Summary
MLLT11, an MLL fusion partner, is a poor prognostic biomarker for paediatric acute myeloid leukaemia (AML), adult normal cytogenetics AML, and adult myelodysplastic syndrome. MLLT11 is highly regulated during haematopoietic progenitor differentiation and development but its regulatory mechanisms have not been defined. In this study, we demonstrate by transfection experiments that MIR29B directly regulates MLLT11 expression in vitro. MIR29B expression level was also inversely related to MLLT11 expression in a cohort of 56 AML patients (P < 0·05). AML patients with low MIR29B/elevated MLLT11 expression had poor overall survival (P = 0·038). Therefore, MIR29B may be a potential prognostic biomarker for AML patients.
Keywords: AML prognostic marker, MLLT11, MLL fusion gene, MIR29, AML survival
The MLLT11 gene, located on chromosome 1 band q21, was initially identified as a mixed-lineage leukaemia (MLL) fusion partner from acute myeloid leukaemia (AML) patients whose leukaemic cells carried a t(1; 11) (q21; q23) chromosomal abnormality (Tse et al, 1995). MLLT11 expression is highly regulated in normal lineage committed haematopoietic progenitor cells (HPC) (Tse et al, 1995). Elevated MLLT11 expression is widely seen in acute myeloid and lymphoid leukaemias (Tse et al, 1995). We have consistently shown that high MLLT11 expression is a poor prognostic biomarker for paediatric AML (Tse et al, 2004), adult normal cytogenetics AML (NC-AML) (Strunk et al, 2009), and adult myelodysplastic syndrome (MDS) (Tse et al, 2005). However, the underlying mechanism(s) of MLLT11, regulation in normal and abnormal haematopoiesis remain unclear.
MicroRNAs are small RNAs that play important roles in the post-transcriptional regulation of genes, such as oncogenes and tumour suppressor genes that are mostly related to cell differentiation. They are located throughout the genome but are often found at fragile sites (Ambros, 2001). We hypothesized that MLLT11 expression may be regulated by micro-RNAs and the MLLT11 3′-UTR, like other genomic regions, is involved in the development of cancers. Through web-based search and analysis, we found and confirmed that MIR29B, a member of the MIR29 family was the strongest candidate to potentially regulate MLLT11. In this report, we demonstrate that MIR29B specifically interacts with the MLLT11 3′-UTR and directly regulates MLLT11 expression and may be a coordinate biomarker with MLLT11 in myeloid leukaemias.
Materials and methods
MicroRNA and gene expression profile from AML patients, human control samples, and statistical analysis
The information, sample collection, and preparation have been previously described (Li et al, 2008) for the 56 patients included in this study. As there is molecular heterogeneity within specific cytogenetic groups that can lead to variation in overall survival (OS), we grouped these patients’ OS according to their MIR29B expression levels instead of cytogenetics. Clinical variables across the groups were compared by using the chi-square or a two-sided Fisher's exact test for categorical variables. P values < 0·05 were considered as statistically significant. OS was calculated by using the method of Kaplan–Meier, and log-rank test was used to assess the differences between survival curves.
Computational predictions of microRNA binding
The web-based “TargetScanHuman” (http://www.targetscan.org/vert_50/) and miRGen (http://www.diana.pcbi.upenn.edu/cgi-bin/miRGen/v3/Targets.cgi#Results) were used to identify microRNA candidates that may potentially regulate MLLT11. The MIR29A/B/C required binding sequence matches of 100% in the MLLT11 3′UTR.
Cell lines and growth conditions
Two human lung cancer cell lines, H157 and SKMES1, and a leukaemic cell line REH were chosen because they have higher MLLT11 expression and acceptable transfection efficiency. Cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum at 37°C and 5% CO2.
Establishment of stable transfectants of green fluorescent protein (GFP), GFP- MLLT11 3′-UTR (GFP-A3U) and GFP- MLLT11 3′-UTR mutation at the MIR29 binding site (GFP-A3U-Mutant)
H157, SKMES1, and REH cells were transfected with 10 nmol/l miRIDIAN MIR29B mimic or miRIDIAN mimic negative control (Dharmacon, Chicago, IL, USA), using Lipofectamine2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA). H157 cells were transfected with GFP, GFP-A3U and GFP-A3U-mutant (mis-matched at 2-nucleotides, S2B) by using Lipofectamine2000 transfection reagent. After transfection, neomycin-resistant clones were selected in the presence of 900 μg/ml G418 sulfate (Mediatech, Manassa, VA, USA). GFP-positive cells were sorted by flow cytometry. Cells were imaged under the Nikon eclipse Ti microscope using the NIS-Elements image system.
