Summary
Nutrient deprivation is a stimulus shared by both autophagy and the formation of primary cilia. The recently discovered role of primary cilia in nutrient sensing and signaling motivated us to explore the possible functional interactions between this signaling hub and autophagy. Here we show that part of the molecular machinery involved in ciliogenesis also participates in the early steps of the autophagic process. Signaling from the cilia, such as that from the Hedgehog pathway, induces autophagy by acting directly on essential autophagy-related proteins strategically located in the base of the cilium by ciliary trafficking proteins. While abrogation of ciliogenesis partially inhibits autophagy, blockage of autophagy enhances primary cilia growth and cilia-associated signaling during normal nutritional conditions. We propose that basal autophagy regulates ciliary growth through the degradation of proteins required for intraflagellar transport. Compromised ability to activate the autophagic response may underlie the basis of some common ciliopathies.
Keywords: primary cilia, intraflagellar transport proteins, lysosomes, autophagosomes, vesicular trafficking
Introduction
Autophagy is a cellular catabolic process that contributes to quality control and maintenance of the cellular energetic balance through the turnover of proteins and organelles in lysosomes1. Induction of autophagy recruits proteins and lipids from different intracellular membranes2 to initiate the formation of autophagosomes, double-membrane vesicles that sequester cytoplasmic material and deliver it through vesicular fusion to lysosomes for degradation3.
The primary cilium is a non-motile signaling organelle that grows in a specific region of the plasma membrane and senses, among others, changes in the enrichment of nutrients in the environment4. Cargo trafficking along the ciliary axoneme (intraflagellar transport; IFT) is maintained through motor proteins (kinesins and dyneins) and two large multiprotein complexes (IFT particles A and B)5. Some subunits of these complexes can be found in other cellular compartments such as Golgi, from where they facilitate mobilization of specific cargo to the basal body and cilium for ciliogenesis and ciliary signaling6. The primary cilium coordinates a variety of signaling pathways including the Hedgehog (Hh) pathway which requires IFT-mediated recruitment to the basal body and axoneme of smoothened (Smo) and the transcription factors Gli1 and Gli27,8.
In many types of cultured cells, deprivation of serum from the media induces almost linear growth of primary cilia up to 2 days9. Induction of autophagy also occurs in the first hours that follow serum removal, and can be sustained for the full duration of the starvation period10. Despite the temporal coincidence of the formation of autophagosomes and primary cilia, any possible relation between the autophagic and ciliogenesis machineries remains unknown. Likewise, the possible involvement of autophagy in ciliary dynamics has not been analyzed.
In this study we investigate the functional interaction between primary cilia and autophagic induction. We have found that maximum activation of autophagy in response to nutrient deprivation requires the strategic location of components of the autophagic machinery at the ciliary base in a IFT and Hh signaling-dependent manner. In contrast, when autophagy is compromised, ciliogenesis is enhanced and cilia grow longer. We conclude that ciliary signaling pathways, such as Hh, may specify a cilia-mediated autophagy, closely related to autophagosome assembly at the plasma membrane and that in turn, basal autophagy negatively modulates ciliary growth through turnover of essential ciliogenesis proteins (Extended Data Fig. 1).
Results
Disruption of IFT compromises autophagy
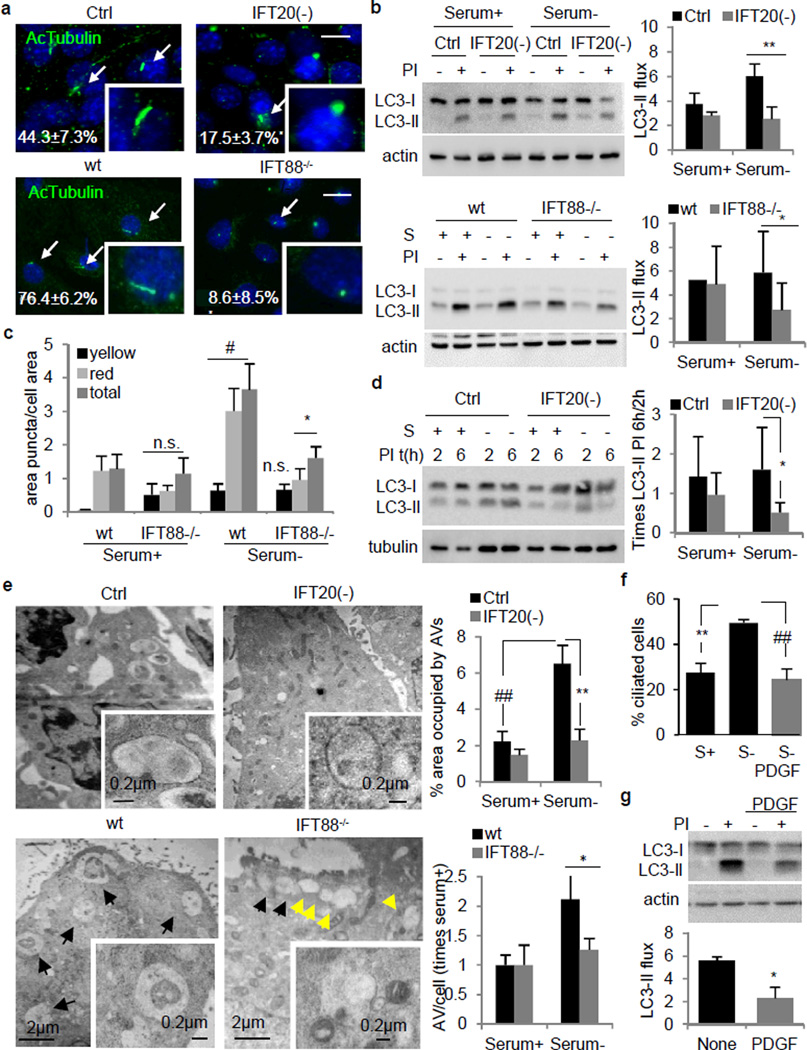
To determine the role of primary cilia on autophagy, we used two cellular models with compromised ciliogenesis: mouse embryonic fibroblast (MEFs) stably knocked-down for IFT20 (IFT20(−)) and kidney epithelial cells (KECs) from mice with a hypomorphic mutant IFT88 (IFT88−/−), two IFT-B complex components required for ciliogenesis6 (Extended Data Fig. 2a). After 24h of serum removal, cells with detectable primary cilia were reduced by 60% in IFT20(−) MEFs and almost absent in the IFT88−/− KECs (Fig. 1a).
Figure 1. Blockage of IFT reduces autophagic activity.
(a) Ciliated control and IFT20(−) MEFs (*p=0.030, n=3), and wt and IFT88−/− KECs (**p=0.003, 25 cell each, n=3). Arrows: cilia. (b) LC3 flux. Ctrl and IFT20(−) MEFs (**p=0.0003, n=7). Wt and IFT88−/− KECs (*p=0.035, n=9). (c) Autophagic flux by mCherry-GFP-LC3. Puncta: Yellow, autophagosomes; red autophagolysosomes; total, both together (#p=0.008, *p=0.025). (d) Autophagosome formation. S: serum; PI: protease inhibitors (*p=0.048, n=4). (e) Electron microscopy and quantification of autophagic vacuoles (AV) in Ctrl and IFT20(−) MEFs (##p=0.001, **p=0.001, 10 fields in 2 experiments) and KECs wt and IFT88−/− (*p=0.037, 5 fields). Arrows: AV (black) and endosomal compartments (yellow). (f) Ciliated MEFs after 50ng/ml PDGF. (**p=0.001, ##p=0.0009, 121, 230 and 162 cells, n=5). (g) LC3 flux upon PDGF (*p=0.005, n=3). Bars: 10µm. n.s., statistically non-significant. Mean±s.d. in b, d and mean±s.e.m rest.
