Abstract
The molecule known as SF2575 from Streptomyces sp. is a tetracycline polyketide natural product that displays antitumor activity against murine leukemia P388 in vivo. In the SF2575 biosynthetic pathway, SsfS6 has been implicated as the crucial C-glycosyltransferase (C-GT) that forms the C-C glycosidic bond between the sugar and the SF2575 tetracycline-like scaffold. Here, we report the crystal structure of SsfS6 in the free form and in complex with TDP, both at 2.4 Å resolution. The structures reveal SsfS6 to adopt a GT-B fold wherein the TDP and docked putative aglycon are consistent with the overall C-glycosylation reaction. As one of only a few existing structures for C-glycosyltransferases, the structures described herein may serve as a guide to better understand and engineer C-glycosylation.
Keywords: natural product, antitumor, GT-B fold, X-ray diffraction, molecular docking, biosynthesis, carbohydrate
INTRODUCTION
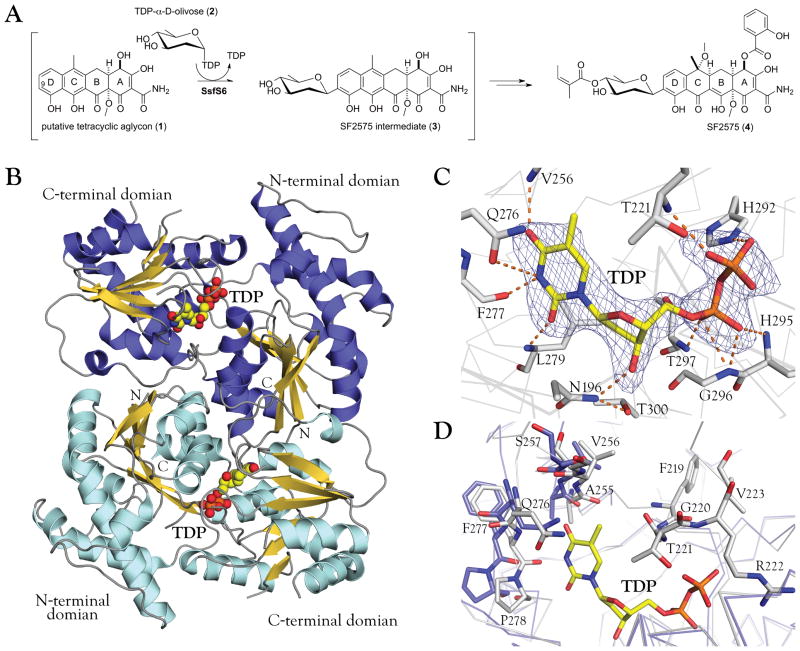
Natural products and their derivatives are an important resource for the discovery of antibiotics and anticancer drugs.1 The bioactivity of such natural products can be influenced by enzyme-catalyzed glycosylation and include natural products bearing O-, N-, S- or even C-glycosides.2,3 An example of the latter is SF2575, a tetracycline polyketide isolated by Hatsu et al. from Streptomyces sp. SF2575 that displays potent cytotoxicity against P388 leukemia cells in vitro (IC50 = 7.5 ng/ml) and increases the life span of P388 leukemia cell ip-implanted mice in vivo.4,5 The biosynthesis of SF2575 has received considerable attention culminating in the work by Tang and co-workers on the cloning, genetics and annotation of the SF2575 biosynthetic gene cluster and subsequent biochemical study of key enzymes encoded by this newly identified biosynthetic locus.6–8 Specifically, several SF2575 intermediates including 3 [Fig. 1(A)] have been identified via proton NMR and HRMS,6 and sequence analysis of ssf biosynthetic gene cluster detected only one putative glycosyltransferase, SsfS6, which has high sequence homology to a C-GT HedJ from hedamycin biosynthetic pathway.9 Based upon this impressive body of work, SsfS6 has been annotated as the unique C-C-bond forming C-GT that catalyzes the transfer of D-olivose from TDP-D-olivose to C9 of the tetracycline-like aglycon [Fig. 1(A)].
Figure 1.
A: Putative biosynthetic pathway of SF2575 highlighting the proposed reaction catalyzed by SsfS6. SsfS6 is postulated to catalyze the transfer of D-olivose from the sugar donor dTDP-D-olivose to C9 of the D-ring of a tetracycline-like aglycon, the structure of which has not been fully elucidated.
B: Quaternary structure of the SsfS6 dimer as predicted by PISA.33 N-terminal and C-terminal in each monomer are indicated and TDP is illustrated as spheres.
