Abstract
Objective
Allelic variation (rs738409C→G) in adiponutrin (patatin-like phospholipase domain-containing protein 3, PNPLA3) has been associated with hepatic steatosis and liver fibrosis. The physiologic impact of the PNPLA3 G allele may be exacerbated in patients with severe obesity. In this study, we investigated the interactions of PNPLA3 rs738409 with a broad panel of metabolic and histologic characteristics of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis (NASH) in patients with medically complicated obesity.
Design and Methods
Consecutive patients undergoing bariatric surgery were selected for a prospective study. They underwent extensive laboratory and histologic (liver biopsy) assessment, as well as evaluation of rs738409 polymorphism by TaqMan assay.
Results
Only 12 (8.3%) of the 144 patients had normal liver histology, with 72 (50%) NASH, of whom 15 (10.4% of total patients) had fibrosis stage 2–3. PNPLA3 GG genotype correlated positively (P < 0.05) with serum levels of alanine aminotransferase (ALT), asparate aminotransferase (AST), glucose, fibrinogen, and insulin-dependent diabetes mellitus, homeostasis model assessment—insulin resistance, and presence of NASH. Multivariate analysis indicated that PNPLA3 rs738409 G versus C allele remained an (independent) risk factor for NASH, in addition to CK-18 >145 IU/l, glucose >100 mg/dl, and C-reactive protein (CRP) >0.8 mg/dl. The probability of NASH increased from 9% (no risk factor) to 82% if all four risk factors were present.
Conclusions
In this cohort of patients with medically complicated obesity, PNPLA3 rs738409 G allelic expression is associated with hepatic (NASH) and nonhepatic complications of obesity, such as insulin resistance. These novel findings may be related to a greater impact of PNPLA3 variant in magnitude and scope in patients with severe obesity than in less obese populations. Further studies are needed to characterize the nature of these associations.
Introduction
Nonalcoholic fatty liver disease (NAFLD), considered to be a hepatic manifestation of the metabolic syndrome, is associated closely with insulin resistance and obesity (1–3). The World Health Organization estimated in 2005 that more than 1.6 billion adults are overweight, and at least 400 million are obese. According to the most recent National Health and Nutrition Examination Survey (4), the prevalence of obesity (BMI ≥ 30) in the United States is 34.4%, of which about half of patients (a total of 14.4%) had obesity class II (BMI 35–39.9) or class III (BMI ≥ 40). The increase in obesity will lead to an attendant increase in the frequency of complications of obesity. A large, population-based study (5) reported hepatic steatosis measured by magnetic resonance spectroscopy in one of three US adults. The physiologic basis of the histological diversity of NAFLD, e.g., why some patients with NAFLD develop steatohepatitis with cirrhosis while others do not progress beyond simple steatosis (SS), is poorly understood. There is increasing evidence that the development of NAFLD and nonalcoholic steatohepatitis (NASH) is affected by genetic factors. Romeo et al. (6) in a large multiethnic North American study reported that allelic variation of the patatin-like phospholipase domain-containing protein 3 (PNPLA3) gene is associated strongly with hepatic fat content. A large, European, genome-wide association study (GWAS) (7) identified the same allelic PNPLA3 variation (rs738409 G) being associated with increases in serum liver enzyme elevations. Since the identification of this allelic variation of PNPLA3, other studies have confirmed the importance of rs738409 G in influencing the fat content of liver (6,8–12) and the histologic severity of NASH (8,9,13,14). Recently, a meta-analysis (15) evaluated previous studies and indicated a clear association between PNPLA3 allelic variation with steatosis and fibrosis in NAFLD. Several other studies have indicated similar associations in patients with alcoholic liver disease (16,17) and hepatitis C (18).
In addition to steatosis and NASH, PNPLA3 and its allelic variance may play a role in nonhepatic metabolic disturbances, such as obesity and insulin resistance (8,19,20). An association of rs738409 with obesity and insulin resistance has been variable. It is possible, as with other genetic predispositions such as alpha-1 antitrypsin deficiency, that the phenotypic manifestations of genetic variants in PNPLA3 are modified by nongenetic susceptibility factors, such as degree of obesity and exposure to saturated fats and cholesterol.
In this study, we investigated the associations of PNPLA3 rs738409 with a broad panel of metabolic parameters and histologic characteristics of NAFLD and NASH in patients with medically complicated obesity.
Methods
Patients
From November 2006 to April 2009 data and specimens were collected prospectively on consecutive patients with medically complicated obesity scheduled to undergo bariatric surgery. All participants gave written informed consent for participation in medical research. The study was approved by the Institutional Review Board.
All patients were assessed before bariatric surgery by an endocrinologist, psychiatrist, dietician, and bariatric surgeon to evaluate comorbidities such as hypertension, diabetes mellitus (DM), dyslipidemia, obstructive sleep apnea syndrome (OSAS), hypothyroidism, or psychiatric diseases, including chemical dependency. For the purpose of this study, comorbidities, medications, alcohol consumption, laboratory values (lipid profile and liver enzymes), and anthropometrics were recorded. The metabolic syndrome was diagnosed according to criteria of the Adult Treatment Panel III (ATP III) published in 2004 by the National Institutes of Health, meeting three or more of the following five criteria: (1) fasting blood glucose > 110 mg/dl or known DM, (2) blood pressure > 135/85 mmHg or under pharmacologic treatment, (3) serum HDL-C < 40 mg/dl in men or <50 mg/dl in women, (4) serum triglycerides > 150 mg/dl, and (5) waist circumference in men > 102 cm or in women > 88 cm. Instead of waist circumference, we used a BMI > 35 kg/m2.
