Abstract
The aryl hydrocarbon receptor (AHR) is a transcription factor and environmental sensor that regulates expression of genes involved in drug-metabolism and cell cycle regulation. Chromatin immunoprecipitation analyses, Ahr ablation in mice and studies with orthologous genes in invertebrates suggest that AHR may also play a significant role in embryonic development. To address this hypothesis, we studied the regulation of Ahr expression in mouse embryonic stem cells and their differentiated progeny. In ES cells, interactions between OCT3/4, NANOG, SOX2 and Polycomb Group proteins at the Ahr promoter repress AHR expression, which can also be repressed by ectopic expression of reprogramming factors in hepatoma cells. In ES cells, unproductive RNA polymerase II binds at the Ahr transcription start site and drives the synthesis of short abortive transcripts. Activation of Ahr expression during differentiation follows from reversal of repressive marks in Ahr promoter chromatin, release of pluripotency factors and PcG proteins, binding of Sp factors, establishment of histone marks of open chromatin, and engagement of active RNAPII to drive full-length RNA transcript elongation. Our results suggest that reversible Ahr repression in ES cells holds the gene poised for expression and allows for a quick switch to activation during embryonic development.
Keywords: Ah receptor, Embryonic stem cells, Pluripotency factors, PcG-mediated repression, RNA polymerase II pausing
1. Introduction
AHR is a member of the bHLH/PAS family of transcription factors and the main mediator of teratogenic and carcinogenic toxicities resulting from exposure to planar polycyclic and halogenated aromatic hydrocarbons present in the environment (Hankinson, 1995). Activation by ligand causes AHR to translocate to the nucleus, dissociate from its cytosolic chaperones and heterodimerize with its ARNT partner, also a member of the bHLH/PAS family (Reyes et al., 1992). Binding of AHR-ARNT complexes to AHR response motifs in the promoters of target genes recruits transcription cofactors and associated chromatin remodeling proteins and signals initiation of gene transcription (Hestermann and Brown, 2003; Schnekenburger et al., 2007). Increasing evidence indicates that in addition to the well-known xenobiotic metabolism genes in the Cyp1 family of cytochromes P450, there are other AHR transcriptional targets, including genes involved in cell cycle regulation and morphogenetic processes that may play a vital function during embryonic development (Gasiewicz et al., 2008; Sartor et al., 2009). Such a developmental role may be an evolutionarily conserved primary function of the AHR, a notion supported by the finding that unlike their vertebrate counterparts, AHR orthologs in invertebrates like the fruit fly D. melanogaster and the nematode C. elegans are not activated by xenobiotic ligands but control expression of homeotic genes involved in neuronal specification during development (Emmons et al., 1999; Hahn, 2002; Qin and Powell-Coffman, 2004; Kim et al., 2006). In mice, Ahr ablation leads to impaired vasculature in kidney, liver sinusoid, and eyes of the neonates (Lahvis et al., 2000) with an ensuing cardiovascular disease that might be directly or indirectly the principal cause of other Ahr deficit phenotypes, such as reduced liver size, patent ductus venosus, cardiac hypertrophy, hypertension, and fibrosis (Fernandez-Salguero et al., 1995; Fernandez-Salguero et al., 1997; Lund et al., 2003; Lahvis et al., 2005; Lund et al., 2008).
Several studies have shown a complex pattern of Ahr expression during early mouse embryogenesis. Fertilized eggs at the 1-cell stage show detectable levels of Ahr mRNA (Dey and Nebert, 1998; Wu et al., 2002) and high levels of AHR activity, as determined by an elevated constitutive mRNA level of the AHR target gene Cyp1a1 (Dey and Nebert, 1998). Thereafter, Ahr mRNA expression is completely silenced between the 2- and 8-cell stages and afterwards increases to a detectable level by late pre-implantation blastocysts (Peters and Wiley, 1995; Dey and Nebert, 1998; Wu et al., 2002). In the post-implantation embryo, Ahr mRNA can be demonstrated as early as gestational day 9.5, followed by widespread expansion into almost all developing organs (Abbott et al., 1995; Jain et al., 1998).
Correct reprogramming of the epigenome during embryonic preimplantation stages is essential for the acquisition of pluripotency to ensure the concerted completion of development. The above findings suggest that, concurrent with the time of reprogramming of the embryonic epigenome and establishment of pluripotency in the inner cell mass blastocysts, embryos show low or undetectable levels of Ahr expression. It is reasonable to hypothesize that, although needed for post-implantation developmental stages, a functional AHR might be detrimental to the preimplantation process and needs to be silenced during this period.
In ES cells, the pluripotency factors OCT3/4, NANOG and SOX2 form a transcriptional network that controls the expression of several hundred target genes, either by activating the promoters of self-renewal genes or by silencing the promoters of differentiation associated genes (Christophersen and Helin, 2010). The specificity of this silencing resides in the quick regulatory reversibility requiring the interplay between core pluripotency factors, numerous chromatin remodeling complexes, and paused RNAPII molecules, that primes target genes and allows them to be ready for fast activation when required by morphogenetic signals (Medvedev et al., 2012). The promoters of these transcription factors are simultaneously marked by active and repressive histone modifications (i.e., H3K4me3 and H3K27me3, respectively) (Mikkelsen et al., 2007) and are repressed by Polycomb Group-mediated mechanisms, including recognition by Polycomb repressive complexes PRC-1 and -2 which further block transcript elongation by RNAPII (Stock et al., 2007; Endoh et al., 2012). In this study, we examine Ahr expression during in vitro non-directed differentiation of mouse ES cells. We find that Ahr is silent in these cells, but its expression is quickly restored upon differentiation. ChIP analyses indicate that expression is silenced by the binding of core pluripotency factors and PcG proteins as well as pausing of RNAPII on the Ahr promoter. These results are consistent with the concept that Ahr silencing is required in ES cells and its expression needed for the completion of subsequent morphogenetic events during differentiation.
2. Materials and Methods
2.1 Antibodies and primers
Lists of primary antibodies and primers used in this work are shown in Supplemental Tables S1 and S2.
2.2 Culture of embryonic stem cells and in vitro differentiation
C57BL/6N-C2 mouse ES cells (Gertsenstein et al. 2010) bearing the Ahrb-1 allele coding for the high ligand-affinity Ah receptor, were used throughout this study. Cells were cultured in Dulbecco’s Modified Essential Medium (Gibco, Grand Island, NY) supplied with 15% (vol/vol) Knock-Out Serum Replacement (KO-SR, Invitrogen, Grand Island, NY), 1000 units/ml ESGRO Leukemia Inhibitory Factor (LIF, Millipore, Billlerica, MA), 50 units/ml penicillin 50 μg/ml streptomycin, 2 mM L-glutamine, 0.1 mM 2-mercaptoethanol, 0.1 mM MEM non-essential amino acids (NEAA, Invitrogen), 1 mM sodium pyruvate in a 5% CO2 humidified incubator at 37°C. Tissue culture plates used for ES cells were coated with 0.1% gelatin at room temperature for 15 min. Plates with feeder cells were prepared with mouse embryonic fibroblasts pre-treated with 10 μg/ml mytomycin C (Sigma-Aldrich, St. Louis, MO) plated at density of 5×104 cells/cm2. ES cells were cultured either on gelatin coated plates or feeder plates and passaged every second or third day. In vitro non-directed differentiation (hereafter referred to as simply differentiation) was initiated by forming embryoid bodies (EB) in differentiation media either in 25-μl hanging drops (6×104 cells/ml) or in rotary cultures (1×106 cells/ml, 40 rpm) at 37°C for 3 days. Differentiation medium was ES medium minus LIF and with FBS instead of KO-SR. After 3 days incubation, 100 EBs were transferred per 10-cm tissue culture plate for use at various time intervals.
