Significance
Early-life exposure to dogs is protective against allergic disease development, and dog ownership is associated with a distinct milieu of house dust microbial exposures. Here, we show that mice exposed to dog-associated house dust are protected against airway allergen challenge. These animals exhibit reduced Th2 cytokine production, fewer activated T cells, and a distinct gut microbiome composition, highly enriched for Lactobacillus johnsonii, which itself can confer airway protection when orally supplemented as a single species. This study supports the possibility that host–environment interactions that govern allergic or infectious airway disease may be mediated, at least in part, by the impact of environmental exposures on the gastrointestinal microbiome composition and, by extension, its impact on the host immune response.
Keywords: house environment, airway adaptive immunity, gastrointestinal bacterial community, Lactobacilliaceae
Abstract
Exposure to dogs in early infancy has been shown to reduce the risk of childhood allergic disease development, and dog ownership is associated with a distinct house dust microbial exposure. Here, we demonstrate, using murine models, that exposure of mice to dog-associated house dust protects against ovalbumin or cockroach allergen-mediated airway pathology. Protected animals exhibited significant reduction in the total number of airway T cells, down-regulation of Th2-related airway responses, as well as mucin secretion. Following dog-associated dust exposure, the cecal microbiome of protected animals was extensively restructured with significant enrichment of, amongst others, Lactobacillus johnsonii. Supplementation of wild-type animals with L. johnsonii protected them against both airway allergen challenge or infection with respiratory syncytial virus. L. johnsonii-mediated protection was associated with significant reductions in the total number and proportion of activated CD11c+/CD11b+ and CD11c+/CD8+ cells, as well as significantly reduced airway Th2 cytokine expression. Our results reveal that exposure to dog-associated household dust results in protection against airway allergen challenge and a distinct gastrointestinal microbiome composition. Moreover, the study identifies L. johnsonii as a pivotal species within the gastrointestinal tract capable of influencing adaptive immunity at remote mucosal surfaces in a manner that is protective against a variety of respiratory insults.
The emerging field of human microbiome research has demonstrated the key role microbial communities play in a variety of critical mammalian processes including ancillary mucosal barrier function (1) and metabolism (2, 3), as well as development and modulation of host immune responses (4, 5). This is particularly evident in the gastrointestinal (GI) tract where the composition of the microbiome in this niche and, specifically, the presence of particular bacterial species such as segmented filamentous bacteria and those belonging to Clostridium clades IV and XIV, have been shown to induce specific T-cell repertoires, i.e., Th17 and CD4+ FoxP3+ T-regulatory cells, respectively (4, 6). These studies demonstrate that despite the complexity of the GI microbiome, the presence or absence of specific bacterial species can dramatically alter the adaptive immune environment.
Human studies appear to support this concept. A large European birth cohort study demonstrated that a significant increase in the number of Escherichia coli or Clostridium difficile in fecal samples from 3-wk-old infants was associated with a greater risk of developing a spectrum of childhood allergic diseases (7), commonly characterized by overactive Th2 adaptive immune response. Early-life exposures, including those known to impact GI microbiome composition, e.g., antibiotic administration and caesarian section delivery, have also been associated with increased risk for childhood asthma (8, 9). Conversely, exposure to livestock or pets, particularly dogs during this early-life period, significantly decreases the risk for disease development (10, 11). Conceivably, the mechanism by which animal exposures mediate their protective effect is through their impact on local environmental microbial exposures, which in turn influence microbiome membership and the immune response of the human host. Because GI microbiome composition clearly impacts immune function, and early GI colonization patterns are linked to allergic disease development, it is necessary to understand whether and how distinct environmental microbial exposures associated with allergy-protective factors influence GI microbiome composition and airway disease outcomes.
Results
House Dust Exposure Affords Airway Protection.
House dust was collected from two residences: one possessed an indoor/outdoor dog (D), and the other had no pet (NP) present. The total weight of dust collected from the D house was approximately fourfold greater than that of the NP house. DNA extraction of 0.1 g of each dust and 16S rRNA amplification under identical conditions resulted in no detectable PCR product from the NP sample, whereas the D sample produced >250 ng of amplicon. This is consistent with our previous study in which NP dust samples exhibited low bacterial burden, with 40% of samples failing to produce a 16S rRNA PCR product (12). Nonmetric multidimensional scaling (NMDS) analysis based on a Canberra distance matrix confirmed that the microbial composition of the D dust used for these experiments was more similar to indoor/outdoor dog-associated dust samples than to any other type of house dust (cat-owning or no-pet homes) examined in our previous study (12) (Fig. S1A). Canberra distances between this sample and various distinct dust sample types previously analyzed confirmed this observation (Fig. S1B).