C: TDP in the binding site of SsfS6 with the simulated annealing Fo-Fc omit map of TDP (contoured at 3.0σ). Residues in the donor-binding site form hydrogen bonds with the thymine base, deoxyribose and the pyrophosphate of TDP.
D: Active site comparison between SsfS6 bound to TDP (grey) and SsfS6 in the free form (light blue). Ligand binding leads to the shift of several residues and a stabilization of the region between G220 and L225 (a region which lacks electron density in apo SsfS6).
Glycosyltransferases comprise 94 diverse sequence-based families of enzymes,10 but they are known to adopt only three different folds (GT-A, GT-B and predicted trans-membrane fold GT-C).11–13 The few C-GT structures elucidated to date adopt a GT-B fold,14,15 which consists of two spatially distant β/α/β Rossmann-like domains. As the novel C-glycoside products synthesized by C-GTs are chemically unique because of their remarkable stability to chemical and enzymatic hydrolysis,9 there remains notable interest in the synthesis and application of C-glycoside-containing O-glycoside surrogates.16,17 In contrast to O-GTs,12 mechanistic, in vitro biochemical and/or structural studies of C-GTs are still limited, which severely restricts both our level of understanding, and ability to engineer and/or exploit,14 these remarkable catalysts.
Here we report the crystal structures of the putative C-GT SsfS6 in both apo and thymidine diphosphate (TDP) bound forms. The structure of TDP-bound SsfS6 elucidates the TDP-enzyme interactions and reveals that the loop residue N196 impedes binding of UDP-sugars by sterically occluding the ribose 2′-OH group.18 Also, two potential active site bases implicated for C-glycosyl catalysis were identified. They are consistent with docking studies that highlight potential binding modes of the putative tetracycline-like aglycon. Cumulatively, this study is an additional structural blueprint for the further general study of C-glycosylation and specifically, SF2575.
MATERIALS AND METHODS
Cloning, expression and purification
The SsfS6 gene (NCBI accession: 292659130) was cloned into NdeI/BamHI-digested pET28a to enable production of recombinant N-His6-SsfS6. For protein production, the corresponding pET28a-SsfS6 construct was transformed into the E. coli methionine auxotroph strain B834 (DE3) on a LB agar plate containing 50 μg/mL kanamycin. A single colony of the plate was then used to inoculate 20 mL MDAG liquid medium19 supplemented with 50 μg/mL kanamycin and cultured at 37°C overnight with shaking at 250 rpm, and the starter culture was then transferred to 1 L of 5SM auto-inducing medium19 in the presence of 50 μg/mL kanamycin and grown at 25°C for 30 h. The cells were harvested by centrifugation at 4200 × g for 30 min at 4°C.
The cell pellet was resuspended in buffer 20 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 7.8, and lysed by sonication (VirSonic 475, Virtis; 100 W, 4×45 s pulses, ~2 min between pulses) on ice. Subsequently, N-His6-SsfS6 was purified via Ni-NTA chelating column (GE Healthcare) following a protocol with a linear imidazole (10–500 mM) elution gradient (50 mM NaH2PO4, 300 mM NaCl, pH 8.0). Then the protein was concentrated and desalted through PD-10 column (GE Healthcare) with an elution buffer of 50 mM Tris, pH 8.0. The purified protein was determined to be 11.4 mg/mL by Bradford assay and flash frozen in liquid nitrogen for storage at −80°C. Unless otherwise indicated, the term SsfS6 herein refers to SeMet-labeled N-His6-SsfS6.
Crystallization, diffraction and structure determination
General screens were performed with PEGRx HT, Crystal Screen HT, Index HT, and SaltRx HT (Hampton Research) utilizing a Mosquito® dispenser (TTP labTech) by the sitting drop method. Crystal growth was monitored by Bruker Nonius Crystal Farms at 20 °C. Apo SsfS6 and SsfS6 with TDP crystals were obtained by mixing 2 μL of protein solution and 2 μL of reservoir solution, 1.13~1.40 M sodium phosphate monobasic monohydrate/potassium phosphate dibasic, pH 8.2, using the sitting drop method. For crystals of SsfS6-TDP, 10 mM TDP was mixed with protein solution in a 1:10 volume ratio. Diffraction quality hexagonal crystals were best obtained by mixing the protein solution 1:1 with buffer solutions and centrifuging before setting up the crystallizations. All crystals were protected by Paratone-N and flash frozen in liquid nitrogen.