Patients were excluded from participation if there was any clinical, laboratory, or histological suspicion of a chronic liver disease other than NAFLD by means of history of alcohol consumption, medication use, viral serologies for hepatitis C and B, iron studies, alpha1-antritrypsin phenotype, ceruloplasmin concentration, and antimitochondrial (AMA), antinuclear (ANA), and anti-smoothmuscle antibodies (ASMA); all data were available within 6 months of a subject entering the study.
Biochemical parameters
Patients underwent fasting morning blood samples within 24 h of liver biopsy. Samples were analyzed for a broad panel of endocrine, metabolic, and immune markers including sulfated dehydroepiandrosterone (DHEA-S), growth hormone (GH), free insulin-like growth factor (IGF-1), insulin, adiponectin, interleukin-6 (IL-6), C-peptide, glucose, leptin, CRP, free IGF, and tumor necrosis factor alpha (TNF-α). Other variables include cytokeratin-18 (CK-18) fragments (M30-Apoptosense® ELISA, Peviva AB, Sweden), complement C4 (cC4—AssayMax Himan complement C4 ELISA, Assaypro, MO), and human resistin (Human Resistin ELISA, Millipore, MA). Blood glucose concentrations were measured enzymatically with an autoanalyzer (Beckman Instruments, Fullerton, CA). All patients were screened for other causes of liver disease by measurement of ceruloplasmin levels, hepatitis C virus (HCV) and hepatitis B virus (HBV) serologies, and ANA, AMA, and ASMA.
Insulin resistance was determined using the homeostasis model assessment—insulin resistance (HOMA-IR) and the quantitative insulin sensitivity check index (QUICKI). HOMA-IR was calculated by fasting plasma glucose (mmol/l) × fasting serum insulin (µU/ml)/22.5. QUICKI was calculated by 1/(log fasting serum glucose in mg/dl + log fasting serum insulin in µU/ml). Insulin resistance was defined as QUICKI < 0.33 and HOMA-IR > 2.6.
Histologic assessment of liver biopsies
Liver biopsies were performed at the time of bariatric surgery. All histological results are based on the interpretation of these single time-point biopsies. A liver specimen with at least six portal tracts was considered adequate for histological evaluation. Liver biopsy specimens were reviewed in a blinded fashion by experienced hepatopathologists and judged on grade/stage of steatosis, necroinflammation, and fibrosis according to the Brunt criteria (21). NASH was defined as a necroinflammatory grade ≥1 (mild: steatosis involving up to 66% of biopsy; occasional ballooned zone 3 hepatocytes may be present; scattered rate intra-acinar polymorphonuclear ± intra-acinar lymphocytes; and no or mild chronic inflammation). All of the patients with NASH in our study had ballooning. Patients who met these criteria for NASH were then divided into those with mild (stage 0–1) or advanced (stage ≥2) fibrosis. Patients classified as having SS in our study had steatosis in the absence of necroinflammation and fibrosis. Patients were thus categorized prospectively into four groups according to liver histology: (1) normal, (2) SS, (3) NASH with fibrosis stage (FS) 0–1 (NASH mild), and (4) NASH with FS 2–3 (NASH advanced).
PNPLA3 rs738498 genotyping
SNP rs738409 was genotyped by TaqMan assay (Applied Biosystems, Foster City, CA) and used according to the manufacturer’s instructions. Briefly, TaqMan Master Mix, water, and oligonucleotides specific for the TaqMan Allelic discrimination assays were added to sample DNA, and analyzed for 40 cycles (92.0° for 15 s followed by 60.0° for 1 min) using an ABI 7900HT analyzer (Applied Biosystems). Genotypes were assigned using the software tools provided with the analyzer.
Statistical analysis
Quantitative patient characteristics are summarized using means ± standard deviations, whereas categorical patient factors are summarized using N’s and percentages. All reported P-values are nonparametric. In the univariate analysis, patients were classified according to their genotype and phenotype and the resulting groups were compared using either the nonparametric Kruskal-Wallis test or the Pearson χ2 test for continuous or categorical factors, respectively. Logistic regression was used to determine the association between NASH and patient risk factors. Patients were classified as either NASH (coded 1) or non-NASH (coded 0 = normal liver histology and SS). To analyze odds ratios (ORs) for the individual risk factors for NASH, patients were divided into normal versus abnormal based on the upper (or lower) limit of normal, or based on a median split. A median split was used if values were within the normal range or if normal ranges were not well validated (GH, IL-6, adiponectin, and C-peptide). In the multivariate analysis, all the statistically significant patient risk factors from the univariate analysis were entered into a logistic regression model. The factor with the greatest P-value was eliminated from the model and this process was repeated until the remaining factors were all statistically significant. Our study had a sample size of 144 patients, with a genotype distribution of approximately 60% homozygous wild type, 30% heterozygous, and 10% homozygous variant. Among the wild type the incidence of NASH was approximately 40%, whereas the incidence of NASH among the heterozygous was 65% and among the homozygous variants was 67% (a distribution very similar to that reported by others (6)).
