Abstract
Inorganic polyphosphate (PolyP) is a biological polymer that plays important roles in the cell physiology of both prokaryotic and eukaryotic organisms. Among the available methods for PolyP localization and quantification, a 4’,6-diamidino-2-phenylindole(DAPI)-based assay has been used for visualization of PolyP-rich organelles. Due to differences in DAPI permeability to different compartments and/or PolyP retention after fixation, a general protocol for DAPI-PolyP staining has not yet been established. Here, we tested different protocols for DAPI-PolyP detection in a range of samples with different levels of DAPI permeability, including subcellular fractions, free-living cells and cryosections of fixed tissues. Subcellular fractions of PolyP-rich organelles yielded DAPI-PolyP fluorescence, although those with a complex external layer usually required longer incubation times, previous aldehyde fixation and/or detergent permeabilization. DAPI-PolyP was also detected in cryosections of OCT-embedded tissues analyzed by multiphoton microscopy. In addition, a semi-quantitative fluorimetric analysis of DAPI-stained fractions showed PolyP mobilization in a similar fashion to what has been demonstrated with the use of enzyme-based quantitative protocols. Taken together, our results support the use of DAPI for both PolyP visualization and quantification, although specific steps are suggested as a general guideline for DAPI-PolyP staining in biological samples with different degrees of DAPI and PolyP permeability.
Key words: DAPI, polyphosphate, fluorescence, fluorimetry
Introduction
Inorganic polyphosphate (PolyP) is a widely distributed biological polymer composed of phosphate residues linked by high-energy phosphoanhydride bonds, and found in every tested organism.1 Several biological functions have been described for PolyP, including a role as phosphate reservoir, metal homeostasis, regulation of transcription factors and transcription fidelity, as well as regulation of several enzymatic activities.2 More recently, studies on the role of PolyP in vertebrate models revealed the participation of PolyP stores in platelet activation and intrinsic coagulation,3-6 cancer metastasis,7,8 activation of the fibroblast growth factor (FGF) signaling and a role in stem cell differentiation.9,10
Although extensively demonstrated in several subcellular compartments such as nuclei, mitochondria, as well as in the cytoplasm of different cells, millimolar concentrations of PolyP are commonly found inside PolyP-rich organelles. A wide variety of PolyP-rich organelles has been described, but recent studies have pointed that at least some of these compartments probably derive from a common evolutionary step and are similar to the protozoan acidocalcisome,11 the best studied PolyP-rich organelle. It has been suggested that PolyP stored in acidocalcisomes have a role in protozoan virulence12,13 as well as in the maintenance of osmotic balance14 and metal homeostasis.15 Together with protozoan acidocalcisomes, bacterial volutin granules are also considered one of the most studied types of PolyP-rich structures. Although volutin granules have been known for a longer period, it was only after the first description of the acidocalcisome in trypanosomes that the concept that this organelle might have a membrane unit containing specific transporters was established in bacteria.16,17 After these initial studies, several other organisms were shown to contain PolyP-rich acidocalcisome-like organelles, including eggs of invertebrates such as insects18,19 and sea urchin,20 and vertebrates (chicken),21 models where the isolation of subcellular fractions of acidocalcisomes was readily demonstrated.
The significant biological relevance of PolyP in cell metabolism has lead to the establishment of different protocols for PolyP visualization and quantification, an important step towards a better comprehension of the roles played by this polymer in different biological models. In this regard, the use of 4’,6-diamidino-2-phenylindole (DAPI), first developed as a trypanocide agent and later largely used for the detection of nucleic acids,22 has progressively become a widely accepted tool for the detection of PolyP-rich compartments. When used as a nucleic acid sensor, both in spectrofluorimetric and microscopic analysis, DAPI-DNA complexes exhibit a maximum fluorescence emission at around 450 nm (blue), whereas when bound to PolyP, a bathochromic shift in the emission to 525-550 nm take place, leading to a greenish-yellowish color.23 DAPI-PolyP interaction was shown to be remarkably specific and to generate a proper quantum yield for microscopic observation.21,24-26 Nevertheless, reports concerning the feasibility of using DAPI as a PolyP probe have remained fragmented among several publications that suggested a number of possible routines for DAPI-PolyP labeling and visualization.23,27,28
Information available in most catalogs and product data sheet classify DAPI (different varieties) as compounds slightly permeable to cell membranes, although it has been generally recognized that it may permeate cell membranes less efficiently in living cells, making them less susceptible to DAPI-staining.29 DAPI permeability to cell walls has been reported,23 although not systematically studied in different cell types. The degree of permeability presumably depends on the structural characteristics of the different cell surfaces in different organisms. In this regard, although a few studies have shown the spectroscopic properties of analytical grade sodium PolyP-DAPI fluorescence for quantification purposes,28,30 quantitative analysis of PolyP mobilization using DAPI as a probe in biological samples has not yet been reported. In the present study, we evaluated the effect of different routines of DAPI incubation in subcellular fractions, whole cells (containing or not cell walls), and OCT-embedded semi-thin sections in order to ascertain a unified guideline for PolyP labeling and detection. A semi-quantitative fluorimetric assay for the analysis of PolyP mobilization in PolyP-enriched extracts obtained from biological samples is also shown and provides a fast protocol for PolyP quantification
Materials and Methods
Animals, cell culture and strains
Trypanosoma cruzi epimastigote forms (Y strain) were grown axenically in liver infusion tryptose (LIT) medium supplemented with 10% fetal calf serum as previously described.31 Cells were collected by centrifugation at 350 g after four days of cultivation. Trypanosoma brucei procyclic forms (strain 427) were maintained at 27-28°C in SDM-79 medium supplemented with 10% fetal bovine serum.32 Cells were harvested by centrifugation at 350 g after two days of cultivation.
The obtainment of Eimeria spp oocysts from chickens was approved by the Ethics Committee of the Biology Institute of the State University of Campinas (UNICAMP) under the protocol number 1084-1, according to the Brazilian federal law (11.794/2008, Decree n. 6.899/2009) that is based on the Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences, USA, and the Australian Code of Practice for Care and Use of Animal for Scientific Purpose. All animals received humane care in compliance with the above-mentioned guides used by the Ethics Committee to approve the protocol. Briefly, four-week-old chickens were inoculated with 1x104 sporulated oocysts of Eimeria acervulina (Cu strain) and 5×103 Eimeria tenella (Pa strain) oocysts, as previously described.33 Oocysts were isolated from feces and sporulated after one week of inoculation. Sporozoites were obtained from oocysts by mechanic rupture, enzymatic treatment, and purification by anion-exchange chromatography using DE-52 cellulose columns.