Mouse hepatoma Hepa-1c1c7 (Hepa-1) cells from the American Type Culture Collection were cultured in DMEM supplemented with 5% (vol/vol) FBS in a 5% CO2 humidified atmosphere at 37°C. Cells were used for experiments when they reached 70 to 80% confluence.
2.3 Whole cell protein extraction and immunoblotting
Cells were harvested after washing twice with PBS. NETN lysis buffer contained 100 mM NaCl, 20 mM Tris-HCl pH 8, 1 mM EDTA, 0.5% NP-40, and 1X complete protease inhibitor cocktail (Roche, Indianapolis, IN). After lysis on ice for 10 min, lysates were sonicated for 2 min and centrifuged at 14,000 rpm at 4°C for 10 min. Protein concentrations were determined by the Bradford assay (Bio-Rad Protein Assay, Bio-Rad, Hercules, CA). Twenty-five micrograms of protein extract were subjected to SDS-polyacrylamide gel electrophoresis and transferred to a 0.45-μm polyvinylidene fluoride membrane (Millipore). Immunoblotting was carried out in TNT buffer containing 20 mM Tris-HCl pH 7.6, 137 mM NaCl, and 0.1% Tween-20. After blocking with 5% non-fat dry milk at room temperature for 2 hours, membranes were probed with the pertinent antibodies followed by species-specific horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Dallas, TX). After washing, immunoblotting signals were visualized with chemiluminescence ECL Western Blotting Substrate (Pierce, Rockford, IL).
2.4 Total RNA isolation, Reverse Transcription, and real-time PCR reactions
Cells were lysed in Iso-RNA lysis reagent (5-Prime, Gaithersbourg, MD) and RNAs were extracted according to the manufacturer’s specifications. Twenty micrograms of total RNA and 20 pmoles of random hexamer primer were denatured at 70°C for 5 minutes and allowed to cool. cDNA was prepared with reverse transcription mix containing 1X reverse transcriptase buffer, 15 μM dNTP, 10 mM DTT, 500 units of Superscript III Reverse Transcriptase (Invitrogen), and 20 unit of RNAsin (Promega, Madison, WI) at 42°C for 2 hours. RNA was removed by treatment in 0.05 N NaOH, and after neutralization cDNA was precipitated with ethanol in the presence of 10 mg glycogen carrier and 300 mM sodium acetate pH 5.2. Precipitates were dissolved in 200 μl ddH2O and 1-μl aliquots were used per PCR reaction. Real-time PCR reactions were done in duplicate in a total volume of 20 μl of SYBR Green PCR Master mix (Applied Biosystems, Grand Island, NY) and 250 nM specific primers. Amplification used an ABI 7500 (Applied Biosystems) with 1-cycle of 95°C for 10 minutes and 40 cycles of 95°C for 30 seconds and 60°C for 1 min. PCR efficiencies were verified by examination of the corresponding melting curves for each primer pair. Data were normalized to Gapdh mRNA standard relative to ES cell expression and shown as ΔΔCt in log2 scale, where ΔΔCt = (CtGene − CtGapdh)Assay − (CtGene − CtGapdh)ESC control.
2.5 Immunofluorescence staining
Day 13 differentiated EB were cultured on 10-mm glass coverslips, fixed with 4% paraformaldehyde in PBS for 20 min, permeabilized with 0.1% triton X-100 for 20 min, blocked with 1% BSA for 1 hour, and incubated with first antibody at 4°C overnight. After washing, coverslips were stained with Alexa 488- or Alexa 567-labeled secondary antibodies and Hoechst solution. The cells were examined and images were captured using a Zeiss Axio microscope.
2.6 Chromatin immunoprecipitation assay
Chromatin immunoprecipitation was done with minor modifications of our previously described procedures (Schnekenburger et al., 2007). Cells were cross-linked with 1% formaldehyde at room temperature for 10 minutes; after quenching the reaction by adding 125 mM glycine at room temperature for 5 minutes, cells were washed twice with cold PBS then scraped and harvested. All solutions except the elution buffer contained 1X complete protease inhibitor cocktail. Cell pellets were lysed in Cell-Lysis-Buffer (5 mM PIPES pH 8.5, KCl 85 mM, and 0.5% NP-40) on ice for 10 minutes; nuclei were pelleted and lysed in Nuclei-Lysis-Buffer (50 mM Tris-HCl pH 8, 10 mM EDTA, and 1% SDS) at an approximate density of 107 nuclei/ml followed by a 10-minute incubation on ice. Chromatin was sheared by sonication with a Bioruptor (Diagenode, Denville, NJ) in a crushed-ice water bath for thirty 30-seconds bursts of 200 W with a 30-second interval between bursts. Supernatant was collected after centrifugation and diluted 6X with IP-Dilution-Buffer (16.7 mM Tris-HCl pH 8, 167 mM NaCl, 1.2 mM EDTA, 1.1% Triton X-100, and 0.01% SDS) and distributed into aliquots corresponding to 2×106 cell-equivalents for each ChIP assay. After pre-cleaning with a 50:50 mix of protein-A/G-agarose (Upstate), 3 μg of the pertinent antibody was added to each ChIP assay tube and incubated on a rotating platform at 4°C overnight. Specific immune complexes were recovered by a 2-hour incubation at 4°C with 30 μl 50:50 protein-A/G-agarose. Pelleted protein-A/G-agarose was washed three times with 1X-Dialysis-Buffer (50 mM Tris-HCl pH 8, 2mM EDTA, 0.2% sarkosyl (omitted if the antibody was a mouse monoclonal) and three times with IP-Wash-Buffer (100 mM Tris-HCl pH 9 (pH 8 for mouse monoclonal antibody), 500 mM LiCl, 1% NP-40, 1% deoxycholic acid) on ice for 10 minutes. Chromatin complexes were released by washing twice with 60 μl Elution-Buffer (50 mM NaHCO3 and 1% SDS) by vigorous shaking in a vortex mixer for at least 20 minutes. Formaldehyde cross-links were reversed and residual RNA removed by addition of 300 mM NaCl and 20 μg RNAse A (Sigma) at 65°C overnight. ChIP-enriched DNA was purified by chromatography in a QIAquick column (QIAGEN, Valencia, CA) after proteinase K treatment at 45°C for 1.5 hours. The concentration of the purified DNA was determined with PicoGreen dsDNA dye (Invitrogen) and an aliquot was used for each PCR reaction. DNA enrichment is represented as the percent of total input after subtracting the values of the relevant control antibodies.
2.7 Induced expression of reprogramming factors OCT3/4, SOX2, KLF4, and MYC in Hepa-1 cells
Hepa-1 cells at 70% confluence were transfected at 37°C for 3 hours with plasmids pBabe-Puro, to provide puromycin resistence, FUW-M2rtTA, to provide a source of tetracycline-activated Tetracycline receptor, and TetO-FUW-OSKM (Addgene, Cambridge, MA) (Carey et al. 2009), to provide Tet-receptor dependent expression of OCT3/4, SOX2, KLF4, and MYC. Transfection was by lipofectamine 2000 and PLUS reagent (Invitrogen) according to the manufacturer’s specifications. Transfected cells were incubated for 3 hours in medium containing 10% FBS and selected with 3 μg/ml puromycin for 3 days. To activate the expression of reprogramming factors coded for by the TetO-FUW-OSKM vector, 5 μg/ml doxycycline was added to the cultures. Samples for mRNA and protein expression analyses were collected after 3 days of puromycin selection and doxycycline treatment.