On the basis of our literature review, we would expect that the PNPLA3 SNP rs738409C>G would have a minor allele frequency (MAF) of approximately 20–30% in a population of European descent. With 144 observations available for genotyping, we estimated the expected genotype frequencies under the assumption of independence of alleles assuming some plausible MAF scenarios (e.g., 20, 25, and 30%).
Our minimum detectable difference in order to have 80% power (α = 0.05) while allowing for unequal sample sizes, based upon the expected genotype frequencies we would obtain assuming 144 subjects for genotyping and MAF ranging from 0.20 to 0.30. For quantitative traits, we determined we would be adequately powered to detect a minimum standardized effect (δ) of 0.50 using a two-group (CC vs. CG/GG) t-test. For categorical traits we determined we would be adequately powered to detect an OR of 3.0 for traits with a frequency greater than or equal to 20% in the CC genotype group using a two-group continuity-corrected χ2 test of equal proportions. If the trait frequency in the CC genotype group was to drop as low as 5 (10)%, the minimum detectable OR would be 5.5 (4.0).
Because of the large of number of hypotheses being tested, the statistical standard for significance was increased, using a Bonferroni adjustment of 0.05/40 tests, to P ≤ 0.025 for univariate analyses. Finally, all two-way interactions among the final patient risk factors were calculated and these interaction terms were evaluated for statistical significance using the likelihood ratio test. The traditional P-value of 0.05 was used to determine statistical significance. Statistical analyses were calculated using Proc. Logistic (SAS) and the JMP software (SAS institute, NC).
Results
Patients
The study included 144 severely obese patients of whom 122 (84.7%) were females. Mean age was 47.9 ± 10.7 years, with a mean BMI of 46.6 ± 7.7 kg/m2. The range of the BMI was 34.8–72.6 kg/m2 with 50% of the BMI values between 41.1 and 50.9 kg/m2. BMI and age did not differ between female and male patients. Comorbidities were common; 56 of the total patients had type 2 DM (38.9%) of whom 15 used insulin, 87 had hypertension (60.4%), 99 (68.7%) had OSAS, and 102 (72.9%) fulfilled the ATP III criteria for metabolic syndrome.
Biochemical and histologic assessment
All patients underwent liver biopsy at the time of bariatric surgery. Evaluation of these liver biopsies showed that 12 patients (8.3%) had normal liver histology, 60 (41.7%) had SS, and 72 (50%) had NASH. Of the 72 patients with NASH, 57 (41.7% of total patients and 79.2% of NASH patients) had early-stage NASH (F 0–1) and 15 (10.4% of total patients and 26.3% of NASH patients) had advanced stage NASH (F 2–3).
Evaluation of NASH versus non-NASH patients
In this severely obese population, sex, BMI, and age did not differ significantly between patients with NASH (n = 72) or without NASH (n = 72) (Supporting Information Table). Patients with NASH had significantly higher serum levels of glucose (123.3 ± 42.0 vs. 115.1 ± 52.9, P = 0.005), insulin (20.7 ± 11.6 vs. 17.5 ± 15.5, P = 0.009), and insulin-resistance scores HOMA-IR (6.7 ± 5.6 vs. 5 ± 5.5, P = 0.003) and QUICKI (0.30 ± 0.03 vs. 0.32 ± 0.03, P = 0.004) than patients without NASH. Analysis of biomarkers indicated that, when compared to patients without NASH, NASH patients had significantly higher mean serum values of IL-6 (11.8 ± 25 vs. 5 ± 4.4, P = 0.02), CK-18 (265.7 ± 139 vs. 225.8 ± 126, P = 0.04), CRP (1.9 ± 0.2 vs. 0.8 ± 0.7, P = 0.005), and decreased values of GH (0.30 ± 0.45 vs. 0.39 ± 0.45, P = 0.02) and adiponectin (7165.8 ± 2942.1 vs. 9579.4 ± 3923.1, P < 0.001). There were no differences in parameters of lipid metabolism (including serum cholesterol, LDL, HDL, and triglycerides). Analysis did not reveal significant different frequencies of comorbidities between the two groups.
PNPLA3 variants
The distribution of PNPLA3 rs738409 genotypes was CC in 88 patients (61.1%), CG in 44 patients (30.6%), and GG in 12 patients (8.3%) (Table 1).
TABLE 1.
Demographics—liver histology and comorbidities—and variation with PNPLA3 genotype
| Variables | PNPLA3 genotyping |
P-values— overall |
||
|---|---|---|---|---|
| CC | CG | GG | ||
| Patients (n, % of all patients) | 88 (61.1%) | 44 (30.6%) | 12 (8.3%) | |
| BMI (kg/m2) | 45.8 ± 7.4 | 47.0 ± 6.6 | 50.7 ± 12.0 | 0.13 |
| Age (years) | 48.3 ± 11.0 | 46.8 ± 10.2 | 49.0 ± 10.4 | 0.69 |
| Liver histology (n, %) | ||||
| Simple steatosis or normal | 52 (59%) | 16 (36%) | 4 (33%) | 0.01 |
| NASH | 36 (41%) | 28 (64%) | 8 (67%) | 0.01 |
| NASH with FS 0–1 | 29 (33%) | 23 (52%) | 5 (42%) | 0.10 |
| NASH with FS 2–4 | 7 (8%)⊤ | 5 (11%) | 3 (25%)⊤ | 0.17 |
| Type 2 DMA total patients (n, %) | 28 (32%) | 21 (48%) | 7 (58%) | 0.07 |
| IDDM (n, %) | 5 (6%) | 6 (14%) | 4 (33%) | 0.009 |
| OSAS (n, %) | 61 (69%) | 31 (71%) | 8 (67%) | 0.96 |
| Metabolic syndrome (n, %) | 62 (71%) | 32 (76%) | 9 (75%) | 0.83 |
| HOMA-IR > 2.6 (n, %)* | 63 (73%) | 37 (88%) | 8 (80%) | 0.15 |
| Hypertension (n, %) | 47 (54%) | 31 (71%) | 9 (75%) | 0.09 |
| Anticholesterol medication | 23 (26%) | 17 (39%) | 6 (50%) | 0.13 |
P-values derived using a Pearson χ2 test of the 2 × 3 table (e.g., IDDM vs. rs738409C>G genotype).