X-ray diffraction data were collected at the Life Science Collaborative Access Team (LS-CAT) with an X-ray wavelength of 1.12715 Å (for both apo SsfS6 and SsfS6-TDP) at the Advanced Photon Source at Argonne National Laboratory. Datasets were indexed and scaled by HKL2000.20 For structure solution of apo SsfS6, phenix.HySS was used for determination of selenium atom sub-structure, AutoSol for phasing and phenix.autobuild for model building.21 For the structure of SsfS6 with bound TDP, molecular replacement was utilized with the apo SsfS6 structure as a starting model. The structures including several double conformations were manually rebuilt in several rounds by Coot22 and further refined by phenix.refine with default parameters.21 The quality of coordinates was verified by MolProbity.23 All structural figures in this paper were generated using PyMOL.24 All docking experiments were done using AutoDock 4.2.25
RESULTS AND DISCUSSION
Overall structure and structure quality
X-ray diffraction data were collected to a resolution of 2.4 Å for both apo SsfS6 and the SsfS6-TDP complex [Table I]. SsfS6 crystals belong to space group P6122 with a packing parameter of 2.66 Å3/Da, having one SsfS6 molecule of 42,712.2 Da (N-terminal His6-tagged protein sequence) per asymmetric unit.
Table I.
Data Collection and Refinement Statistics for SsfS6 and SsfS6-TDP complex
| SsfS6 | SsfS6-TDP | |
|---|---|---|
| Data collection | ||
| Resolution range (Å) | 50-2.40 | 50-2.40 |
| Wavelength (Å) | 1.13 | 1.13 |
| Space group | P6122 | P6122 |
| a, b, c (Å) | 82.61, 82.61, 230.52 | 81.14, 81.14, 229.91 |
| No. of molecules per asymmetric unit | 1 | 1 |
| Measured reflections | 218,101 | 288,940 |
| Unique reflections | 18,943 | 18,077 |
| Rmerge | 0.094 (1.00) | 0.20 (0.88) |
| Completeness | 99.04 (96.18) | 98.94 (93.87) |
| Redundancy | 11.5 (11.8) | 15.9 (16.0) |
| Mean I/sigma(I) | 11.24 (1.54) | 8.24 (2.41) |
| Refinement | ||
| Rcryst/Rfreea | 0.224/0.262 | 0.244/0.269 |
| No. of protein atoms | 2,642 | 2,687 |
| No. of ligand atoms | - | 25 |
| No. of solvent atoms | 110 | 67 |
| Average B factor (Å2) | ||
| Protein | 39.7 | 32.3 |
| Water | 36.5 | 26.3 |
| TDP | - | 26.7 |
| RMSD from ideal | ||
| Bond length (Å) | 0.004 | 0.005 |
| Bond angles (deg) | 0.861 | 0.780 |
| Ramachandran plot (%) | ||
| Favored regions | 96.8 | 99.1 |
| Allowed regions | 2.9 | 0.9 |
| Outliers | 0.3 | 0 |
| PDB ID | 4FZR | 4G2T |
Values in parentheses are for the highest resolution shell.
Rfree was calculated as Rcryst using 5.0% of randomly selected unique reflections (in thin resolution shells) that were omitted from the structure refinement.
Although only one SsfS6 monomer was detected in the asymmetric unit, the chain forms a C2-symmetric dimer with another SsfS6 chain in the unit cell [Fig. 2(B)], with a buried interface area of 1574 Å2 as calculated by PISA.26 Other members of the GT1 sequence family, such as UrdGT2, CalG3 and SpnG,18,27,28 adopt similar homodimer architectures. The dimeric interface comprises residues within both the N-terminal and the C-terminal domains, including intertwined hydrophobic interactions with a contact center generated by two stacking tryptophan residues (W22). Thus we propose that SsfS6 is a dimer in solution. The SsfS6 monomer consists of two β/α/β Rossmann-like domains that face each other and form an active site cleft. The N-terminal domain (residues 1–198) comprises seven parallel β-sheets surrounded by six α-helices while the C-terminal domain (residues 214–383) consists of six parallel β-sheets surrounded by seven α-helices. A long loop (resides 184–213) containing a small conserved α-helix connects the two domains. Several loops are not assignable in the electron density maps of either apo SsfS6 or SsfS6-TDP. A number of SsfS6 structurally related proteins were identified by Dali29 from which the top five hits (27%~31% sequence identity with SsfS6) are listed as aligned sequences [Fig. S1]. The best of these was the C-GT UrdGT2 (PDB ID 2P6P, Z = 46.1), while the remaining four were O-GTs. Superposition of SsfS6 and these five comparators confirms common folding features of GT-B family proteins.
