Abstract
Optimizing multivalency: Clusters of Man9 glycans which are recognized by broadly-neutralizing anti-HIV antibody 2G12 have potential as HIV vaccines. However, optimal recognition by 2G12 requires optimal clustering of glycans. Using our recently described SELMA technique (SELection with Modified Aptamers), we have developed Man9 clusters in which glycans are supported by DNA sequences selected from among 2 × 1013 variants.
Keywords: carbohydrates, cluster effect, directed evolution, glycoconjugates, multivalency
Carbohydrate-protein interactions mediate numerous important biological events, such as cell signalling, recognition and adhesion, and host-pathogen recognition.[1] The affinity of individual glycans for individual protein binding sites is normally very low, with Kd’s in the mM range. High avidity binding is achieved by multivalency, wherein multiple glycans within a cluster bind cooperatively to multiple sites in a single protein or in several clustered proteins.[2] This concept has been extensively applied to the design of synthetic glycan clusters that bind to interesting biological targets.[3]
However, the number of glycans (valency) is not the only important feature in multivalent ligand design. For applications in which the carbohydrate binding sites are separated by a fixed distance, avidity is also greatly affected by the spacing and orientation of glycans within the cluster. Although glycans attached by long flexible linkers can easily span target binding sites, the resulting avidity is suboptimal due to entropic costs, as the unbound ligand has many more possible conformations than the bound one.[2b] Moreover, flexible glycan clusters may not be specific for a single target. Design of more conformationally-defined glycocluster ligands is desirable due to the potential to achieve high avidity and specificity, but is challenging, as small deviations from ideal glycan arrangement may completely prevent the desired binding interaction. For these rigid glycoclusters, rational design of the optimal structure may require extensive trial and error as well as structural insight.
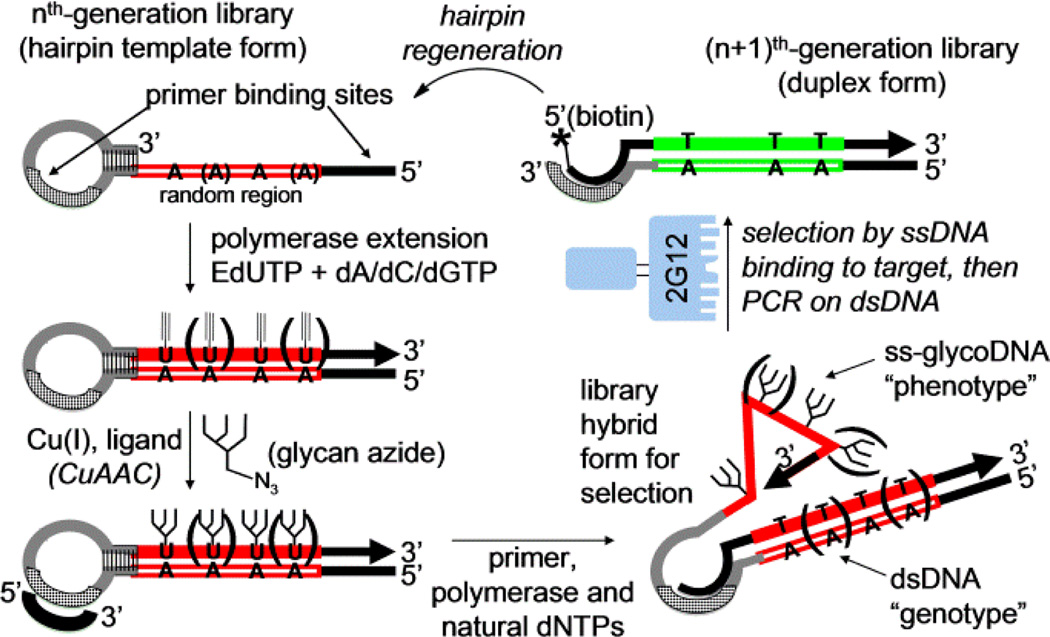
As an alternative to rational glycocluster design, we have developed SELMA (SELection with Modified Aptamers, Scheme 1).[4] This technique enables evolution of DNA aptamers bearing large covalent modifications such as oligosaccharides.[5] The process begins with a library template of synthetic DNA hairpins, containing a single-stranded random sequence region and a 3’-terminal self-priming patch of self-complementary sequence. Polymerase extension with EdU (alkynyl U) substituted for T installs alkynes at every position in which the single stranded template contains an A, and then CuAAAC chemistry[6] attaches glycans (or potentially other large modifications) to each of these alkynes. The glycosylated strand is then displaced by extension of a small primer inside the hairpin loop, using all-natural dNTPs. After this step, the single-stranded glycosylated DNA can adopt a sequence-dependent fold, whereas the covalently-attached dsDNA contains a copy of the same sequence, though with all natural bases. This “genotype” sequence can readily be amplified by PCR if the “phenotype” portion enables survival during selection. In our original report, we applied SELMA to the evolution of decavalent clusters of Man4 oligosaccharides, which bound to HIV broadly-neutralizing antibody 2G12 with Kd’s of ~250–500 nM. In the present paper, we describe further developments of this method to afford Man9 glycoclusters of higher avidity, smaller size, and lower valency.