RNA isolation, cDNA synthesis, quantitative reverse transcription polymerase chain reaction (qRT-PCR), and Western blotting
Total RNA isolation, cDNA synthesis, qRT-PCR, and cell lysates were performed as previously reported (Tse et al, 2004). qRT-PCR was used to determine the MLLT11, GFP, and B2M (internal control) expression. Corresponding MLLT11 protein levels in three tested cell lines were confirmed by Western blot using the rabbit monoclonal anti-MLLT11 (Epitomics, Burlingame, CA, USA). Samples were tested in triplicate fashion. Primers for each gene amplification were as follows:
MLLT11 Forward: 5′-GCACTCCCTCCATCTTTGGA-3′
MLLT11 Reverse: 5′- CAGCTCCGACAGATCCAGTTC-3′
GFP Forward: 5′-CGACAAGCAGAAGAACGGCATCAA-3′
GFP Reverse: 5′-AACTCCAGCAGGACCATGTGAT-3′
B2M Forward: 5′-ATGAGTATGCCTGCCGTGTGA-3′
B2M Reverse: 5′-GGCATCTTCAAACCTCCATG-3′
Construction of GFP reporter vectors
The MLLT11 3′-UTR was subcloned into pEGFP-C1 (Clontech, Mountain View, CA, USA) at the EcoRI/BamHI sites. Mutation of the MIR29B binding site in the MLLT11 3′UTR was created by the Stratagene QuickChange method with the following primers:
MLLT11-3UTR (forward): 5′-TACTGTGTTGGTGGTTCGGATGAATCTG-3′
MLLT11-3UTR (reverse): 5′-CAGATTCATCCGAACCACCAACACAGTA-3′
Results and discussion
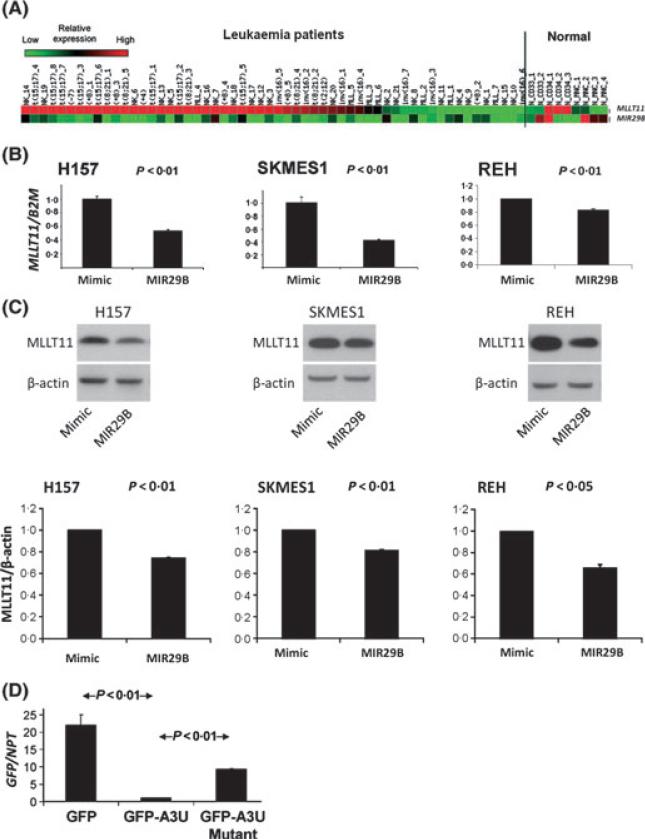
Through using a web-based scoring system, MIR29 was identified as a potential regulator of MLLT11 expression. This observation was further supported by our subsequent finding that MLLT11 and MIR29A/B/C expression were inversely related in the tested cell lines and AML patient samples. We chose to study MIR29B in this report because only MIR29B expression had significant predictive power for OS in a cohort of AML patients reported here. It was confirmed that most of the 56 AML patients with elevated MLLT11 expression had depressed MIR29B expression (Fig 1A). The prevalence of elevated MLLT11 expression among this cohort of AML patients (66%) was similar to our previous reports (Tse et al, 2004; Strunk et al, 2009). To test whether MIR29B could specifically and directly regulate MLLT11 expression by interacting with the MLLT11 3′-UTR, MIR29B was transfected into 3 cell lines (H157, SKMES1, and REH) and found to knockdown MLLT11 mRNA and MLLT11 protein (Fig 1B, C). This regulatory relationship between MIR29B and MLLT11 was direct and specific because the knockdown effect on MLLT11 mRNA and MLLT11 protein could be ablated by mutation of the MIR29B binding site in the MLLT11 3′-UTR (Fig 1D).
Fig 1.
