Significance
Environmental stresses, such as drought and high salinity, adversely affect plant growth and productivity. Although various phytohormones are known to be involved in regulation of plant stress responses, the role of strigolactone (SL) in this important process remains elusive. By using different molecular and physiological approaches, we provide compelling evidence that, in Arabidopsis, SL acts as positive regulator of plant responses to drought and salt stress, which was associated with shoot- rather than root-related traits. Comparative transcriptome analysis suggests that plants integrate multiple hormone-response pathways—at least SL, abscisic acid, and cytokinin pathways—for adaptation to environmental stress. Our findings demonstrate that genetic modulation of SL content/response could provide a new approach for development of crops with improved stress tolerance.
Keywords: hormonal regulation, plant adaptation, transcriptome analysis
Abstract
This report provides direct evidence that strigolactone (SL) positively regulates drought and high salinity responses in Arabidopsis. Both SL-deficient and SL-response [more axillary growth (max)] mutants exhibited hypersensitivity to drought and salt stress, which was associated with shoot- rather than root-related traits. Exogenous SL treatment rescued the drought-sensitive phenotype of the SL-deficient mutants but not of the SL-response mutant, and enhanced drought tolerance of WT plants, confirming the role of SL as a positive regulator in stress response. In agreement with the drought-sensitive phenotype, max mutants exhibited increased leaf stomatal density relative to WT and slower abscisic acid (ABA)-induced stomatal closure. Compared with WT, the max mutants exhibited increased leaf water loss rate during dehydration and decreased ABA responsiveness during germination and postgermination. Collectively, these results indicate that cross-talk between SL and ABA plays an important role in integrating stress signals to regulate stomatal development and function. Additionally, a comparative microarray analysis of the leaves of the SL-response max2 mutant and WT plants under normal and dehydrative conditions revealed an SL-mediated network controlling plant responses to stress via many stress- and/or ABA-responsive and cytokinin metabolism-related genes. Our results demonstrate that plants integrate multiple hormone-response pathways for adaptation to environmental stress. Based on our results, genetic modulation of SL content/response could be applied as a potential approach to reduce the negative impact of abiotic stress on crop productivity.
Strigolactones (SLs), a small class of carotenoid-derived compounds, were first characterized more than 45 y ago as seed germination stimulants in root parasitic plants, such as Striga, Orobanche, and Phelipanche species (1, 2). SL was later reported as a root-derived signal that can enhance symbiosis between plants and arbuscular mycorrhizal fungi (AMF), possibly through its ability to induce AMF hyphal branching (3). More recently, SL was reported to play an important role in the suppression of shoot branching by inhibiting the outgrowth of axillary buds (4, 5).
In the nonmycotrophic Arabidopsis, more axillary growth (MAX) genes, namely MAX1/AT2G26170, MAX3/AT2G44990, and MAX4/AT4G32810, have been identified to encode enzymes that are involved in the SL-biosynthetic pathway. Additionally, two genes, MAX2/AT2G42620 and BRC1/AT3G18550 (branched 1), have been reported to play a role in SL responses (2). MAX3 and MAX4 encode carotenoid cleavage dioxygenase 7 (CCD7) and CCD8, respectively, which catalyze sequential carotenoid cleavage reactions to produce an apocarotenone called carlactone, a proposed SL precursor (6). MAX1 is a cytochrome P450 monooxygenase that is presumably involved in a catalytic step downstream of MAX3 and MAX4 (2). MAX2 encodes an F-box leucine-rich repeat protein that acts as the substrate-recruiting subunit of an Skp, Cullin, and F-box (SCF)-type ubiquitin E3 ligase complex. Given the structural similarity between MAX2 and the auxin receptor transport inhibitor response 1 (TIR1), MAX2 has been suggested to be involved in SL perception (2). Arabidopsis max3 and max4 mutants have a 70–75% reduction in SL content as determined by Striga germination assays (4). Unlike max3 and max4, whose branching phenotype is rescued by exogenous application of SL, the branching phenotype of max2 is not. These data provide further evidence that max2 is a SL-signaling and not a SL-biosynthetic mutant (4). The BRC1 is induced by SL and encodes a TCP-type transcription factor (TF) that acts downstream of MAX2 in the regulation of shoot branching (2).
Environmental stresses, such as drought and high salinity, adversely affect plant growth and productivity. Various phytohormones, such as abscisic acid (ABA), brassinosteroid, and cytokinin (CK), have been shown to cooperatively regulate adaptive responses to these stressors (7–9). Currently, the role of SL in plant stress responses, if any, remains unknown. In the present study, various SL-deficient and SL-signaling mutants were functionally analyzed to determine the involvement of SL in regulating drought and salt stress responses. Results demonstrate that SL acts as a positive regulator of stress signaling networks and diverse ABA signaling pathways by regulating the expression of many stress and/or ABA-responsive genes involved in plant development and abiotic stress response. Furthermore, impaired SL signal transduction also led to the down-regulation of CK oxidase/dehydrogenase (CKX) encoding genes that are required for CK degradation (7). Collectively, these results indicate that coordinated cross-talk between SL, ABA, and CK signaling networks regulates plant adaptation to adverse environmental conditions.
Results
SL-Deficient and SL-Response max Mutants Exhibit Hypersensitivity to Drought and Salt Stress.