Rates of lysosomal protein degradation after serum deprivation, a stimulus that activates autophagy, were reduced in the two ciliogenesis-impaired models (Extended Data Fig. 2b). To test if autophagy was defective we analyzed flux through the autophagic pathway using lysosomal proteolysis inhibitors and measuring changes in levels of LC3-II, an autophagosome component degraded with the cargo in lysosomes11. The characteristic upregulation of the autophagic flux after serum removal was reduced in IFT20(−) MEFs whereas basal autophagic flux (rich media) was only discretely reduced (Fig. 1b). Although a high percentage of KECs are ciliated under basal conditions and serum removal has small effect on ciliogenesis (Extended Data Fig. 2c), we still observed a reduction in autophagic flux upon serum removal in IFT88−/− KECs when compared to wild type (wt) (Fig. 1b), suggesting that primary cilia are not necessary to maintain the high basal autophagic flux of these cells (Extended Data Fig. 2d), but are essential to sustain proper autophagic flux during nutrient deprivation.
To directly analyze autophagosome formation and their clearance by lysosomes, we transfected cells with a pH-sensitive reporter (mCherry-GFP-LC3) that highlights autophagosomes as yellow puncta and autophagolysosomes (post-lysosomal fusion) as red puncta12. Basal levels of autophagic vacuoles were comparable in wt and IFT88−/− KECs, but upon removal of serum IFT88−/− KECs displayed a significantly lower content of autophagolysosomes (Fig. 1c and Extended Data Fig. 2e). This reduction in autophagolysosomes was not paralleled by an increase in autophagosomes, and the overall content of LC3-positive vesicles in serum deprived IFT88−/− KECs was lower than in wt cells (Extended Data Fig. 2f) suggesting that reduced autophagic flux was mainly due to lower autophagosome synthesis rather than blockage in their degradation. Autophagosome formation (assessed as the increase in LC3-II levels at two time points after inhibition of lysosomal proteolysis) was also reduced in IFT20(−) MEFs (Fig. 1d). Ultrastructural analysis confirmed that despite normal autophagosome morphology, IFT20(−) and IFT88−/− cells failed to expand the autophagic compartment in response to serum deprivation (Fig. 1e).
Contrary to the restricted location of IFT88 to the ciliary base13, IFT20 is also present in other cellular compartments6. However the effect of IFT20 on autophagy seems to be primarily related to its function in ciliogenesis since additional knock-down for IFT20 in IFT88 knock-down cells did not result in further reduction of autophagy (Extended Data Fig. 3a–d), supporting that blockage of anterograde IFT (aIFT) compromises activation of inducible autophagy. Consistent with this notion, cilia resorption induced by treatment with platelet-derived growth factor (PDGF)14 (Fig. 1f) markedly decreased starvation-induced autophagic flux in MEFs, KECs and in RGC-5 cells, a retinal ganglion cell line with robust induction of ciliogenesis in response to serum removal (Fig. 1g and Extended Data Fig. 3e).
Overall these findings support a reduced ability for autophagosome synthesis in cells with compromised ciliogenesis that limits their capacity to upregulate autophagy in response to nutrient deficiency.
Ciliary Hh signaling induces autophagy
Altered aIFT blocks ciliogenesis and prevents the recruitment of essential signaling molecules to the ciliary region6. We hypothesized that malfunctioning of ciliary signaling may be behind the defective starvation-induced autophagy in cells with compromised IFT. Although primary cilia downregulates signaling through the mammalian Target of Rapamycin (mTOR) pathway15, a well-known negative regulator of autophagy, treatment of IFT20(−) and IFT88−/− cells with the mTOR inhibitor rapamycin failed to restore normal autophagic activity in these cells (data not shown); thereby reduced autophagy in ciliogenesis-defective cells was not due to enhanced mTOR signaling.
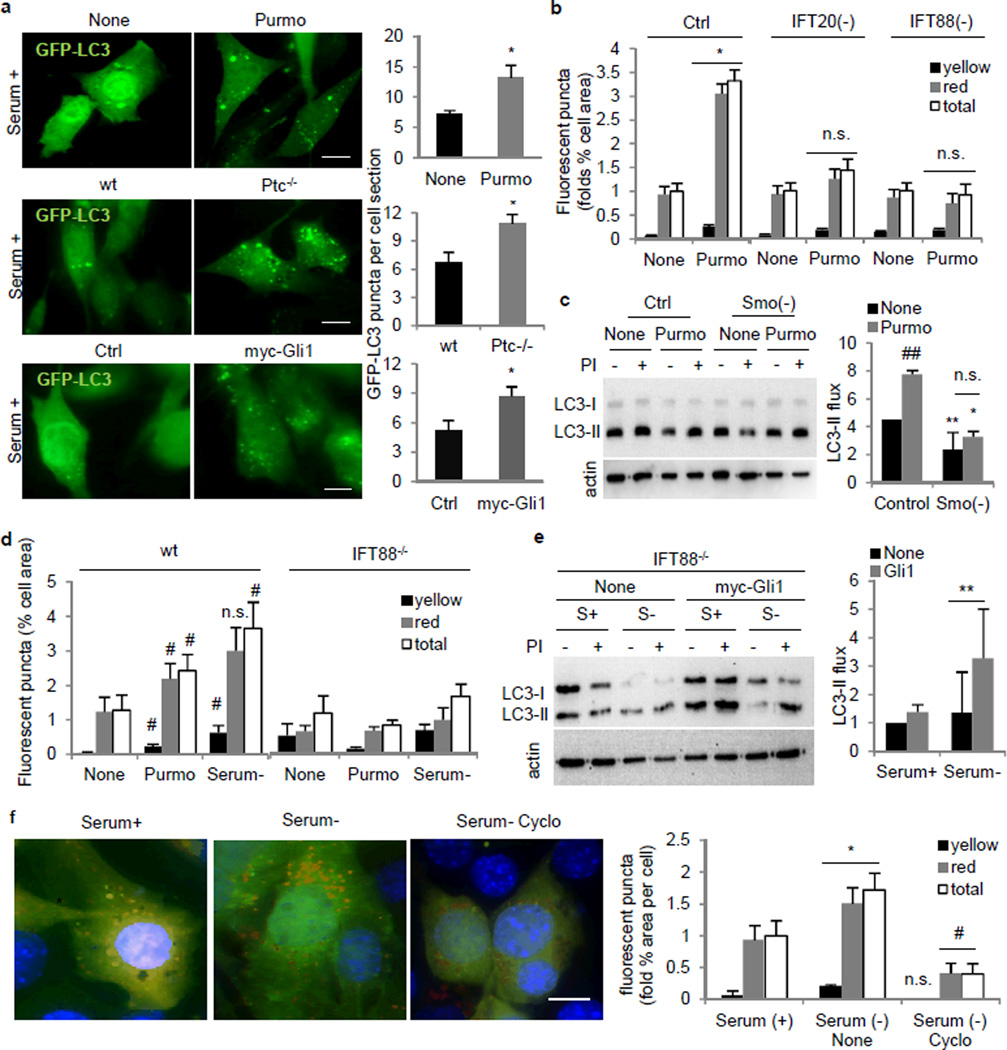
We next focused on the Hedgehog (Hh) signaling pathway because of its dependence on the primary cilium and intact IFT7,8. Treatment of serum-supplemented MEFs or KECs with purmorphamine (Purmo), a smoothened (Smo) agonist that activates expression of Hh downstream factors (Extended Data Fig. 4a,c), returned content of LC3-positive compartments and autophagic flux to values observed upon serum removal (Fig. 2a–d and Extended Data Fig. 4d–f). Purmo failed to induce autophagy in IFT20(−) or IFT88(−) MEFs and in IFT88−/− KECs supporting dependence on IFT (Fig. 2b,d and Extended Data Fig. 4f) and in Smo(−) cells (Fig. 2c) which as expected showed reduced Purmo-mediated upregulation of Gli1 and Gli2 (Extended Data Fig. 4c). Two other conditions that activate Hh signaling, Patched-1 receptor knockout (Ptc−/−; constitutive activation of Hh signaling and ciliogenesis8) and overexpression of Gli1 also upregulated autophagy (Fig. 2a and Extended Data Fig. 4d,g–j), and in fact, overexpression of Gli1 is sufficient to partially rescue the autophagic defect in IFT88−/− cells (Fig. 2e). Conversely, two interventions that reduce Hh signaling, knock-down of Smo and treatment with Hh antagonist cyclopamine, reduced starvation-induced autophagy (Fig. 2c,f and Extended Data Fig. 4k).
