scholar]
- 34.Jackson JB, Kirby MX, Berger WH, Bjorndal KA, Botsford LW, Bourque BJ, Warner RR. 2001. Historical overfishing and the recent collapse of coastal ecosystems. Science 293, 629–637 (doi:10.1126/science.1059199) [DOI] [PubMed] [Google Scholar]
- 35.Amorosi T, McGovern TH, Perdikaris S. 1994. Bioarchaeology and cod fisheries: a new source of evidence . ICES J. Mar. Sci. 198, 31–48 [Google Scholar]
- 36.Gunnarsson G. 1983. Monopoly trade and economic stagnation. Studies in the foreign trade of Iceland 1602–1787. Lund: Skrifter Utgivna av Ekonomisk-Historiska Föreningen 38 [Google Scholar]
- 37.Ogilvie AE, Jónsson T. 2001. ‘Little Ice Age’ research: a perspective from Iceland. Clim. Change 48, 9–52 (doi:10.1023/A:1005625729889) [Google Scholar]
- 38.Patterson WP, Dietrich KA, Holmden C, Andrews JT. 2010. Two millennia of North Atlantic seasonality and implications for Norse colonies. Proc. Natl Acad. Sci. USA 107, 5306–5310 (doi:10.1073/pnas.0902522107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller GH, Geirsdóttir Á, Zhong Y, Larsen DJ, Otto-Bliesner BL, Holland MM, Thordarson T. 2012. Abrupt onset of the Little Ice Age triggered by volcanism and sustained by sea-ice/ocean feedbacks. Geophys. Res. Lett. 39, L02708 (doi:10.1029/2011GL050168) [Google Scholar]
- 40.Patterson WP. 2013. Climate change in the north atlantic. In Histories from the north: environments, movements, and narratives (eds Ziker JP, Stammler F.), pp. 139–143 Dubuque, IA: Kendall Hunt Publishing Company [Google Scholar]
- 41.O'Brian CM, Fox CJ, Planque B, Casey J. 2000. Climate variability and North Sea cod. Nature 404, 142 (doi:10.1038/35004654) [DOI] [PubMed] [Google Scholar]
- 42.Planque B, Frédou T. 1999. Temperature and the recruitment of Atlantic cod (Gadus morhua). Can. J. Fish. Aquat. Sci. 56, 2069–2077 (doi:10.1139/f99-114) [Google Scholar]
- 43.Mourier T, Ho SY, Gilbert MTP, Willerslev E, Orlando L. 2012. Statistical guidelines for detecting past population shifts using ancient DNA. Mol. Biol. Evol. 29, 2241–2251 (doi:10.1093/molbev/mss094) [DOI] [PubMed] [Google Scholar]
- 44.Trippel EA. 1995. Age at maturity as a stress indicator in fisheries. Bioscience 45, 759–771 (doi:10.2307/1312628) [Google Scholar]
- 45.Pauly D, Christensen V, Dalsgaard J, Froese R, Torres F. 1998. Fishing down marine food webs. Science 279, 860–863 (doi:10.1126/science.279.5352.860) [DOI] [PubMed] [Google Scholar]
- 46.Johansen S, Bakke I. 1996. The complete mitochondrial DNA sequence of Atlantic cod (Gadus morhua): relevance to taxonomic studies among codfishes. Mol. Mar. Biol. Biotech. 5, 203–214 [PubMed] [Google Scholar]
- 47.Paradis E. 2010. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26, 419–420 (doi:10.1093/bioinformatics/btp696) [DOI] [PubMed] [Google Scholar]
- 48.R Core Team 2012. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0/. See http://www.r-project.org/.
- 49.Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (doi:10.1093/bioinformatics/btp187) [DOI] [PubMed] [Google Scholar]
- 50.Petit RJ, El Mousadik A, Pons O. 1998. Identifying populations for conservation on the basis of genetic markers. Conserv. Biol. 12, 844–855 (doi:10.1046/j.1523-1739.1998.96489.x) [Google Scholar]
- 51.Goudet J. 2005. Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 5, 184–186 (doi:10.1111/j.1471-8286.2004.00828.x) [Google Scholar]
- 52.Minin VN, Bloomquist EW, Suchard MA. 2008. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol. Biol. Evol. 25, 1459–1471 (doi:10.1093/molbev/msn090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (doi:10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 (doi:10.1093/molbev/msn083) [DOI] [PubMed] [Google Scholar]
- 55.Suchard MA, Weiss RE, Sinsheimer JS. 2001. Bayesian selection of continuous-time Markov chain evolutionary models. Mol. Biol. Evol. 18, 1001–1013 (doi:10.1093/oxfordjournals.molbev.a003872) [DOI] [PubMed] [Google Scholar]
- 56.Beaumont MA, Zhang WY, Balding DJ. 2002. Approximate Bayesian computation in population genetics. Genetics 162, 2025–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cornuet JM, Santos F, Beaumont MA, Robert CP, Marin JM, Balding DJ, Estoup A. 2008. Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics 24, 2713–2719 (doi:10.1093/bioinformatics/btn514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cornuet JM, Ravigné V, Estoup A. 2010. Inference on population history and model checking using DNA sequence and microsatellite data with the software DIYABC (v1.0). BMC Bioinform. 11, 401 (doi:10.1186/1471-2105-11-401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marine Research Institute 1996. Marine Research Technical Report no 54. Marine Research Institute, Reykjavik: See http://www.hafro.is/Bokasafn/Timarit/fjolrit-054.pdf [Google Scholar]