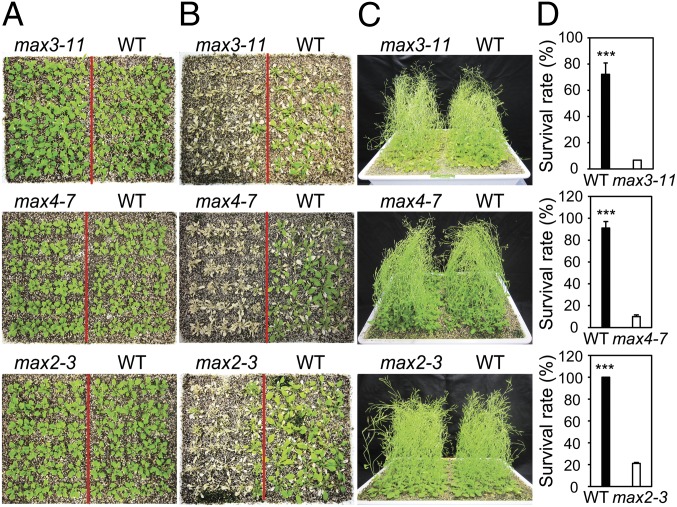
To determine the potential involvement of SL in the response of Arabidopsis to abiotic stress, the ability of the Arabidopsis max mutant and WT plants to survive drought and high salinity was examined. Two independent lines from each of the SL-biosynthetic max3 and max4 mutants, as well as the SL-signaling max2 mutant, were subjected to a drought tolerance assay in which water was withheld from 3-wk-old plants growing in soil. WT and all max mutant plants displayed similar plant size and growth rate without the application of drought stress (Fig. 1 A and C and Fig. S1 A and C). A significantly lower number of SL-deficient or SL-response max mutant plants, however, survived the drought stress compared with WT plants (Fig. 1 B and D and Fig. S1 B and D). For the salt tolerance test, 3-wk-old plants were irrigated with 200 mM NaCl instead of water for 6 d. Plants were subsequently supplied with water for 4 d, and the effect of salt stress on plants was then evaluated. A significantly higher number of SL-deficient and SL-signaling max mutant plants died compared with WT plants in response to salinity stress (Fig. S2A). A germination assay conducted on germination medium (GM) agar plates amended with 100 mM NaCl also resulted in reduced germination rates for the max mutants compared with WT plants (Fig. S2B). These data indicate that max mutants are hypersensitive to salt stress at the germination and vegetative stages of growth, and further establish that a reduction in endogenous SL content or impairment in SL signaling compromises the plant’s ability to tolerate drought and salt stress. Thus, SL plays an important role in the regulation of plant responses to abiotic stress.
Fig. 1.
Hypersensitivity of SL-deficient and SL-signaling max mutant plants to drought stress. (A) Three-wk-old WT and SL-deficient max3-11 and max4-7 and SL-signaling max2-3 mutant plants before being subjected to a drought stress. (B) WT and mutant plants subjected to a drought stress and then rewatered for 3 d. Inflorescences were removed from the surviving plants before photographing. (C) Unstressed (control) WT and max plants grown in parallel with the drought test. (D) Percent survival rates of WT and mutant plants. Data represent the mean and SE from data pooled from three independent experiments (n = 30 per genotype per experiment). Asterisks indicate significant differences as determined by a Student t test (***P < 0.001).
Exogenous Application of SL Rescues the Drought-Sensitive Phenotype of SL-Deficient Mutants and Enhances the Drought Tolerance of WT Plants.
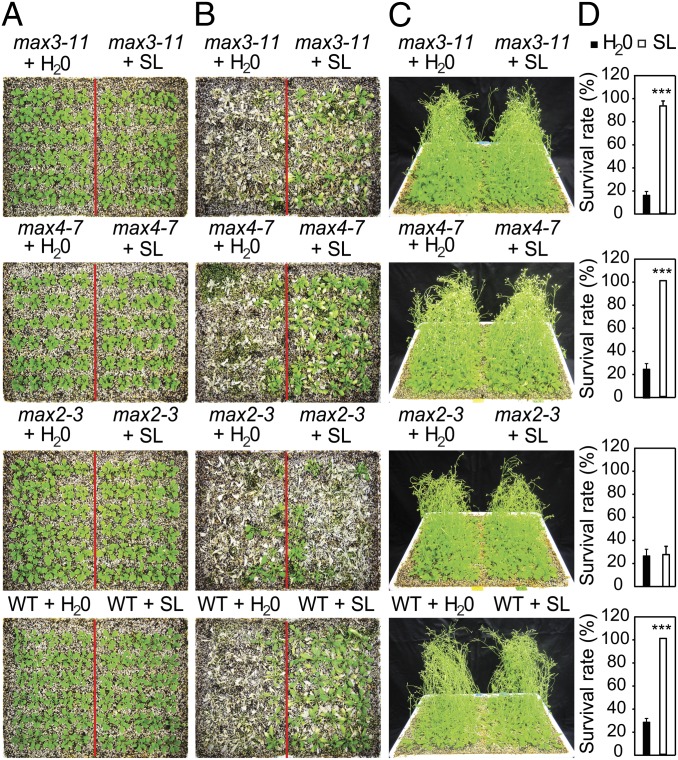
To further confirm the role of SL in drought stress, the effect of exogenous SL on the phenotype of the SL-deficient and SL-response max mutant and WT plants subjected to drought stress was determined. A comparison of the max mutants subjected to drought stress with or without exogenous application of SL revealed that the drought-sensitive phenotype of the SL-deficient max3 and max4 mutants could be rescued when sprayed with SL to almost the same level of WT plants (Figs. 1 and 2), whereas no significant effect of SL application was observed on SL-response max2 plants (Fig. 2). Furthermore, SL-treated WT plants were much more tolerant to drought than the untreated WT plants, as evidenced by their higher survival rate (100% survived among SL-treated plants vs. ∼29% survived among water-treated plants; Fig. 2). These data further support the role of SL as a positive regulator of plant response to drought stress.