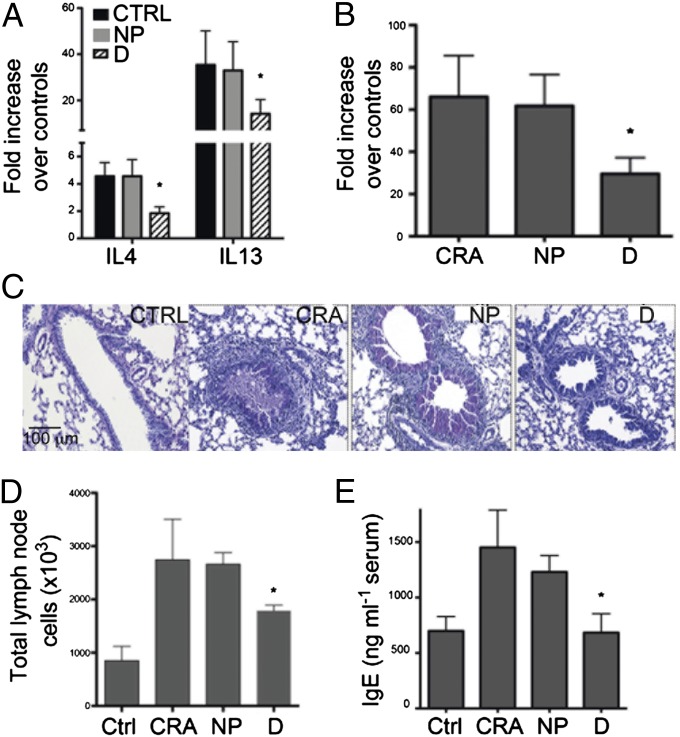
House dust bacterial burden and diversity likely play a significant role in defining environmental microbial exposures. Therefore, in an effort to faithfully replicate the microbial exposures present in these houses, the weight of D dust used to gavage mice was four times greater than that used to gavage mice receiving NP dust. Four treatment groups [D or NP dust-gavaged, cockroach allergen (CRA)-challenged mice; unsupplemented CRA-challenged; or control, unsupplemented, unchallenged animals; Fig. S2A] were used in a CRA airway challenge model described in SI Materials and Methods. Following final CRA challenge, all animals in each treatment group (n = 5) were assessed for airway pathophysiology and immune responses. Animals exposed to D dust exhibited a significant reduction in lung Th2 cytokine mRNA expression levels (IL-4 and IL-13) compared with those treated with dust from NP houses (Fig. 1A). This abrogated Th2 response was accompanied by significantly reduced expression of the mucus-associated gene, gob5 (chloride channel calcium activated 3; Fig. 1B). In addition, airway histology demonstrated an overall reduction in both the level of inflammation and goblet cell metaplasia, which was relatively intense in CRA and NP+CRA-exposed animals but virtually absent from the D dust-exposed mice (Fig. 1C). Enumeration of total mediastinal (lung-draining) lymph node cells, which serve as an indicator of immune activation and lymphocyte expansion, revealed that animals treated with D dust demonstrated a significant reduction in total lymph node cell numbers (Fig. 1D). Because epidemiologic studies have demonstrated an association between the presence of a pet in the home and reduced levels of serum IgE in infants (13), we also examined serum IgE levels in animals treated with NP or D dust and demonstrated a significant reduction in D dust-supplemented, compared with NP dust-treated animals (Fig. 1E). Given their role in controlling proinflammatory responses, we also examined total T-reg cell numbers in the lung, lymph nodes, and Peyer’s patch. However, no significant difference in the total number of these regulatory cells were identified.
Fig. 1.
Exposure of animals to dust from homes with dogs attenuates the development of allergen-induced airways disease and serum IgE. (A) Whole-lung mRNA analysis by Q-PCR demonstrates a significant decrease in IL-4 and IL-13 in D dust-supplemented, but not in NP-supplemented animals compared with controls. (B) Accompanying the Th2 cytokine reduction was significantly reduced expression of the mucus-associated gene, gob5. (C) Reduced airway mucus secretion and goblet cell metaplasia is observed in the D dust-supplemented animals, as depicted by PAS staining in lung histology. (D) A reduction in draining lymph node numbers in D dust-supplemented animals was also observed. (E) Serum IgE levels reflected the reduction in the development of the Th2 environment in the D dust-supplemented animals. Data represent the mean ± SE from five mice per group; *P < 0.05.
Because it could be argued that the lack of protection by NP dust was simply due to a lower level of inoculation, we performed a separate experiment using newly collected dust samples, exposing animals to equal weights of either D- or NP-associated house dusts. Irrespective of the weight of dust administered, D-associated house dust consistently demonstrated a significant reduction in airway Th2 cytokine expression compared with NP dust exposure (Fig. S3), although decreased gob5 expression was lost upon reduced exposure (Fig. S3). Collectively, these data demonstrate that exposure to D-associated house dust alters allergen-induced airway immune responses, via down-regulation of Th2 responses and serum total IgE levels and that this protective phenomenon persisted even when D dust exposure is reduced to levels equivalent to that encountered in NP-owning households.
To determine whether the alteration of pulmonary immunity in D dust-exposed animals extended to other allergens, we used an antigen-specific ovalbumin (OVA) murine model that involves transfer of carboxyfluorescein succinimidyl ester (CFSE)-stained splenic naïve T cells from DO.11 OVA protein-specific animals before airway OVA challenge (detailed in SI Materials and Methods; Fig. S2B). DO.11 T-cell proliferation in the mediastinal lymph nodes was assessed across treatment groups and indicated that animals treated with dust from homes with dogs displayed significantly lower proliferative responses, as assessed by reduced dilution of the CFSE-labeled OVA-specific DO.11 T cells in these animals (Fig. S4A). The number of OVA-specific DO.11 T cells was also lower in these mice, suggesting a reduction in expansion and/or recruitment of the responding T-cell population (Fig. S4B). Impressively, when airway histology was examined, a dramatic reduction in overall inflammation and airway pathology was observed in animals exposed to D dust (Fig. S4C). Collectively, these studies demonstrate that exposure to D-associated house dust alters host responses to two distinct antigen systems via modulation of both T-cell numbers and activity.