Figure 2. Hedgehog signaling requires the primary cilia to regulate autophagy.
(a) GFP-LC3 in MEFs with Purmorphamine (Purmo; *p=0.028, n=4), Ptc−/− (*p=0.015, n=4) and myc-Gli1 overexpression (*p=0.012, n=3). (b) mCherry-GFP-LC3 upon Purmo treatment in MEFs. Yellow, autophagosomes; red, autophagolysosomes; total, both. (*p=0.0323, 40 fields). (c) LC3 flux in Ctrl (**p=0.001) and Smo(−) MEFs (**p=0.0001) upon Purmo (*p=0.009). (n=4). (d) mCherry-GFP-LC3 in wt and IFT88−/− KECs upon Purmo. (wt Purmo, #p=0.046; wt Serum-, #p=0.008, 40 fields). (e) LC3 flux in IFT88−/− KECs overexpressing myc-Gli1 (**p=0.00006, n=3). (f) mCherry-GFP-LC3 in MEFs with Cyclopamine (Cyclo). (*p=0.011, #p=0.013, 25 fields). Bars: 10µm. n.s., statistically non-significant. Mean±s.d in c, f and mean±s.e.m rest.
Altogether, these data reveal a positive regulatory effect of Hh signaling on autophagy, and support that the inability to activate autophagy in cells with defective IFT originates, at least in part, from the loss of Hh signaling.
Autophagic machinery localizes at the cilia
Besides ciliogenesis, IFT also participates in recruitment of non-ciliary proteins to the plasma membrane16, a site of autophagosome formation17, which made us hypothesize an involvement of aIFT in the delivery of the autophagic machinery to this membrane.
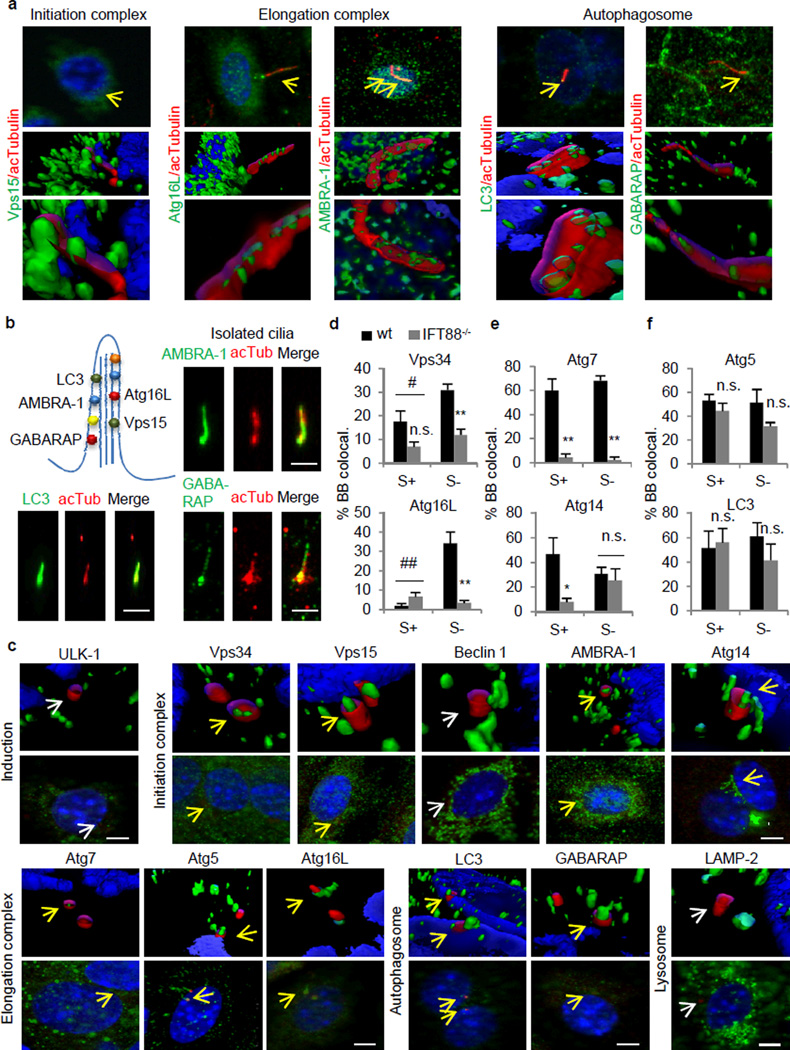
Co-immunostaining for different Atgs (green) and acetylated tubulin (red) to highlight the ciliary axoneme in serum-deprived KECs, revealed that of the twelve Atgs analyzed, the five lipid-binding Atgs2,3 (Atg16L, AMBRA-1, LC3, GABARAP and Vps15) localized as discrete puncta along the ciliary axoneme, clearly distinguishable from the surrounding cytosol signal with deconvolution immunofluorescence and 3D wire modeling (Fig. 3a, Extended Data Fig. 5a,b and Videos 1–5). Similar colocalization was observed using fluorescent tagged versions of the ciliary protein inversin18 or of Atgs (Extended Data Fig. 5c,d). Other more abundant Atgs, such as Atg14, were not detectable in the axoneme, and knock-down against cilia-associate Atgs abolished their staining (Extended Data Fig. 5b,d) supporting specificity of the staining. These Atgs were also detectable in primary cilia isolated by the “peeling off” procedure (Fig. 3b).