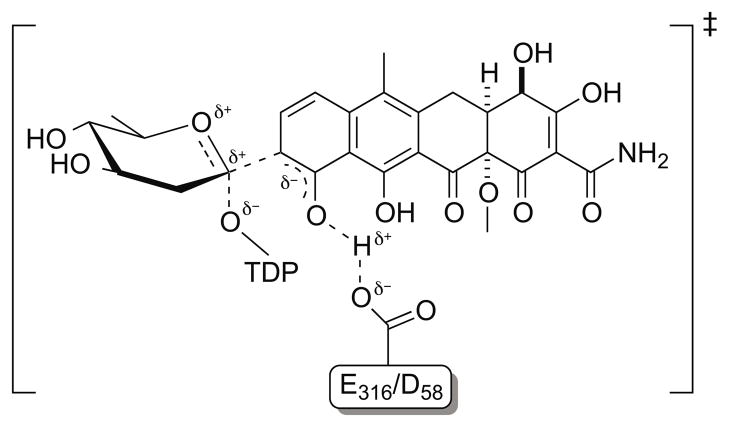
Figure 2.
The proposed mechanism of C-glycosylation catalyzed by SsfS6.
TDP active site
The SsfS6-TDP complex structure reveals TDP bound to the C-terminal domain of SsfS6 with its pyrophosphate pointing toward the active site [Fig. 2(C)]. Interactions between TDP and SsfS6 are multi-faceted and include interactions with thymine, deoxyribose and the pyrophosphate of TDP. Thymine interactions include hydrogen bonding between thymine O2 and L279 main chain NH, thymine N3 and F277 main chain NH, and thymine O4 and V256 main chain NH. For the deoxyribose, the NH2 of N196 forms a hydrogen bond with the TDP 3′-OH group and also likely excludes UDP due to anticipated steric infringement with the 2′-OH group. TDP specificity of SsfS6 may also derive from the substitution of T300 for a conserved glutamate within UDP-utilizing enzymes (such as UGT71G1, 2ACW),30 which, in the latter, is important for hydrogen bonding interactions with the 2′-OH and 3′-OH groups of UDP ribose, further stabilized by a conserved glutamine. In SsfS6, T300 also coordinates N196 to help select for the TDP-sugar and L279 substitutes for the above-mentioned glutamine.
In most GT-B enzymes, the NDP-sugar pyrophosphate is stabilized by a neighboring “H-X3-G-T loop”31 which corresponds to SsfS6 292HGGHGT297. Important contributors to pyrophosphate hydrogen bonding include the side chain NE2 group of H292 and the main chain NH groups of H295, G296 and T297. A comparison of the apo and TDP-bound forms of SsfS6 reveals a shift of several residues (255AVS257 and 276QFP278) near the active site upon ligand binding. In addition, the loop 220GTRVPL225 became ordered upon ligand binding wherein main chain NH group of T221 engaged hydrogen bonding interactions with the TDP pyrophosphate [Fig. 2(D)].
Proposed mechanism
Several research groups have proposed that inverting C-glycosylation is achieved by a single nucleophilic displacement at the anomeric carbon, reminiscent of an inverting O-glycosyltransferase mechanism.9,15,32 In C-glycosylation, activation of the aromatic carbon nucleophile is believed to derive from deprotonation of an adjacent hydroxyl (i.e., the C-10 hydroxyl of the SsfS6 D ring). Indirect evidence to support this mechanism includes the ability of the C-GT UrdGT2 to glycosylate surrogate substrates which contain a heteroatom nucleophile at the C-glycosylation site15 (e.g., analogous to C-9 of the SsfS6 D ring) and recent engineering studies which demonstrated the swapping of glycosidic bond-type specificity between C-GTs and O-GTs.14,33
Based upon the determined structure of the SsfS6-TDP complex, we postulate either D58 or E316 may serve as the active-site base and the side chain of H292 may be used to stabilize the β-phosphate during catalysis. While the nature of the actual SsfS6 aglycon remains to be elucidated,6,34 and co-crystallization experiments with a range of tetracycline analogs were unsuccessful, molecular docking was employed in an attempt to glean additional information.25 Four docked tetracycline-like aglycon orientations were identified as potential poses in which the protein SsfS6 could productively interact [Fig. S2]. These four poses were given to three independent scoring functions35–37 for validation of the docked poses [Table S1]. This analysis was consistent with the putative role of D58 or E316 as the active site base and also revealed the side chain of R222 to occupy the proposed space for olivose (within the product glycoside) and thereby be involved in product release. In summary, while the unknown nature of the SsfS6 aglycon prohibits biochemical study of this intriguing catalyst, the structural studies presented herein are consistent with a putative canonical C-GT mechanism [Fig. 2] and highlight key residues that may impact upon catalysis and/or substrate specificity. As such, this work may advance the study of SF2575 biosynthesis and also set the stage for future protein engineering work on SsfS6.