Airway Protection Is Associated with GI Microbiome Restructuring.
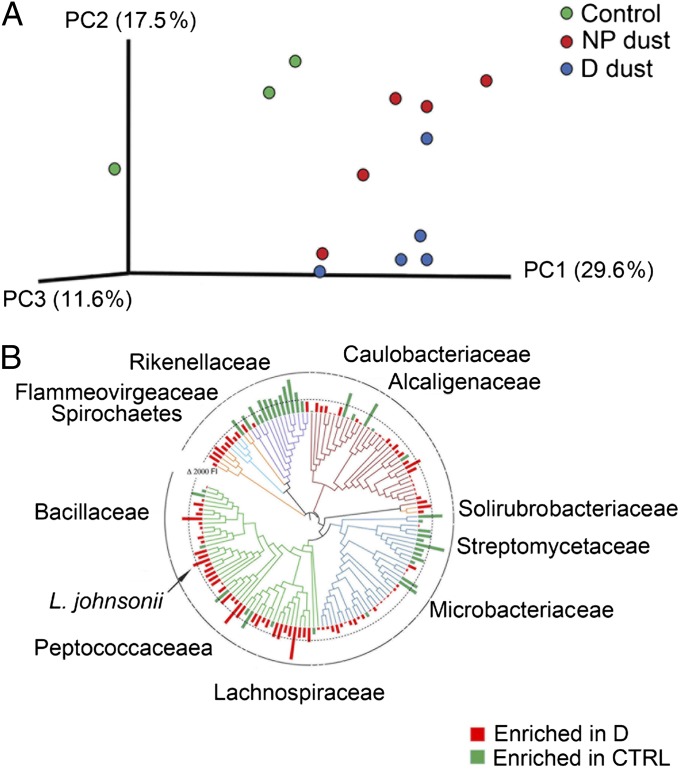
We next examined the cecal microbiota of control mice and those gavaged with either D or NP dust in the OVA challenge experiment, using a phylogenetic microarray. This platform was used rather than next-generation sequencing to generate a high-resolution microbiome profile for comparative analyses across treatment groups and identify specific taxa that exhibit significantly increased or decreased relative abundance, irrespective of their rank (dominance) in the community. Relative measures of community richness, Pielou’s evenness, and Faith’s phylogenetic diversity were not significantly altered across control and treatment groups (P ≤ 0.13, P ≤ 0.21, P ≤ 0.15, respectively; Kruskal–Wallis ANOVA; Fig. S5). Fast UniFrac (14), which considers and weights phylogenetic relationships when producing a distance matrix, was used to analyze microbiota profiles, and data were visualized using principal coordinate analysis (PCoA) to determine whether microbiota composition differed across treatment groups and whether there was a phylogenetic signal associated with such compositional differences. Control animals exhibited compositionally distinct microbiota compared with animals who received either D or NP dust before airway challenge, indicating orally gavaged house dust impacts the composition of the lower GI microbiota (Fig. 2A). Moreover, the majority of mice exposed to D dust exhibited distinct cecal microbiota from animals exposed to NP dust, indicating that exposure to dusts with distinct microbial content elicited a differential effect on GI microbiome membership.
Fig. 2.
(A) Exposure to house dust alters cecal microbiome composition. UniFrac-based cluster analysis of cecal microbiota of control, D dust- and NP dust-supplemented animals reveals distinct microbiota compositions in each treatment group. (B) Phylogenetic tree displaying all taxa that exhibited significant (P < 0.05; q < 0.15) relative enrichment (red bars) or depletion (green bars) in airway-protected mice supplemented with D-associated house dust compared with unprotected control animals. Phyla are indicted by color: Acidobacteria (light blue), Actinobacteria (teal), Bacteriodetes (purple), Firmicutes (green), Proteobacteria (red), and other (orange). Family designation of highly enriched or depleted taxa are indicated.
In an effort to identify candidate members of the GI microbiota that afforded protection against airway OVA challenge, we performed a between-group comparison, focusing on organisms significantly enriched in the ceca of protected mice. Following Welch’s t testing and false-discovery rate correction for multiple observations (15), we identified 104 taxa significantly (P ≤ 0.05, q ≤ 0.15) enriched in protected animals. Of these, taxa exhibiting the greatest enrichment in protected animals were represented by multiple distinct genera primarily belonging to the Firmicutes (classes Clostridia and Bacilli), including taxa belonging to the genus Lactobacillus (operational taxonomic unit 7028), which ranked among those taxa most highly enriched in protected animals (Table S1; Fig. 2B). Using quantitative PCR (Q-PCR), we confirmed the array-reported relative abundance of taxon 7028, the most highly enriched Lactobacillus in these communities (Fig. S6). Based on these findings, and the capacity to selectively culture Lactobacillus species on de Man, Rogosa, and Sharpe (MRS) agar, we enriched the cecal contents of four additional animals gavaged with D dust for members of this genera, a process that yielded several hundred morphologically identical colonies. Bidirectional sequencing of the full-length 16S rRNA gene of six colonies per animal, resulted in 21 high-quality almost full-length (1,462 bp) reads, all of which exhibited at least 99% coverage and 99% homology to Lactobacillus johnsonii (BLAST bit scores: 2,685 ± 28; E values: 0.0). This species resides in taxon 7028 (represented by Lactobacillus gasseri), which exhibited the most significant Lactobacillus enrichment in D dust-exposed animals (Table S1), indicating a high degree of concordance between molecular and culture-based assessments of the cecal microbiome of these animals.