Low MIR29B/high MLLT11 expression in AML patient samples and demonstration of direct regulation of MLLT11 by direct interaction of MIR29B with the MLLT11 3-UTR. (A) MLLT11 and MIR29B expression heat map: columns and rows represent patients and MLLT11/MIR29Bexpression, respectively. Red to green indicates high to low expression of MLLT11/MIR29B, respectively. (B) Assessment of expression of MLLT11 mRNA by qRT-PCR and its response to the transfection of miR-29b: transfection of MIR29B into H157 lung cancer cells, SKMES1 lung cancer cells, REH leukaemic cells and a negative control for 24 h, respectively. (C) Assessment of MLLT11 protein level by Western blots in samples corresponding to those of B. (D) GFP reporter stable transfectants in H157 cells showing GFP empty vector (control), GFP-A3U (wild-type), and GFP-A3U-Mutant as well as their images taken by the NIS-Elements image system. Assessment of expression of GFP mRNA by qRT-PCR normalized to neomycin phosphotransferase (NPT) mRNA that also digitally confirmed the image observation. Data in B/C/D are representative of three independent experiments.
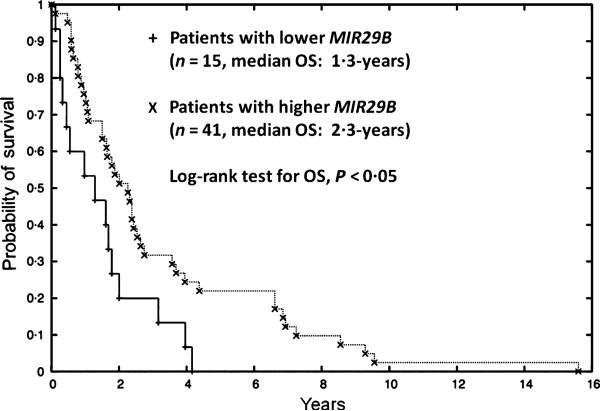
We next wanted to determine whether MIR29B would offer the same prognostic power as the high MLLT11 expression quartile in AML patients (Strunk et al, 2009). It was found that 15 patients with low MIR29B expression (2·6-fold), corresponding to the highest quartile of high MLLT11 expression, had significantly poorer OS (P < 0·05, Fig 2) and a stronger trend toward an association with adverse cytogenetics (P = 0·06) compared to the remaining AML patients. Therefore, low MIR29B expression could be a similar poor prognostic marker as high MLLT11 expression and adverse cytogenetics in AML patients as we reported (Tse et al, 2004; Strunk et al, 2009), and vice versa. Notably among 56 AML patients, there was only one patient with −7q, one of the coding regions for MIR29B, who had very low MIR29B expression but very high MLLT11 expression. This indicative case demonstrated that expression of mature MIR29B in AML might probably come from the chromosomal locus 7q23. The observation on this specific case potentially provides a hint that deletion of the tumour suppressive MIR29B coding region may be mechanistically responsible for the pathogenesis of MDS/AML patients with −7q/monosomy 7.
Fig 2.
Lower MIR29B expression is a poor prognosis marker for AML patients. Kaplan-Meier survival curves for 56 AML patients are stratified by MIR29B expression. AML patients with lower MIR29B expression had a strong trend of adverse cytogenetics (<2·6-fold; P = 0·06) and significantly poorer OS (median OS 1·3-year vs. 2·3-year; P < 0·05) compared to AML patients with higher MIR29B expression (>2·6-fold).
The MIR29 family comprises a group of small non-coding RNAs (MIR29A/B/C) that are actively involved in regulating many developmental and differentiation related genes (Ambros, 2001). MIR29B is considered a tumour suppressor and has been found to directly regulate MCL1 (Mott et al, 2007), TCL1A (TCL1) (Pekarsky et al, 2006), and DNMT3A/B (DNMT3) (Garzon et al, 2009; Takada et al, 2009). It is believed that through targeting p85α and CDC42, MIR29 can activate TP53 as part of its biological function as a tumour suppressor (Park et al, 2009). MIR29 also regulates lung (Williams et al, 2007), myoblast (Wang et al, 2008), and osteoblast (Li et al, 2009) differentiation and development that may contribute to a role in lung cancer and rhadomyosarcoma.
Despite that the biological function of MLLT11 is unclear, we have demonstrated that it is tightly regulated in HPC differentiation and development (Tse et al, 1995). We also have consistently shown that elevated MLLT11 expression is a poor prognostic biomarker for paediatric AML (Tse et al, 2004), adult NC-AML (Strunk et al, 2009), and adult MDS (Tse et al, 2005). Another series of studies also showed similar observations, that naturally elevated MIR29B expression appears to associate with certain lower risk AML patients such as those NC-AML patients with NPM1 mutation (Garzon et al, 2008). Given the fact that MIR29B directly regulates MLLT11 expression, the observations of Garzon et al (2008) are consistent with our previous study that high MLLT11 expression is a poor prognostic marker for NC-AML (Strunk et al, 2009). However, our current study further shows that MIR29B expression levels may also have OS predictive value for AML patients with different cytogenetics and this observation needs to be confirmed in a larger AML cohort. Biologically, over-expression of pre- MIR29B reduces global DNA hypomethylation and restores expression of the hypermethylated tumour suppressors ESR1 and CDKN2B (p15INK4b) in AML patients. These changes may explain why these AML patients have better outcomes (Garzon et al, 2009). Another group of investigators also observed that the MIR29 family is down-regulated in AML patients with balanced 11q23 translocations targeting the oncogene TCL1A, which represents poor prognosis AML (Pekarsky et al, 2006).