Figure 3. Autophagy-related proteins associate with ciliary structures in a serum-dependent manner.
(a) Co-immunostaining and 3D reconstruction for Atgs (green) and acetylated tubulin (red). Yellow arrows: colocalization. (b) Atgs associated with the axoneme. Co-immunostaining for Atgs and acetylated tubulin in isolated cilia. (c) Co-immunostaining and 3D reconstruction for Atgs (green) and gamma tubulin (red). Arrows: colocalization (yellow), no colocalization (white). (d–f) Cells with colocalizing Atgs in the basal body (BB). (d) Serum- and IFT-dependent (Vps34, #p=0.04, **p=0.002, n=4; Atg16L, ##p=0.0003, **p=0.0009, 15 cells each per experiment, n=7), (e) IFT-dependent but serum-independent (Atg7, S+ **p=0.001, S− **p= 9.33768E-06, n=4; Atg14, *p=0.018, 15 cells each per experiment, n=5), and (f) BB association independent on serum or IFT (Atg5, n=5; LC3, n=8; 15 cells each per experiment). Bars: 10µm. n.s., statistically non-significant. Mean±s.e.m.
A larger number of Atgs associated with the basal body (highlighted with anti-gamma tubulin). In addition to the five cilia-associate Atgs, we also found Atg14, Vps34, Atg7 and Atg5, but not Beclin 1 or ULK-1 at the base of the axoneme (Fig. 3c; LAMP-2 is used a negative control). Similar colocalization experiments in serum-supplemented cells, to repress autophagy, and in cells with compromised ciliogenesis (IFT88−/− cells) revealed three types of association of Atgs with the basal body: serum- and aIFT-dependent, such as Vps34 and Atg16L; serum-independent but aIFT-dependent, as Atg7 and Atg14; and serum- and aIFT-independent, as Atg5 and LC3 (Fig. 3d–f and Extended Data Fig. 6).
Disruption of IFT also altered the overall intracellular distribution of Atgs that localize at the basal body in an aIFT-dependent manner. Thus, Atg7 was normally observed in KECs as discrete cytosolic puncta that markedly increased upon starvation, but it adopted a diffuse reticular pattern unchanged by starvation in IFT88−/− KECs (Extended Data Fig. 7a). Disruption of IFT also reduced the basal association of Atg14, key for autophagy initiation3, with the Golgi and its mobilization into discrete cytosolic puncta upon serum removal (Extended Data Fig. 7a).
Interestingly, IFT88-deficiency also blunted the starvation-induced changes in the intracellular location of Atgs that localize to the basal body independently of IFT88, such as LC3 and Atg5 (Fig. 3f). The association with the plasma membrane of LC3 and GABARAP (both involved in autophagosome membrane formation/elongation3), clearly noticeable in wt cells upon serum removal was no longer observed in the IFT88-deficient cells (Extended Data Fig. 7a). Likewise, the often “hook-like” organization of Atg5 (involved in autophagosome membrane elongation)at the basal body was also abrogate in IFT88−/− KECs (Extended Data Fig. 7b,c). The abundance of Atg5 hook-like structures under basal conditions in cells with constitutive Hh signaling (Ptc−/−; ; Extended Data Fig. 7d) suggests the need of a functional cilium for Atg5 clustering in this location.
These findings support a role for aIFT in the cellular relocation of the autophagic machinery to sites of autophagosome ..
IFT-dependent trafficking of Atg16L
To gain a better understanding of the mechanisms behind the ciliary regulation of starvation induced autophagy, we focused on Atgs that associated with ciliary structures preferentially upon serum removal. We focused on Atg16L because it was almost undetectable in the basal body in serum-supplemented cells (Fig. 3d), starvation induced its IFT-dependent association with the basal body and its presence in this location increased in two paradigms with enhanced cilliary Hh signaling (Extended Data Fig. 8a,b). We proposed that active recruitment of Atg16L to the cilliary base during starvation could be the trigger for ciliary-induced autophagy.
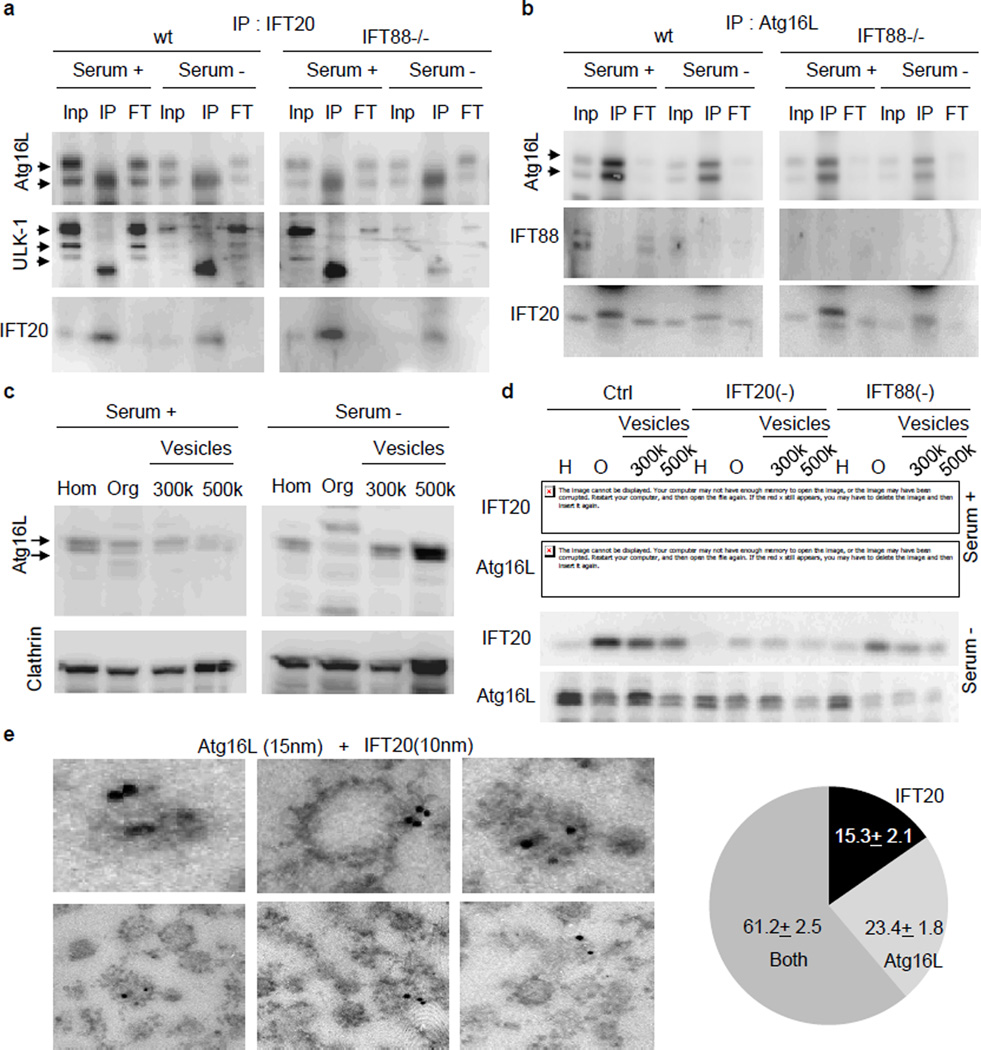
We first investigated the contribution of IFT20 to ciliary delivery of Atg16L since added to aIFT, IFT20 also participates in trafficking of ciliary membrane proteins from the Golgi to the base of the cilium6. To separately analyze both functions of IFT20, we compared its association with Atg16L in cells with intact or disrupted aIFT (IFT88−/− KECs). We found that IFT20 and Atg16L co-localized in small cytosolic vesicles that become more abundant upon serum removal and that this colocalization was only partially reduced when aIFT was disrupted (Extended Data Fig. 8c). Both proteins co-immunoprecipitated independently of the presence of IFT88 (Fig. 4a); and IFT20, but not IFT88, could be pulled-down with Atg16L (Fig. 4b). Starvation did not change the association between Atg16L and IFT20 but changed the location of the IFT20-Atg16L-positive vesicles from the proximity of the Golgi towards a more cytosolic location (Extended Data Fig. 8c), suggesting that the same signal that enhances IFT20-mediated trafficking of proteins from the Golgi to the cilia may also be responsible for cilliary delivery of Atg16L. Other Atgs (Vps15, Atg7) display minimal colocalization with IFT20, or colocalize only at the Golgi (Atg14) (Extended Data Fig. 8d) and none of them binds directly to IFT20 (Extended Data Fig. 8e–g).