L. johnsonii Oral Supplementation Mediates Airway Protection.
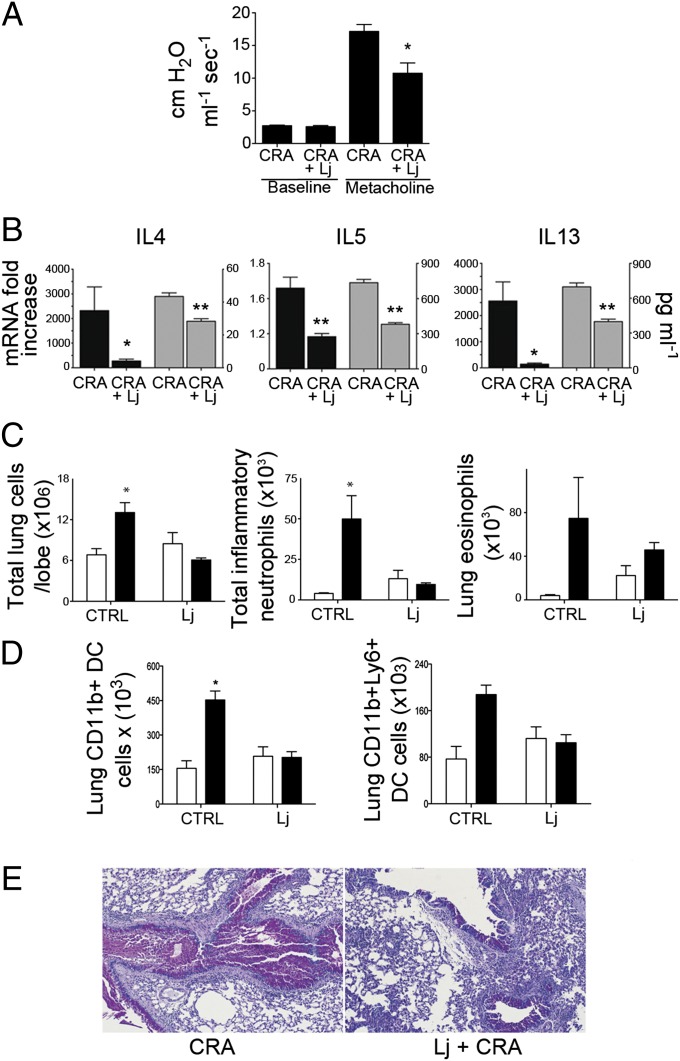
L. johnsonii has recently been shown to delay or inhibit the onset of type 1 diabetes in diabetes-prone rats (16) and protects against atopic dermatitis in mice if introduced during the weaning period (17), supporting its role as a protective species in mammalian systems. Hence, to determine as proof of principle whether this member of the protective microbiome played a role in modulating adaptive immune responses associated with airway protection, we generated standardized supplements of this species that were used to gavage animals in a similar experimental design as the CRA airway challenge model. Administration of this single Lactobacillus species resulted in significantly reduced bronchial responsiveness (Fig. 3A) as well as abrogated pulmonary mRNA levels and restimulated lymph node production of Th2 cytokines, IL-4, IL-5, and IL-13 (Fig. 3B). Likewise, when the inflammatory cell infiltrates were assessed in enzyme-dispersed lungs of control or L. johnsonii-supplemented, allergen-sensitized and -challenged animals, there was a significant reduction in overall inflammation, with neutrophils, but not eosinophils, reduced in the lungs of L. johnsonii-supplemented animals (Fig. 3C). In addition, although control animals exhibited an increase in distinct inflammatory DC population migration into the lungs upon CRA challenge, L. johnsonii-treated animals did not (Fig. 3D). This overall reduction in inflammation was evident in histopathological sections, which demonstrated that, although L. johnsonii-treated animals did display minor signs of allergic inflammation, both inflammatory cell infiltration and goblet cell metaplasia were substantially reduced compared with unsupplemented animals (Fig. 3E). L. johnsonii was not detected in the airways of these animals (data not shown), indicating that GI L. johnsonii plays a pivotal (albeit not as entirely comprehensive as that afforded by D-associated dust) role in airway protection.
Fig. 3.
Supplementation of mice with L. johnsonii attenuates the development of allergic airways disease. (A) Examination of allergen-induced airway hyperactivity (AHR) following methacholine (250 μg/kg, i.v.) exposure demonstrated reduced responses in L. johnsonii-supplemented animals. (B) Th2 cytokine mRNA in the lungs (black bars) and protein expression in allergen-restimulated lymph node cells (gray bars) indicated a significant attenuation in L. johnsonii-supplemented mice. (C) Upon CRA exposure (black bars), significant increases in total leukocytes, granulocytes (neutrophils and eosinophils), and (D) in total and inflammatory (Ly6c+) DC populations were only observed in the control but not the L. johnsonii-supplemented animals (CRA-unexposed controls represented by white bars). (E) Histologic staining with PAS stain revealed a distinct reduction in the inflammatory and mucogenic responses in L. johnsonii-supplemented animals. Data represent mean ± SE from five mice per group. *P < 0.05, **P < 0.01.