2.8 Statistical analyses
All experiments were done in biological duplicates or triplicates; data are presented as means ± SD. Group comparisons were made by two-way analysis of variance (ANOVA) followed by Bonferroni post test. A p-value equal to or less than 0.05 was considered statistically significant.
3. Results
3.1 Ahr is silent in ES cells and its expression is up-regulated upon differentiation
Previous work has shown that Ahr is expressed in fertilized eggs (Dey and Nebert, 1998; Wu et al., 2002) but becomes silent at the 2-cell stage, remaining completely silenced until the late preimplantation stage, at what time blastocysts begin to show detectable de novo Ahr expression (Peters and Wiley, 1995; Dey and Nebert, 1998; Wu et al., 2002). An expression pattern of quick reversibility such as this is consistent with a developmental role for AHR that might be harmful to the early preimplantation embryo but needed for the late preimplantation and postimplantation embryos. To investigate the mechanisms responsible for this pattern of expression and identify what these functions might be, we followed the temporal sequence of Ahr expression in mouse ES cells, a ready source of in vitro pluripotent preimplantation blastocysts and their differentiated progeny. Relative to Gapdh, Ahr mRNA levels were low but detectable in ES cells and increased with time of differentiation, reaching maximal levels 9 days after initiation of differentiation (Fig. 1A, upper panel). Starting on day 7, significant mRNA levels of the prototypical AHR target gene Cyp1a1 were present (Fig. 1A, lower panel), suggestive of endogenous AHR activation, independent of activation by a xenobiotic ligand. AHR protein levels followed a similar pattern of expression as the mRNA, reaching a maximum after 7 days of differentiation (Fig. 1B).
Fig. 1. Ahr expression is silent in pluripotent mouse ES cells and activated during non-directed differentiation.
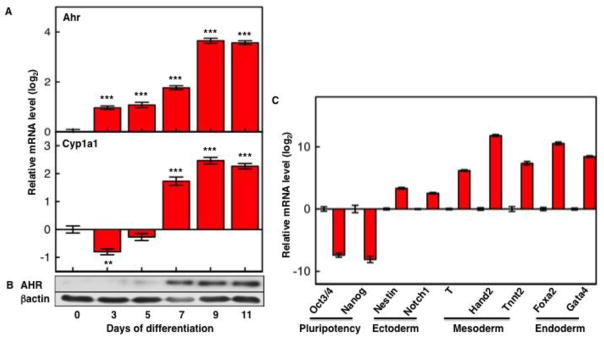
After 3-days of EB formation, differentiation was followed for 11 or 13 days on cover slips. Ahr and Cyp1a1 mRNA levels (A) were normalized to Gapdh and are shown relative to the levels in ES cells (day 0). Immunoblots with AHR and β–actin specific primary antibodies (B) were visualized with chemiluminescence ECL Western Blot Substrate. (C) mRNA expression levels of pluripotency and germ cell layer marker genes were examined by real time PCR, normalized to Gapdh and are shown relative to the levels in ES cells. The asterisk (*) indicates significant differences to values in ES cells: (*) p<0.05; (**) p<0.01; (***) p<0.001.
Determination of mRNA levels of germ layer marker genes (Fig. 1C) indicated that our non-directed differentiation protocol generated a mixed population of differentiated progeny in which all three cellular lineages, i.e. ectoderm, mesoderm, and endoderm, were present, albeit possibly in different proportions. Expression of AHR and its co-localization with germ layer markers was also demonstrated by immunofluorescence staining of 13-day EBs, which showed that AHR independently co-localized with ectoderm marker keratin 14, cardiac mesoderm marker troponin-T, and endoderm marker Gata4 (Supplemental Fig. S1).
These data indicate that mechanisms exists to silence Ahr transcription in ES cells and that differentiation in vitro faithfully recapitulates the regulatory events of AHR expression described earlier and previously observed in vivo.
3.2 Recruitment of Sp1 and Sp3 factors to the proximal promoter up-regulates Ahr transcription in differentiated cells
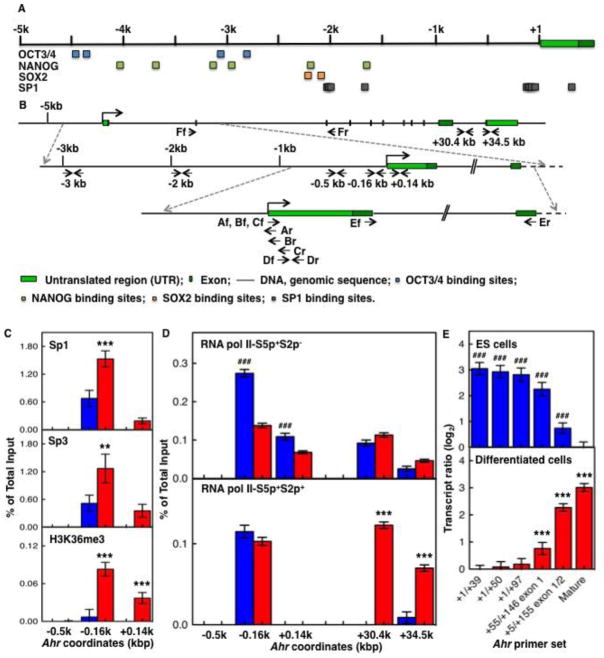
Constitutive Ahr expression depends on binding of Sp factors to four Sp1 binding sites located between coordinates −174 and +70 from the TSS of the Ahr promoter (Fitzgerald et al., 1998) (Fig. 2A,B). Ahr transcriptional silence may simply be due to lack of functional expression of Sp-factors or to impairment of chromatin binding in the ES cells but not in the differentiated cells. To test this hypothesis, we used ChIP analyses to measure binding of Sp-factors to their sites in the Ahr proximal promoter and to compare pluripotent ES cells to cells on day 9 of non-directed differentiation, at a time when Ahr expression is maximal. Binding of Sp1 and Sp3, already high in ES cells, doubled in the day-9 samples (Fig. 2C, upper and middle panels), suggesting that the promoter might be transcriptionally active under both experimental conditions. However, this was clearly not the case, since there was little if any Ahr mRNA in the ES cells. In agreement with gene silencing in ES cells, H3K36me3, a histone mark associated with actively transcribed genes (Bannister et al., 2005), was practically undetectable in ES cells and significantly high in the differentiated cells (Fig. 2C, lower panel), confirming that the Ahr promoter proceeds from being silent to activated in parallel to the differentiation of the ES cells.
Fig. 2. Sp-factors and RNAPII control the transition of silent to activated Ahr.
(A) Schematic representation of the mouse Ahr gene and location of core pluripotency factors OCT3/4, SOX2, and NANOG and Sp1 binding sites along 5 kb of the Ahr promoter. MatInspector software from Genomatrix was used to screen binding sites of transcription factors. Blue, green, amber, and gray squares represent OCT3/4, SOX2, NANOG, and Sp1 binding sites respectively. (B) Location of primers used in this study. Upper level: structure of the Ahr gene from 5 kb upstream of the TSS to the TES region. Middle level: expansion of the sequence from 3 kb upstream of the TSS to the 2nd exon region. Lower level: expansion of the sequence from 1 kb upstream of the TSS to the 2nd exon. Arrows indicate primers and target DNA strands, forward (f)/reverse (r) versus sense/antisense. (C) ES cells (blue bars) and cells differentiated for 9 days (red bars) were compared for binding of Sp-factors (upper and middle panels) and active transcription histone mark H3K36me3 (lower panel) on the Ahr proximal promoter region. (D) binding of RNAPII-S5p+S2p− (upper panel) and RNAPII-S5p+S2p+ (lower panel) on both the Ahr proximal promoter and the TES region. The asterisk (*) and the pound (#) denote significant difference to ES or to day-9 differentiation, respectively: (*,#) p<0.05; (**,##) p<0.001; (***,###) p<0.001. (E) The ratio of Ahr short transcripts to mature mRNA was determined as described in the text; the upper panel shows the ratio of transcripts of various lengths in ES cells relative to mature mRNA in day-9 samples; the bottom panel shows the ratio of transcripts of various lengths in day-9 samples relative to mature mRNA in ES cells.