scheme 1.
Overview of SELMA (SELection with Modified Aptamers)
2G12 is a target of interest because it neutralizes a broad range of HIV strains[7] and confers sterilizing immunity in rhesus macaque models of HIV infection.[8] As 2G12 binds to several Man9 glycans on the HIV envelope protein gp120,[9] there has been considerable effort to design Man9 glycoclusters which mimic the 2G12 epitope and might re-elicit 2G12-like antibodies.[10] Immunogenicity studies with Man9 glycoclusters designed so far have been unsuccessful at eliciting a 2G12-like antibody response, so design of new constructs which might mimic the true Man9 presentation on gp120 are desirable.
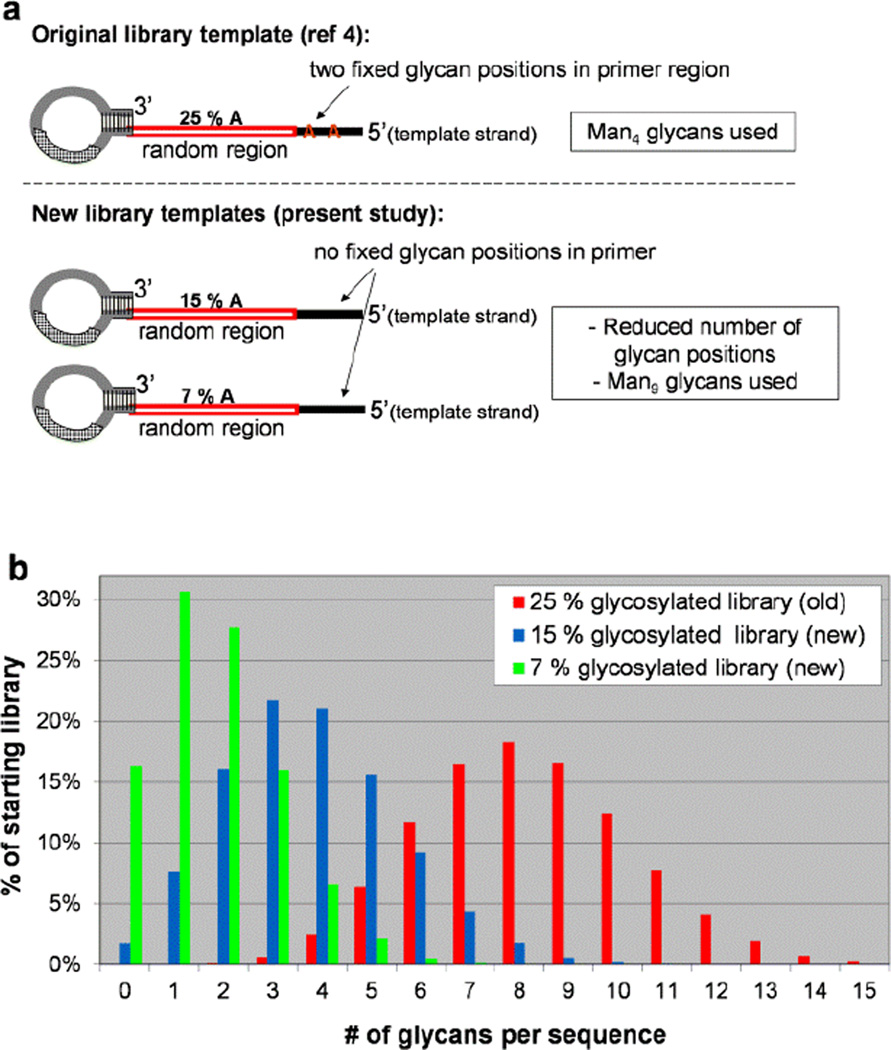
For the present study (Figure 1), we designed our SELMA selection to differ from the original study[4] in 2 respects: 1) glycoclusters would contain the full Man9 oligosaccharide rather than the Man4 fragment which was used previously,[11] and 2) we would begin selection with libraries biased to contain species with fewer glycans. As depicted in Scheme 1, the positions of glycans in library sequences are dictated their CuAAAC attachment to “alkynyl-U” (EdU) bases, which are in turn located opposite A bases in the template strand.