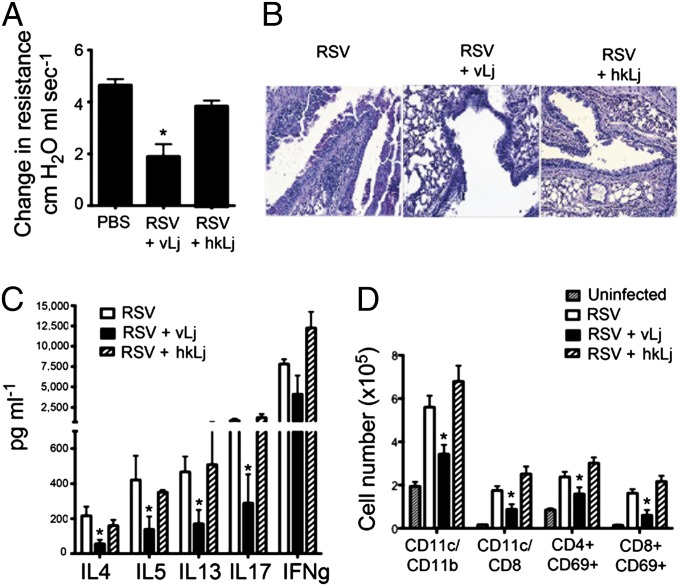
To further examine whether this protective effect was more generalized, we used a primary viral infection model. Respiratory syncytial virus (RSV) infections during infancy are especially problematic, and early and severe RSV infection is both the leading cause of childhood hospitalizations and a risk factor for development of childhood asthma (18–20). Using the same murine L. johnsonii supplementation strategy (Fig. S1), animals were infected with RSV (line 19 strain, 1 × 105 pfu/mouse). In these studies, an additional group of animals received autoclaved heat-killed (H.K.) L. johnsonii to determine whether viable organisms were necessary for airway protection. Animals were examined for pathologic and immunologic outcomes that phenocopy many of the clinical outcomes found in severely infected infants, i.e., airway hyperresponsiveness (AHR), mucus hypersecretion, and T-cell cytokine production. Those supplemented with live L. johnsonii displayed significantly decreased AHR (Fig. 4A) and histologically presented with less inflammation and reduced numbers of periodic acid-Schiff (PAS)-positive mucus-producing goblet cells (Fig. 4B), whereas the H.K. L. johnsonii-supplemented animals were not significantly different from untreated, RSV-infected animals. Similar to observations made in the allergen model, a significant reduction in draining lymph node T-cell production of IL-4, IL-5, IL-13, and IL-17 was also observed in only the live L. johnsonii-supplemented animals (Fig. 4C). Finally, analyses of immune populations infiltrating the lymph nodes demonstrated a reduction in total numbers of cells. A reduction in number of DC subsets, CD11c+/CD11b+ and CD11c+/CD8+, as well as CD69+ activated CD4 and CD8 T cells was also observed (Fig. 4D). Thus, these studies confirm that L. johnsonii supplementation significantly reduced the RSV-induced pulmonary responses and that viable organisms are necessary to transduce the altered responses.
Fig. 4.
Viable L. johnsonii is necessary to attenuate RSV-induced airway responses. (A) Viable (vLj) but not heat-killed (hkLj) L. johnsonii supplementation protects animals from RSV-induced airway hyperreactivity (AHR) assessed at 8 d postinfection. (B) Histologic examination of lungs from RSV-infected animals demonstrates reduced inflammation and PAS-stained airway mucus only in the treatment group who received viable organisms. (C) RSV-restimulated lymph node cell-induced cytokine responses are significantly lower in animals who received viable L. johnsonii supplements. (D) Significant reductions in the number of various leukocyte subsets in the lungs are only observed in animals who received viable L. johnsonii. Data represent mean ± SE from five mice per group. *P < 0.05.
L. johnsonii Supplementation Impacts GI Microbiome Composition.
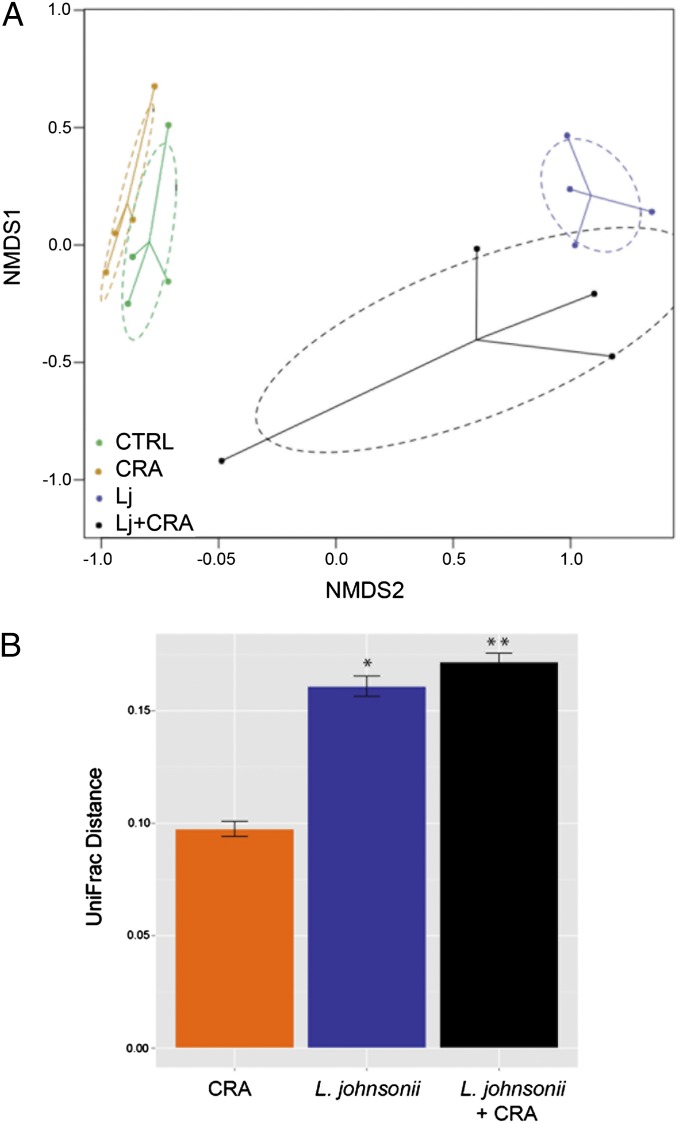
To determine whether supplementation of animals with L. johnsonii impacted the microbiome, cecal contents of animals in each treatment group from the CRA challenge model were subjected to microbiome profiling. Using Fast UniFrac (14) PCoA with ordilipse to impose a 95% confidence ellipse around samples in each treatment group demonstrated that samples clustered, irrespective of CRA challenge, in discrete groups based on L. johnsonii treatment (Fig. 5A). Examination of β-diversity (based on between-group UniFrac distances compared with the control) confirmed that L. johnsonii-supplemented communities exhibited the greatest phylogenetic distance from supplemented, control animals (Fig. 5B), indicating that supplementation with this species elicited a significant change in community phylogeny. Because the airway protective effect observed is likely due to the combined activities of L. johnsonii and other cecal cocolonizers, we next identified the specific taxa enriched in these phylogenetically distinct and protective cecal communities. Compared with the number of taxa exhibiting significant enrichment in D dust-supplemented animals, L. johnsonii-supplemented mice possessed relatively fewer taxa significantly increased in relative abundance. Those taxa significantly enriched in L. johnsonii-supplemented mice included members of the Rikenellaceae II (primarily organisms of GI origin), whereas unprotected animals were characterized by enrichment of Lachnospiraceae and Ruminococceaceae, including Clostridium and Bacteroides species (Table S2).