Fig. 2.
Effect of SL treatment on survival of SL-deficient and SL-response mutants and WT plants. (A) Three-wk-old WT and SL-deficient max3-11 and max4-7 and SL-signaling max2-3 mutant plants before drought stress. (B) Three-wk-old plants sprayed with 5 mL of 5 µM SL or water [sprayed once at 4:00 PM (the first day and from the 7th to the 13th days) and twice at 10:00 AM and 4:00 PM (from the second day to the sixth day) during water withholding period] and subjected to a drought stress. Plants were photographed 3 d subsequent to rewatering and after removal of inflorescences from the surviving plants. (C) Nonstressed WT and max plants sprayed with 5 mL of 5 µM SL or water as shown in B. (D) Percent survival of mutant and WT plants sprayed with SL or water and subjected to a drought stress as described earlier. Data represent the mean and SE from data pooled from three independent experiments (n = 30 plants per genotype per experiment). Asterisks indicate significant differences as determined by a Student t test (***P < 0.001).
SL-Deficient and SL-Signaling max Mutants Are Less Sensitive to Exogenous ABA Than WT Plants.
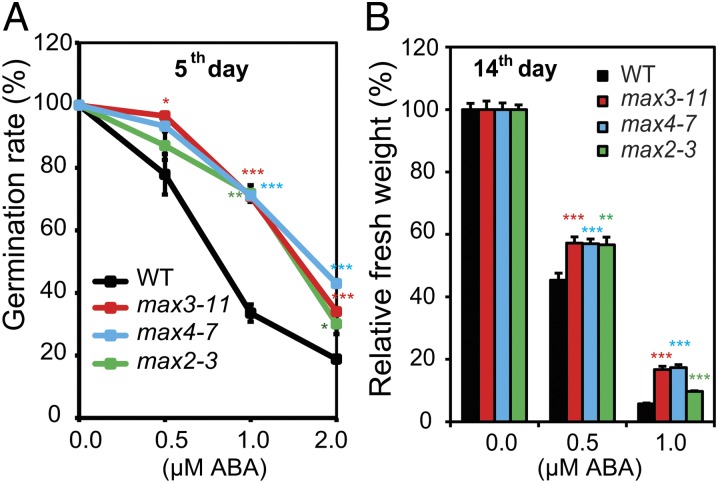
Plant responses to ABA and abiotic stresses are interrelated. ABA is induced by abiotic stresses and ABA signaling plays a pivotal role in controlling plant adaptation to many types of abiotic stress (9). To determine if ABA is involved in SL-mediated plant responses to stress, we analyzed responsiveness of the max mutants to various concentrations of ABA during germination and postgermination developmental stages. We observed that exogenous ABA more severely inhibited germination and postgermination growth of WT seedlings compared with that of the max mutants, indicating that the max mutants have reduced sensitivity to ABA than WT at the stages examined (Fig. 3 and Fig. S3). These results suggest the existence of cross-talk between SL and ABA signaling pathways in the regulation of plants stress responses.
Fig. 3.
Response of SL-deficient and SL-signaling max mutant plants to exogenous ABA treatment. (A) Percent germination of SL-deficient max3-11 and max4-7 mutant, SL-signaling max2-3 mutant, and WT seeds treated with different levels of exogenous ABA. Data represent the mean plus SE of data pooled from three independent experiments (n = 50 seeds per genotype per experiment). (B) Relative fresh weight of SL-deficient max3-11 and max4-7 mutant, SL-signaling max2-3 mutant, and WT seedlings to application of different concentrations of exogenous ABA. Relative fresh weights of all seedlings were determined after 14 d of incubation at 22 °C. Data represent the mean and SE (n = 6, where each replicate is composed of seven pooled plants). Asterisks indicate significant differences as determined by a Student t test (*P < 0.05, **P < 0.01, and ***P < 0.001).
Root Growth of max and WT Plants Under High Salinity and Osmotic Stress.
One of the successful strategies exhibited by plants to deal with osmotic stress is to alter root-related traits, particularly root growth (10, 11). To gain insight into mechanisms that render max mutant plants more sensitive to abiotic stress, we examined root growth in max and WT plants under salt and osmotic stresses. In our experimental design, various concentrations of mannitol were used to induce osmotic stress. Root growth in the max mutant and WT plants was inhibited to similar extents by treatments with different concentrations of NaCl and mannitol (Fig. S4), indicating that the stress-sensitive phenotype of max plants is not associated with a differential effect on root growth or development, at least to 11 d of growth.
Comparison of Dehydration-Induced Water Loss Rates, ABA-Mediated Stomatal Closure, and Stomatal Density in the max Mutant and WT Plants.