Euglena gracilis wild strain was cultivated axenically at room temperature and constant illumination as previously described.34 Rhodnius prolixus Stahl, 1859 (Hemiptera, Reduviidae) were reared in a colony maintained at 28°C and 70-80% relative humidity. The insects were fed with rabbit blood in an artificial apparatus as described before.35 Total egg homogenates (TEH) were prepared by disrupting the laid eggs and a fraction enriched in acidocalcisomes was obtained as described by Ramos et al.19 The final pellet was used as acidocalcisome-enriched fraction (acidocalcisome fraction). Anticarsia gemmatalis specimens were obtained as described;36 briefly, a colony was kept about 27°C and 70% relative humidity. Adults were maintained in a plastic cage and paper sheets were added for eggs deposition. After 24 h, eggs were transferred to a plastic box and left for egg hatching and larvae development. Larvae were fed as previously described,37 until they reached the fifth instar around the 10th or 11th day after hatching as detected by visual inspection.
PolyP staining in whole cells
For PolyP detection, living cells were incubated with 50 μg/mL DAPI for 30 min, or different times where indicated, at room temperature. Alternatively, Eimeria sporocysts were first fixed with 4% formaldehyde for 5-20 min at room temperature and permeabilized with 0.3% Triton X-100 for 5 min prior to DAPI incubation. Mechanically disrupted sporocysts were also prepared as described.33 Following DAPI incubation, samples were mounted on glass slides and observed with a Zeiss Axioplan epifluorescence microscope using a personalized filter set with a bandpass excitation maximum at 350 nm and a 500 nm long pass emission filter, combined with the appropriate dichroic mirror or a FITC-optimized filter set. Following image acquisition, deconvolution was performed using a no-neighbor algorithm.
Isolation of PolyP-rich organelles and PolyP staining
Acidocalcisome-like organelles from Rhodnius prolixus were fractionated and enriched as described.19,21,38 Isolated acidocalcisomes were incubated with 2-10 μg/mL DAPI at room temperature, mounted on a glass slides and observed under a Zeiss Axioplan epifluorescence microscope using a personalized filter set as above.
Midgut optimum cutting temperature embedding and PolyP staining in semi-thin sections
Midguts from 5th instar larvae of the insect Anticarsia gemmatalis were dissected and fixed for 3 h at room temperature using 4% formaldehyde, 0.1% glutaraldehyde, 0.1 M sodium cacodylate pH 7.2. The tissue was cryoprotected in 10% sucrose overnight and in 30% sucrose for 24 h before Tissue-Tek optimum cutting temperature (OCT) immersion and freezing in LN2. Semi-thin sections were obtained in a cryostat and left at room temperature to dry for 60 min, then incubated with 0.1 μg/mL DAPI, washed with PBS, and mounted on slides using n-propyl gallate. The sections were observed under a Zeiss Axioplan epifluorescence microscope using a fluorescein filter set. After image acquisition, deconvolution was performed using a no-neighbor algorithm. Alternatively, samples were observed with a Zeiss LSM 510 Meta NLO multiphoton microscope using a two photon fluorescence excitation of 780 nm. A wavelength scan was performed in order to detect the most intense fluorescence wavelengths.
Fluorimetric analysis of PolyP
Time-lapse analysis of PolyP fluorescence was evaluated by washing cell suspensions with PBS and incubating with 100 μg/mL DAPI. After addition of DAPI, samples were immediately transferred to a microplate reader and the fluorescence intensity was registered over the course of 60 min, using excitation/emission wavelength of 350/450 (nucleic acid signal) and 350/550 (PolyP signal). Water-soluble PolyP fractions (WSF) were prepared from Periplaneta americana and R. prolixus eggs as described26 and both WSF, sodium phosphate glass - PolyP75+ (Sigma Aldrich, St. Louis, MO, USA) – and plasmidial DNA were incubated with 20 μg/mL DAPI for 30 min at room temperature. Samples were analyzed with a spectrofluorometer at the excitation wavelength of 354 nm. Relative quantification of PolyP in R. prolixus eggs was performed as described.26 For comparison, PolyP samples were extracted as described and incubated with an excess of scPPX for 15 minutes at 37oC.39 Resulting Pi levels were analyzed with malachite green.
Results
PolyP-DAPI staining of subcellular isolated compartments and single cells
Subcellular fractions of PolyP-rich organelles were obtained as previously described19 and verified by electron microscopy analysis of whole mounts adhered to formvar-coated grids, showing the typical electrondense characteristic of acidocalcisomes (Figure 1A). Additional confirmation was obtained by electron-probe X-ray microanalysis where high amounts of phosphorus and oxygen, bound to a number of cations, were detected (Figure 1B). PolyP stores in these fractions were rapidly stained (2-3 min at room temperature) using a range of 2-10 μg/mL DAPI and observed using a filter set optimized for DAPI-PolyP visualization (Figure 1C). Similar results were obtained using acidocalcisome fractions isolated from other organisms, such as PolyP-rich granules of chicken eggs (data not shown) and are consistent with those obtained from iodixanol differential sedimentation from other models.19-21,25,40
Figure 1.
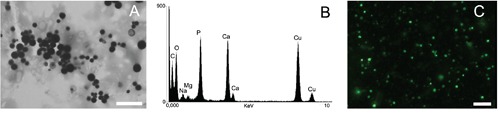
PolyP detection on subcellular stores in fraction of PolyP-rich organelles obtained from R. prolixus eggs. A) Electron micrograph where the stores appear as electron dense granules. B) X-ray microanalysis elemental composition. C) PolyP-rich granules after DAPI staining as observed by epifluorescence microscope with a long pass 500 nm emission filter. Scale bars: A, 2 μm; C, 2.5 μm.