3.3 Unproductive RNA polymerase II phosphorylated in CTD-Ser-5 but not Ser-2 is paused on the Ahr proximal promoter of ES cells
Changes in the phosphorylation of the RNAPII carboxyl-terminal repeat domain (CTD) are associated with transcriptional initiation, elongation and termination (Phatnani and Greenleaf, 2006; Buratowski, 2009). Transcription initiation without elongation leads to RNAPII pausing at transcriptional start domains, a phenomenon first described in Drosophila (Gilmour and Lis, 1986) and now recognized as a critical mechanism to poise downstream target genes for rapid and precise gene activation. While poised genes may lack RNAPII binding or may bind RNAPII phosphorylated in Ser-5 (RNAPII-S5p+S2p−), active transcriptional elongation is generally characterized by RNAPII phosphorylated in both Ser-5 and Ser-2 (RNAPII-S5p+S2p+) (Stock et al., 2007; Brookes et al., 2012). Studies in human and mouse ES cells have revealed the prevalence of RNAPII-S5p+S2p− paused at genes encoding proteins critical for developmental functions, stimulus-responsiveness, and signaling components that regulate ES pluripotency and self-renewal (Guenther et al., 2007; Min et al., 2011; Gilchrist et al., 2012). Consistent with our findings with Sp-factors and H3K36me3, binding of RNAPII-S5p+S2p− on the Ahr proximal promoter was higher in ES cells than in day-9 differentiated cells (Fig. 2D, upper panel), suggesting that silent Ahr expression in ES cells might be the consequence of an unproductive RNAPII form paused on the proximal promoter domains. To test this hypothesis, we measured the binding of the active form RNAPII-S5p+S2p+ to the Ahr gene proximal promoter and transcription end-site (TES) domains of ES and day-9 differentiated cells. Binding to the proximal promoter was similar in ES and day-9 differentiated cells, but only the differentiated cells showed significant binding of the active RNAPII-S5p+S2p+ form to the TES domain (Fig. 2D, lower panel), indicative of transcriptional elongation.
3.4 Paused RNA polymerase II generates Ahr short transcripts in ES cells
Increasing evidence from studies in a variety of systems, including Drosophila and ES cells, indicate that the main developmental checkpoint that directs differential gene expression during differentiation is the regulation of transcription elongation by release of tethered RNAPII from the proximal promoter of the affected genes (Levine, 2011). The paused RNAPII produces only short aborted transcripts, while its release from the promoter results on full-length transcript elongation (Rasmussen and Lis, 1995; Guenther et al., 2007). To determine if the silent Ahr in ES cells produced short transcripts absent in differentiated cells, we used real-time RT-PCR to measure the number of increasingly longer transcripts from the 5′-end of the mRNA (Fig. 2E) relative to the number of full-length molecules. We used PCR primer pairs A – E (Fig. 2B and Table S2) to amplify, A: the 5′-most 39, B: 50, and C: 97 nucleotides of Ahr mRNA; D: a 91-nucleotide segment from coordinates +55 to +146 from exon 1; and, E: a 150-nucleotide segment bracketing exons 1 and 2, from +5 to +155. The number of amplified molecules for each of these amplicons was determined relative to the number of amplified products of amplicon F, the 93-nucleotide segment bracketing exons 2 and 3 used to assess the level of full-length transcription. In ES cells, short Ahr transcripts were 8-fold more abundant than full-length and their abundance decreased as the location of the PCR amplicon proceeded 3′-ward, with much less transcripts reaching position +155 (Fig. 2E, upper panel). In contrast, in day-9 differentiated cells most transcripts were elongated beyond +155 and hardly any short transcript were detected, with 8-fold more full-length transcripts than in ES cells (Fig. 2E lower panel). These results confirm that pausing of unproductive RNAPII on the Ahr proximal promoter generates short transcripts and that the transition from pluripotent ES cell to differentiated cells is accompanied by a switch in Ahr expression from repressed to active state.
3.5 The core pluripotency factors, OCT3/4, NANOG, and SOX2 bind to the Ahr distal promoter domain
In ES cells, the three transcription factors OCT3/4, NANOG, and SOX2 establish a regulatory network critical to maintain pluripotency and self-renewal ability (Christophersen and Helin, 2010). On the basis of their transcriptional status, binding of these three factors characterizes promoters that are active or primed for expression but repressed. On the one hand, regulatory complexes of OCT3/4, NANOG, and SOX2 activate and maintain the expression of pluripotency genes, including themselves (Kim et al., 2008; Chambers and Tomlinson, 2009); on the other hand, the same complexes cooperate with Polycomb Group-mediated repressive mechanisms to keep developmentally regulated genes in a repressed state (Boyer et al., 2006; Loh et al., 2006). By scanning the 5 kb of Ahr promoter upstream of the TSS we identified two clusters of pluripotency factor binding sites. One cluster, located in the minus-3 kb region, comprises 2 NANOG and 2 OCT3/4 binding motifs; the second, located in the minus-2 kb region, contains 1 NANOG and 2 SOX2 motifs (Fig. 2A). The presence of these motifs suggested the possibility that binding of OCT3/4-NANOG-SOX2 complexes to these sites could be actively involved in repressing Ahr expression in ES cells, a prediction that was confirmed by ChIP analyses. Binding of OCT3/4 and SOX2 was observed primarily on the minus-2 kb region and was significantly higher in ES cells than in day-9 differentiated cells (Fig. 3A,B). NANOG bound to both minus-2 kb and minus-3 kb clusters, at levels significantly higher in ES cells than in day-9 differentiated cells (Fig. 3C). These results suggest that binding of pluripotency factors to the Ahr promoter correlates with and might significantly contribute to its silencing in ES cells.
Fig. 3. Binding of pluripotency factors to the Ahr distal promoter domain.
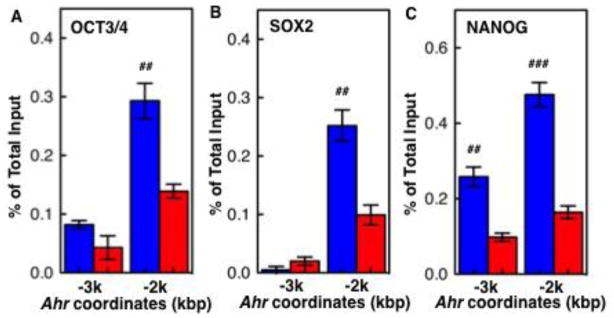
As shown in Fig. 2, clusters of pluripotency factor binding sites are located in the minus-3 kb, and minus-2 kb Ahr domains upstream of the TSS. OCT3/4 (A), SOX2 (B), and NANOG (C) bind to these sites to a significantly greater extent in ES cells than in day-9 differentiated cells. The pound (#) denotes significant differences between ES cells and day-9 differentiation: (#) p<0.05; (##) p<0.01; (###) p<0.001.