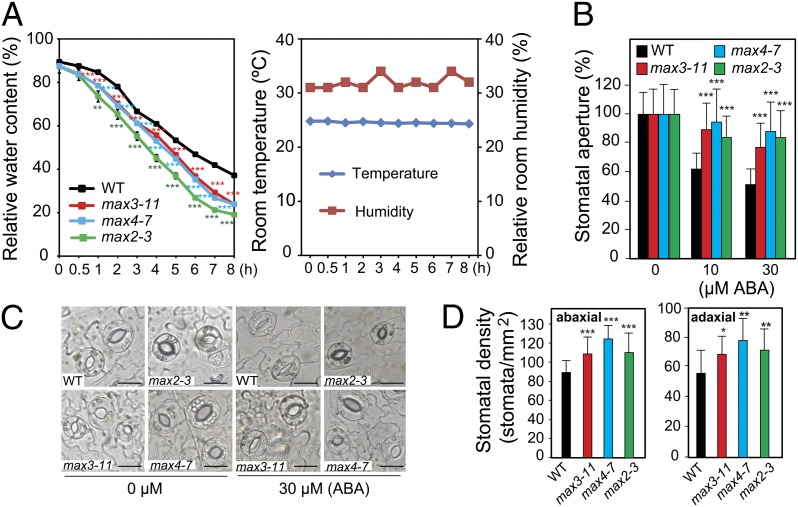
It was of interest to determine whether an alteration in shoot-related traits was the cause of the stress-sensitive phenotype observed in max plants. Leaf water status and water loss rates of WT and max mutant plants exposed to dehydration were compared. Seventeen-day-old plants were subjected to dehydration by removing them from their growth medium and placing them on a paper towel. The plants were then periodically weighed to determine leaf relative water content (RWC) and rate of water loss. By using this dehydration assay, it was observed that SL-deficient and SL-signaling max mutant plants lost water faster than WT plants (Fig. 4A), suggesting that an altered transpiration rate might be responsible for the lower tolerance of max plants to water deficit stress.
Fig. 4.
RWC, relative size of the stomatal aperture and stomatal density of the WT and SL- deficient and SL-signaling max mutant plants. (A) Time course of RWC of WT and SL-deficient max3-11 and max4-7 and SL-signaling max2-3 plants exposed to dehydration stress. Data represent the mean and SE (n = 5, where each replicate represents the weight of six plants). Room temperature and relative room humidity data recorded during the course of the experiment are also presented. (B) Average size of the stomatal aperture of rosette leaves from 3-wk-old WT and max mutant plants in the presence or absence of ABA presented as a percentage relative to the size of stomatal aperture in WT and mutant plants not exposed to ABA, which was defined as 100%. Epidermal peels were treated with ABA for 1 h after stomatal preopening under light conditions. Data represent the mean and SD (n = 200). (C) Guard cells of 3-wk-old WT and max mutant plants exposed to 30 µM ABA for 1 h or left unexposed. (Scale bars: 20 µm.) (D) Average stomatal density on the abaxial and adaxial sides of rosette leaves from 3-wk-old WT and max mutant plants. Data represent the mean and SD (n = 30). Asterisks indicate significant differences as determined by a Student t test (*P < 0.05, **P < 0.005, and ***P < 0.001).
It is well established that ABA regulates turgor pressure in guard cells of leaves, inducing stomatal closure when water stress is perceived, and that alteration in ABA-mediated stomatal closure and/or density can affect transpiration rates during water stress (12). Because WT and max mutant plants displayed different degrees of ABA responsiveness (Fig. 3 and Fig. S3), ABA-mediated stomatal movement in WT and max plants was analyzed. As shown in Fig. 4 B and C, stomatal cells of the SL-deficient and SL-signaling max mutants closed more slowly than in WT plants in response to ABA treatment. Additionally, stomatal density was higher in max mutant lines than in WT plants (Fig. 4D). These data indicate that the slower ABA-mediated stomatal closure and higher stomatal density observed in the max mutants contribute to an increased water loss rate, rendering the max mutants more sensitive to drought than WT.
Comparative Transcriptome Analysis of Leaves of the SL-Response max2-3 and WT Plants Under Well-Watered and Dehydrative Conditions.
A comparative transcriptome analysis of leaves of WT and SL-signaling max2-3 plants under normal and dehydrative conditions was conducted by using the Arabidopsis 44K DNA oligo microarrays using the experimental design illustrated in Fig. S5 A and B. This was done to identify genes involved in the downstream pathways affected by SL-mediated responses to abiotic stress. The microarray data can be accessed through Gene Expression Omnibus (accession no. GSE48949). Results of the microarray analyses are summarized in Dataset S1. A comparison of the max2-3 and WT leaf transcriptomes under nonstress conditions revealed 231 up-regulated and 262 down-regulated genes in max2-3 with respect to the WT, by using the criteria of fold change at least two and a false discovery rate-corrected P value (i.e., q-value) <0.05 (max2-3 well-watered control 0 h vs. WT well-watered control 0 h; Fig. S5C and Datasets S2 and S3). Among the 262 down-regulated genes in this comparison, 50 genes are drought-inducible (Fig. S5 D, i, and Dataset S4), among which nine are also ABA-inducible in at least five independent ABA treatment data sets among the 13 ABA treatment datasets contained in Genevestigator (Dataset S5).
A greater number of stress-inducible and/or ABA-inducible genes that are down-regulated in the max2 mutant were expected to be identified when plants were exposed to water stress. To verify this assumption, transcriptomes of leaves of max2 and WT plants exposed to drying for 2 or 4 h were compared. Approximately 1,022 and 2,767 genes were down-regulated (with a ratio ≥2) in max2 plants dehydrated for 2 and 4 h, respectively, in comparison with similarly treated WT plants (max2-3 dehydrated vs. WT dehydrated at 2 h and 4 h; Fig. S5C and Dataset S3). A Venn diagram constructed from the two down-regulated gene sets (2, 4) identified 491 and 955 genes, respectively, as dehydration-inducible (Fig. S5 D, ii and iii and Dataset S4), many of which are also ABA-inducible (Dataset S5), suggesting that ABA signaling-mediated dehydration-responsive gene expression is affected in max mutants. This significant overlap was further corroborated by the high Z-scores obtained by using Genesect at VirtualPlant (version 1.3; Dataset S6). The relatively lower expression of a number of drought- and/or ABA-inducible genes in max mutants under nonstress and drought stress conditions may be correlated with the drought-sensitive phenotype of these plants. A significant proportion (43.89%) of the down-regulated genes identified in unstressed max2 vs. unstressed WT overlapped with down-regulated genes recorded in dehydrated max2 vs. dehydrated WT comparisons (Fig. S5 D, iv, and Dataset S4). The reliability of our microarray data were validated by examining the expression of several genes by using real-time quantitative PCR (RT-qPCR; Fig. S6).