DAPI-PolyP was also detected in whole cells of Trypanosoma cruzi (Figure 2A-B), a protozoan where both the size and number of T. cruzi PolyP-rich organelles (acidocalcisomes) are well known.41,42 Preparations of whole cell mounts seen in TEM showed electron dense organelles in the intracellular milieu (Figure 2A), presenting morphological similarities with the acidocalcisome-rich fractions depicted in Fig. 1A. X-ray microanalysis spectra confirmed the high content of phosphorus, oxygen and several cations (Figure 2A, inset), as previously described.43,44 When DAPI-PolyP staining was carried out in whole cells, higher concentrations of DAPI and longer incubation times were usually required. Incubation of intact cells with DAPI at the same conditions used for staining subcellular fractions (2-10 μg/mL or 5 min) did not result in satisfactory staining (data not shown). In contrast, 50 μg/mL DAPI for 10-20 min showed an intense staining pattern compatible with the presence and distribution of acidocalcisomes (Figure 2B) in these cells. This result suggested that the presence of external membranes or additional layers with different degrees of DAPI permeability may potentially diminish the penetration of the dye, therefore requiring slight modifications in the staining protocol. To verify whether these additional layers would actually interfere in PolyP-DAPI staining, we performed a PolyP detection assay under similar conditions in a range of unicellular organisms, i) containing plasma membrane with different properties, or ii) presenting additional cell or cyst walls. We first analyzed detection of PolyP stores in Euglena gracillis, known to present unique plasma membrane morphology supported by a dense fibrillar layer, the pellicular cortex.45 In Euglena cells, intracellular PolyP stores (acidocalcisome-like) were usually detected using higher concentrations of DAPI (50-200 μg/mL), following a 30 min incubation period at room temperature, without any further modifications in the protocol (Figure 3A). To verify whether the presence of a cell wall would diminish DAPI penetration into the cells, we incubated Candida albicans cells with different concentrations of DAPI and followed the staining pattern over time. In general, although the additional cell wall layer seemed to reduce DAPI penetration in some cells, PolyP stores were readily visible using 50 μg/mL DAPI (Figure 3B). Eimeria parasites have also been shown to contain PolyP-rich organelles.33 These cells provided an interesting model for testing PolyP detection with DAPI, since the sporozoites (infective cells that contain only a limiting plasma membrane as a surface barrier) are found inside sporocysts that contain a cyst wall (each sporocyst contain two zoites). In contrast to yeast cells, PolyP detection in Eimeria sporozoites protected by a cyst wall inside sporocysts was not possible using the above mentioned protocols. In an attempt to enhance the access of DAPI to the intracellular milieu, sporocysts were submitted to i) gentle mechanical disruption/permeabilization as previously reported46 (Figure 3 C,D) or ii) Triton X-100 permeabilization followed by aldehyde fixation (Figure 3 E,F), before incubation with DAPI. Results showed an intense staining of PolyP-rich organelles (acidocalcisomes) inside the sporozoites. Although a previous step of aldehyde fixation has not significantly altered PolyP staining properties of DAPI in Eimeria cells, the same procedure seemed to impair DAPI-PolyP staining in other organisms, either by excluding DAPI penetration across the fixed cell wall or causing leaking of PolyP from the target organelles after chemical fixation (Figure 3B, inset). Nevertheless, it is important to note that in other yeast cell types such as the pathogenic fungi Cryptococcus neoformans, successful
Figure 2.
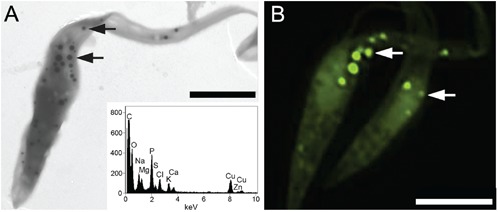
PolyP staining of T. cruzi whole cells. A) transmission electron micrograph of the electron dense acidocalcisomes whose elemental composition was determined by X-ray microanalysis (A, inset). B) PolyP-rich composition of acidocalcisomes was confirmed by fluorescence DAPI staining (50 μg/mL DAPI for 10-20 min). Arrows: acidocalcisomes. Scale bars: A, 3 μm; B, 4 μm.
Figure 3.
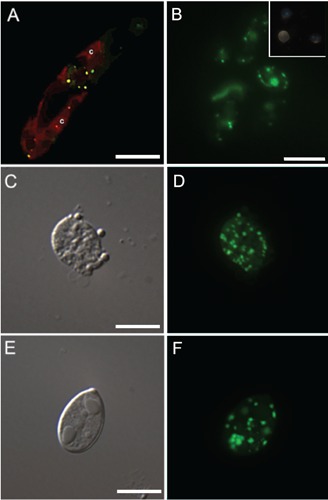
Neither prior fixation nor permeabilization was needed for detecting PolyP by DAPI staining (50 μg/mL for 30 min) in intracellular stores in living cells from (A) E. gracillis presenting c: chlorophyll autofluorescence, or (B) C. albicans. Formaldehyde fixation prevented DAPI staining of the PolyP granules in C. albicans (B, inset). On the other hand, formaldehyde fixation followed by either mechanical lysis (C,D) or Triton X-100 permeabilization (E, F) was needed to visualize subcellular PolyP stores in Eimeria cells by DAPI staining. Scale bars: A, 10 μm; B, 10 μm; C, 5 μm; E, 5 μm.
PolyP-DAPI staining has been carried out without the previous step of aldehyde fixation (data not shown), suggesting that the structural organization of the cell wall may be important for the permeation of the dye. Additional time-lapse analyses showed that the staining intensity was time-depended and varied with the cell type and the number and type of surface barriers. Fluorescence microscopy analysis of C. albicans incubated for different times showed acidocalcisomes became visible after 30 min and fluorescence intensity reached a plateau within 50 min (Figure 4A). Time-scale spectrofluorimetric analysis showed that in contrast to C. albicans, this plateau was reached in T. cruzi and T. brucei within 15-20 min, both for nuclei and PolyP staining (Figure 4 B,C).
Figure 4.