3.6 H3K27ac, H3K9ac, H3K4me2 and H3K4me3 open Ahr proximal promoter chromatin for expression during differentiation
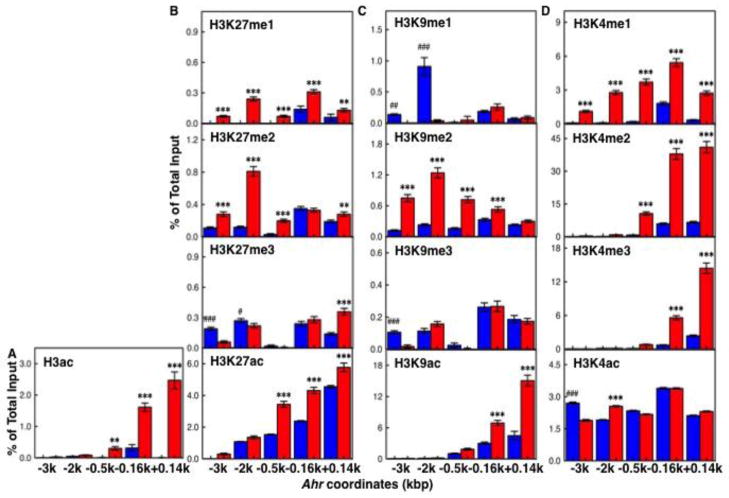
In addition to be repressed and primed for expression through control of pluripotency factors, promoters of developmental transcription factors are in a bivalent state, simultaneously marked by active and repressive methylation and acetylation marks in Lys-4, Lys-9 and Lys-27 (Mikkelsen et al., 2007). To determine if the Ahr promoter had a characteristic pattern of bivalent marks in ES cells that resolved after differentiation, we examined the degree of acetylation and methylation of H3K4, H3K9 and H3K27 in both distal and proximal Ahr promoter domains. Total histone H3 acetylation and H3K9ac was greatly increase in the day-9 differentiated cell population relative to ES cells, particularly in the proximal promoter, while H3K4ac and H3K27ac levels were similar in both sets of cells (Fig. 4A–D, lower panels). In contrast to acetylation, H3K27me1 and H3K27me2 were higher in the differentiated cells, while H3K27me3 was not different in ES and day 9 cells (Fig. 4B). Monomethylation of Lys-9 was very high in ES cells, while dimethylation was higher in differentiated cells and both sets of cells had similar levels of trimethylated Lys-9 (Fig. 4C), a hallmark of heterochromatin structure. Consistent with an active transcription state, H3K4me3, H3K4me2 and H3K4me1 levels were much higher in differentiated cells that in ES cells, especially in the proximal promoter domains (Fig. 4D).
Fig. 4. Comparison of histone marks in the Ahr promoter in ES and day-9 differentiated cells.
Levels of total histone H3 acetylation (panel A) and of mono-/di-/tri-methylated and acetylated H3K27, H3K9 and H3K4 (panels B, C and D, respectively) were determined by chromatin immunoprecipitation in ES cells and day-9 differentiated cells. The asterisk (*) and the pound (#) denote significant difference to ES or to day-9 differentiation: (*,#) p<0.05; (**,##) p<0.001; (***,###) p<0.001.
3.7 Binding of PcG proteins characterizes the bivalent state of the Ahr promoter in ES cells
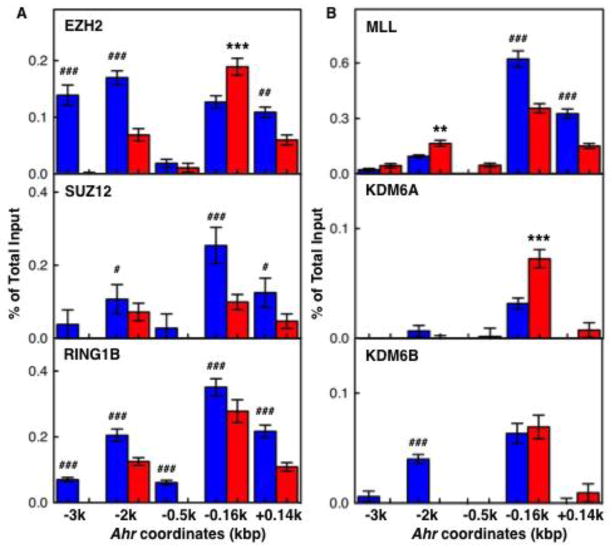
As described earlier, pluripotency factors cooperate with reversible PcG-mediated mechanisms in the transcriptional repression of differentiation associated genes in ES cells (Bernstein et al., 2006). The reversibility of PcG-mediated repression is provided by interactions between multiple protein complexes, including PRC1, PRC2, and the Trithorax Group (TxG) proteins, which are recruited to antagonize PcG effects (Morey and Helin, 2010; Schuettengruber et al., 2011). To further characterize Ahr repression in ES cells, we examined the binding of several PcG and TxG proteins on the Ahr promoter. Binding of, (i), the PRC2 components EZH2, a H3K27 histone methyltransferase, and SUZ12; (ii) the PRC1 component RING1B, a H2AK119 E3 ubiquitin ligase; and (iii) MLL, a SET domain TxG protein and H3K4 histone methylatransferase, was higher in ES cells than in day-9 differentiated cells in all Ahr promoter locations analyzed (Fig. 5A and top panel of 5B). The same was true for the H3K27me3 demethylase KDM6B but not for KDM6A (Fig. 5B middle and bottom panels). It appears that PcG/TxG interactions at the promoter are key to maintain the Ahr gene in a repressed state in pluripotent ES cells.
Fig. 5. Binding of PcG and TxG proteins to the Ahr promoter.
Binding profiles of PcG proteins EZH2, SUZ12, and RING1B (panel A) and TxG proteins MLL, KDM6A and KDM6B (panel B) on both Ahr distal and proximal promoter regions. The asterisk (*) and the pound (#) denote significant differences to ES or to day-9 differentiated cells respectively: (*,#) p<0.05; (**,##) p<0.001; (***,###) p<0.001.
3.8 Ectopic expression of reprogramming factors OCT3/4, SOX2, KLF4, and MYC represses Ahr expression in hepatoma cells
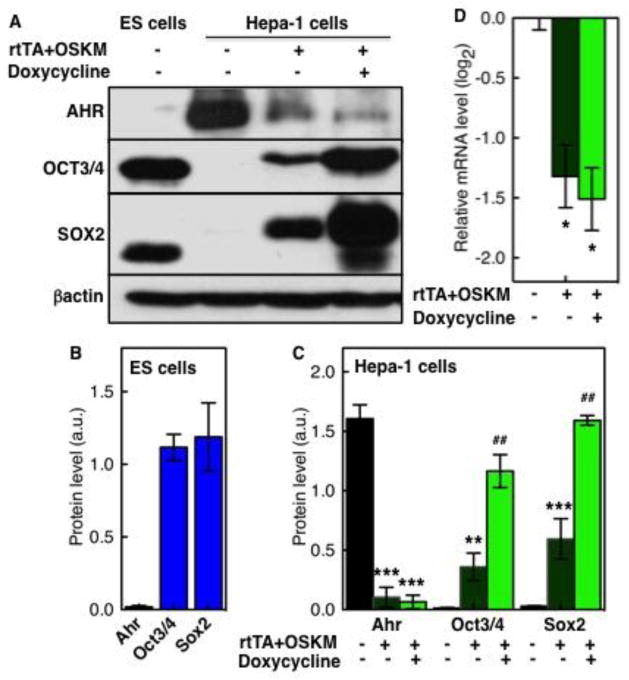
Major progress in stem cell biology has resulted from studies that directly return somatic cells to the pluripotent state by introduction of the so-called Yamanaka factors, namely OCT3/4, SOX2, KLF4, and MYC (OSKM), which give rise to induced pluripotent stem (iPS) cells from embryonic and/or adult fibroblasts (Takahashi and Yamanaka, 2006). To determine whether expression of pluripotency factors could be responsible for Ahr repression in ES cells, we induced the ectopic expression of a doxycycline-inducible OSKM expression vector (Hockemeyer et al., 2008; Carey et al., 2009) in mouse hepatoma Hepa-1 cells, which express high levels of AHR protein, and measured the effect of OSKM expression on AHR expression in the transfected cells. Three days after transfection, Tet receptor dependent induction of ectopic OCT3/4 and SOX2 was readily detectable in transfected cells, possibly due to leakiness in the system, as already described (Carey et al., 2009), and more so in cells treated with doxycycline, in which expression reached levels comparable to those in ES cells (Fig. 6A). In contrast, in the presence of OCT3/4 and SOX2, reduced levels of AHR protein, high in untransfected cells, were detected; considerably lower in transfected cells and less so in cells treated with doxycycline (Fig. 6A). When immunoblot bands were quantified and normalized to β–actin levels, it was evident that the low AHR and high OCT3/4-SOX2 levels observed in ES cells (Fig. 6B) were reversed in the transfected Hepa-1 cells, in which even in the absence of doxycycline treatment, AHR expression was inhibited by greater than 15-times and more so after doxycycline treatment (Fig. 6C). Consistent with the protein data, Ahr mRNA levels were 3 times lower in transfected cells than in wild-type cells (Fig. 6D). These data suggest that the pluripotency factors OCT3/4 and SOX2 can repress Ahr expression and thus, may be directly responsible for Ahr repression in ES cells.