Several dehydration- and/or ABA-inducible genes down-regulated in max2 mutant plants encode regulatory and functional proteins, such as AP2-, myeloblastosis (MYB)-, and NAC-type TFs, CIPKs and leucine-rich repeat kinases, heat shock proteins, ATP-binding cassette (ABC) transporters, LEA (late embryogenesis abundant) proteins, α/β-hydrolases, glycosyltransferases, and glycoside hydrolases (Fig. S6 and Dataset S3). A subset of these genes is related to metabolism categories (Figs. S7 and S8), in particular regulatory and functional genes involved in flavonoids biosynthesis, such as production of anthocyanin pigment 1 (PAP1)/MYB75, PAP2/MYB90, CHS, FLS1, DFR, and LDOX (13, 14) (Dataset S7). Flavonoids are known to protect plants against various environmental stresses (15). The majority of the flavonoid biosynthesis-related genes identified in the present study are inducible by drought in an ABA-independent manner, suggesting that SL also modulates stress response independently of ABA (Dataset S7). Genes involved in CK degradation, namely CKX1, CKX2, CKX3, and CKX5, were repressed in max2 mutant under normal and dehydrative conditions (Dataset S7). These genes have previously been correlated with stress and ABA responses (16–18). Thus, the down-regulation of a number of genes with known links to stress-, ABA-, and CK-signaling pathways is concomitant with reduced drought and salt tolerance of max mutant plants, clearly establishing the link of SL with plant stress responses.
A closer look at the up-regulated gene sets identified an interesting phenomenon. A significant number of photosynthesis-related genes, almost all of which were down-regulated by dehydration in an ABA-independent mechanism, were up-regulated in max2 vs. WT plants, especially under dehydration stress conditions (Fig. S9 and Dataset S8). These data suggest a correlation between misregulation of photosynthesis-related genes, which would result in an enhanced photosynthesis and reduced drought tolerance of the max plants. In addition, microarray and RT-qPCR analyses indicated that, among the three known SL-biosynthetic genes, MAX3 and MAX4 were significantly induced by dehydration in leaves of WT plants (Fig. S10A and Dataset S2), further supporting that modulation of SL synthesis is important for plant adaptation to stresses.
Discussion
Considerable research has been carried out in the past 20 y to elucidate the functional role of various hormones in plant responses to environmental stress and the molecular events involved in hormone-mediated adaptation to abiotic stress (7–9). Little information is available, however, regarding the function of SL in plant abiotic stress responses despite the fact that this hormone was discovered more than 45 y ago. In the present study, a loss-of-function approach was used to explore the regulatory role of SL in plant response and adaptation to abiotic stress. Results demonstrated that all the examined max mutants displayed increased sensitivity to drought and salt stress (Fig. 1 and Figs. S1 and S2), thus implicating the involvement of SL as a positive regulator in abiotic stress responses. This premise was further supported by the rescue of the drought-sensitive phenotype of SL-deficient max3 and max4 mutants but not of the SL-response max2 mutant, and the enhancement of drought tolerance of WT plants by exogenous application of SL to the plants (Fig. 2). It is worthy to note that no remarkable change in shoot branching phenotype of the WT plants was observed during exogenous SL treatment, which is in agreement with previous findings (4, 5).
Consistent with the drought-sensitive phenotype, leaves of max mutant plants were found to lose water more rapidly and to a greater extent than leaves of WT plants when they were subjected to dehydration (Fig. 4A). Importantly, no significant differences in root growth were observed in nutrient medium grown mutant and WT plants subjected to salinity and osmotic stress treatments (Fig. S4). These data indicate that the higher transpiration rate may contribute to the reduced tolerance of the max mutants against drought stress. The increased transpiration rate in max mutants was associated with increased stomatal density and alterations in ABA-mediated stomatal closure (Fig. 4 B–D). Additionally, all max mutants tested here exhibited a lower sensitivity to various concentrations of ABA compared with WT plants during the germination and young seedling stages of growth (Fig. 3 and Fig. S3). The reduced sensitivity to ABA may be responsible for the slower stomatal closure, leading to greater loss of water and the overall reduced drought tolerance of max mutants. Several studies have demonstrated a close relationship between drought tolerance, leaf water status, stomatal density, and rates of stomatal closure. For instance, mutations in the Arabidopsis K+ uptake transporter 6 (KUP6), KUP8, and GORK (guard cell outward rectifying K+ channel) resulted in reduced ABA sensitivity, impaired ABA-mediated stomatal closure, and decreased plant survival under drought stress (19). Conversely, overexpression of EDT1 (enhanced drought tolerance1) in Arabidopsis conferred enhanced drought tolerance that was associated with a lower transpiration rate and lower stomatal density (20). In the present study, the ABA-insensitive phenotype and increased stomatal density and water loss noted in SL biosynthesis and SL signaling impaired mutants (Figs. 3 and 4) all point to SL positively regulating abiotic stress responses, at least in part through ABA signaling.