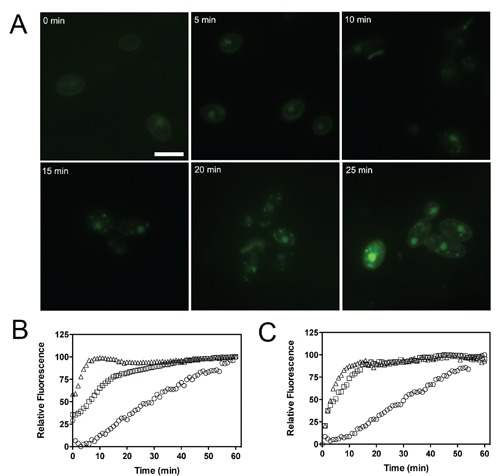
Time-lapse analysis of PolyP DAPI fluorescence. (A) Development of PolyP store labeling in C. albicans after staining with 50 μg/mL DAPI in PBS. (B,C) The influence of cell permeability to DAPI for the signal yield was compared over time using samples of T. cruzi (squares), T. brucei (triangles) and C. albicans (circles) incubated with 100 μg/mL DAPI at room temperature. Normalized fluorescence intensities were recorded by a microplate fluorimeter using an excitation/emission wavelength of 350/450 B: nuclear staining) or 350/500 (C: PolyP staining). Scale bar: 10 μm.
DAPI staining in tissues
Several studies have shown that the inorganic content of PolyP stores is usually washed during different steps used in sample preparation for electron microscopy, resulting in empty organelles observed in transmission electron microscopy images from several models.24,47,48 This suggested that the preparation of semi-thin sections of PolyP-containing tissues for light microscopy would potentially require protocol adaptations to preserve the PolyP content. In this regard, preparation of semi-thin cryosections of OCT-embedded tissues allowed on one hand minimal sample handling and, on the other, the obtainment of sections containing the whole diameter of the PolyP-rich organelles, avoiding the leaking of PolyP from sectioned organelles, as observed in TEM preparations. Analysis of semi-thin sections of the midgut of the insect A. gemmatalis showed the presence of PolyP stores in some apocrine vesicles at the apical region of columnar cells (Figure 5A) as well as in vesicles occurring mainly around the goblet cell cavities (Figure 5B), as previously described.36,49
Figure 5.
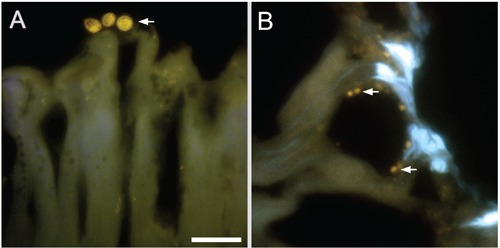
In situ detection of PolyP in midgut cells of A. gemmatali. Cryostat sections after incubation with 0.1 μg/mL DAPI. Apocrine secretions containing PolyP stores were detected emerging from columnar cells (arrows in A). PolyP granules concentrate around goblet cells (arrows in B). Scale bar: 25 μm.
Results provided by different groups have shown that in situ DAPI-PolyP staining of PolyP granules may result in a slight variation of fluorescence emission peaks or colors (from green to yellow) (Figure 3).50,51 To check whether different organelles in the same organism might yield different emission peaks, a localized in situ wavelength scan of different PolyP-rich apocrine secretions in the midgut of A. gemmatalis was carried out using multiphoton microscopy. Results showed that different organelles generated similar fluorescence emission profiles, ranging from 516 to 592 nm, with a maximum around 528-549 nm (Figure 6, lines 1-3), which in general corresponded to DAPI-bound commercial sodium PolyP75 emission wavelengths (Figure 7A). Wavelength scans of control areas did not generate similar fluorescence spectra (Figure 6, line 4).
Figure 6.
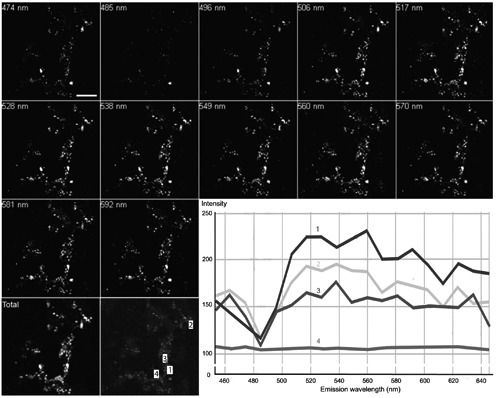
Spectral analysis of PolyP detection in situ. Cryostat sections of the midgut of A. gemmatalis after incubation with 0.1 μg/mL DAPI as observed by a multiphoton microscope at 780 nm excitation wavelength. PolyP stores fluorescence was more intense around 520-550 nm (lines 1- 3). The background signal was negligible and did not show wavelength specificity (line 4). Scale bar: 100 μm.
Figure 7.
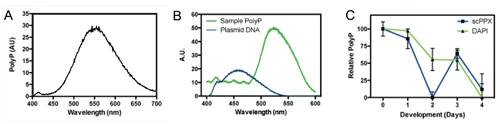
Spectroscopic analysis of PolyP fluorescence after staining with 20 μg/mL DAPI for 30 min at room temperature. The fluorescence intensities were measured by a fluorimeter at 354 nm excitation wavelength. Spectra were obtained from (A) 50 ug PolyP75 on (B) the water soluble fraction from P. americana (green line); plasmid DNA was also measured (blue line). (C) normalized DAPI fluorescence at 550 nm for PolyP samples of R. prolixus eggs at different development days (green line); the results were compared against an exopolyphosphatase (scPPX)-based assay (blue line), as a control.
Quantitative fluorimetric analysis of DAPI-PolyP staining in dynamic events
Spectrofluorimetric analysis of analytical grade sodium polyphosphate (PolyP75) (Figure 7A) and PolyP extracted from P. americana WSF (Figure 7B) confirmed fluorescence spectra qualitatively different from DNA-rich (plasmid) samples (Figure 7B), supporting the use of fluorimetric methods for the quantitative analysis of DAPI-PolyP staining. Although some reports have suggested similar approaches with minor variations,28,30,52 a semi-quantitative measurements of PolyP mobilization in biological samples using DAPI as a probe were yet to be compared against results from more widely accepted methodologies such as scPPX quantification following PolyP binding onto glassmilk particles.14,39 Here, we measured PolyP mobilization by following DAPI-PolyP fluorescence levels in samples containing PolyP extracted from eggs of the insect R. prolixus during the early stages of development. Results showed that a strong mobilization of PolyP takes place during this period (Figure 7C) and were validated after a comparison against the well-established protocol for PolyP quantification based on the PolyP binding to silica powder in suspension followed by enzymatic PolyP hydrolysis into Pi by a recombinant yeast exopolyphosphatase,14,26,39,53 which showed similar profiles of PolyP mobilization during the early days of development.