Fig. 6. Doxycycline-induced expression of reprogramming factors in Hepa-1 cells represses AHR expression.
Hepa-1 cells were transfected with a rtTA-regulated doxycycline-inducible expression system of reprogramming factors (Oct3/4/SOX2/KLF4/MYC) (see Materials and Methods). Uninduced and 3-day doxycycline-induced transfected Hepa-1 cells and control ES cells were collected and subjected to mRNA and protein expression analyses. Protein levels of AHR, OCT3/4 and SOX2 were determined in Western blots (panel A) and quantified relative to β-actin levels for ES cells and Hepa-1 cells (panels B and C, respectively). Ahr mRNA levels in uninduced and doxycycline-induced OSKM-transfected Hepa-1 cells are shown in panel D relative to the levels in untransfected cells. The asterisk (*) and the pound (#) denote significant difference to untransfected or to uninduced Hepa-1 cells, respectively: (*,#) p<0.05; (**,##) p<0.001; (***,###) p<0.001.
4. Discussion
The results from this study show that Ahr expression is maintained in a repressed but poised state in pluripotent ES cells that allows quick activation as the cells differentiate. Repression results from the binding of a pluripotency factor complex of OCT3/4-NANOG-SOX2 on the Ahr distal promoter region in cooperation with other reversible regulatory mechanisms, including PcG-mediated repression and pausing of unproductive RNAPII-S5p+S2p− molecules on the TSS proximal promoter region. The switch of Ahr expression from repression to activation follows the release of pluripotency factors from their binding sites, withdrawal of PcG proteins, recruitment of Sp-factors, acquisition of histone marks characteristic of open chromatin on the proximal promoter region, and engagement of phosphorylated RNAPII-S5p+S2p+ to drive full-length transcript elongation.
Our observations are consistent with previous findings in vivo that Ahr expression is silenced at the 2-cell stage, becoming again detectable in late blastocysts (Peters and Wiley, 1995; Dey and Nebert, 1998; Wu et al., 2002) and in most developing organs during post-implantation development (Abbott and Probst, 1995; Jain et al., 1998). Such an expression pattern suggests that AHR is developmentally bivalent and that its functions during post-implantation must be silenced if pluripotency is maintained. Studies on the OCT3/4-NANOG-SOX2 regulatory network in ES cells have shown that pluripotency factors maintain ES cell properties by simultaneously activating expression of pluripotency-related genes, including their own (Kim et al., 2008; Chambers and Tomlinson, 2009) and repressing key differentiation transcription factors (Boyer et al., 2006; Loh et al., 2006). Among the latter, OCT3/4 was previously implicated on Ahr repression (Loh et al., 2006). In good agreement, we find that, not just OCT3/4, but NANOG and SOX2 as well are actively involved in Ahr repression in ES cells through binding to their cognate sites on the Ahr minus-2 kb distal promoter region. Ectopic OSKM expression in mouse hepatoma Hepa-1 cells confirmed this conclusion. Endogenous Ahr expression is very high in these cells, but it was significantly repressed by the transfected OSKM proteins both at the protein and mRNA levels. This finding confirms that pluripotency factors are the main agents of Ahr repression in pluripotent cells and suggests that Ahr repression may be required to maintain pluripotency. In this context, it may be significant that AHR regulates the multipotency of hematopoietic stem cells, since its inhibition promotes their greater expansion (Boitano et al., 2010), while deletion of the Ahr gene increases the proliferative capacity of hematopoietic progenitors (Lindsey and Papoutsakis, 2012; Gasiewicz et al., 2010; Singh et al., 2011).
Pluripotency factors do not act alone; they share overlapping networks of target genes with PcG proteins that further silence their targets (Bernstein et al., 2006; Boyer et al., 2006; Loh et al., 2006). In ES cells, differentiation associated genes are subject to flexible epigenetic controls that repress and prime their expression but do not fully silence them. Genes encoding transcription factors that play an essential role during differentiation carry bivalent promoter domain marks in ES cells, consisting of active H3K4me3 and repressive H3K27me3 (Mikkelsen et al., 2007). These marks, through their interplay with PcG and TxG proteins, represses transcription but keep the genes in a primed state (Morey and Helin, 2010; Schuettengruber et al., 2011). The Ahr promoter does not show bivalent domains in ES cells; in fact, in our hands and in good agreement with previous studies (Mikkelsen et al., 2007; Ku et al., 2008), H3K27me3 is low in both ES and differentiated cells and H3K4me3 is low in ES cells and reaches high levels in differentiated cells. Conversely, consistent with recent work that has classified Ahr as a PcG-silenced gene (Brookes et al., 2012), our ChIP analyses show that PcG-mediated mechanisms are strongly involved in Ahr repression in ES cells. Several PcG proteins, including EZH2, SUZ12, RING1B, and TxG proteins MLL and KDM6B showed higher binding on the Ahr promoter in ES than in differentiated cells. The E3 ubiquitin ligase activity of the PRC1 component RING1B mediates pausing of unproductive RNAPII through ubiquitination of histone H2A, contributing a key element to gene repression (Endoh et al., 2012; Stock et al., 2007). We conclude that, in addition to the inhibitory effect resulting from direct binding of pluripotency factors, Ahr is also repressed by PcG-mediated mechanisms. Binding of the two TxG proteins, the H3K4 methyltransferase MLL and the H3K27 demethylase KDM6B, suggests that their role is to quickly activate the Ahr upon reception of morphogenetic signals. The main features of the transition of Ahr repression from undifferentiated ES cells to activation in differentiated cells are schematically presented in Fig. 7.
Fig. 7. Schematic model of the transition of Ahr expression from the repressed state in pluripotent ES cells to the activated state in differentiated cells.