Figure 4. IFT20 regulates trafficking of Atg16L to the cilium.
(a,b) Reciprocal co-immunoprecipitation of IFT20 (a) and Atg16L (b). Inp: 1/10 input; IP: immunoprecipitate; FT: 1/10 flow-through. (c) Immunoblot for Atg16L and Clathrin in MEFs homogenate (Hom), and the pellets from 1h centrifugation at 100,000 g (organelles (Org)), 300,000 g and 500,000 g (Vesicles). (d) Immunoblot for IFT20 and Atg16L in the same fractions isolated from Ctrl, IFT20(−) and IFT88 (−) MEFs. (e) Immunogold electron microscopy for Atg16L (15nm particles) and IFT20 (10nm particles) in isolated vesicles from MEFs. Bottom: IFT20 and Atg16L presence in the vesicles (11 fields). Mean±s.e.m.
The amount of of Atg16L in isolated clathrin-enriched vesicles19 was very low under basal conditions, but it markedly increasedupon serum removal (Fig. 4c). Although the vesicular content of IFT20 did not change with starvation (Fig. 4d), knock-down for IFT20 reduced starvation-induced loading of Atg16L in the vesicles(Fig. 4d). Interestingly, Atg16L content was also reduced in vesicles isolated from serum-deprived cells knocked down for IFT88 (Fig. 4d) explaining the reduced colocalization between IFT20 and Atg16L in IFT88−/− cells. Electron microscopy and double immunogold labeling for IFT20 and Atg16L confirmed the vesicle nature of the fractions and the coincidence of both proteins in more than 60% of the labeled vesicles (Fig. 4e and Extended Data Fig. 8h).
We propose that during starvation Atg16L reaches the base of the cilia through the Golgi-to-cilia shuttling function of IFT20, and then it access the ciliary region by IFT88-dependent mechanisms20. Overall, our results support a novel role for the centrioles as organization center for the autophagic machinery and of IFT proteins as common components shared by both ciliogenesis and starvation-induced autophagosome formation.
Autophagy activation reduces cilia growth
Although inducible autophagy relies on IFT and cilia function, we found that autophagy is not required for cilia formation. In fact, Atg5−/− MEFs formed longer cilia and faster than wt MEFs upon serum removal, and a higher percentage of them grew cilia even in basal conditions (Fig. 5a–d). Knock-down of two other Atgs and even chemical inhibition of autophagy were sufficient to induce ciliogenesis in different cell types under basal conditions, whereas upregulation of autophagic activity with rapamycin did not affect either basal or inducible ciliary growth (Extended Data Fig. 9).
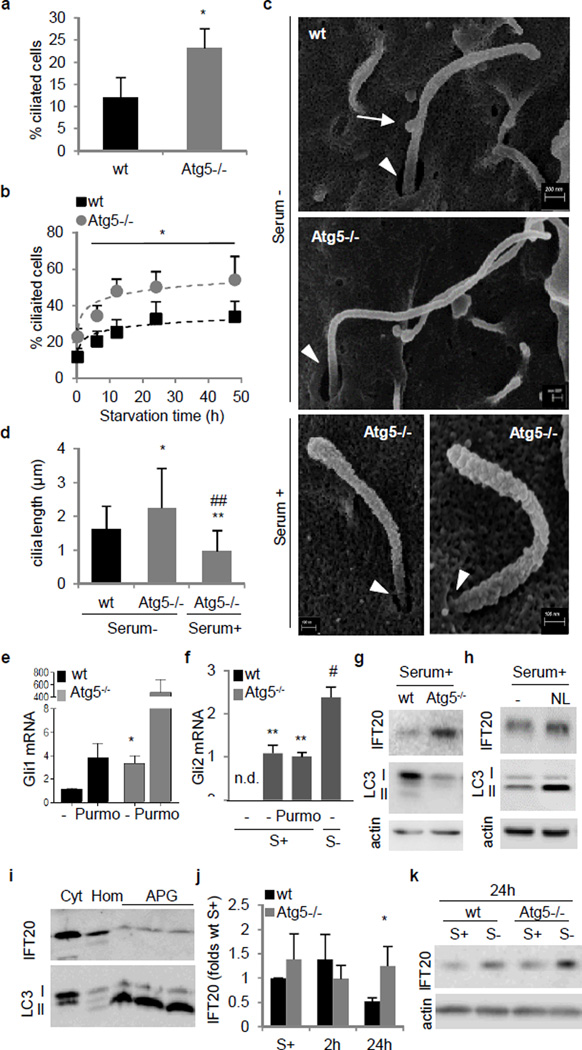
Figure 5. Ciliogenesis is enhanced in autophagy-defective cells.
(a,b) Ciliated Atg5−/− MEFs in serum+ (*p=0.038, 25 cells each per experiment, n=3) (a) or serum− (*p=0.006, non-linear fit regression, 25 cells each per experiment, n=3) (b). (c) Scanning electron microscopy (SEM) of primary cilia. Arrows: cilia-associated vesicles. Arrowheads: ciliary pocket. (d) Cilia length quantification from SEM (*p=0.027 n=28; **p=0.001, ##p=0.0001, n=23). (e,f) Gli1 (*p=0.04, n=3) (e) and Gli2 (S+ **p=0.0005, S+ Purmo **p=0.0046, S− #p=0.0132, n=3) (f) mRNA expression in Atg5−/− MEFs. (g,h) IFT20 immunoblot in Atg5−/− (g) and wt MEFs with lysosomal inhibitors (NL) (h). (i) IFT20 immunoblot in cytosol (Cyt), homogenate (Hom) and autophagosomes (APG). (j) IFT20 protein levels in Atg5−/− MEFs (*p=0.038, n=6). (k) IFT20 immunoblot in Atg5−/− MEFs. n.s., statistically non-significant. Mean±s.d. in d and j, Mean±s.e.m rest.