Supplementary Material
Acknowledgments
This material is based upon work supported by the Protein Structure Initiative project, Enzyme Discovery for Natural Product Biosynthesis (NATPRO) with NIH/NIGMS grant number U01GM098248 and a NIH MERIT Award (AI52218) to JST. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. The authors would also like to thank the Center for Eukaryotic Structural Genomics for various contributions, Dr. Guojong Wang (University of Kentucky) for providing premithramycinone, and Mr. Heng Keat Tam (University of Freiburg) and Dr. Hongnan Cao (Rice University) for their helpful comments on this paper.
References
- 1.Walsh CT, Fischbach MA. Natural products version 2. 0: connecting genes to molecules. J Am Chem Soc. 2010;132(8):2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu HW, Thorson JS. Pathways and mechanisms in the biogenesis of novel deoxysugars by bacteria. Annu Rev Microbiol. 1994;48:223–256. doi: 10.1146/annurev.mi.48.100194.001255. [DOI] [PubMed] [Google Scholar]
- 3.Weymouth-Wilson AC. The role of carbohydrates in biologically active natural products. Nat Prod Rep. 1997;14(2):99–110. doi: 10.1039/np9971400099. [DOI] [PubMed] [Google Scholar]
- 4.Hatsu M, Sasaki T, Gomi S, Kodama Y, Sezaki M, Inouye S, Kondo S. A new tetracycline antibiotic with antitumor activity. II. The structural elucidation of SF2575. J Antibiot (Tokyo) 1992;45(3):325–330. doi: 10.7164/antibiotics.45.325. [DOI] [PubMed] [Google Scholar]
- 5.Hatsu M, Sasaki T, Watabe H, Miyadoh S, Nagasawa M, Shomura T, Sezaki M, Inouye S, Kondo S. A new tetracycline antibiotic with antitumor activity. I. Taxonomy and fermentation of the producing strain, isolation and characterization of SF2575. J Antibiot (Tokyo) 1992;45(3):320–324. doi: 10.7164/antibiotics.45.320. [DOI] [PubMed] [Google Scholar]
- 6.Pickens LB, Kim W, Wang P, Zhou H, Watanabe K, Gomi S, Tang Y. Biochemical analysis of the biosynthetic pathway of an anticancer tetracycline SF2575. J Am Chem Soc. 2009;131(48):17677–17689. doi: 10.1021/ja907852c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pickens LB, Tang Y. Decoding and engineering tetracycline biosynthesis. Metab Eng. 2009;11(2):69–75. doi: 10.1016/j.ymben.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P, Gao X, Chooi YH, Deng Z, Tang Y. Genetic characterization of enzymes involved in the priming steps of oxytetracycline biosynthesis in Streptomyces rimosus. Microbiology. 2011;157(Pt 8):2401–2409. doi: 10.1099/mic.0.048439-0. [DOI] [PubMed] [Google Scholar]
- 9.Bililign T, Hyun CG, Williams JS, Czisny AM, Thorson JS. The hedamycin locus implicates a novel aromatic PKS priming mechanism. Chemistry & Biology. 2004;11(7):959–969. doi: 10.1016/j.chembiol.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 10.Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328(2):307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 11.Bojarová P, Kren V. Glycosidases: a key to tailored carbohydrates. Trends Biotechnol. 2009;27(4):199–209. doi: 10.1016/j.tibtech.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 13.Palcic MM. Glycosyltransferases as biocatalysts. Curr Opin Chem Biol. 2011;15(2):226–233. doi: 10.1016/j.cbpa.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 14.Härle J, Günther S, Lauinger B, Weber M, Kammerer B, Zechel DL, Luzhetskyy A, Bechthold A. Rational design of an aryl-C-glycoside catalyst from a natural product O-glycosyltransferase. Chemistry & Biology. 2011;18(4):520–530. doi: 10.1016/j.chembiol.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Dürr C, Hoffmeister D, Wohlert SE, Ichinose K, Weber M, Von Mulert U, Thorson JS, Bechthold A. The glycosyltransferase UrdGT2 catalyzes both C- and O-glycosidic sugar transfers. Angew Chem Int Ed Engl. 2004;43(22):2962–2965. doi: 10.1002/anie.200453758. [DOI] [PubMed] [Google Scholar]