A comparative transcriptome analysis of the SL-response max2 mutant and WT plants under well-watered and dehydrative conditions was conducted to gain insight into the molecular mechanisms involved in SL-mediated drought stress responses. The results indicate that the down-regulation of many stress- and/or ABA-inducible genes encoding a wide range of stress-related regulatory and functional proteins (21, 22) may be responsible for the decreased levels of stress tolerance observed in the max mutant plants. For instance, dehydration-, high salinity-, and ABA-inducible AtNAC2/AT5G39610 (Fig. S6 and Dataset S3), which was previously reported as a positive regulator of salt stress response (23), was down-regulated in max2 plants relative to WT under normal and dehydration conditions. Similarly, expression of CIPK1/AT3G17510, a positive regulator of ABA and osmotic stress responses (24), was repressed in unstressed max2-3 compared with unstressed WT plants (Dataset S3). More importantly, down-regulation of ABCG22/AT5G06530 and ABCG40/AT1G15520 ABA importer genes in well-watered and dehydrated max2 leaves, respectively, compared with the respective WT may explain the impaired stomatal function observed in the max mutants and thus the drought-sensitive phenotype (Figs. 1 and 4 and Dataset S3). This viewpoint is supported by data indicating that abcg22 and abcg40 mutants are drought-sensitive as a result of higher transpirational water loss resulted from diminished stomatal closure compared with WT plants (25, 26). Additionally, recent studies reported that the constitutive overexpression of CKX genes, namely CKX1, CKX2, CKX3, and CKX4, resulted in higher ABA sensitivity, enhanced ABA response and improved drought and salt tolerance in transgenic Arabidopsis and tobacco plants (17, 18). The transcriptome analysis conducted in the present study revealed that, among seven Arabidopsis CKX genes, four (CKX1, CKX2, CKX3, and CKX5) were down-regulated in the max2 mutant with and/or without being subjected to dehydration (Dataset S7). The down-regulation of the four CKX genes may have contributed to the decreased stress tolerance of max plants by reducing their ABA response. Furthermore, many genes involved in flavonoid biosynthesis (13, 14) were significantly repressed by the dehydration stress treatment in max2-3 plants relative to the level of expression in WT plants (Dataset S7). This suppression may result in a decrease in the biosynthesis of flavonoid compounds, which are known to ameliorate the injury to plant cells induced by various abiotic stresses (15). In addition, the drought-sensitive phenotype of the max mutant plants might also be attributed to an enhanced photosynthesis, as suggested by the up-regulation of a subset of photosynthesis-related genes, which are negatively regulated under dehydration conditions, in the stressed max2 mutant relative to the stressed WT (Fig. S9 and Dataset S8). The enhancement of photosynthesis in the max plants was also supported by the increased transpiration rate observed in these plants, which resulted, at least in part, from enhanced stomatal activity observed during dehydration treatment (Fig. 4). An increased rate of the highly energy demanding photosynthesis could make the plant more vulnerable to stressful environments as it consumes resources that are limitedly available during stress. Thus, one of the strategies plants use to adapt to stresses is growth reduction by decreasing photosynthesis, as supported by the observed down-regulation of photosynthesis-related genes in plants in response to stresses (Dataset S8) (27, 28), to reallocate limited energy resources from the developmental programs toward efficient stimulation of defense pathways against harmful stresses (7, 29). Furthermore, a large portion of the identified SL signaling-mediated photosynthesis- (Dataset S8) and flavonoid biosynthesis-related genes (Dataset S7) were down- and up-regulated, respectively, by dehydration in an ABA-independent mechanism, suggesting that SL controls stress response not only in an ABA-dependent (Figs. 3 and 4 and Dataset S5) but also in an ABA-independent manner.
In accordance with a positive regulatory role for SL in drought responses, among the three known biosynthetic MAX genes, MAX3 and MAX4 were found to be significantly induced by dehydration treatment in leaves of WT plant (Fig. S10A). The induced expression of MAX3 and MAX4 by stress may trigger an efficient activation of SL biosynthesis, leading to the activation of SL signaling, thereby enabling plants to better adapt to adverse conditions. The observed up-regulation of MAX3 and MAX4 by high salinity and ABA treatments in WT plants further supports this mechanism (Fig. S10B). The expression of MAX3 and MAX4 was strongly induced after 2 h of salt treatment, whereas that of MAX1 was mostly unchanged. All the SL-biosynthesis MAX1, MAX3, and MAX4 genes exhibited a tendency to be induced by longer periods of ABA treatment, especially MAX3 and MAX4, which were induced more than four- and eightfold after 10 h of treatment with ABA (Fig. S10B). Furthermore, repression of MAX3 as well as MAX1 in the SL-response max2 mutant by dehydration relative to the dehydrated WT plants further supports the premise that stress-regulated SL homeostasis is essential for plant adaptation to abiotic stress (Dataset S3).
Overall, results of the present study demonstrated that SL positively modulates stress responses in Arabidopsis through ABA-dependent and ABA-independent pathways. Biological processes responsible for plant adaptation to adverse environmental conditions are complex, requiring the involvement of multiple hormone regulatory pathways. This complexity is reflected, for instance, in the functions of ABA, CK, and SL in the regulation of stomatal closure and leaf senescence, two traits that are closely related to stress responses and adaptation. Whereas ABA and SL promote the senescence of leaves, CK delays leaf senescence (30, 31). On the other hand, ABA (32) and SL (the present study) act as positive regulators of stomatal closure and thus stress response, whereas CK acts as a negative regulator of the same process (18, 33). Importantly, results of the present study demonstrate the potential for the use of genetic engineering to improve the stress tolerance of crop plants by manipulating SL biosynthesis and/or SL signaling as suggested by the enhanced drought tolerance exhibited by WT plants sprayed with SL (Fig. 2).
Materials and Methods
Plant Materials and ABA and High Salinity Treatments.