Scanning electron microscopy revealed that starvation induced in Atg5−/− MEFformation s of long and narrow cilia that come out of smalle ciliary pocketsand confirmed that Atg5−/− MEFs grow primary cilia in serum-supplementd media, although shorter and with less surface-adhered vesicles (presumably exosomes) than the cilia grown during starvation (Fig. 5c and Extended Data Fig. 10). Activation of ciliary Hh signaling by starvation or treatment with Purmo showed efficient recruitment of Smo to the cilia in Atg5−/− cells and increased expression of the downstream transcription factors Gli1 and Gli2 in these cells even under basal conditions, supporting full functionality of the cilia induced by autophagic blockage (Fig. 5e,f and Extended Data Fig. 11a).
To start investigating the basis for this inhibitory effect of autophagy on ciliogenesis, we compared levels and distribution of IFT proteins in wt and Atg5−/− MEFs and found that basal levels of IFT20, but not IFT88, were markedly higher in Atg5−/− MEFs (Fig. 5g). Since chemical inhibition of lysosomal proteolysis in control cells reproduces a similar increase in IFT20 levels (Fig. 5h), and IFT20 could be detected in isolated autophagic vacuoles (Fig. 5i), we propose that basal levels of IFT20, and consequently ciliary growth, are regulated at least in part, through IFT20 degradation by basal autophagy. Although most IFT20 associates with Golgi, the basal increase in cellular IFT20 levels in Atg5−/− MEFs seems to occur in the small cytosolic vesicles only visible in control cells during starvation-induced ciliogenesis (Extended Data Fig. 11b). Interestingly, the differences in the IFT20 content between wt and Atg5−/− MEFs were no longer observed early after serum removal, but become apparent again after 24h in serum-deprived media (Fig. 5j,k). Studies with lysosomal proteolysis inhibitors in wt MEFs confirmed absence of lysosomal degradation of IFT20 during the first 12h of serum removal, when inducible autophagy is maximally activated (Extended Data Fig. 11c,d). In light of these findings, we propose that basal autophagy contributes to regulate ciliogenesis and the length of an already formed cilium, at least in part through the degradation of IFT20. The switch from basal to induced autophagy seems to spare cytosolic IFT20 from degradation and allow its active engagement in the vesicular trafficking required for ciliogenesis.
Discussion
In this work, we have identified a previously unknown reciprocal relationship between primary cilium and autophagy whereby components of the ciliary machinery are shared with autophagy for starvation-induced autophagosome biogenesis. In contrast, basal autophagy inhibits ciliogenesis by limiting trafficking to the cilium of components required for ciliary growth. The temporal coincidence of activation of ciliogenesis and autophagy at the early times of nutritional deprivation may thus serve as a self-regulatory brake for each of these processes (Extended Data Fig. 1).
The localization of autophagy initiating Atgs at the ciliary base and the active recruitment of Atg16L to this location upon starvation suggests that sensing of nutrient deficiency and activation of signaling from the cilium could initiate a cilia-mediated autophagic program (see additional Discussion 1 in SI). The presence along the cilium axoneme of the pre-autophagosomal Atg16L and integral autophagosome membrane Atgs – e.g. LC3 and GABARAP – and their IFT-dependent enrichment at the plasma membrane suggest that cilia-mediated autophagy may induce autophagosome formation from this location, making the ciliary pocket, characterized by high vesicular activity21, an attractive place for autophagosome formation.
The negative regulatory role of basal autophagy on ciliogenesis identified here may take place through different mechanisms. We have found that basal autophagy controls trafficking of ciliary proteins by limiting the amount of IFT20 accessible for shuttling between the Golgi and the ciliary base (see additional Discussion 3 in SI). Since induction of autophagy depends on the same IFT20 protein that autophagy degrades, this could represent a novel mechanism for self-containment of the autophagic process.
The primary cilium is constitutively present in most tissues, but the fact that ciliogenesis blockage in cells with permanent cilia compromises their starvation-induced autophagy, supports that it is not the mere presence of a cilium but the activation of ciliary signaling by starvation that contributes to autophagic induction. We have also identified the regulatory effect of the ciliary Hedgehog signaling pathway, for which an emerging role in autophagy regulation has been proposed22–25 (see additional Discussion 4 in SI). The possible participation of the cilium in the integration of other autophagy-inducing signals and the effect of autophagy in IFT20 functions beyond ciliogenesis16 need further investigation.
Methods
Cells and reagents
Mouse embryonic fibroblasts (MEFs) were prepared as previously described26. Atg5−/− MEFs were a gift from Dr. N. Mizushima (Tokyo University), Wt (Tg737+/+), IFT88−/− (Tg737−/−) mouse kidney epithelial cells were from Dr. G. Pazour (University of Massachusetts), Ptc−/− MEFs were from Dr. M.P. Scott (Stanford University) and RGC-5 the cell line from Dr. Boya (CIB, Madrid, Spain). Inversin-GFP 3T3s were from Drs. Veland and Christensen (University of Copenhagen, Denmark). All cells were cultured at 37°C and 5% CO2 in Dulbecco’s Modified Eagle’s Medium (Sigma) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin-fungizone. Serum removal was performed by thoroughly washing the cells with Hanks’ Balanced Salt Solution (Invitrogen) and placing them in serum-free medium. Where indicated, cells were treated with the macroautophagy inhibitor 3-methyladenine (10mM) (Sigma) or with NH4Cl (20mM) and leupeptin (100µM) (Fisher BioReagents) to inhibit lysosomal proteolysis or with rapamycin (100µM) (Calbiochem) to activate autophagy. For cilia resorption experiments Cytochalasin D (0.5µM) (Tocris) and PDGF (50ng/ml) (R&D systems) were used. Purmophamine was from Calbiochem and Cyclopamine from Sigma. The plasmid for EGFP-LC3 was from Addgene, and Inversin-GFP was from Drs. Veland and Christensen (University of Copenhagen, Denmark). The antibodies against acetylated tubulin, beta-tubulin, gamma-tubulin (mouse), and ULK-1 were from Sigma; against IFT20 and IFT88 from Proteintech (rabbit) and Abcam (mouse); against AMBRA-1, Atg5/12, Beclin, IFT88 and Vps15 from Novus; against Atg7 (immunoblot) and LC3 from Cell Signaling; against Atg7 (immunofluorescence) from Serotec; against GABARAP and gamma-tubulin (rabbit) from Santa Cruz; against LC3 (immunogold), Atg14, and Atg16(L) from MBL; against actin and detyrosinated alpha tubulin (Glu-tubulin) from AbCam; against Vps34 from Invitrogen; against Smo from AbCam, against LAMP-2 from the Iowa Hybridoma Bank and against clathrin Heavy Chain (mouse) from BD Biosciences. The rest of chemicals were from Sigma.