Discussion
PolyP-rich organelles comprise the major PolyP reservoir in several eukaryotic models such as yeast and protozoans.11,54-56 PolyP stores were first detected with metachromatic reactions using basic dyes such as toluidine blue, methylene blue or neutral red, and were, thus, named metachromatic granules.57-62 These reactions generally depend on a previous step of specimen fixation, thus limiting experiments in live cells. Polymeric compounds other than PolyPs, such as nucleic acids, have also been shown to be stained with basic dyes,63,64 dragging the attention to the poor specificity of some dyes and the necessity for the use of markers with higher specificity. Tetracycline has been alternatively used to indirectly stain bacterial PolyP.65 Nevertheless, fluorescence mechanism involves binding of tetracycline to divalent cations (usually chelated by PolyP) within PolyP-rich organelles, providing
signals of low specificity. Alternative methods such as the use of PolyP-binding domain of the E. coli exopolyphosphatase (PPBD) fused with an Xpress tag66 for the indirect detection of PolyP with the use of secondary antibodies have been reported.67,68 As yeast exopolyphosphatase (scPPX) is highly specific to PolyP, the method provides both specificity and sensitivity, although as PPBD is not commercially available, in lab production of PPBD requires protein engineering, bacterial strain transformation and protein expression and purification. In the last years, the use of DAPI as a PolyP-rich organelle sensor has gained popularity.21,24-26 It yields fluorescence at 525-550 nm after PolyP binding, displays a greenish-yellowish color23 and is both commercially available and present a fairly good sensitivity and specificity for the detection of PolyP-rich stores. In the present report, we evaluated the effect of different routines for PolyP detection and demonstrated the feasibility of using DAPI as a probe for the detection of PolyP-rich organelles. We further demonstrated that a set of straightforward treatments steps allows a wide range of biological samples to be analyzed following sample-optimized protocols. DAPI was used to detect PolyP stores in biological models after a relatively simple incubation step. Nevertheless, DAPI concentrations needed for PolyP detection were generally above those used for nuclei staining and usually demanded longer incubation times. Whether those conditions might not be optimal for in vivo studies, they are still the least invasive technique for PolyP visualization at present. Nevertheless, it was observed that DAPI intake could not be performed in vivo in some models. This might be the result of both the inability of DAPI to surpass complex cell layers or outward DAPI pumping mechanism of living cells. In those cases, we have been able to observe PolyP stores by using both pre-incubation cell fixation and permeabilization routines. Nevertheless, special care must be taken as we have observed that aldehyde fixation could either prevent or allow visualization of PolyP granules by DAPI.
Subcellular fractionation has been the method of choice for the isolation and posterior characterization of PolyP storage compartments.20,25,40,48 In those cases, a fast and reliable method of PolyP detection is essential as it allows identification of PolyP-enriched granules among the several resulting fractions. The subcellular compartments we tested did not need previous treatments, and smaller DAPI concentrations (2-10 μM DAPI) were successfully used. Additionally, DAPI staining in semi-thin sections have not been widely used and scarcely reported.49 In part, this is due to a loss of inorganic components during sample preparation, resulting in empty compartments. In this work, we have used an OCT-embedding protocol avoiding extensive sample manipulation and the steps of alcohol dehydration and medium substitution found in paraffin embedding protocols.
Several microscope settings have been previously used for in situ detection of PolyP stores by DAPI likely resulting in a confusing scenario for the outcomer researcher. As PolyP shifts DAPI emission from 450 nm to 525-550 nm,23 the fluorescence hue changes to a yellowish-green color. Thus, fluorescence emitted from nuclei can theoretically be discerned from fluorescence from PolyP using regular DAPI filters (data not shown), if color sensitive cameras are used. Nevertheless, emissions from nuclei often become too intense to enable differentiation from the weaker PolyP emissions. The DAPI filter system has thus to be modified and set to block wavelengths shorter than 500-515 nm. This will not prevent nuclei greenish emission area from being detected, yet it should block the most intense blue wavelengths and even allow detection by a monochromatic detection system. During our analyses, these settings more commonly provided optimal results suggesting that it should be used as a default condition for DAPI-PolyP
detection. Alternatively, previous reports suggested that the use of excitation wavelengths around 400-415 nm could similarly favor PolyP signals.28 A similar adjustment was used with OCT-embedded sections with good results. Nevertheless, care must be taken as DAPI excitation is performed over the maximum 350 nm excitation wavelength and low signals might be generated, hindering proper detection.
Quantification of PolyP levels have been done either with the use of its metachromatic properties69 or using enzymatic assays mainly based on the hydrolysis of PolyP by scPPX14,70 or incorporation of Pi from PolyP into ADP and synthesis of ATP by a polyphosphate kinase (PPK).39 Again, the same issues described for detection of PolyP stores are present and the methods either lack specificity and sensitivity, or require recombinant enzymes that are not commercially available. Nevertheless, biotechnological applications of PolyP such as the removal of phosphorous and heavy metals from wastewater demands a faster and cheaper method of quantification that still maintain its reliability.50 In this regard, some authors have approached on a method for DAPI-PolyP quantification.26,28,30,50,71 Here we performed an evaluation of relative PolyP levels to ascertain PolyP mobilization on a time scale using fluorescence levels generated by DAPI as a probe. Again, fluorescence levels were measured using the maximum DAPI excitation wavelength (~350 nm). It has been suggested that DAPI-DNA and unbound DAPI signal are significantly removed by using 415 nm excitation, which is imperative to discern PolyP specific signals in conditions of low PolyP content. Our present results were compared with the scPPX-based routine, which remains the most widely accepted PolyP quantification method, and demonstrated no major discrepancy between both methods. This suggests that when significant levels of PolyP are present, therefore giving a proper specific PolyP signal when compared to unbound or DNA/RNA-bound DAPI signals, quantification using excitation wavelengths around 350 nm are allowed. Nevertheless, it has been demonstrated that smaller polymers (<5 Pi residues) do not yield DAPI fluorescence.30 Interference by DAPI binding to contaminants may also account for imprecise quantification. In this regard, DAPI binding to RNA has been described,64 and also shown that treatment of PolyP extracts with RNAse into has a strong influence on DAPI-PolyP signals under specific circumstances.72 Analysis of the level of interference of DAPI-RNA on the samples should help to establish how absolute quantification of PolyP by DAPI can be accomplished.