[6] Thus, to create starting libraries with less glycosylation, we made two modifications which reduce the number of A’s in the original library template sequences (Figure 1a). Whereas the original library contained two fixed A’s in a primer region and 25% A content in the random sequence region, our two new libraries contained no A’s in the constant primer regions, and a lower percentage of A’s (7% or 15%) in the random sequence region. Figure 1b shows the statistically predicted percentages of different valencies within each starting library; whereas species containing 6–10 glycans were prevalent in the original library, the new starting libraries contain larger percentages of species with fewer glycans.
Figure 1.
(a) new library templates, containing reduced A’s, leading to reduced glycosylation sites (b) predicted multivalency profile of each library (% of sequences with 0, 1, 2, 3, …, 15 glycans)
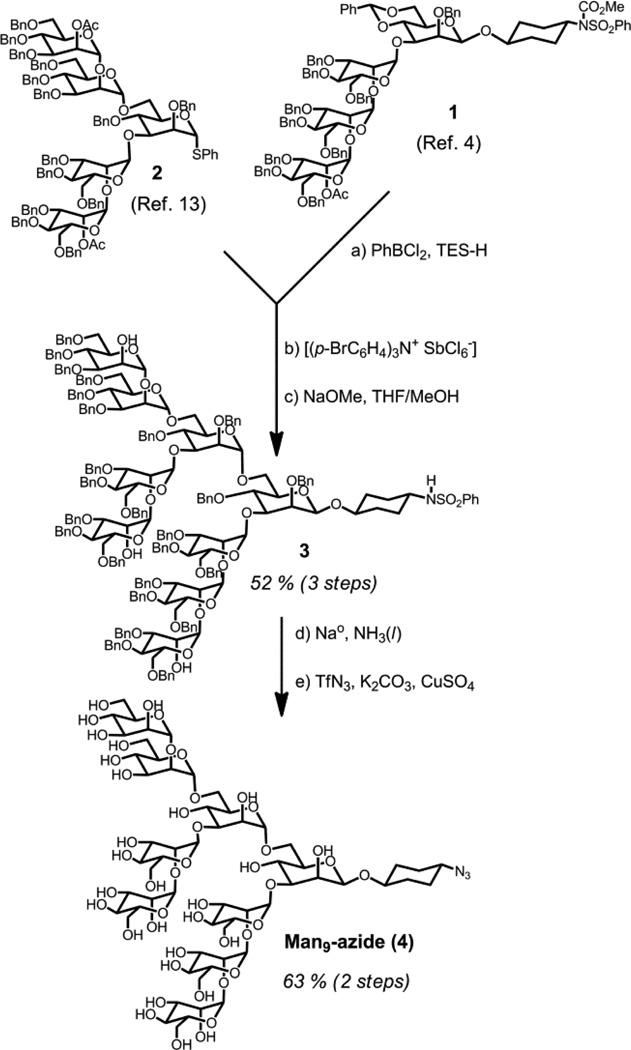
To enable selection with Man9 oligosaccharides, we prepared Man9 azide by the convergent block assembly method shown in Scheme 2. Man4 tetrasaccharide 1, containing a reducing-terminal β-mannose and cyclohexyl linker was prepared as in our previous study.[4] Deprotection of the benzylidene 6-hydroxyl, followed by Sinay glycosylation[12] with Man5 donor 2[13] and partial deprotection afforded Man9 derivative 3 in 52 % yield over three steps. Global deprotection,[14] followed by Wong’s azidation protocol,[15] afforded 4 in 63 % yield (nearly 100 mg) over two steps.