- 16.Lee DY, He M. Recent advances in aryl C-glycoside synthesis. Curr Top Med Chem. 2005;5(14):1333–1350. doi: 10.2174/156802605774643042. [DOI] [PubMed] [Google Scholar]
- 17.Bililign T, Griffith BR, Thorson JS. Structure, activity, synthesis and biosynthesis of aryl-C-glycosides. Nat Prod Rep. 2005;22(6):742–760. doi: 10.1039/b407364a. [DOI] [PubMed] [Google Scholar]
- 18.Isiorho EA, Liu HW, Keatinge-Clay AT. Structural studies of the spinosyn rhamnosyltransferase, SpnG. Biochemistry. 2012;51(6):1213–1222. doi: 10.1021/bi201860q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sreenath HK, Bingman CA, Buchan BW, Seder KD, Burns BT, Geetha HV, Jeon WB, Vojtik FC, Aceti DJ, Frederick RO, Phillips GN, Jr, Fox BG. Protocols for production of selenomethionine-labeled proteins in 2-L polyethylene terephthalate bottles using auto-induction medium. Protein Expr Purif. 2005;40(2):256–267. doi: 10.1016/j.pep.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Terwilliger TC, Adams PD, Read RJ, Mccoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, Hung LW. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallographica Section D-Biological Crystallography. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeLano WL. Use of PYMOL as a communications tool for molecular science. Abstracts of Papers of the American Chemical Society. 2004;228:U313–U314. [Google Scholar]
- 25.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 27.Chang A, Singh S, Helmich KE, Goff RD, Bingman CA, Thorson JS, Phillips GN., Jr Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity. Proc Natl Acad Sci U S A. 2011;108(43):17649–17654. doi: 10.1073/pnas.1108484108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mittler M, Bechthold A, Schulz GE. Structure and action of the C-C bond-forming glycosyltransferase UrdGT2 involved in the biosynthesis of the antibiotic urdamycin. J Mol Biol. 2007;372(1):67–76. doi: 10.1016/j.jmb.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao H, He X, Achnine L, Blount JW, Dixon RA, Wang X. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell. 2005;17(11):3141–3154. doi: 10.1105/tpc.105.035055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulichak AM, Lu W, Losey HC, Walsh CT, Garavito RM. Crystal structure of vancosaminyltransferase GtfD from the vancomycin biosynthetic pathway: interactions with acceptor and nucleotide ligands. Biochemistry. 2004;43(18):5170–5180. doi: 10.1021/bi036130c. [DOI] [PubMed] [Google Scholar]
- 32.Fischbach MA, Lin H, Liu DR, Walsh CT. In vitro characterization of IroB, a pathogen-associated C-glycosyltransferase. Proc Natl Acad Sci U S A. 2005;102(3):571–576. doi: 10.1073/pnas.0408463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutmann A, Nidetzky B. Switching between O- and C-Glycosyltransferase through Exchange of Active-Site Motifs. Angew Chem Int Ed Engl. 2012;51(51):12879–12883. doi: 10.1002/anie.201206141. [DOI] [PubMed] [Google Scholar]
- 34.Pickens LB, Sawaya MR, Rasool H, Pashkov I, Yeates TO, Tang Y. Structural and biochemical characterization of the salicylyl-acyltranferase SsfX3 from a tetracycline biosynthetic pathway. J Biol Chem. 2011;286(48):41539–41551. doi: 10.1074/jbc.M111.299859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang R, Lai L, Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J Comput Aided Mol Des. 2002;16(1):11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- 36.Neudert G, Klebe G. DSX: a knowledge-based scoring function for the assessment of protein-ligand complexes. J Chem Inf Model. 2011;51(10):2731–2745. doi: 10.1021/ci200274q. [DOI] [PubMed] [Google Scholar]
- 37.Fan H, Schneidman-Duhovny D, Irwin JJ, Dong G, Shoichet BK, Sali A. Statistical potential for modeling and ranking of protein-ligand interactions. J Chem Inf Model. 2011;51(12):3078–3092. doi: 10.1021/ci200377u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.