FS, fibrosis stage; OSAS, obstructive sleep apnea.
Histologic associations with PNPLA3 variants
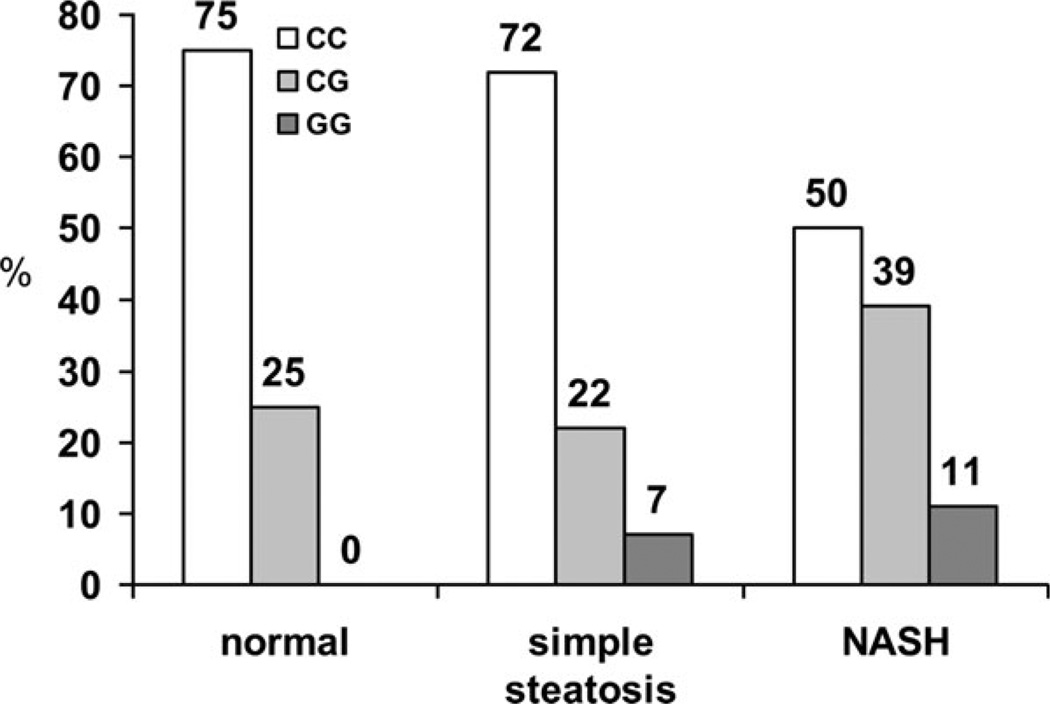
The prevalence of NASH varied significantly with PNPLA3 genotypes (P = 0.01) (Table 1 and Figure 1). Of the 12 patients with GG genotype, eight (66.7%) had NASH, including three patients (25%) with progressive NASH (F2–3). Twenty-eight patients (63.6%) of the CG genotype had NASH, including five (11.4%) with fibrosis stages 2–3, whereas 36 patients (40.9%) of the CC genotype had NASH, including seven patients (8%) with fibrosis stage 2–3.
FIGURE 1.
Variation in PNPLA3 genotype with severity of liver disease (a) and variation severity of liver disease with PNPLA3 genotype (b). The distribution of PNPLA3 rs738409 genotypes was CC in 88 patients (61.1%), CG in 44 patients (30.6%), and GG in 12 patients (8.3%). For histological findings, 12 patients (8.3%) had normal liver histology, 60 patients (41.7%) had simple steatosis, and 72 patients (50%) had NASH. The prevalence of NASH was significantly different with the PNPLA3 genotypes, with analysis of the subpopulations indicating that CG patients have a higher prevalence of NASH (P < 0.05) when compared with CC patients.
Nonhepatic associations with PNPLA3 variants
Analysis of the comorbidities (Table 1) indicated that the prevalence of insulin-dependent DM (IDDM) differed (P < 0.009) between the three populations. Other differences (P < 0.025) between the three PNPLA3 genotypes were serum levels of AST and trends (P < 0.1) for HOMA-IR, IL-6, and serum fibrinogen (Table 2).
TABLE 2.