scheme 2.
Synthesis of Man9 azide. a) PhBCl2, TES-H, 4Å MS, 80 %; b) 2, Sinaÿ Reagent [(p-BrC6H4)3N+ SbCl6ȡ], 4Å MS, 0 °C, MeCN, 81 %; c) NaOMe, MeOH/THF, 81 %; d) Na metal, THF, −78 °C, NH3(l); e) TfN3, K2CO3, CuSO4, DCM/MeOH/H2O, 63 % (2 steps).
With Man9 derivative 4 in hand, we then initiated SELMA selections with 40 pmol each (2 × 1013 sequences) of the 7 % and 15 % libraries shown in Figure 1. Briefly, libraries generated as outlined in Scheme 1 (see Supporting Information for details) were incubated with mAb 2G12, and the bound complexes were captured with protein A magnetic beads. A counterselection with only protein A beads was run in rounds 2, 4 and 6 to remove potential protein A binders from the library. Stringency was increased by decreasing 2G12 concentration from 50 nM in rounds 1–4 to 10, 5 and 1.4 nM in rounds 5, 6 and 7, respectively. Library enrichment was monitored by observing the decrease in the number of PCR cycles required to regenerate the library after selection. After seven rounds, enrichment levelled off and the libraries were cloned, yielding 31 unique sequences (among 93 sequence reads, see SI Table 3).
These sequences contained from 3 to 12 glycosylation sites, with an average of 7.9, as compared to 10.5 in the sequences obtained from our original 25 % glycosylated library selection. Filter binding studies on nine representative sequences (Table 1) showed that these glycoclusters were recognized by 2G12 with Kd’s ranging from 310 nM to > 1µM. Although these sequences exhibited moderate reductions in Kd and valency compared with those from our original library (compare with entry 9), CLUSTAL sequence alignment[16] (SI Table 3) revealed several interesting features. Most sequences resulting from the new selections contained a consensus motif (Table 1, in yellow), and strikingly, this sequence was present in clones from both libraries. Moreover, the motif was located at various positions, either early or late, within different sequences. This suggests that the multiple clones containing this motif resulted from convergent evolution, and were not derived from mutations of a single clone or cross-contamination between libraries.
Table 1.
Representative 5‘->3‘ sequences after 7 rounds of SELMA selection, Kd for binding to 2G12, and consensus sequence.
| Clone ID | Sequence ( = glycosylated EdU, yellow = consensus, blue=constant primer regions) = glycosylated EdU, yellow = consensus, blue=constant primer regions) |
Kd | Bmax |
|---|---|---|---|
| 7A8-4 | > 1 µM | NDa | |
| 7A8-9 | > 1 µM | NDa | |
| 15A8-B | > 1 µM | NDa | |
| 15A8-F | 410±90 | 0.85±0.10 | |
| M3-HH | 320±80 | 0.65±0.08 | |
| 7A8-1 | 310±110 | 0.73±0.13 | |
| 7A8-8 | 320±10 | 0.86±0.02 | |
| 15A8-J | n.b.b | 0 | |
| M3-30 | n.b.b | 0 | |
| F1 controlc | 430±130 | 0.33±0.06 |
Not determined; binding curve was linear up to highest 2G12 concentration tested.
no binding.