Autophagic measurements
Intracellular protein degradation was measured by metabolic labeling and pulse chase experiments as described before27. Briefly, confluent cells labeled with [3H]leucine (2µCi/ml) (NENPerkinElmer Life Sciences) for 48h at 37°C were extensively washed and maintained in medium supplemented or not with serum with an excess of unlabeled leucine (2.8mM). Aliquots of the medium taken at different time points were precipitated in trichloroacetic acid and proteolysis was measured as the amount of acid-precipitable radioactivity (proteins) transformed in acid-soluble (amino acids and small peptides) at each time. This assay quantifies the degradation of the pool of long-lived proteins that become radiolabeled during the 48h pulse. Lysosomal degradation was determined as the percentage of degradation sensitive to inhibition by ammonium chloride and leupeptin. Autophagic flux was measured by immunoblot as changes in levels of LC3-II upon inhibition of lysosomal proteolysis (net flux) and autophagosome formation as the increase in LC3-II levels at two consecutive times during lysosomal proteolysis inhibition11. Autophagosome content was evaluated as the number of fluorescent puncta in cells transfected with GFP-LC3 or after immunostaining with antibodies against endogenous LC3. Autophagic flux was also tested by transient transfection of the mCherry-GFP-LC3 plasmid, which was a gift from Dr. Johansen. Constructs were transfected using either Lipofectamine™ 2000 or Optifect reagent (Invitrogen) according to manufacturer's instructions. Quantification of the yellow and red puncta was performed by Green and Red Puncta Colocalization Macro for Image J (D. J. Swiwarski modified by R.K. Dagda).
Lentivirus-mediated shRNA
The transfer vector plasmids containing the IFT20 and IFT88 shRNA sequence were purchased from Sigma Mission Library (Clone ID NM_018854.4-417s21c1 and NM_009376.1-762s1c1). Lentiviral particles were generated by co-transfection with the third-generation packaging constructs pMDLg/pRRE and pRSV-REV, and as envelope the G glycoprotein of the VSV (pMD2.G) into HEK293T cells as described before26. Cultured cells were transduced by addition of packed virus at a titer of 2.63×106 units/ml.
Immunocytochemistry and morphometric analysis
Cells grown on coverslips were fixed either in 4% paraformaldehyde or 100% cold methanol, blocked and permeabilized, and incubated with primary antibodies (1:200 except for acetylated tubulin that was 1:1000) and fluorophore-conjugated secondary antibodies (1:200) sequentially. Mounting medium contained DAPI (4′,6-diamidino-2-phenylindole) to highlight the nucleus. Images were acquired with an ApoTome.2 system using an Axiovert 200 fluorescence microscope (Carl Zeiss) equipped with a ×63 1.4 NA oil objective lens and red (ex. 570/30 nm, em. 615/30 nm), cyan (ex. 365/50 nm and em. 530/45 nm) and green (ex. 475/40 nm and em. 535/45 nm) filter sets (Chroma), and prepared using Adobe Photoshop 6.0 software (Adobe Systems). 3D reconstruction images were modeled as mixed renderings using the Inside4D module for AxioVision Rel. 4.8. after applying the Nyquist sampling criteria. For quantification of the number of ciliated cells Z-stack sections were acquired and orthogonal views generated using AxioVision Rel. 4.8. Cilia length was measured using AxioVision Rel. 4.8 and number of fluorescent puncta using ImageJ (NIH) after thresholding of the images. Quantification of co-localization was performed using Image J (NIH) in individual frames after thresholding, and co-localization was calculated with the JACoP plugin of the same program in merged images.
Transmission and scanning electron microscopy and morphometric analysis
Cells grown to confluence were pelleted and fixed in 2.5% glutaraldehyde in 100mM sodium cacodylate (SC) pH 7.43 at room temperature for 45min. The pellet was then rinsed in SC, post-fixed in 1% osmium tetroxide in SC followed by 1% uranyl acetate, dehydrated through a graded series of ethanol, and embedded in LX112 resin (LADD Research Industries). Ultrathin sections were cut on a Reichert Ultracut E, stained with uranyl acetate followed by lead citrate. Immunogold labeling was performed in ultrathin sections of isolated cytosolic vesicles fixed in 4% paraformaldehyde/0.1% glutaraldehyde in sodium cacodylate, dehydrated and embedded in Lowicryl. Grids were washed in 50mM glycine in phosphate buffered saline, blocked, either single labeled with Atg16L or IFT20 or double labeled with Atg16L and IFT20 antibody for 2h, washed extensively and incubated with the gold-conjugated secondary antibodies (1:100) for 2 h. Control grids were incubated with the secondary antibody alone or with an irrelevant immunoglobulin G. After extensive washing, samples were fixed a second time for 5min in 2% glutaraldehyde, washed and negatively stained with 1% uranyl acetate for 15min. All grids were viewed on a JEOL 1200EX transmission electron microscope at 80kV Scanning electron microscopy was performed by the Analytical Imaging facility at the Albert Einstein College of Medicine, following standard protocols30. Images were acquired on a Zeiss Supra 40 Field Emission Scanning Electron Microscope at 5kV. Morphometric analysis of transmitted and scanning electron micrographs was done using Image J (NIH).
Isolation of subcellular factions
Cilia was isolated by peel-off technique as previously described28. Briefly, a poly-L-lysine-coated coverslip was placed on top of kidney epithelial cells cultured in absence of serum. After application of light pressure, the poly-L-lysine coated coverslip containing the isolated cilia was lifted up and processed for immunofluorescence. Autophagosomes and autophagolysosomes were isolated by differential centrifugation and floatation in metrizamide gradients as previously described29. The cytosolic fraction was obtained by centrifugation for 1h at 100,000×g of the supernatant obtained after separating the autophagosome-autophagolysosome-enriched fraction.
Isolation of cytosolic vesicles
Vesicles were isolated from 10-cm confluent plates of MEF cells as described before19. Briefly, cells collected in PBS were disrupted in homogenizing buffer (0.25M sucrose, 20mM MOPS, pH 7.3) by nitrogen cavitation (nitrogen bomb; Parr Instrument Company, Moline, IL) applying a pressure of 3500psi for 7min. Cell homogenate was centrifuged at 2500×g for 15min to pellet the cell debris (heavy particles, or fractions of cells) and supernatant was centrifuged at 100,000×g for 60min in a TLA 110 rotor (Beckman) to pellet all major organelles and was designated as 100K. The supernatant of this 100K pellet was further subjected to two successive centrifugations; 300,000×g, and 500,000×g and the pellets from each of these centrifugations were collected as the 300K and 500K crude vesicular fractions, respectively.
Co-immunoprecipitation assays
Cytosol was resuspended in a buffer containing 0.5% NP-40, and incubated over-night with the desired antibody at 4°C. The fraction bound to antibodies was precipitated using either ProteinA or ProteinG coated beads, eluted and subjected to SDS-PAGE as described in the general methods.
mRNA quantification
Semi-quantitative real-time PCR was performed after extracting total RNA using the RNeasy Protect Mini Kit (Qiagen) following the manufacturer's indications. The first strand cDNA was synthesized from 0.5µg of the total RNA with the SuperScript II RNase H Reverse Transcriptase (Invitrogen) and oligo(dT)12–18 primers. The expression levels were normalized to levels of β-actin in the same samples after amplification using the SYBR Green PCR kit (PE Biosystems) and the following primers from the QuantiTect Primer Assay (Quiagen): Gli1 (QT00173537) Gli2 (QT01062236); Ptc1 (QT00149135) and mouse β-actin 5’-GGCTGTATTCCCCTCCATCG-3’. Differences between samples were calculated using the ΔΔCt Method.