Fig. 5.
(A) L. johnsonii-supplemented animals that exhibit airway protection exhibit altered cecal microbiome composition. Nonmetric dimensional scaling based on a UniFrac distance matrix reveals that microbial communities of mice supplemented with L. johnsonii are compositionally and phylogenetically distinct from unsupplemented animals. Ellipses constructed around each treatment group indicate the 95% confidence intervals. (B) Compared with unsupplemented control animals, communities supplemented with L. johnsonii or L. johnsonii followed by CRA exposure exhibit the greatest phylogenetic distance (*P < 0.0005, **P < 0.0001, respectively).
Predicted Cecal Community Function Using PICRUSt.
To identify microbiome functions that characterized airway-protected animals, we used PICRUSt to predict (based on 16S rRNA data) the metagenome of cecal microbiomes present in D dust or L. johnsonii-supplemented animals. Compared with L. johnsonii-supplemented animals, D dust-exposed animals were enriched in a substantially wider range of predicted KEGG pathways, including sphingolipid, arachadonic, and carbohydrate metabolism, among others (Fig. S7). In comparison, L. johnsonii-supplemented animals exhibited comparatively fewer functions, although common predicted functions shared by these protected animals, included N-glycan biosynthesis (Fig. S7), a KEGG pathway very recently shown to be down-regulated in severe asthmatics (21).
Discussion
Humans are constantly exposed to environmental microbes, a salient point given recent microbiome studies that have underscored the reliance of mammalian systems on appropriate microbial colonization to support metabolic and immune homeostasis. In Western nations, the burden of chronic inflammatory disease, including allergic asthma, have increased dramatically over the past several decades and humans spend ∼92% of their time indoors (22), therefore a large portion of environmental microbial exposure originates from the household living environment. Hence this study was devised to test the hypothesis that distinct indoor microbial exposures impact allergic airway disease outcomes in mice and that outcome is predicated on GI microbiome composition. Moreover, the study aimed to identify key microbial species within these complex GI consortia that are critical to airway protection.
The importance of the GI microbiome in defining both the local and systemic immune environment has been demonstrated in a number of recent studies, indicating that strategies to manipulate gut microbiome membership and function may have far-reaching implications for extraintestinal niches, such as the skin or joints (23). In addition, several studies have demonstrated the importance of the GI microbiome in airway response to pathogenic agents. Ichinohe et al. (24) recently demonstrated that the GI microbiota composition critically regulates the generation of CD4 and CD8 T cells and antibody responses to respiratory influenza virus infection. Here, we not only demonstrate that distinct house dust exposures lead to differential airway allergic outcomes in pre-adult mice, but that protected animals possess a compositionally distinct cecal microbiome enriched for L. johnsonii, which can, when provided as a single species supplement, promote airway protection against allergen challenge. What is even more compelling is that airway protection, which is associated with significant reductions in total and activated T-cell numbers as well as total IgE levels, also extends to respiratory syncytial viral infection, indicating a common protective mechanism against distinct airway pathological agents and suggesting that exposure to D-associated house dust may be prophylactic particularly in the earlier phases of life as the microbiome and the immune response are developing in parallel.