Laboratory findings—variation with PNPLA3 genotype
| Variables | PNPLA3 genotyping |
P-values— overall |
||
|---|---|---|---|---|
| CC | CG | GG | ||
| ALT (IU/l) | 30.9 ± 15.1 | 33.0 ± 16.4 | 47.3 ± 31.9 | 0.16 |
| AST (IU/l) | 27.2 ± 8.3 | 26.7 ± 11.2 | 37.8 ± 15.9 | 0.02 |
| Cholesterol (mg/dl) | 195.0 ± 39.4 | 186.7 ± 34.7 | 184.6 ± 33.6 | 0.60 |
| LDL (mg/dl) | 112.9 ± 32.5 | 108.8 ± 32.3 | 107.3 ± 32.1 | 0.81 |
| HDL (mg/dl) | 47.4 ± 12.0 | 48.4 ± 12.0 | 44.6 ± 10.0 | 0.80 |
| Triglycerides (mg/dl) | 165.0 ± 73.3 | 155.26 ± 2.7 | 163.1 ± 70.1 | 0.85 |
| Glucose (mg/dl) | 112.5 ± 35.6 | 124.0 ± 54.7 | 153.8 ± 86.9 | 0.36 |
| Insulin (µIU/ml) | 18.3 ± 14.8 | 19.6 ± 11.4 | 23.2 ± 14.1 | 0.20 |
| HOMA-IR | 5.2 ± 5.2 | 6.3 ± 5.0 | 9.9 ± 9.2 | 0.09 |
| IL-6 (pg/ml) | 5.8 ± 6.7 | 13.5 ± 30.4 | 7.1 ± 4.3 | 0.08 |
| CK-18 (IU/l) | 247 ± 137 | 242 ± 127 | 225 ± 86 | 0.96 |
| CRP (mg/dl) | 0.96 ± 1.20 | 1.39 ± 1.93 | 1.07 ± 0.77 | 0.49 |
| cC4 (µg/ml) | 419 ± 126 | 401 ± 145 | 412 ± 65 | 0.35 |
| Free IGF-1 (ng/ml) | 0.94 ± 0.28 | 0.97 ± 0.30 | 0.97 ± 0.43 | 0.75 |
| GH (ng/ml) | 0.28 ± 0.35 | 0.42 ± 0.53 | 0.56 ± 0.67 | 0.54 |
| DHEA-S (µg/dl) | 97.9 ± 74.0 | 92.8 ± 86.5 | 62.4 ± 39.4 | 0.36 |
| C-peptide (nmol/l) | 1.3 ± 0.5 | 1.4 ± 0.9 | 1.1 ± 0.6 | 0.27 |
| Leptin (ng/ml) | 47.6 ± 25.1 | 50.0 ± 24.9 | 60.3 ± 44.3 | 0.63 |
| Adiponectin (ng/ml) | 8435 ± 3693 | 8375 ± 3815 | 8317 ± 3243 | 0.997 |
| Resistin (ng/ml) | 16.3 ± 5.5 | 17.1 ±8.4 | 17.0 ± 5.9 | 0.89 |
| TNF-α (pg/ml) | 1.57 ± 0.60 | 1.58 ± 0.76 | 1.79 ± 0.68 | 0.37 |
| Fibrinogen | 441 ± 93 | 468 ± 108 | 521 ± 104 | 0.07 |
Comparison of CC versus non-CC.
Odds ratios and multivariate model for predicting NASH
Odds ratios were calculated for significant risk factors of NASH in order to compare these risk factors. Analysis (Table 3) indicated that the ORs of these predictive variables were more or less similar, with PNPLA3 genotype having similar ORs as for glucose (>100 mg/dl) and CK-18 (>145 IU/l), and just less than other, well-established risk factors such as insulin resistance (HOMA-IR > 2.6) and abnormal ALT (>29 IU/l upper limit of normal).
TABLE 3.
Odds ratios of variables significantly different in patients with NASH
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Variables (normal value/or median) |
Odds ratios (95% CI) |
P-values | Odds ratios (95% CI) |
P-values |
| HOMA-IR > 2.6 | 3.2 (1.3–7.8) | 0.001 | A | A |
| ALT > 29 IU/l | 3.0 (1.5–5.9) | 0.002 | A | A |
| CRP > 0.80 mg/l | 2.9 (1.4–5.8) | 0.02 | 2.4 (1.1–5.3) | 0.02 |
| PNPLA3 G | 2.8 (1.4–5.5) | 0.004 | 2.5 (1.1–5.3) | 0.03 |
| Glucose > 100 mg/dl | 2.7 (1.3–5.6) | 0.006 | 2.5 (1.1–5.4) | 0.03 |
| CK-18 > 145 IU/l | 2.7 (1.1–6.7) | 0.03 | 3.1 (1.2–8.2) | 0.02 |
| AST > 31 IU/l | 2.5 (1.2–5.5) | 0.02 | A | A |
| Insulin > 14 µIU/l | 2.3 (1.2–4.6) | 0.02 | A | A |
| Adiponectin < 7,943 ng/ml | 2.3 (1.2–4.6) | 0.02 | A | A |
| Interleukin-6 > 4.3 pg/ml | 1.9 (0.9–4.0) | 0.09 | A | A |
| GH > 0.17 ng/ml | 0.6 (0.3–1.1) | 0.10 | A | A |
| C-peptide > 1.2 nmol/l | 1.2 (0.6–2.3) | 0.64 | A | A |
The importance of PNPLA3 genotype in this severely obese study population was further shown by multivariate analysis. Analysis indicated that the final model of independent risk factors for NASH included CRP > 0.80 mg/dl (OR 2.43 [1.12–5.31]), glucose > 100 mg/dl (OR 2.45 [1.12–5.38]), CK-18 > 145 IU/l (OR 3.11 [1.18–8.20]), and PNPLA3 G allele (OR 2.43 [1.12–5.31]). This analysis also indicated that after adjusting PNPLA3 genotype for the other significant risk factors the value of the OR only decreased slightly.