Statistical analysis
Results are shown as mean ± s.e.m. or mean ± s.d., and represent data from a minimum of three independent experiments unless otherwise stated. Student's t-test for unpaired data was used for statistical analysis and one-way analysis of variance (ANOVA) was used for multiple comparisons. For cilia growing curves Nonlinear Regression was performed applying the Least Squares fitting method with Sigma Plot software. A value of p< 0.05 was considered statistically significant.
General methods
Cells were solubilized in RIPA buffer (150mM NaCl, 50mM Tris pH=8, 1% NP-40, 0.5% NaDoc, 0.1% SDS) and protein concentration was determined by the Lowry method. After SDS-PAGE and immunoblot, the proteins recognized by the specific antibodies were visualized by chemiluminescent HRP substrate from Pierce in a Fujifilm Las-300 Imager. Densitometric quantification of the bands was done using the square method with ImageJ (NIH), and all values were corrected by actin or tubulin as labeled in the figures.
Supplementary Material
Acknowledgements
We thank Dr. Mizushima and Dr. Pazour for providing MEFs and Drs. Veland and Christensen for providing the plasmid for expression of GFP-inversin. We also thank the personnel at the Analytical Imaging Facility for their technical assistance with TEM, Ms Jennifer Kraut and Dr. Natalia Rodriguez for their early assistance in cilia staining optimization and Dr. Kaushik for critically reviewing this manuscript. This work was supported by grants NIH/NIA AG031782 and AG038072 (to AMC), NIH/NIDKK DK412960 and DK098408 (to AMC and PS), INSERM (to PC and IB), InCa&ANR (to PC), and the generous support of Robert and Renee Belfer. OP was supported by a Basque Government Postdoctoral Fellowship.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
OP designed and performed most of the experiments, analyzed the data and contributed to writing the paper; IO and IB performed and analyzed the experiments related to Hh signaling; BP performed the electron microscopy studies and morphometric analysis; SS performed the cytosolic vesicle experiments; ADC assisted with cell culture; PC conceived the part of the study related to Hh signaling, provided interpretation to the data and contributed to the writing and revision of the paper; BHS and PS set the bases for the rationale of the study, provided feedback in the interpretation of the data and revised the written manuscript; AMC coordinated the study, designed experiments, analyzed data and contributed to the writing and revision of the paper.
Competing Financial Interest
The authors declare that they have no competing interests.
References
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamasaki M, Shibutani ST, Yoshimori T. Up-to-date membrane biogenesis in the autophagosome formation. Curr Opin Cell Biol. 2013;25:455–460. doi: 10.1016/j.ceb.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satir P, Pedersen LB, Christensen ST. The primary cilium at a glance. J Cell Sci. 2010;123:499–503. doi: 10.1242/jcs.050377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taschner M, Bhogaraju S, Lorentzen E. Architecture and function of IFT complex proteins in ciliogenesis. Differentiation. 2012;83:S12–S22. doi: 10.1016/j.diff.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Follit JA, Xu F, Keady BT, Pazour GJ. Characterization of mouse IFT complex B. Cell Motil Cytoskeleton. 2009;66:457–468. doi: 10.1002/cm.20346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liem KF, et al. The IFT-A complex regulates Shh signaling through cilia structure and membrane protein trafficking. The Journal of Cell Biology. 2012;197:789–800. doi: 10.1083/jcb.201110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rohatgi R, Milenkovic L, Scott MP. Patched1 Regulates Hedgehog Signaling at the Primary Cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 9.Kiprilov EN, et al. Human embryonic stem cells in culture possess primary cilia with hedgehog signaling machinery. J Cell Biol. 2008;180:897–904. doi: 10.1083/jcb.200706028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- 11.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal Turnover, but Not a Cellular Level, of Endogenous LC3 is a Marker for Autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 12.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pazour GJ, San Agustin JT, Follit JA, Rosenbaum JL, Witman GB. Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Current Biology. 2002;12:R378–R380. doi: 10.1016/s0960-9822(02)00877-1. [DOI] [PubMed] [Google Scholar]
- 14.Schneider L, et al. PDGFRalphaalpha signaling is regulated through the primary cilium in fibroblasts. Curr Biol. 2005;15:1861–1866. doi: 10.1016/j.cub.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Boehlke C, et al. Primary cilia regulate mTORC1 activity and cell size through Lkb1. Nat Cell Biol. 2010;12:1115–1122. doi: 10.1038/ncb2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finetti F, et al. Intraflagellar transport is required for polarized recycling of the TCR/CD3 complex to the immune synapse. Nat Cell Biol. 2009;11:1332–1339. doi: 10.1038/ncb1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veland IR, et al. Inversin/Nephrocystin-2 is required for fibroblast polarity and directional cell migration. PLoS One. 2013;8:e60193. doi: 10.1371/journal.pone.0060193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sridhar S, et al. The lipid kinase PI4KIIIbeta preserves lysosomal identity. EMBO J. 2013;32:324–339. doi: 10.1038/emboj.2012.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richey EA, Qin H. Dissecting the sequential assembly and localization of intraflagellar transport particle complex B in Chlamydomonas. PLoS One. 2012;7:e43118. doi: 10.1371/journal.pone.0043118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satir P. The new biology of cilia: review and annotation of a symposium. Dev. Dynam. 2012;241:426–430. doi: 10.1002/dvdy.23713. [DOI] [PubMed] [Google Scholar]
- 22.Li H, et al. Sonic hedgehog promotes autophagy of vascular smooth muscle cells. Am J Physiol Heart Circ Physiol. 2012;303:H1319–H1331. doi: 10.1152/ajpheart.00160.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jimenez-Sanchez M, et al. The Hedgehog signalling pathway regulates autophagy. Nat Commun. 2012;3:1200. doi: 10.1038/ncomms2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Han C, Lu L, Magliato S, Wu T. Hedgehog signaling pathway regulates autophagy in human hepatocellular carcinoma cells. Hepatology. 2013 doi: 10.1002/hep.26394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petralia RS, et al. Sonic hedgehog promotes autophagy in hippocampal neurons. Biology open. 2013;2:499–504. doi: 10.1242/bio.20134275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auteri JS, Okada A, Bochaki V, Dice JF. Regulation of intracellular protein degradation in IMR- 90 human diploid fibroblasts. J Cell Physiol. 1983;115:159–166. doi: 10.1002/jcp.1041150210. [DOI] [PubMed] [Google Scholar]
- 28.Huang BQ, et al. Isolation and characterization of cholangiocyte primary cilia. Am J Physiol Gastrointest Liver Physiol. 2006;291:G500–G509. doi: 10.1152/ajpgi.00064.2006. [DOI] [PubMed] [Google Scholar]
- 29.Marzella L, Ahlberg J, Glaumann H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J Cell Biol. 1982;93:144–154. doi: 10.1083/jcb.93.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wada K, et al. Application of photoacoustic microscopy to analysis of biological components in tissue sections. Chemical & pharmaceutical bulletin. 1986;34:1688–1693. doi: 10.1248/cpb.34.1688. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.