Acknowledgments
This work was supported by grants from the following Brazilian agencies: Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, INCT de Entomologia Molecular and INCT de Biologia Estrutural e Bioimagem), Coordenação de Aperfeiçoamento do Pessoal de Nível Superior (CAPES), Fundação Carlos Chagas Filho de Amparo à Pesquisa de Estado do Rio de Janeiro (FAPERJ) and CAPES-Petrobrás. We are grateful to Dr Urara Kawazoe (UNICAMP) and Dr Renato Damatta (UENF) for providing Eimeira cells, and for Dr. Eduardo Fox for critical opinion during the revision of the manuscript.
References
- 1.Kulaev I, Kulakovskaya T.Polyphosphate and phosphate pump. Annu Rev Microbiol. 2000;54:709-34. [DOI] [PubMed] [Google Scholar]
- 2.Rao NN, Gomez-Garcia MR, Kornberg A.Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem. 2009;78:605-47 [DOI] [PubMed] [Google Scholar]
- 3.van der Meijden P, Heemskerk J.Polyphosphates: a link between platelet activation, intrinsic coagulation and inflammation? Expert Rev Hematol. 20103:269-72 [DOI] [PubMed] [Google Scholar]
- 4.Muller F, Mutch NJ, Schenk WA, Smith SA, Esterl L, Spronk HM, et al. Platelet polyphosphates are proinflammatory and procoagulant mediators in vivo. Cell. 2009;139:1143-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith SA, Morrissey JH.Polyphosphate enhances fibrin clot structure. Blood. 2008;112:2810-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith SA, Mutch NJ, Baskar D, Rohloff P, Docampo R, Morrissey JH.Polyphosphate modulates blood coagulation and fibrinolysis. Proc Natl Acad Sci USA. 2006;103:903-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Fraley CD, Faridi J, Kornberg A, Roth RA.Inorganic polyphosphate stimulates mammalian TOR, a kinase involved in the proliferation of mammary cancer cells. Proc Natl Acad Sci USA. 2003;100:11249-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han KY, Hong BS, Yoon YJ, Yoon CM, Kim YK, Kwon YG, et al. Polyphosphate blocks tumor metastasis via anti-angiogenic activity. Biochem J. 2007;406:49-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawazoe Y, Katoh S, Onodera Y, Kohgo T, Shindoh M, Shiba T.Activation of the FGF signaling pathway and subsequent induction of mesenchymal stem cell differentiation by inorganic polyphosphate. Int J Biol Sci. 2008;4:37-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiba T, Nishimura D, Kawazoe Y, Onodera Y, Tsutsumi K, Nakamura R, et al. Modulation of mitogenic activity of fibroblast growth factors by inorganic polyphosphate. J Biol Chem. 2003;278:26788-92 [DOI] [PubMed] [Google Scholar]
- 11.Docampo R, de Souza W, Miranda K, Rohloff P, Moreno SN.Acidocalcisomes - conserved from bacteria to man. Nat Rev Microbiol. 2005;3:251-61 [DOI] [PubMed] [Google Scholar]
- 12.Espiau B, Lemercier G, Ambit A, Bringaud F, Merlin G, Baltz T, et al. A soluble pyrophosphatase, a key enzyme for polyphosphate metabolism in Leishmania. J Biol Chem. 2006;281:1516-23 [DOI] [PubMed] [Google Scholar]
- 13.Luo S, Ruiz FA, Moreno SNJ.The acidocalcisome Ca2+-ATPase (TgA1) of Toxoplasma gondii is required for polyphosphate storage, intracellular calcium homeostasis and virulence. Mol Microbiol. 2005;55:1034-45 [DOI] [PubMed] [Google Scholar]
- 14.Ruiz FA, Rodrigues CO, Docampo R.Rapid changes in polyphosphate content within acidocalcisomes in response to cell growth, differentiation, and environmental stress in Trypanosoma cruzi. J Biol Chem. 2001;276:26114-21 [DOI] [PubMed] [Google Scholar]
- 15.Docampo R, Scott DA, Vercesi AE, Moreno SN.Intracellular Ca2+ storage in acidocalcisomes of Trypanosoma cruzi. Biochem J. 1995;310:1005-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seufferheld MJ, Kim KM, Whitfield J, Valerio A, Caetano-Anolles G.Evolution of vacuolar proton pyrophosphatase domains and volutin granules: clues into the early evolutionary origin of the acidocalcisome. Biol Direct. 2011;6:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seufferheld M, Lea C, Vieira M, Oldfield E, Docampo R.The H+-pyrophosphatase of Rhodospirillum rubrum is predominantly located in polyphosphate-rich acidocalcisomes. J Biol Chem. 2004;279:51193-202 [DOI] [PubMed] [Google Scholar]
- 18.Motta LS, Ramos IB, Gomes FM, de Souza W, Champagne DE, Santiago MF, et al. Protonpyrophosphatase and polyphosphate in acidocalcisome-like vesicles from oocytes and eggs of Periplaneta americana. Insect Biochem Mol Biol. 2009;39:198-206 [DOI] [PubMed] [Google Scholar]
- 19.Ramos I, Gomes F, Koeller CM, Saito K, Heise N, Masuda H, et al. Acidocalcisomes as calcium- and polyphosphate-storage compartments during embryogenesis of the insect Rhodnius prolixus Stahl. PLOS One. 2011;6:e27276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramos IB, Miranda K, Pace DA, Verbist KC, Lin FY, Zhang Y, et al. Calcium-and polyphosphate-containing acidic granules of sea urchin eggs are similar to acidocalcisomes, but are not the targets for NAADP. Biochem J. 2010;429:485-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramos IB, Miranda K, Ulrich P, Ingram P, LeFurgey A, Machado EA, et al. Calcium- and polyphosphate-containing acidocalcisomes in chicken egg yolk. Biol Cell. 2010;102:421-34 [DOI] [PubMed] [Google Scholar]
- 22.Kapuscinski J.DAPI: a DNA-specific fluorescent probe. Biotech Histochem. 1995; 70:220-33 [DOI] [PubMed] [Google Scholar]