Probability of NASH
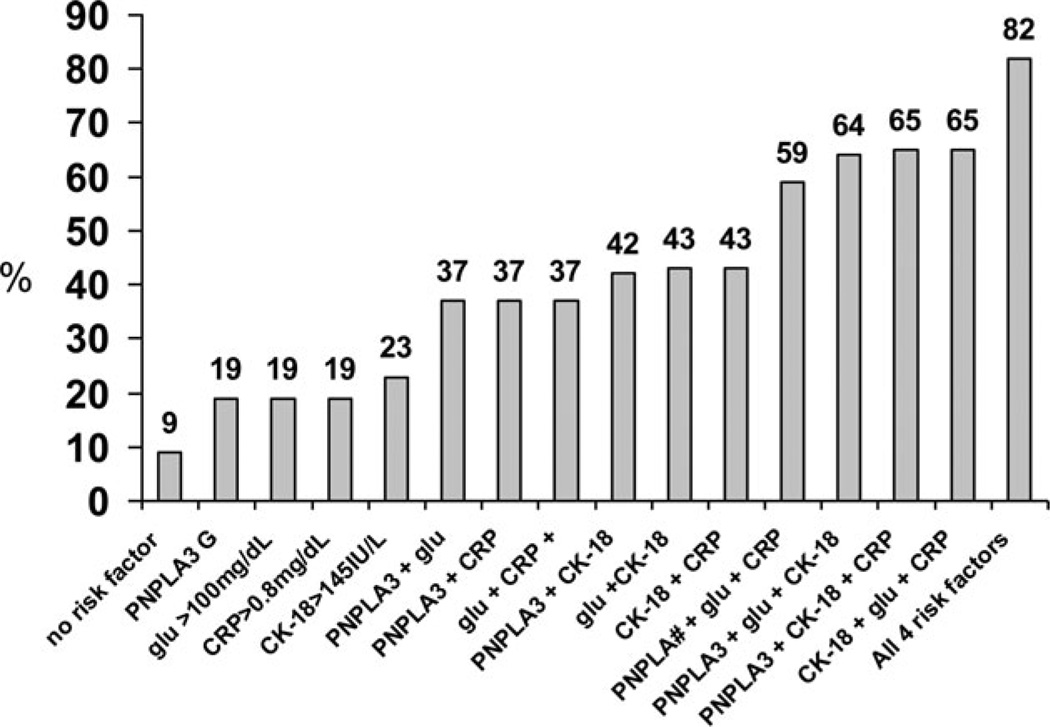
Based on the four risk factors independently predictive of NASH (PNPLA3 G, CRP > 0.80 mg/dl, glucose > 100 mg/dl, and CK-18 > 145IU/l), the probability of NASH was calculated. (Figure 2) This calculation revealed that in patients with no risk factor the probability of NASH was 9% (95% confidence intervals: 3–24%), whereas if all four risk factors were present the probability of NASH increased to 82% (65–91%). Only three patients in our study had no risk factor, whereas nine patients had all four risk factors. The remaining distribution of risk factors was as follows: 32 patients with one risk factor, 45 patients with two risk factors, and 44 patients with three risk factors. The calculated probabilities of NASH for these combinations are shown in Figure 2.
FIGURE 2.
The prevalences of NASH based on the presence of four risk factors found to be independently predictive of NASH (PNPLA3 G, CRP > 0.80 mg/dl, glucose > 100 mg/dl, and CK-18 > 145IU/l). In patients with no risk factor the probability of NASH was 9% (95% confidence intervals: 3–24%), whereas if all four risk factors were present the probability of NASH increased to 82% (65–91%). Only three patients in our study had no risk factor, 32 had one risk factor, 45 had two risk factors, 44 had three risk factors, and nine patients had all four risk factors.
Discussion
Allelic variation in PNPLA3 (rs738409 G) has been repeatedly shown to correlate with transaminases, hepatic steatosis (6–12), and also with histologic severity of NASH in both adults and children (8,9,13,14). The impact of risk factors for more histologically severe NAFLD (NASH) is likely to vary with the magnitude of established risk factors, such as the degree and severity of medical complications of obesity. Patients with medically complicated obesity have a high prevalence of comorbidities (e.g., insulin resistance and type 2 DM), which may negate or exacerbate the effects of genetic variation of PNPLA3 on the development of steatosis and NASH. The impact of genetic variation in PNPLA3 may, for example, be different for an individual with a BMI of 28 kg/m2 than for an individual with a BMI of 48 kg/m2. For this reason we felt it was important to determine the interactions of a broad panel of biologic parameters with PNPLA3 genotype and histologic characteristics of NAFLD and NASH in a high-risk population—patients with medically complicated obesity. Our study population was enriched with obesity-related liver disease, with only 10% of studied patients having normal liver histology and 50% meeting histologic criteria for NASH. The high prevalence of NAFLD and NASH occurred in concert with other obesity-related complications, including hypertension, dyslipidemia, and insulin resistance. We made several important observations with regard to PNPLA3 rs738409 genotype in this study population.
Similar to analyses in less severely obese patients (22), we saw an association of PNPLA3 genotype with the presence of NASH. Specifically, we found that the G allele is associated with an OR of 2.75 for NASH when compared with the C allele. We also noted a new association of the PNPLA3 genotype with IDDM and insulin resistance scores. The frequency of insulin requiring type 2 DM was 33% of patients with GG genotype vs. 14 and 6%, respectively, for CG and CC genotypes.