The Arabidopsis max2-3 (SALK_092836), max2-4 (SALK_028336), max3-11 (SALK_023975), max3-12 (SALK_015785), max4-7 (SALK_082552), and max4-8 (SALK_072750) mutants used in this study are in Columbia genetic background and were obtained as in a previous study (4). For ABA and high salinity treatments, Arabidopsis WT plants were grown on GM agar plates for 14 d (22 °C, 16 h light/8 h dark cycle, 60 µmol⋅m–2⋅s–1 photon flux density) and treated with water (hydroponic control), 100 µM ABA, or 250 mM NaCl for the indicated time periods.
Assessment of Drought, Salt, and Osmotic Stress Tolerance.
Drought stress tolerance assay.
The same tray method was used to evaluate drought tolerance as previously described (18). Seedlings were grown in soil by using Dio Propagation Mix no. 2 for Professional soil (Dio Chemicals). When an SL treatment was applied, 30 plants grown in soil trays were sprayed with 5 mL of 5 µM SL (rac-GR24; Chiralix) or water control once (on the first day and from the 7th to 13th days) at 4:00 PM and twice at 10:00 AM and 4:00 PM (from the second day to the sixth day). Rewatering was performed when the greatest difference in appearance was discerned between the control and mutant plants. Trays were photographed 3 d subsequent to rewatering and after removal of inflorescences from the survived plants.
Salt stress tolerance assay.
WT and mutant plants were grown in soil for 3 wk, as they were in the drought tolerance test. Each tray was then irrigated at the bottom of the tray with a total of 2 L of 200 mM NaCl instead of water for 6 d. The plants were then irrigated with water from the seventh day onward. Plant survival was recorded 4 d later (day 10).
Germination assay.
Seeds were sown on GM medium containing 1% sucrose with or without 100 mM NaCl. Those with expanded cotyledons were recorded as germinated seeds.
Root growth assay.
Four-day-old plants germinated and grown on vertical GM plates were transferred onto 0.5× Murashige–Skoog plates containing 1.2% (wt/vol) agar with and without the indicated concentration of NaCl or mannitol. Root growth of vertically grown seedlings was examined 7 d after transfer.
Stomatal Closure Assay and Measurement of Stomatal Density.
ABA-induced stomatal closure was assessed as previously described (19) with slight modifications. Epidermal peels from leaves of 3-wk-old plants grown on GM plates were incubated for 4 h in a solution containing 10 mM KCl, 0.2 mM CaCl2, and 10 mM Mes⋅KOH (pH 6.15) under white light (300 μmol⋅m−2⋅s−1). The peeled strips were subsequently incubated in a solution containing the same buffer plus ABA. Guard cells were photographed by using a light microscope equipped with a digital camera. Two hundred stomatal apertures were measured for each mutant and WT plant. Stomatal density was determined by counting the number of stomates on the abaxial and adaxial sides of leaves of 3-wk-old mutant and WT seedlings. Leaves were cleared with chloral hydrate and visualized by the light microscopy. All stomates from 30 images of the central region of the leaves from eight individual plants were counted for each mutant and WT.
Assay for Sensitivity to ABA.
Germination and growth inhibition assays were performed on GM medium containing 1% sucrose and various concentrations of ABA as described previously (18).
Expression Analyses.
Total RNA was extracted with an RNeasy Plant Mini Kit (Qiagen). Procedures described previously (34) were used for cDNA synthesis and RT-qPCR. Polyubiquitin 10 (UBQ10) was used as a reference gene for expression analysis. Primer pairs used to examine the expression levels of specific genes are listed in Dataset S9.
Dehydration Treatment and Microarray Analysis.
WT and max2-3 plants (30 plants each) were grown in soil as described previously (18) and in the drought tolerance assay. Aerial portions of 24-d-old plants were detached and exposed to dehydration by placing them on paper towels on a laboratory bench. At the indicated time points, RWC of treated samples was measured (n = 5). Rosette leaves of three independent WT and SL-signaling max2-3 mutant plants treated for 0, 2, 4, and 6 h were then collected to make three biological replicates for microarray and expression analyses. Microarray analysis by using the Arabidopsis Oligo 44K DNA microarray (version 4.0; Agilent Technology) was performed as described previously (35, 36). The microarray data and a detailed protocol were deposited in the Gene Expression Omnibus database (accession no. GSE48949). MapMan (http://mapman.gabipd.org) and VirtualPlant (http://virtualplant.bio.nyu.edu/cgi-bin/vpweb/) were used to analyze the data. In some cases, ABA and stress-responsive gene expression was analyzed by using Genevestigator (www.genevestigator.com) or the Arabidopsis eFP browser (http://bar.utoronto.ca/efp_arabidopsis/cgi-bin/efpWeb.cgi).
Supplementary Material
Acknowledgments
C.V.H. was supported by a PhD fellowship from the International Program Associate of RIKEN. This work was supported in part by Howard Hughes Medical Institute Grant 55005946 (to L.H.-E.) and a grant (Project Code 03/2012/HĐ-ĐTĐL) from the Vietnam Ministry of Science and Technology to the Research Group of N.V.D.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE48949).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322135111/-/DCSupplemental.