- 23.Allan RA, Miller JJ.Influence of S-adenosylmethionine on DAPI-induced fluorescence of polyphosphate in the yeast vacuole. Can J Microbiol. 1980;26:912-20 [DOI] [PubMed] [Google Scholar]
- 24.Marchesini N, Ruiz FA, Vieira M, Docampo R.Acidocalcisomes are functionally linked to the contractile vacuole of Dictyostelium discoideum. J Biol Chem. 2002;277:8146-53 [DOI] [PubMed] [Google Scholar]
- 25.Ruiz FA, Marchesini N, Seufferheld M, Govindjee, Docampo R.The polyphosphate bodies of Chlamydomonas reinhardtii possess a proton-pumping pyrophosphatase and are similar to acidocalcisomes. J Biol Chem. 2001;276:46196-203 [DOI] [PubMed] [Google Scholar]
- 26.Gomes FM, Ramos IB, Motta LM, Miranda K, Santiago MF, de Souza W, et al. Polyphosphate polymers during early embryogenesis of Periplaneta americana. J Insect Physiol. 2008;54:1459-66 [DOI] [PubMed] [Google Scholar]
- 27.Gomes FM, Carvalho DB, Peron AC, Saito K, Miranda K, Machado EA.Inorganic polyphosphates are stored in spherites within the midgut of Anticarsia gemmatalis and play a role in copper detoxification. J Insect Physiol. 2012;58:211-9 [DOI] [PubMed] [Google Scholar]
- 28.Aschar-Sobbi R, Abramov AY, Diao C, Kargacin ME, Kargacin GJ, French RJ, et al. High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J Fluoresc. 2008;18:859-66 [DOI] [PubMed] [Google Scholar]
- 29.Zink D, Sadoni N, Stelzer E.Visualizing chromatin and chromosomes in living cells. Methods. 2003;29:42-50 [DOI] [PubMed] [Google Scholar]
- 30.Diaz JM, Ingall ED.Fluorometric quantification of natural inorganic polyphosphate. Environ Sci Technol. 2010;44:4665-71 [DOI] [PubMed] [Google Scholar]
- 31.Camargo EP.Growth and differentiation in Trypanosoma cruzi. I. Origin of metacyclic Trypanosomes in Liquid media. Rev Inst Med Trop Sao Paulo. 1964;6:93-100 [PubMed] [Google Scholar]
- 32.de Jesus TC, Tonelli RR, Nardelli SC, da Silva Augusto L, Motta MC, Girard-Dias W, et al. Target of rapamycin (TOR)-like 1 kinase is involved in the control of polyphosphate levels and acidocalcisome maintenance in Trypanosoma brucei. J Biol Chem. 2010;285:24131-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soares Medeiros LC, Gomes F, Maciel LR, Seabra SH, Docampo R, Moreno S, et al. Volutin granules of Eimeria parasites are acidic compartments and have physiological and structural characteristics similar to acidocalcisomes. J Eukaryot Microbiol. 2011;58:416-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Einicker-Lamas M, Mezian A.Euglena gracilis as a model for the study of Cu2+ and Zn2+ toxicity and accumulation in eukaryotic cells. Environ Pollut. 2002;120:779-86 [DOI] [PubMed] [Google Scholar]
- 35.Garcia ES, Macarini JD, Garcia ML, Ubatuba FB.Feeding of Rhodnius prolixus in the laboratory. An Acad Bras Cienc. 1975;47:537-45 [PubMed] [Google Scholar]
- 36.Gomes FM, Carvalho DB, Peron AC, Saito K, Miranda K, Machado EA.Inorganic polyphosphates are stored in spherites within the midgut of Anticarsia gemmatalis and play a role in copper detoxification. J Insect Physiol. 2012;58:211-9 [DOI] [PubMed] [Google Scholar]
- 37.Hoffmann-Campo C, Oliveira E, Moscardi F. Criação massal da lagarta da soja Anticarsia gemmatalis. Londrina, EMBRAPA-CNPSo, 1985. [Google Scholar]
- 38.Rohloff P, Miranda K, Rodrigues JC, Fang J, Galizzi M, Plattner H, et al. Calcium uptake and proton transport by acidocalcisomes of Toxoplasma gondii. PLOS One. 2011;6:e18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ault-Riché D, Fraley CD, Tzeng CM, Kornberg A.Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J Bacteriol. 1998;180:1841-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruiz FA, Lea CR, Oldfield E, Docampo R.Human platelet dense granules contain polyphosphate and are similar to acidocalcisomes of bacteria and unicellular eukaryotes. J Biol Chem. 2004;279:44250-7 [DOI] [PubMed] [Google Scholar]
- 41.Miranda K, Benchimol M, Docampo R, de Souza W.The fine structure of acidocalcisomes in Trypanosoma cruzi. Parasitol Res. 2000;86:373-84 [DOI] [PubMed] [Google Scholar]
- 42.Scott DA, Docampo R, Dvorak JA, Shi S, Leapman RD.In situ compositional analysis of acidocalcisomes in Trypanosoma cruzi. J Biol Chem. 1997;272:28020-9 [DOI] [PubMed] [Google Scholar]
- 43.Souza W, Carreiro I, Miranda K, Silva N.Two special organelles found in Trypanosoma cruzi. An Acad Bras Cienc. 2000;72:421-32 [DOI] [PubMed] [Google Scholar]
- 44.Miranda K, Docampo R, Grillo O, de Souza W.Acidocalcisomes of trypanosomatids have species-specific elemental composition. Protist. 2004;155:395-405 [DOI] [PubMed] [Google Scholar]
- 45.Lefort-Tran M, Bre MH, Ranck JL, Pouphile M.Euglena plasma membrane during normal and vitamin B12 starvation growth. J Cell Sci. 1980;41:245-61 [DOI] [PubMed] [Google Scholar]
- 46.Miranda K, Rodrigues CO, Hentchel J, Vercesi A, Plattner H, de Souza W, et al. Acidocalcisomes of Phytomonas francai possess distinct morphological characteristics and contain iron. Microscopy and Microanalysis. 2004;10:647-55 [DOI] [PubMed] [Google Scholar]
- 47.Miranda K, Docampo R, Grillo O, Franzen A, Attias M, Vercesi A, et al. Dynamics of polymorphism of acidocalcisomes in Leishmania parasites. Histochem Cell Biol. 2004;121:407-18 [DOI] [PubMed] [Google Scholar]
- 48.Scott DA, Docampo R.Characterization of Isolated Acidocalcisomes of Trypanosoma cruzi. J Biol Chem. 2000;275:24215-21 [DOI] [PubMed] [Google Scholar]