Implications and the potential basis of these observations merit detailed consideration. The PNPLA3 gene encodes for a protein product that is part of the patatin-like family of proteins containing a phospholipase domain. Within this family of proteins, PNPLA3 has most resemblance with PNPLA2, which is the major triglyceride lipase in adipose tissue and skin. Prior studies (23,24) have illustrated that PNPLA3 also has triglyceride lipase activity in addition to acyl-CoA-independent transacylation of acylglycerols. Based on the observation of no effect of PNPLA3 knockout mice on liver steatosis (25), the effect of rs738409 G allele is probably not related to loss of function of the wild-type protein but to the differential expression and function of variant PNPLA3. The effect of variant PNPLA3 protein may relate to diminished mobilization or hydrolysis of triglycerides (26). Nonhepatic effects of PNPLA3 genotype might be expected because expression of PNPLA3 is also seen in adipocyte and skin tissue (27). In addition, studies have shown that PNPLA3 expression is rapidly induced by caloric intake and by insulin (27,28). It is therefore not surprising that the expression of PNPLA3 has been shown to be increased in obese individuals (29), which also implies a greater expression of its G allele-associated protein. This was further illustrated by Davis et al. (30) in obese children, who observed that high dietary carbohydrate and sugar intake correlated with hepatic fat, but only in patients with the GG genotype (independent of covariates). It is therefore possible that patients with severe obesity have increased (negative) effects of PNPLA3 G allele variant protein due to increased expression when compared with less severely obese patients. Study selection is thus key in finding some or no clinical associations with PNPLA3 G genotype; the earliest findings of PNPLA3 G genotype in the Dallas heart study (3) did not reveal an association of PNPLA3 G with transaminases in relatively lean European and African Americans (with low prevalences of NAFLD), whereas this association was shown in Hispanics (with high prevalence of NAFLD) as well as in subsequent studies in populations with a high prevalence of NAFLD and NASH (8–11). With 144 total participants, our study was not powered to detect any differential effect of ethnicity. It is possible that the results may have been confounded by cryptic stratification on the basis of ethnicity. In support of a possible association between the PNPLA3 G allelic variant and insulin resistance are the findings of Johansson et al. (19) who reported that insulin resistance in a large Swedish population-based sample (n = 1,811) increased more rapidly with increasing BMI in carriers of the rs738409 G genotype when compared with wild-type carriers. In addition, two other SNPs of the PNPLA3 gene were associated with insulin sensitivity in a large population of Swedish children (31). Despite multiple comorbidities in our study patients and a high prevalence of other NASH-related risk factors, PNPLA3 remained an independent risk factor, even after adjusting for all the other risk factors. The four independent risk factors of NASH in this study population were PNPLA3 G allele, CK-18 > 145 IU/l, glucose > 100 mg/dl, and CRP > 0.8 mg/dl. In patients without any of these risk factors the probability of NASH was only 9%, increasing to 82% in patients with all four risk factors.
In summary, we report that PNPLA3 G allelic expression is an independent risk factor for NASH in patients with severe obesity with a high prevalence of insulin resistance and several indices of insulin resistance. We report novel associations of PNPLA3 G genotype with nonhepatic complications, such as insulin resistance. The nature and mechanism of the association of PNPLA3 genotype with nonhepatic complications of obesity warrant future studies.
Acknowledgments
Funding agencies: This work has been supported by Public Health Service grants NIDDK RO1 DK41876, DK069757-05 (to M. R. C.), and GCRC RR00585.
Footnotes
Author contributions: MMJ Guichelaar: acquisition of data, analysis and interpretation of data, and drafting of the manuscript; S Gawrieh, M Oliver, M Viker, A Krishnan, KD Watt, JM Swain, and M Sarr: acquisition of data and critical revision of the manuscript; S Sanderson: acquisition of data and critical revision of the manuscript for important intellectual content; M Malinchoc: statistical analyses and critical revision of the manuscript for important intellectual content; MR Charlton: acquisition of data, analysis and interpretation of data, drafting of the manuscript, and critical revision of the manuscript for important intellectual content.