References
- 1.Xie X, Yoneyama K, Yoneyama K. The strigolactone story. Annu Rev Phytopathol. 2010;48:93–117. doi: 10.1146/annurev-phyto-073009-114453. [DOI] [PubMed] [Google Scholar]
- 2.Ruyter-Spira C, Al-Babili S, van der Krol S, Bouwmeester H. The biology of strigolactones. Trends Plant Sci. 2013;18(2):72–83. doi: 10.1016/j.tplants.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Akiyama K, Matsuzaki K, Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. 2005;435(7043):824–827. doi: 10.1038/nature03608. [DOI] [PubMed] [Google Scholar]
- 4.Umehara M, et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455(7210):195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- 5.Gomez-Roldan V, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455(7210):189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- 6.Alder A, et al. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science. 2012;335(6074):1348–1351. doi: 10.1126/science.1218094. [DOI] [PubMed] [Google Scholar]
- 7.Ha S, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. Cytokinins: metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 2012;17(3):172–179. doi: 10.1016/j.tplants.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Choudhary SP, Yu JQ, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. Benefits of brassinosteroid crosstalk. Trends Plant Sci. 2012;17(10):594–605. doi: 10.1016/j.tplants.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 9.Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res. 2011;124(4):509–525. doi: 10.1007/s10265-011-0412-3. [DOI] [PubMed] [Google Scholar]
- 10.Manavalan LP, Guttikonda SK, Tran LS, Nguyen HT. Physiological and molecular approaches to improve drought resistance in soybean. Plant Cell Physiol. 2009;50(7):1260–1276. doi: 10.1093/pcp/pcp082. [DOI] [PubMed] [Google Scholar]
- 11.Galvan-Ampudia CS, Testerink C. Salt stress signals shape the plant root. Curr Opin Plant Biol. 2011;14(3):296–302. doi: 10.1016/j.pbi.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 12.Casson SA, Hetherington AM. Environmental regulation of stomatal development. Curr Opin Plant Biol. 2010;13(1):90–95. doi: 10.1016/j.pbi.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Saito K, et al. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol Biochem. 2013;72:21–34. doi: 10.1016/j.plaphy.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53(5):814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 15.Pourcel L, Routaboul JM, Cheynier V, Lepiniec L, Debeaujon I. Flavonoid oxidation in plants: From biochemical properties to physiological functions. Trends Plant Sci. 2007;12(1):29–36. doi: 10.1016/j.tplants.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Werner T, et al. Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell. 2010;22(12):3905–3920. doi: 10.1105/tpc.109.072694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macková H, et al. Enhanced drought and heat stress tolerance of tobacco plants with ectopically enhanced cytokinin oxidase/dehydrogenase gene expression. J Exp Bot. 2013;64(10):2805–2815. doi: 10.1093/jxb/ert131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishiyama R, et al. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell. 2011;23(6):2169–2183. doi: 10.1105/tpc.111.087395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osakabe Y, et al. Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell. 2013;25(2):609–624. doi: 10.1105/tpc.112.105700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu H, et al. Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell. 2008;20(4):1134–1151. doi: 10.1105/tpc.108.058263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartels D, Sunkar R. Drought and salt tolerance in plants. Crit Rev Plant Sci. 2005;24(1):23–58. [Google Scholar]
- 22.Jogaiah S, Govind SR, Tran LSP. Systems biology-based approaches toward understanding drought tolerance in food crops. Crit Rev Biotechnol. 2013;33(1):23–39. doi: 10.3109/07388551.2012.659174. [DOI] [PubMed] [Google Scholar]
- 23.He XJ, et al. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005;44(6):903–916. doi: 10.1111/j.1365-313X.2005.02575.x. [DOI] [PubMed] [Google Scholar]
- 24.D’Angelo C, et al. Alternative complex formation of the Ca-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J. 2006;48(6):857–872. doi: 10.1111/j.1365-313X.2006.02921.x. [DOI] [PubMed] [Google Scholar]
- 25.Kang J, et al. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci USA. 2010;107(5):2355–2360. doi: 10.1073/pnas.0909222107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuromori T, Sugimoto E, Shinozaki K. Arabidopsis mutants of AtABCG22, an ABC transporter gene, increase water transpiration and drought susceptibility. Plant J. 2011;67(5):885–894. doi: 10.1111/j.1365-313X.2011.04641.x. [DOI] [PubMed] [Google Scholar]
- 27.Matsui A, et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 2008;49(8):1135–1149. doi: 10.1093/pcp/pcn101. [DOI] [PubMed] [Google Scholar]
- 28.Le DT, et al. Differential gene expression in soybean leaf tissues at late developmental stages under drought stress revealed by genome-wide transcriptome analysis. PLoS ONE. 2012;7(11):e49522. doi: 10.1371/journal.pone.0049522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaves MM, Flexas J, Pinheiro C. Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot (Lond) 2009;103(4):551–560. doi: 10.1093/aob/mcn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim PO, Kim HJ, Nam HG. Leaf senescence. Annu Rev Plant Biol. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- 31.Czarnecki O, Yang J, Weston DJ, Tuskan GA, Chen JG. A dual role of strigolactones in phosphate acquisition and utilization in plants. Int J Mol Sci. 2013;14(4):7681–7701. doi: 10.3390/ijms14047681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka Y, et al. Cytokinin and auxin inhibit abscisic acid-induced stomatal closure by enhancing ethylene production in Arabidopsis. J Exp Bot. 2006;57(10):2259–2266. doi: 10.1093/jxb/erj193. [DOI] [PubMed] [Google Scholar]
- 34.Le DT, et al. Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Res. 2011;18(1):17–29. doi: 10.1093/dnares/dsq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishiyama R, et al. Transcriptome analyses of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS ONE. 2012;7(2):e32124. doi: 10.1371/journal.pone.0032124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishiyama R, et al. Arabidopsis AHP2, AHP3, and AHP5 histidine phosphotransfer proteins function as redundant negative regulators of drought stress response. Proc Natl Acad Sci USA. 2013;110(12):4840–4845. doi: 10.1073/pnas.1302265110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.