- 49.Gomes FM, Carvalho DB, Machado EA, Miranda K.Ultrastructural and functional analysis of secretory goblet cells in the midgut of the lepidopteran Anticarsia gemmatalis. Cell Tissue Res. 2013;352:313-26 [DOI] [PubMed] [Google Scholar]
- 50.Kulakova AN, Hobbs D, Smithen M, Pavlov E, Gilbert JA, Quinn JP, et al. Direct quantification of inorganic polyphosphate in microbial cells using 4’-6-diamidino-2- phenylindole (DAPI). Environ Sci Technol. 2011;45:7799-803 [DOI] [PubMed] [Google Scholar]
- 51.Omelon S, Omelon S, Georgiou J, Georgiou J, Henneman ZJ, Henneman ZJ, et al. Control of Vertebrate Skeletal Mineralization by Polyphosphates. PLOS One. 2009;4:e5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pavlov E, Aschar-Sobbi R, Campanella M, Turner RJ, Gomez-Garcia MR, Abramov AY.Inorganic polyphosphate and energy metabolism in mammalian cells. J Biol Chem. 2010;285:9420-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gomes FM, Oliveira DMP, Motta LS, Ramos IB, Miranda KM, Machado EA.Inorganic polyphosphate inhibits an aspartic protease-like activity in the eggs of Rhodnius prolixus (Stahl) and impairs yolk mobilization in vitro. J Cell Physiol. 2010;222:606-11 [DOI] [PubMed] [Google Scholar]
- 54.Miranda K, de Souza W, Plattner H, Hentschel J, Kawazoe U, Fang J, et al. Acidocalcisomes in Apicomplexan parasites. Exp Parasitol. 2008;118:2-9 [DOI] [PubMed] [Google Scholar]
- 55.Tijssen JP, Beekes HW, van Steveninck J.Localization of polyphosphates in Saccharomyces fragilis, as revealed by 4’,6-diamidino- 2 -phenylindole fluorescence. Biochim Biophys Acta. 1982;721:394-8 [DOI] [PubMed] [Google Scholar]
- 56.Urech K, D¸rr M, Boller T, Wiemken A, Schwencke J.Localization of polyphosphate in vacuoles of Saccharomyces cerevisiae. Arch Microbiol. 1978;116:275-8 [DOI] [PubMed] [Google Scholar]
- 57.Ebel J.Recherches sur les polyphosphates contenus dans diverses cellules vivantes. IV. Localisation cytologique et role physiologique des polyphosphates dans la cellule vivante. Bull Soc Chim Biol (Paris). 1952;34:498-505 [PubMed] [Google Scholar]
- 58.Wiame J.Remarque sur la metachromasie des cellules de levure. CR Soc Biol. 1946;140:897-99 [Google Scholar]
- 59.Dmitrieva S, Bekker Z.Some data on the nature of the volutin granules in Penicillium chrysogenum. Tsitologiia. 1961;4:691-95 [PubMed] [Google Scholar]
- 60.Voelz H, Voelz U, Ortigoza RO.The polyphosphate overplus phenomenon in Myxo coccus xanthus and its influence on the architecture of the cell. Arch Microbiol. 1966;53:371-88 [DOI] [PubMed] [Google Scholar]
- 61.Lopez Revilla R, Gomez Dominguez R.Incorporation and Toxicity of 32P Orthophosphate and Occurrence of Polyphosphate in Entamoeba Tropho zoites. J Eukaryot Microbiol. 1985;32:353-5 [DOI] [PubMed] [Google Scholar]
- 62.Suresh N, Warburg R, Timmerman M, Wells J, Coccia M, Roberts M, et al. New strategies for the isolation of microorganisms responsible for phosphate accumulation. Water Science & Technology. 1985;17:99-111 [Google Scholar]
- 63.Kulaev IS, Vagabov V, Kulakovskaya T. Wiley Online Library; 1979. The biochemistry of inorganic polyphosphates. [Google Scholar]
- 64.Tanious FA, Veal JM, Buczak H, Ratmeyer LS, Wilson WD.DAPI (4“, 6-diamidino-2 -phenylindole) binds differently to DNA and RNA: minor-groove binding at AT sites and intercalation at AU sites. Biochemistry. 1992;31:3103-12 [DOI] [PubMed] [Google Scholar]
- 65.Gunther S, Trutnau M, Kleinsteuber S, Hause G, Bley T, Roske I, et al. Dynamics of polyphosphate-accumulating bacteria in wastewater treatment plant microbial communities detected via DAPI (4’, 6’-diamidino-2-phenylindole) and tetracycline labeling. Appl Environ Microbiol. 2009;75:2111-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saito K, Ohtomo R, Kuga-Uetake Y, Aono T, Saito M.direct labeling of polyphosphate at the ultrastructural level in Saccharomyces cerevisiae by Using the affinity of the polyphosphate binding domain of Escherichia coli exopolyphosphatase. Appl Environ Microbiol. 2005;71:5692-701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuga Y, Saito K, Nayuki K, Peterson RL, Saito M.Ultrastructure of rapidly frozen and freeze-substituted germ tubes of an arbuscular mycorrhizal fungus and localization of polyphosphate. New Phytologist. 2008;178:189-200 [DOI] [PubMed] [Google Scholar]
- 68.Saito K, Kuga-Uetake Y, Saito M, Peterson RL.Vacuolar localization of phosphorus in hyphae of Phialocephala fortinii, a dark septate fungal root endophyte. Can J Microbiol. 2006;52:643-50 [DOI] [PubMed] [Google Scholar]
- 69.Chernysheva E, Kritskii M, Kulaev I.[The determination of the degree of polymerization of different inorganic polyphosphate fractions from mycelia of Neurospora crassa]. [Article in Russian]. Biokhimiia. 1971;36:138-42 [PubMed] [Google Scholar]
- 70.Rao NN, Liu S, Kornberg A.Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J Bacteriol. 1998;180:2186-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abramov AY, Fraley C, Diao CT, Winkfein R, Colicos MA, Duchen MR, et al. Targeted polyphosphatase expression alters mitochondrial metabolism and inhibits calciumdependent cell death. Proc Natl Acad Sci USA. 2007;104:18091-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martin P, Van Mooy BA.Fluorometric quantification of polyphosphate in environmental plankton samples: extraction protocols, matrix effects, and nucleic acid interference. Appl Environ Microbiol. 2013;79:273-81 [DOI] [PMC free article] [PubMed] [Google Scholar]