Disclosure: The authors declared no conflict of interest.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Angulo P, Keach JC, Batts KP, Lindor KD. Independent predictors of liver fibrosis in patients with nonalcoholic steatohepatitis. Hepatology. 1999;30:1356–1362. doi: 10.1002/hep.510300604. [DOI] [PubMed] [Google Scholar]
- 2.Luyckx FH, Lefebvre PJ, Scheen AJ. Non-alcoholic steatohepatitis: association with obesity and insulin resistance and influence of weight loss. Diabetes Metab. 2000;26:98–106. [PubMed] [Google Scholar]
- 3.Dixon JB, Bhathal PS, O’Brien PE. Nonalcoholic fatty liver disease: predictors of nonalcoholic steatohepatitis and liver fibrosis in the severely obese. Gastroenterology. 2001;121:91–100. doi: 10.1053/gast.2001.25540. [DOI] [PubMed] [Google Scholar]
- 4.Shields M, Carrol MD, Ogden CL. Adult obesity prevalence in Canada and the United States. NHCS Data Brief. 2011;56:1–8. [PubMed] [Google Scholar]
- 5.Browning JD, Szczepaniak LS, Dobbins R, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 6.Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan X, Waterworth D, Perry JER, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83:520–528. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valenti L, Al-Serri A, Daly AK, et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51:1209–1217. doi: 10.1002/hep.23622. [DOI] [PubMed] [Google Scholar]
- 9.Sookoian S, Castano GO, Burhueno AL, et al. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res. 2009;50:2111–2116. doi: 10.1194/jlr.P900013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kantartzis K, Peter A, Machicao F, et al. Dissociation between fatty liver and insulin resistance in humans carrying a variant of the patatin-like phospholipase 3 gene. Diabetes. 2009;58:2616–2623. doi: 10.2337/db09-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotronen A, Peltonen M, Hakkarainen A, et al. Prediction of non-alcoholic fatty liver disease and liver fat using metabolic and genetic factors. Gastroenterology. 2009;137:865–872. doi: 10.1053/j.gastro.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Kotronen A, Johansson LE, Johansson LM, et al. A common variant in PNPLA3, which encodes adiponutrin, is associated with liver fat content in humans. Diabetologia. 2009;52:1056–1060. doi: 10.1007/s00125-009-1285-z. [DOI] [PubMed] [Google Scholar]
- 13.Rotman Y, Koh C, Zmuda JM, et al. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology. 2010;52:894–903. doi: 10.1002/hep.23759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hotta K, Yoneda M, Hyogo H, et al. Association of the rs738409 polymorphism in PNPLA3 with liver damage and the development of nonalcoholic fatty liver disease. BMC Med Genet. 2010;11:1–10. doi: 10.1186/1471-2350-11-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sokooian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 16.Tian C, Stokowski RP, Kershenobich D, et al. Variant in PNPLA3 is associated with alcoholic liver disease. Nat Genet. 2010;42:21–23. doi: 10.1038/ng.488. [DOI] [PubMed] [Google Scholar]
- 17.Stickel F, Buch S, Lau K, et al. Genetic variation in the PNPLA3 gene with alcoholic liver injury in Caucasians. Hepatology. 2011;53:86–95. doi: 10.1002/hep.24017. [DOI] [PubMed] [Google Scholar]
- 18.Valenti L, Rumi M, Galmozzi E, et al. Patatin-like phospholipase domain-containing 3 I148M polymorphism, steatosis, and liver damage in chronic hepatitis C. Hepatology. 2011;53:791–799. doi: 10.1002/hep.24123. [DOI] [PubMed] [Google Scholar]
- 19.Johansson LE, Lindblad U, Larsson CA, et al. Polymorphisms in the adiponutrin gene are associated with increased insulin secretion and obesity. Eur J Endocrinol. 2008;159:577–583. doi: 10.1530/EJE-08-0426. [DOI] [PubMed] [Google Scholar]
- 20.Steinberg GR, Kemp BE, Watt MJ. Adipocyte triglyceride lipase expression in human obesity. Am J Physiol Endocrinol Metab. 2007;293:E958–E964. doi: 10.1152/ajpendo.00235.2007. [DOI] [PubMed] [Google Scholar]
- 21.Kleiner DE, Brunt EM, Van Natta M, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 22.Romeo S, Sentinelli F, Dash S, et al. Morbid obesity exposes the association between PNPLA3 I148M (rs738409) and indices of hepatic injury in individuals of European descent. Int J Obes (Lond) 2010;34:190–194. doi: 10.1038/ijo.2009.216. [DOI] [PubMed] [Google Scholar]
- 23.Lacke AC, Sun Y, Li JL, et al. Expression, regulation and triglyceride hydroxylase activity of adiponutrin family members. J Lipid Res. 2005;46:2477–2487. doi: 10.1194/jlr.M500290-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins CM, Mancuso DJ, Yan W, et al. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004;279:48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Chang B, Li L, Chan L. PNPLA3/adiponutrin deficiency in mice is not associated with fatty liver disease. Hepatology. 2010;52:1134–1142. doi: 10.1002/hep.23812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He S, McPhaul C, Li JZ, et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem. 2010;285:6706–6715. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polson DA, Thompon MP. Adiponutrin mRNA expression in white adipose tissue is rapidly induced by meal-feeding a high sucrose diet. Biochem Biophys Res Commun. 2003;301:261–266. doi: 10.1016/s0006-291x(02)03027-9. [DOI] [PubMed] [Google Scholar]
- 28.Moldes M, Beauregard G, Faraj M, et al. Adiponutrin gene is regulated by insulin and glucose in human adipose tissue. Eur J Endocrinol. 2006;155:461–468. doi: 10.1530/eje.1.02229. [DOI] [PubMed] [Google Scholar]
- 29.Steinberg GR, Kemp BE, Watt MJ. Adipocyte triglyceride lipase expression in human obesity. Am J Physiol Endocrinol Metab. 2007;293:E958–E964. doi: 10.1152/ajpendo.00235.2007. [DOI] [PubMed] [Google Scholar]
- 30.Davis JN, Le KR, Walker RW, et al. Increased hepatic fat in overweight Hispanic youth influenced by interaction between genetic variation in PNPLA3 and high dietary carbohydrate and sugar consumption. Am J Clin Nutr. 2010;92:1522–1527. doi: 10.3945/ajcn.2010.30185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansson LE, Johansson LM, Danielsson P, et al. Genetic variance in the adiponutrin gene family and childhood obesity. PloS One. 2009;4:e5327. doi: 10.1371/journal.pone.0005327. [DOI] [PMC free article] [PubMed] [Google Scholar]