10-Man4 winner from previously published selection with 25 % glycosylated library (Ref 4).
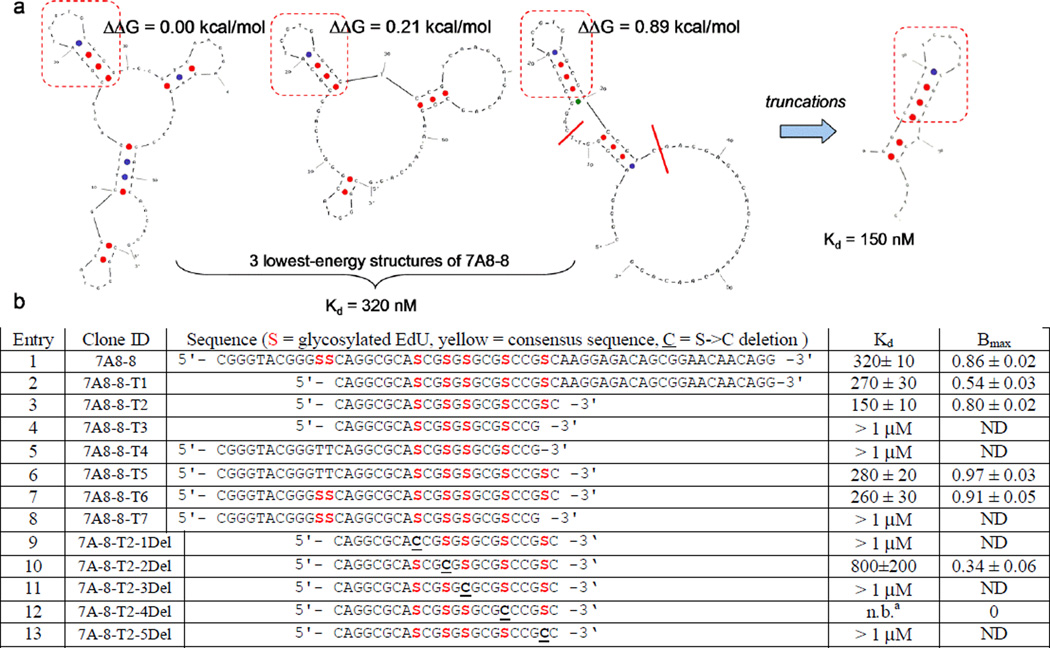
The existence of the consensus motif in most selection winners suggested this region as a common site of 2G12 recognition. To investigate this further, we performed Mfold secondary structure analyses[17] on representative sequences (see Supporting Information). As Mfold does not take into account the potential major effects of our large unnatural base modifications we cautiously regarded the computational output only as a starting point to suggest further experiments. For most sequences, Mfold predicted most stable secondary structures in which the “consensus” motif was present at the end of a stem-loop region. In the case of clone 7A8-8 from the 7 % glycosylated library, we noted that a large region of sequence outside the consensus motif (Figure 2a, regions outside the red boxes) was not predicted to have any predominant structure. We thus prepared truncated versions of 7A8-8, removing regions at the 3’ and 5’ ends. In contrast to our previous work with the 25 % glycosylated library, we found in this case that large truncations were not only tolerated, but beneficial.
Figure 2.
a) M-fold structure predictions of clone 7A8-8 and suggested truncation. b) Effect of truncations/deletions on Kd of 2G12 recognition. [a] no binding
Truncation at the 5’ end (7A-8-T1, Figure 2b, entry 2) improved 2G12 recognition slightly, from 320 to 270 nM, even though the number of glycosylation sites decreased from 7 to 5. Additional truncation at the 3’ end (7A-8-T2, entry 3) further improved binding to 150 nM. Although removal of two more bases from the 3’end appeared to be allowable on the basis of the M-fold structure, this truncation (7A-8-T3, entry 4) eliminated recognition by 2G12, showing that glycosylation at this 5th site in the 7A-8-T2 sequence is important. As expected, binding of these more extreme 3’ truncations was not rescued by lesser truncations at the 5’ end (entries 5 and 8). Finally, truncation only at the 3’ end resulted in Kd’s similar to truncation only at the 5’ end (entries 6 and 7).