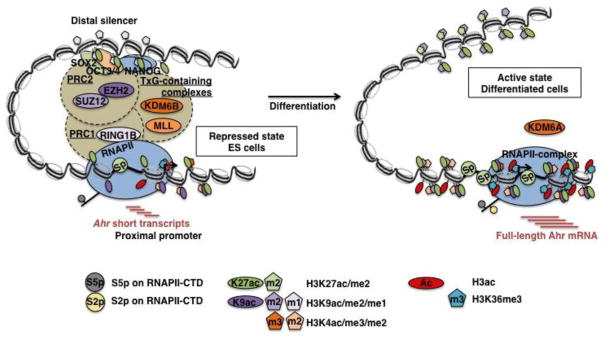
The model integrates the results from this study into a prior knowledge base. In pluripotent ES cells, Ahr is repressed by binding of pluripotency factors OCT3/4, NANOG, and SOX2 on its distal silencer region and pausing of unproductive RNAPII-S5p+S2p− on the proximal promoter region; short Ahr transcripts are produced in these cells. RNAPII pausing is the likely consequence of the cooperation between pluripotency factors and PcG proteins in PRC1 and PRC2 that interact with their antagonists TxG proteins. This active but primed state of the Ahr promoter in ES cells does not show an enrichment of repressive histone marks but maintains a basal level of histone acetylation, tri-/di-methylated H3K4, and binding of Sp-factors on the proximal promoter domain. Differentiation signals resolve the Ahr promoter into an activated state, characterized by the release of pluripotency factors and PcG proteins from their binding sites, acquisition of histone marks for open chromatin, recruitment of Sp-factors on the proximal promoter region, and engagement of active RNAPII-S5p+S2p+ to synthesize mature Ahr mRNA.
5. Conclusions
Our observations are consistent with the concept that Ahr is repressed in ES and conversely, rapid activation from the silent state accompanies progression of differentiation. We can only speculate as to what are the AHR functions that need to be regulated by this concert of repression and activation lest development be disrupted. Two of the most prominent endogenous AHR functions are the inhibition of TGFβ signaling pathways (Fan et al., 2010; Chang et al., 2007) and the cooperation with the retinoblastoma protein RB1 to promote cell cycle arrest (Puga et al., 2005). Given the requirement for TGFβ-related signaling to maintain a uniform population of proliferating undifferentiated ES cells (Galvin-Burgess et al., 2013) and the impairment of pluripotency when all three members of the retinoblastoma family, are ablated (Dannenberg et al., 2000), it is attractive to speculate that TGFβ and RB might be the targets of these functions and that expression of a functional AHR in ES cells might be detrimental to their survival but beneficial to their differentiation.
Supplementary Material
Highlights.
Pluripotency factors repress Ahr gene expression in embryonic stem cells.
Repression depends on Polycomb group proteins and pausing of RNA Polymerase-II.
Paused RNA polymerase-II generates short Ahr transcript in embryonic stem cells.
Ahr expression is quickly initiated upon in vitro differentiation.
Acknowledgments
We thank Drs. Andras Nagy and Marina Gertsenstein, Mount Sinai Hospital and Samuel Lunenfeld Research Institute, Toronto, Canada for the C57BL/6N-C2 ES cells. We also thank Drs. Hisaka Kurita, Francisco-Javier Sánchez-Martín, Vinicius Carreira and Jerry Ovesen for a critical reading of the manuscript. This research was supported by NIEHS grants R01 ES06273, R01 ES10807 and the NIEHS Center for Environmental Genetics grant P30 ES06096.
Abbreviations and Acronyms
- AHR
Aryl hydrocarbon receptor
- AhRE
AHR response element
- ARNT
Ah receptor nuclear translocator
- bHLH/PAS
Basic helix-loop-helix/Per-ARNT-Sim
- ChIP
Chromatin immunoprecipitation
- CTD
Carboxyl-terminal repeat domain
- EB
Embryoid bodies
- EMT
Epithelial-to-mesenchymal transition
- ESC
Embryonic stem cells
- EZH2
Enhancer of zeste homolog 2
- H3ac
Acetylated histone H3
- H3K27ac
Acetylated lysine-27 of histone H3
- H3K27me3/2/1
Tri/Di/Mono-methylated lysine-27 of histone H3
- H3K36me3
Tri-methylated lysine-36 of histone H3
- H3K4me3/2/1
Tri/Di/Mono-methylated lysine-4 of histone H3
- H3K9ac
Acetylated lysine-9 of histone H3
- H3K9me3/2/1
Tri/Di/Mono-methylated lysine-9 of histone H3
- HMT
Histone methyltransferase
- ICM
Inner-cell-mass
- iPSC
Induced pluripotent stem cells
- KDM6A/B
Lysine demethylase 6A and 6B
- KO
Knock out
- MET
Mesenchymal-to-epithelial transition
- MLL
Myeloid/lymphoid or mix-lineage leukemia
- OSKM
OCT3/4, SOX2, KLF4, MYC
- PcG
Polycomb group proteins
- PRC1/2
Polycomb repressive complexes 1 and 2
- RING1B
Ring finger protein 1B
- RNAPII
RNA polymerase II
- RNAPII (S5p+S2p−)
RNA polymerase II phosphorylated in CTD serine-5 but not serine-2
- RNAPII (S5p+S2p+)
RNA polymerase II hyperphosphorylated in CTD serine-5 and serine-2
- SUZ12
Suppressor of zeste 12 homolog
- TxG
Trithorax group proteins
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- TES
Transcription end site
- TSS
Transcription start site
Footnotes
Authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott BD, Birnbaum LS, Perdew GH. Developmental expression of two members of a new class of transcription factors: I. Expression of aryl hydrocarbon receptor in the C57BL/6N mouse embryo. Dev Dynamics. 1995;204:133–143. doi: 10.1002/aja.1002040204. [DOI] [PubMed] [Google Scholar]
- Abbott BD, Probst MR. Developmental expression of two members of a new class of transcription factors: II. Expression of aryl hydrocarbon receptor nuclear translocator in the C57BL/6N mouse embryo. Developmental Dynamics. 1995;204:144–155. doi: 10.1002/aja.1002040205. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boitano AE, Wang J, Romeo R, Bouchez LC, Parker AE, Sutton SE, Walker JR, Flaveny CA, Perdew GH, Denison MS, Schultz PG, Cooke MP. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329:1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brookes E, de SI, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, Kimura H, Ragoussis J, Teichmann SA, Pombo A. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell. 2012;10:157–170. doi: 10.1016/j.stem.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Hanna J, Saha K, Gao Q, Mitalipova M, Jaenisch R. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc Natl Acad Sci U S A. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Tomlinson SR. The transcriptional foundation of pluripotency. Development. 2009;136:2311–2322. doi: 10.1242/dev.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang X, Fan Y, Karyala S, Schwemberger S, Tomlinson CR, Sartor MA, Puga A. Ligand-independent regulation of transforming growth factor beta1 expression and cell cycle progression by the aryl hydrocarbon receptor. Mol Cell Biol. 2007;27:6127–6139. doi: 10.1128/MCB.00323-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophersen NS, Helin K. Epigenetic control of embryonic stem cell fate. J Exp Med. 2010;207:2287–2295. doi: 10.1084/jem.20101438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg JH, van RA, Schuijff L, te RH. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 2000;14:3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Nebert DW. Markedly increased constitutive CYP1A1 mRNA levels in the fertilized ovum of the mouse. Biochem Biophys Res Commun. 1998;251:657–661. doi: 10.1006/bbrc.1998.9519. [DOI] [PubMed] [Google Scholar]
- Emmons RB, Duncan D, Estes PA, Kiefel P, Mosher JT, Sonnenfeld M, Ward MP, Duncan I, Crews ST. The spineless-aristapedia and tango bHLH-PAS proteins interact to control antennal and tarsal development in Drosophila. Development. 1999;126:3937–3945. doi: 10.1242/dev.126.17.3937. [DOI] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Endoh T, Isono K, Sharif J, Ohara O, Toyoda T, Ito T, Eskeland R, Bickmore WA, Vidal M, Bernstein BE, Koseki H. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet. 2012;8:e1002774. doi: 10.1371/journal.pgen.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Boivin GP, Knudsen ES, Nebert DW, Xia Y, Puga A. The aryl hydrocarbon receptor functions as a tumor suppressor of liver carcinogenesis. Cancer Res. 2010;70:212–220. doi: 10.1158/0008-5472.CAN-09-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Salguero P, Pineau T, Hilbert DM, McPhail T, Lee SS, Kimura S, Nebert DW, Rudikoff S, Ward JM, Gonzalez FJ. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science. 1995;268:722–726. doi: 10.1126/science.7732381. [DOI] [PubMed] [Google Scholar]