That community members beyond L. johnsonii are necessary for full airway protection is indicated by our observation that animals supplemented with this one species did not exhibit the same level of airway protection compared with animals supplemented with D-associated house dust. Multiple phylogenetically distinct species significantly enriched in these animals were not enriched in L. johnsonii-supplemented animals, implicating these species in enhanced airway protection, and indicating that GI species that contribute to airway protection originate from environmental sources. Indeed, previous research has demonstrated that, although oral supplementation of animals with a single species can elicit airway protection (25, 26), several studies that supplemented with a phylogenetically distinct multispecies consortia have demonstrated improved efficacy (23, 27). As indicated from our predictive metagenomic analyses, protection is likely a combinatorial effect based on multiple pathways and resulting microbial products encoded by these cocolonizers resulting in presentation of a specific suite of microbial ligands and a distinct profile of microbial metabolites to the innate and acquired components of the host immune system.
The change in the immune environment observed in these studies may be due directly to the specific supplemented bacterial species, the ancillary species that are enriched because of the presence of the supplemented species, or the depletion of organisms outcompeted in this remodeled assemblage, that differentially program host immune responses. Although the fundamental mechanisms that govern this phenomenon remain unclear, the studies reported here provide proof-of-principle that differential environmental exposures result in GI microbiome remodeling that impacts host immune responses associated with both allergic airway response and respiratory viral infection. Moreover, this study identifies a single GI bacterial species, L. johnsonii, that is pivotal to airway protection, and identifies functional gene pathways in protective microbiota that may be responsible for airway protection. These studies suggest that GI microbiome manipulation represents a promising and efficacious therapeutic strategy to protect individuals against both pulmonary infection and allergic airway disease.
Materials and Methods
House Dust Collection.
Dust from homes with or without dogs was collected using a sterile fabric filter sock inserted into a sterile vacuum nozzle immediately before vacuuming a 3′ × 3′ area for 3 min. Further details are provided in SI Materials and Methods.
Murine Models.
BALB/c mice were used for airway allergen challenge and RSV studies; a detailed procedure is provided in SI Materials and Methods. All mouse studies were reviewed and approved by the University of Michigan's University Committee on Use and Care of Animals (UCUCA).
Lung Histology, mRNA Extraction, Reverse Transcription, and RT-PCR.
The left lung was perfused with 4% (vol/vol) formaldehyde for fixation and embedded in paraffin. Further details are provided in SI Materials and Methods. mRNA was isolated from ground lung tissue using TRIzol reagent (Invitrogen) or the RNeasy Mini kit (Qiagen) according to manufacturer’s instructions. Further details are provided in SI Materials and Methods.
Culture and Stimulation of Lymph Node Cells and Flow Cytometry.
Mediastinal lymph nodes were digested mechanically, using 18-gauge needles, and enzymatically, via incubation with 1 mg/mL Collagenase A (Roche) and DNase I (Sigma-Aldrich) in RPMI 1640 with 10% FCS. Further details are provided in SI Materials and Methods. Following FcR blocking, single-cell suspensions of bronchoalveolar lavage, lung, and lymph node cells were stained with anti-CD11c (N418), anti-Ly6C (HK1.4), anti-Ly6G (1A8; Biolegend), anti-CD11b (M1/70), anti-CD103 (2E7) (eBioscience), and anti-MHC-II/IAb (AF6-120.1; BD Biosciences). Inflammatory neutrophils were gated as low autofluorescent, CD11cloCD11bhiLy6C+Ly6G+ with low forward scatter. Inflammatory monocytes were analyzed as low autofluorescent, CD11cloCD11bhiLy6C+Ly6G− cells with low forward scatter. Further details are provided in SI Materials and Methods.
Cecal Microbiome Profiling.
Dust samples were extracted using a cetyltrimethylammonium bromide (CTAB)-PEG protocol as previously described (28). Briefly, 0.5 mL of modified CTAB extraction buffer [1:1 10% CTAB in 1 M NaCl to 0.5 M phosphate buffer (pH 7.5–8) in 1 M NaCl] were added to 0.2 g of dust (when available) in Lysing Matrix E tubes (MP Biomedicals), followed by 500 μL of phenol:cholorform:isoamyl alcohol (25:24:1). Samples were bead-beaten using MPBio FastPrep-24 at 5.5 m/s for 30 s before centrifugation for 5 min at 16,000 × g at 4 °C. Further details of this procedure are provided in SI Materials and Methods.
L. johnsonii Quantification, Isolation, Identification, and Supplement Generation.
Q-PCR was used to validate L. johnsonii relative abundance reported by the array using the QuantiTect SYBR Green PCR kit per the manufacturer’s instructions (Qiagen) and the L. johnsonii-specific primer pair Lj1 and La2 (29). Further details are provided in SI Materials and Methods. Details of L. johnsonii isolation and identification as well as how the strain was prepared for oral supplementation studies are provided in SI Materials and Methods.
Statistical Analyses.
As an exploratory tool to examine community composition dissimilarity, NMDS or PCoA was performed, based on Canberra (30) or UniFrac (14) distance matrices, respectively. PhyloChip fluorescence intensities, normalized to quantitative standards, were log2 × 1,000 transformed before analyses. Further details of ecological and traditional statistical approaches used to analyze the datasets reported in this study are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases P01AI089473-01A1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microbiome data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE52909).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310750111/-/DCSupplemental.