We were interested in whether all five of the glycosylation sites in 7A-8-T2 were necessary for 2G12 recognition, or whether further reductions in valency would be tolerated. To this end, we prepared a series of (S->C) mutants, corresponding to deletion of the glycan at each of the five remaining positions (Figure 2b, entries 9–13). We were surprised to find that deletion of glycan at any of these positions resulted in major loss of binding to 2G12. Based on published 2G12 structural studies, it is unlikely that all five glycans bind to 2G12 simultaneously. However, each glycan likely plays a major structural role, considering that glycans comprise 50% of the molecular weight of this small aptamer.
In conclusion, we have found that SELMA selection of Man9-cluster libraries biased toward low carbohydrate valency leads to selection winners with significantly lower valency than our previous library and modestly better recognition by 2G12.[4] Interestingly, these selection winners contained a conserved 2G12 recognition motif; moreover, it was found that the region outside this motif could be significantly truncated, which actually led to improved 2G12 recognition with fewer glycans. Compared with monovalent Man9 glycan (Kd = 180 µM), these pentavalent glycoclusters exhibit up to a ~1000-fold binding enhancement (Kd = 150 nM), though they are still 30-fold less antigenic than Wong’s nonavalent Man9 dendrimer.[10i, 18] Our truncated oligo, 7A8-8-T2, is relatively short and should be amenable to further structural study by NMR and X-ray crystallography. Most importantly, this construct is a candidate for immunogenicity studies to re-elicit 2G12-like antibodies.
Supplementary Material
Acknowledgments
I.J.K. gratefully acknowledges Brandeis University and the NIH (R01 AI090745). We are grateful to Marcus Long for insightful discussions, and Polymun Scientific for a gift of mAb 2G12.
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the
References
- 1.“Part Four – Protein-Carbohydrate Interactions” Ch 13–20, in Gabius Hans-Joachim., editor. The Sugar Code. Weinheim: Wiley-Blackwell, Wiley-VCH Verlag GmbH & Co. KGaA; 2009. pp. 233–346.
- 2.a) Mammen M, Choi S-K, Whitesides GM. Angew. Chem. Int. Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]; b) Fasting C, Schalley CA, Weber M, Seitz O, Hecht S, Koksch B, Dernedde J, Graf C, Knapp EW, Haag R. Angew. Chem. Int. Ed. 2012;51:10472–10498. doi: 10.1002/anie.201201114. [DOI] [PubMed] [Google Scholar]
- 3.a) Levine PM, Carberry TP, Holub JM, Kirshenbaum K. MedChemComm. 2013;4:493–509. [Google Scholar]; b) Wittmann V, Pieters RJ. Chem. Soc. Rev. 2013;42:4492–4503. doi: 10.1039/c3cs60089k. [DOI] [PubMed] [Google Scholar]
- 4.MacPherson IS, Temme JS, Habeshian S, Felczak K, Pankiewicz K, Hedstrom L, Krauss IJ. Angew. Chem. Int. Ed. 2011;50:11238–11242. doi: 10.1002/anie.201105555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.SELEX has previously been performed with dNTP’s bearing modifications which are tolerated by polymerases. For selected examples, see: Keefe AD, Cload ST. Curr. Opin. Chem. Biol. 2008;12:448–456. doi: 10.1016/j.cbpa.2008.06.028. Li MY, Lin N, Huang Z, Du LP, Altier C, Fang H, Wang BH. J. Am. Chem. Soc. 2008;130:12636–12638. doi: 10.1021/ja801510d. Hollenstein M, Hipolito CJ, Lam CH, Perrin DM. ChemBioChem. 