- Fernandez-Salguero PM, Ward JM, Sundberg JP, Gonzalez FJ. Lesions of aryl-hydrocarbon receptor-deficient mice. Vet Pathol. 1997;34:605–614. doi: 10.1177/030098589703400609. [DOI] [PubMed] [Google Scholar]
- Fitzgerald CT, Nebert DW, Puga A. Regulation of mouse Ah receptor (Ahr) gene basal expression by members of the Sp family of transcription factors 1. DNA Cell Biol. 1998;17:811–822. doi: 10.1089/dna.1998.17.811. [DOI] [PubMed] [Google Scholar]
- Galvin-Burgess KE, Travis ED, Pierson KE, Vivian JL. TGF-beta-superfamily signaling regulates embryonic stem cell heterogeneity: self-renewal as a dynamic and regulated equilibrium. Stem Cells. 2013;31:48–58. doi: 10.1002/stem.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiewicz TA, Henry EC, Collins LL. Expression and activity of aryl hydrocarbon receptors in development and cancer. Crit Rev Eukaryot Gene Expr. 2008;18:279–321. doi: 10.1615/critreveukargeneexpr.v18.i4.10. [DOI] [PubMed] [Google Scholar]
- Gasiewicz TA, Singh KP, Casado FL. The aryl hydrocarbon receptor has an important role in the regulation of hematopoiesis: implications for benzene-induced hematopoietic toxicity. Chem Biol Interact. 2010;184:246–251. doi: 10.1016/j.cbi.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertsenstein M, Nutter LM, Reid T, Pereira M, Stanford WL, Rossant J, et al. Efficient generation of germ line transmitting chimeras from C57BL/6N ES cells by aggregation with outbred host embryos. PLoS One. 2010;5:e11260. doi: 10.1371/journal.pone.0011260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Fromm G, dos SG, Pham LN, McDaniel IE, Burkholder A, Fargo DC, Adelman K. Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes Dev. 2012;26:933–944. doi: 10.1101/gad.187781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986;6:3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn ME. Aryl hydrocarbon receptors: diversity and evolution. Chem Biol Interact. 2002;141:131–160. doi: 10.1016/s0009-2797(02)00070-4. [DOI] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Hestermann EV, Brown M. Agonist and chemopreventative ligands induce differential transcriptional cofactor recruitment by aryl hydrocarbon receptor. Mol Cell Biol. 2003;23:7920–7925. doi: 10.1128/MCB.23.21.7920-7925.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Maltepe E, Lu MM, Simon C, Bradfield CA. Expression of ARNT, ARNT2, HIF1 alpha, HIF2 alpha and Ah receptor mRNAs in the developing mouse. Mech Dev. 1998;73:117–123. doi: 10.1016/s0925-4773(98)00038-0. [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MD, Jan LY, Jan YN. The bHLH-PAS protein Spineless is necessary for the diversification of dendrite morphology of Drosophila dendritic arborization neurons. Genes Dev. 2006;20:2806–2819. doi: 10.1101/gad.1459706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, Adli M, Kasif S, Ptaszek LM, Cowan CA, Lander ES, Koseki H, Bernstein BE. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahvis GP, Lindell SL, Thomas RS, McCuskey RS, Murphy C, Glover E, Bentz M, Southard J, Bradfield CA. Portosystemic shunting and persistent fetal vascular structures in aryl hydrocarbon receptor-deficient mice. Proc Natl Acad Sci U S A. 2000;97:10442–10447. doi: 10.1073/pnas.190256997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahvis GP, Pyzalski RW, Glover E, Pitot HC, McElwee MK, Bradfield CA. The aryl hydrocarbon receptor is required for developmental closure of the ductus venosus in the neonatal mouse. Mol Pharmacol. 2005;67:714–720. doi: 10.1124/mol.104.008888. [DOI] [PubMed] [Google Scholar]
- Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey S, Papoutsakis ET. The evolving role of the aryl hydrocarbon receptor (AHR) in the normophysiology of hematopoiesis. Stem Cell Rev. 2012;8:1223–1235. doi: 10.1007/s12015-012-9384-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Lund AK, Agbor LN, Zhang N, Baker A, Zhao H, Fink GD, Kanagy NL, Walker MK. Loss of the aryl hydrocarbon receptor induces hypoxemia, endothelin-1, and systemic hypertension at modest altitude. Hypertension. 2008;51:803–809. doi: 10.1161/HYPERTENSIONAHA.107.100586. [DOI] [PubMed] [Google Scholar]
- Lund AK, Goens MB, Kanagy NL, Walker MK. Cardiac hypertrophy in aryl hydrocarbon receptor null mice is correlated with elevated angiotensin II, endothelin-1, and mean arterial blood pressure. Toxicol Appl Pharmacol. 2003;193:177–187. doi: 10.1016/j.taap.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Medvedev SP, Pokushalov EA, Zakian SM. Epigenetics of pluripotent cells. Acta Naturae. 2012;4:28–46. [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010;35:323–332. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Peters JM, Wiley LM. Evidence that murine preimplantation embryos express aryl hydrocarbon receptor. Toxicol Appl Pharmacol. 1995;134:214–221. doi: 10.1006/taap.1995.1186. [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Puga A, Tomlinson CR, Xia Y. Ah receptor signals cross-talk with multiple developmental pathways. Biochem Pharmacol. 2005;69:199–207. doi: 10.1016/j.bcp.2004.06.043. [DOI] [PubMed] [Google Scholar]
- Qin H, Powell-Coffman JA. The Caenorhabditis elegans aryl hydrocarbon receptor, AHR-1, regulates neuronal development. Dev Biol. 2004;270:64–75. doi: 10.1016/j.ydbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. Short transcripts of the ternary complex provide insight into RNA polymerase II elongational pausing. J Mol Biol. 1995;252:522–535. doi: 10.1006/jmbi.1995.0517. [DOI] [PubMed] [Google Scholar]
- Reyes H, Reiz-Porszasz S, Hankinson O. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science. 1992;256:1193–1195. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- Sartor MA, Schnekenburger M, Marlowe JL, Reichard JF, Wang Y, Fan Y, Ma C, Karyala S, Halbleib D, Liu X, Medvedovic M, Puga A. Genomewide analysis of aryl hydrocarbon receptor binding targets reveals an extensive array of gene clusters that control morphogenetic and developmental programs. Environ Health Perspect. 2009;117:1139–1146. doi: 10.1289/ehp.0800485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnekenburger M, Peng L, Puga A. HDAC1 bound to the Cyp1a1 promoter blocks histone acetylation associated with Ah receptor-mediated trans-activation. Biochim Biophys Acta. 2007;1769:569–578. doi: 10.1016/j.bbaexp.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12:799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- Singh KP, Garrett RW, Casado FL, Gasiewicz TA. Aryl hydrocarbon receptor-null allele mice have hematopoietic stem/progenitor cells with abnormal characteristics and functions. Stem Cells Dev. 2011;20:769–784. doi: 10.1089/scd.2010.0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Wu Q, Ohsako S, Baba T, Miyamoto K, Tohyama C. Effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) on preimplantation mouse embryos. Toxicology. 2002;174:119–129. doi: 10.1016/s0300-483x(02)00047-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.