References
- 1.Abreu NA, et al. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Science Transl Med. 2012;4(151):151ra124. doi: 10.1126/scitranslmed.3003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nistal E, et al. Differences in faecal bacteria populations and faecal bacteria metabolism in healthy adults and celiac disease patients. Biochimie. 2012;94(8):1724–1729. doi: 10.1016/j.biochi.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 3.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489(7415):242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 4.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen CH, et al. Patterns of early gut colonization shape future immune responses of the host. PLoS One. 2012;7(3):e34043. doi: 10.1371/journal.pone.0034043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Penders J, et al. Gut microbiota composition and development of atopic manifestations in infancy: The KOALA Birth Cohort Study. Gut. 2007;56(5):661–667. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joffe TH, Simpson NA. Cesarean section and risk of asthma. The role of intrapartum antibiotics: A missing piece? J Pediatr. 2009;154(1):154. doi: 10.1016/j.jpeds.2008.08.039. [DOI] [PubMed] [Google Scholar]
- 9.Johnson CC, et al. Antibiotic exposure in early infancy and risk for childhood atopy. J Allergy Clin Immunol. 2005;115(6):1218–1224. doi: 10.1016/j.jaci.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 10.Ownby DR, Johnson CC, Peterson EL. Exposure to dogs and cats in the first year of life and risk of allergic sensitization at 6 to 7 years of age. JAMA. 2002;288(8):963–972. doi: 10.1001/jama.288.8.963. [DOI] [PubMed] [Google Scholar]
- 11.von Mutius E, Vercelli D. Farm living: Effects on childhood asthma and allergy. Nat Rev Immunol. 2010;10(12):861–868. doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- 12.Fujimura KE, et al. Man's best friend? The effect of pet ownership on house dust microbial communities. J Allergy Clin Immunol. 2010;126(2) doi: 10.1016/j.jaci.2010.05.042. 410–412, 412.e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Havstad S, et al. Effect of prenatal indoor pet exposure on the trajectory of total IgE levels in early childhood. J Allergy Clin Immunol. 2011;128(4):880–885.e4. doi: 10.1016/j.jaci.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamady M, Lozupone C, Knight R. Fast UniFrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4(1):17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valladares R, et al. Lactobacillus johnsonii N6.2 mitigates the development of type 1 diabetes in BB-DP rats. PLoS One. 2010;5(5):e10507. doi: 10.1371/journal.pone.0010507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue R, Otsuka M, Nishio A, Ushida K. Primary administration of Lactobacillus johnsonii NCC533 in weaning period suppresses the elevation of proinflammatory cytokines and CD86 gene expressions in skin lesions in NC/Nga mice. FEMS Immunol Med Microbiol. 2007;50(1):67–76. doi: 10.1111/j.1574-695X.2007.00233.x. [DOI] [PubMed] [Google Scholar]
- 18.Régnier SA, Huels J. Association between respiratory syncytial virus hospitalizations in infants and respiratory sequelae: Systematic review and meta-analysis. Pediatr Infect Dis J. 2013;32(8):820–826. doi: 10.1097/INF.0b013e31829061e8. [DOI] [PubMed] [Google Scholar]
- 19.Szabo SM, et al. Elevated risk of asthma after hospitalization for respiratory syncytial virus infection in infancy. Paediatr Respir Rev. 2013;13(Suppl 2):S9–S15. doi: 10.1016/S1526-0542(12)70161-6. [DOI] [PubMed] [Google Scholar]
- 20.Tregoning JS, Schwarze J. Respiratory viral infections in infants: Causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev. 2010;23(1):74–98. doi: 10.1128/CMR.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orsmark-Pietras C, et al. Transcriptome analysis reveals upregulation of bitter taste receptors in severe asthmatics. Eur Respir J. 2013;42(1):65–78. doi: 10.1183/09031936.00077712. [DOI] [PubMed] [Google Scholar]
- 22.Ott WR. 1989. Human activity patterns: A review of the literature for estimating time spend indoors, outdoors and in transit. Proceedings of the Research Planning Conference on Human Activity Patterns, EPA/600/4-89/004 (EPA National Exposure Research Laboratory, Las Vegas), 3.
- 23.Kwon HK, et al. Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc Natl Acad Sci USA. 2010;107(5):2159–2164. doi: 10.1073/pnas.0904055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA. 2011;108(13):5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forsythe P, Inman MD, Bienenstock J. Oral treatment with live Lactobacillus reuteri inhibits the allergic airway response in mice. Am J Respir Crit Care Med. 2007;175(6):561–569. doi: 10.1164/rccm.200606-821OC. [DOI] [PubMed] [Google Scholar]
- 26.Adams VC, et al. Mycobacterium vaccae induces a population of pulmonary CD11c+ cells with regulatory potential in allergic mice. Eur J Immunol. 2004;34(3):631–638. doi: 10.1002/eji.200324659. [DOI] [PubMed] [Google Scholar]
- 27.de Vrese M, et al. Effect of Lactobacillus gasseri PA 16/8, Bifidobacterium longum SP 07/3, B. bifidum MF 20/5 on common cold episodes: A double blind, randomized, controlled trial. Clin Nutr. 2005;24(4):481–491. doi: 10.1016/j.clnu.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 28.DeAngelis KM, et al. Selective progressive response of soil microbial community to wild oat roots. ISME J. 2009;3(2):168–178. doi: 10.1038/ismej.2008.103. [DOI] [PubMed] [Google Scholar]
- 29.Furet JP, Quénée P, Tailliez P. Molecular quantification of lactic acid bacteria in fermented milk products using real-time quantitative PCR. Int J Food Microbiol. 2004;97(2):197–207. doi: 10.1016/j.ijfoodmicro.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 30.Lance GN, Williams WT. Mixed-data classificatory programs, I.) Agglomerative systems. Aust Comput J. 1967;1(1):15–20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.