2009;10:1988–1992. doi: 10.1002/cbic.200900314. Vaught JD, Bock C, Carter J, Fitzwater T, Otis M, Schneider D, Rolando J, Waugh S, Wilcox SK, Eaton BE. J. Am. Chem. Soc. 2010;132:4141–4151. doi: 10.1021/ja908035g. for selected examples of carbohydrates attached to nucleic acids, see: Matsuura K, Hibino M, Yamada Y, Kobayashi K. J. Am. Chem. Soc. 2000;123:357–358. doi: 10.1021/ja001945j. Adinolfi M, De Napoli L, Di Fabio G, Iadonisi A, Montesarchio D. Org. Biomol. Chem. 2004;2:1879–1886. doi: 10.1039/b401819b. Matsuura K, Hibino M, Ikeda T, Yamada Y, Kobayashi K. Chem. Eur. J. 2004;10:352–359. doi: 10.1002/chem.200305465. Yamada Y, Matsuura K, Kobayashi K. Bioorg. Med. Chem. 2005;13:1913–1922. doi: 10.1016/j.bmc.2005.01.021. Matsui M, Nishiyama Y, Ueji SI, Ebara Y. Bioorg. Med. Chem. Lett. 2007;17:456–460. doi: 10.1016/j.bmcl.2006.10.020. D'Onofrio J, Petraccone L, Martino L, Di Fabio G, Iadonisi A, Balzarini J, Giancola C, Montesarchio D. Bioconjugate Chem. 2008;19:607–616. doi: 10.1021/bc7003395. Gorska K, Huang K-T, Chaloin O, Winssinger N. Angew. Chem. Int. Ed. 2009;48:7695–7700. doi: 10.1002/anie.200903328. Moni L, Pourceau G, Zhang J, Meyer A, Vidal S, Souteyrand E, Dondoni A, Morvan F, Chevolot Y, Vasseur JJ, Marra A. ChemBioChem. 2009;10:1369–1378. doi: 10.1002/cbic.200900024. Kiviniemi A, Virta P, Lonnberg H. Bioconjugate Chem. 2010;21:1890–1901. doi: 10.1021/bc100268w. Ciobanu M, Huang KT, Daguer JP, Barluenga S, Chaloin O, Schaeffer E, Mueller CG, Mitchell DA, Winssinger N. Chem. Commun. 2011;47:9321–9323. doi: 10.1039/c1cc13213j. Scheibe C, Bujotzek A, Dernedde J, Weber M, Seitz O. Chem. Sci. 2011;2:770–775. Matsui M, Ebara Y. Bioorg. Med. Chem. Lett. 2012;22:6139–6143. doi: 10.1016/j.bmcl.2012.08.028. Scheibe C, Wedepohl S, Riese SB, Dernedde J, Seitz O. ChemBioChem. 2013;14:236–250. doi: 10.1002/cbic.201200618.
- 6.a) Kolb HC, Finn MG, Sharpless KB. Angew. Chem.-Int. Edit. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]; b) Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew. Chem.-Int. Edit. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]; c) Gierlich J, Burley GA, Gramlich PM, Hammond DM, Carell T. Org. Lett. 2006;8:3639–3642. doi: 10.1021/ol0610946. [DOI] [PubMed] [Google Scholar]; d) Gierlich J, Gutsmiedl K, Gramlich PM, Schmidt A, Burley GA, Carell T. Chem. Eur. J. 2007;13:9486–9494. doi: 10.1002/chem.200700502. [DOI] [PubMed] [Google Scholar]
- 7.Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. J. Virol. 2004;78:13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, Beary H, Hayes D, Frankel SS, Birx DL, Lewis MG. Nat. Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]; b) Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, Forthal DN, Koff WC, Watkins DI, Burton DR. PLoS Pathog. 2009;5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. J. Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, Kwong PD, Moore JP. J. Virol. 2002;76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. J. Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]; e) Calarese DA, Lee H-K, Huang C-Y, Best MD, Astronomo RD, Stanfield RL, Katinger H, Burton DR, Wong C-H, Wilson IA. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13372–13377. doi: 10.1073/pnas.0505763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.For selected approaches, see: Singh S, Ni JH, Wang LX. Bioorg. Med. Chem. Lett. 2003;13:327–330. doi: 10.1016/s0960-894x(02)01025-9. Li HG, Wang LX. Org. Biomol. Chem. 2004;2:483–488. doi: 10.1039/b314565d. Wang LX, Ni J, Singh S, Li H. Chem. Biol. 2004;11:127–134. doi: 10.1016/j.chembiol.2003.12.020. Ni JH, Song HJ, Wang YD, Stamatos NM, Wang LX. Bioconjugate Chem. 2006;17:493–500. doi: 10.1021/bc0502816. Wang J, Li H, Zou G, Wang LX. Org. Biomol. Chem. 2007;5:1529–1540. doi: 10.1039/b702961f. Krauss IJ, Joyce JG, Finnefrock AC, Song HC, Dudkin VY, Geng X, Warren JD, Chastain M, Shiver JW, Danishefsky SJ. J. Am. Chem. Soc. 2007;129:11042–11044. doi: 10.1021/ja074804r. Joyce JG, Krauss IJ, Song HC, Opalka DW, Grimm KM, Nahas DD, Esser MT, Hrin R, Feng MZ, Dudkin VY, Chastain M, Shiver JW, Danishefsky SJ. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15684–15689. doi: 10.1073/pnas.0807837105. Luallen RJ, Lin JQ, Fu H, Cai KK, Agrawal C, Mboudjeka I, Lee FH, Montefiori D, Smith DF, Doms RW, Geng Y. J. Virol. 2008;82:6447–6457. doi: 10.1128/JVI.00412-08. Wang SK, Liang PH, Astronomo RD, Hsu TL, Hsieh SL, Burton DR, Wong CH. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3690–3695. doi: 10.1073/pnas.0712326105. Astronomo RD, Kaltgrad E, Udit AK, Wang SK, Doores KJ, Huang CY, Pantophlet R, Paulson JC, Wong CH, Finn MG, Burton DR. Chem. Biol. 2010;17:357–370. doi: 10.1016/j.chembiol.2010.03.012. Doores KJ, Fulton Z, Hong V, Patel MK, Scanlan CN, Wormald MR, Finn MG, Burton DR, Wilson IA, Davis BG. Proc. Natl. Acad. Sci. U. S. A. 2010;107:17107–17112. doi: 10.1073/pnas.1002717107. Marradi M, Di Gianvincenzo P, Enriquez-Navas PM, Martinez-Avila OM, Chiodo F, Yuste E, Angulo J, Penades S. J. Mol. Biol. 2011;410:798–810. doi: 10.1016/j.jmb.2011.03.042. Clark BE, Auyeung K, Fregolino E, Parrilli M, Lanzetta R, De Castro C, Pantophlet R. Chem. Biol. 2012;19:254–263. doi: 10.1016/j.chembiol.2011.12.019.
- 11.The Man4 fragment used in our previous approach (Ref. 4) is known to be responsible for most of the contacts with 2G12, according to X-ray crystallographic data (Ref. 10d,e)
- 12.Sinay P. Pure and Applied Chemistry. 1991;63:519–528. [Google Scholar]
- 13.Geng X, Dudkin VY, Mandal M, Danishefsky SJ. Angew. Chem. Int. Ed. 2004;43:2562–2565. doi: 10.1002/anie.200353626. [DOI] [PubMed] [Google Scholar]
- 14.Iserloh U, Dudkin V, Wang Z-G, Danishefsky SJ. Tetrahedron Lett. 2002;43:7027–7030. [Google Scholar]
- 15.Nyffeler PT, Liang C-H, Koeller KM, Wong C-H. J. Am. Chem. Soc. 2002;124:10773–10778. doi: 10.1021/ja0264605. [DOI] [PubMed] [Google Scholar]
- 16.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuker M. Nucl. Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The assay used in Wong’s study (ref. 10i), based on competition binding to Man9-printed glass slides, was sensitive to glycan printing density. Given the large differences between our solution-phase assay format and their surface-based assay, caution should be exercised in comparison of the two types of data
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.