Our current study suggests that MIR29B, like MLLT11, can be a potential prognostic marker for AML. Most importantly the MIR29 family regulates a group of genes, such as MCL1,TP53, DNMT3A/B, and now MLLT11 that are related to cell apoptosis, DNA methylation, and differentiation. The MIR29B biological phenomenon warrants further clinical investigation to determine whether the signal transduction profiles related to the MIR29 family and its regulated genes could serve as therapeutic targets for AML patients. This approach alone or in combination with existing targeted apoptotic, methylation, or differentiation therapies might provide novel therapeutic avenues especially for high risk AML patients.
Footnotes
Author contributions
YX, ZL, MJ, JB, JP, and JVT designed and conducted experiments. ACT: analysed microarray data and performed statistical analysis data. KDB, GH, CJ, and JC: analysed data and participated in manuscript writing. LTB, WT designed experiments and wrote manuscript.
References
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C, Volinia S, Liu CG, Schnittger S, Haferlach T, Liso A, Diverio D, Mancini M, Meloni G, Foa R, Martelli MF, Mecucci C, Croce CM, Falini B. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, Havelange V, Volinia S, Blum W, Rush LJ, Perrotti D, Andreeff M, Bloomfield CD, Byrd JC, Chan K, Wu LC, Croce CM, Marcucci G. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, Chen P, Wang Y, Yan M, Qian Z, Neilly MB, Jin J, Zhang Y, Bohlander SK, Zhang DE, Larson RA, Le Beau MM, Thirman MJ, Golub TR, Rowley JD, Chen J. Distinct microRNA expressionprofiling in acute myeloid leukemia with common translocation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. Journal of Biological Chemistry. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott JL, Kobayashi S, Bronk SF, Gores GJ. MiR-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Lee JH, Ha M, Nam JW, Kim VN. MiR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nature Structural & Molecular Biology. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- Pekarsky Y, Santannam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu CG, Rassenti L, Calin GA, Hagan JP, Kipps T, Croce CM. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Research. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- Strunk CJ, Platzbecker U, Thiede C, Schaich M, Illmer T, Kang Z, Leahy P, Li C, Xie X, Laughlin MJ, Lazarus HM, Gerson SL, Bunting KD, Ehninger G, Tse W. Elevated AF1q expression is a poor prognostic marker for adult acute myeloid leukemia patients with normal cytogenetics. American Journal of Hematology. 2009;84:308–309. doi: 10.1002/ajh.21396. [DOI] [PubMed] [Google Scholar]
- Takada S, Berezikov E, Choi YL, Yamashita Y, Mano H. Potential role of miR-29b in modulation of Dnmt3a and Dnmt3b expression in primordial germ cells of female mouse embryos. RNA. 2009;15:1507–1514. doi: 10.1261/rna.1418309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse W, Zhu W, Chen HS, Cohen A. A novel gene, AF1q, fused to MLL in t(1;11) (q21;q23), is specifically expressed in leukemic and immature hematopoietic cells. Blood. 1995;85:650–656. [PubMed] [Google Scholar]
- Tse W, Meshinchi S, Alonzo TA, Stirewalt DL, Gerbing RB, Woods WG, Appelbaum FR, Radich JP. Elevated expression of the AF1q gene, an MLL fusion partner, is an independent adverse prognostic factor in pediatric acute myeloid leukemia. Blood. 2004;104:3058–3063. doi: 10.1182/blood-2003-12-4347. [DOI] [PubMed] [Google Scholar]
- Tse W, Deeg HJ, Stirewalt D, Appelbaum FR, Radich J, Gooley T. Increased AF1q gene expression in high-risk myelodysplastic syndrome. British Journal of Haematology. 2005;128:218–220. doi: 10.1111/j.1365-2141.2004.05306.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, Croce CM, Guttridge DC. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AE, Larner-Svensson H, Perry MM, Barnes PJ, Lindsay MA. Maternally imprinted microRNAs are differentially expressed during mouse and human lung development. Developmental Dynamics. 2007;236:572–580. doi: 10.1002/dvdy.21047. [DOI] [PMC free article] [PubMed] [Google Scholar]