- 60.Geffen AJ, Høie H, Folkvord A, Hufthammer AK, Andersson C, Ninnemann U, Nedreaas K. 2011. High-latitude climate variability and its effect on fisheries resources as revealed by fossil cod otoliths. ICES J. Mar. Sci. 68, 1081–1089 (doi:10.1093/icesjms/fsr017) [Google Scholar]
- 61.Jakobsson J, Stefánsson G. 1998. Rational harvesting of the cod–capelin–shrimp complex in the Icelandic marine ecosystem. Fish. Res. 37, 7–21 (doi:10.1016/S0165-7836(98)00123-4) [Google Scholar]
- 62.Pepin P. 1991. Effect of temperature and size on development, mortality, and survival rates of the pelagic early life history stages of marine fish. Can. J. Fish. Aquat. Sci. 48, 503–518 (doi:10.1139/f91-065) [Google Scholar]
- 63.Drinkwater KF. 2005. The response of Atlantic cod (Gadus morhua) to future climate change. ICES J. Mar. Sci. 62, 1327–1337 (doi:10.1016/j.icesjms.2005.05.015) [Google Scholar]
- 64.Edwards M, Richardson AJ. 2004. Impact of climate change on marine pelagic phenology and trophic mismatch. Nature 430, 881–884 (doi:10.1038/nature02808) [DOI] [PubMed] [Google Scholar]
- 65.Hays GC, Richardson AJ, Robinson C. 2005. Climate change and marine plankton. Trends Ecol. Evol. 20, 337–344 (doi:10.1016/j.tree.2005.03.004) [DOI] [PubMed] [Google Scholar]
- 66.Richardson AJ. 2008. In hot water: zooplankton and climate change. ICES J. Mar. Sci. 65, 279–295 (doi:10.1093/icesjms/fsn028) [Google Scholar]
- 67.Soon W, Baliunas S. 2003. Proxy climatic and environmental changes of the past 1000 years. Clim. Res. 23, 89–110 (doi:10.3354/cr023089) [Google Scholar]
- 68.Bianchi GG, McCave IN. 1999. Holocene periodicity in North Atlantic climate and deep-ocean flow south of Iceland. Nature 397, 515–517 (doi:10.1038/17362) [Google Scholar]
- 69.Rahmstorf S. 2003. Timing of abrupt climate change: a precise clock. Geophys. Res. Lett. 30, 1510 (doi:10.1029/2003GL017115) [Google Scholar]
- 70.Starkey DJ, Holm P, Barnard M. (eds). 2008. Oceans past: management insights from the history of marine animal populations. London, UK: Earthscan [Google Scholar]
- 71.Annales Islandici III. 1933 1, p. 71. Reykjavík, Iceland: Hið Íslenska bókmenntafélag. http://handrit.is/en/manuscript/view/is/JS02-0039.
- 72.Dunlop ES, Enberg K, Jørgensen C, Heino M. 2009. Editorial: toward Darwinian fisheries management. Evol. App. 2, 245–259 (doi:10.1111/j.1752-4571.2009.00087.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang J, Whitlock MC. 2003. Estimating effective population size and migration rates from genetic samples over space and time. Genetics 163, 429–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Therkildsen NO, et al. 2013. Spatiotemporal SNP analysis reveals pronounced biocomplexity at the northern range margin of Atlantic cod Gadus morhua. Evol. App. 6, 690–705 (doi:10.1111/eva.12055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Robichaud D, Rose GA. 2004. Migratory behaviour and range in Atlantic cod: inference from a century of tagging. Fish Fish. 5, 185–214 (doi:10.1111/j.1467-2679.2004.00141.x) [Google Scholar]
- 76.Luikart G, Cornuet JM. 1998. Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conserv. Biol. 12, 228–237 (doi:10.1046/j.1523-1739.1998.96388.x) [Google Scholar]
- 77.Knutsen H, Olsen EM, Jorde PE, Espeland SH, Andre C, Stenseth NC. 2011. Are low but statistically significant levels of genetic differentiation in marine fishes ‘biologically meaningful’? A case study of coastal Atlantic cod. Mol. Ecol. 20, 768–783 (doi:10.1111/j.1365-294X.2010.04979.x) [DOI] [PubMed] [Google Scholar]
- 78.Pitman J. 1999. Coalescents with multiple collisions. Ann. Prob. 27, 1870–1902 (doi:10.1214/aop/1022677552) [Google Scholar]
- 79.Eldon B, Wakeley J. 2006. Coalescent processes when the distribution of offspring number among individuals is highly skewed. Genetics 172, 2621–2633 (doi:10.1534/genetics.105.052175) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ardren WR, Kapuscinski AR. 2003. Demographic and genetic estimates of effective population size (Ne) reveals genetic compensation in steelhead trout. Mol. Ecol. 12, 35–49 (doi:10.1046/j.1365-294X.2003.01705.x) [DOI] [PubMed] [Google Scholar]
- 81.Fraser DJ, Hansen MM, Østergaard S, Tessier N, Legault M, Bernatchez L. 2007. Comparative estimation of effective population sizes and temporal gene flow in two contrasting population systems. Mol. Ecol. 16, 3866–3889 (doi:10.1111/j.1365-294X.2007.03453.x) [DOI] [PubMed] [Google Scholar]
- 82.Belmar-Lucero S, Wood JL, Scott S, Harbicht AB, Hutchings JA, Fraser DJ. 2012. Concurrent habitat and life history influences on effective/census population size ratios in stream-dwelling trout. Ecol. Evol. 2, 562–573 (doi:10.1002/ece3.196) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genbank accession numbers and haplotype frequencies can be found in electronic supplementary material, table S3.