Significance
Studies focused on understanding how transcription factors control gene expression have shown that transcription-factor binding sites generally greatly exceed the number of regulated genes, making it challenging to identify functional binding sites. Using Notch pathway inhibitors, we identified a subset of Notch-binding sites in leukemia cell genomes that are dynamic, changing in occupancy relatively rapidly when Notch signaling is perturbed. Dynamic Notch sites are highly associated with genes that are directly regulated by Notch and mainly lie in large regulatory switches termed superenhancers, which control genes with key roles in development and cancer. This work links Notch signaling to superenhancers and suggests that assessment of transcription factor–genome dynamics can help to identify functionally important regulatory sites.
Keywords: gene regulation, Notch signaling
Abstract
The main oncogenic driver in T-lymphoblastic leukemia is NOTCH1, which activates genes by forming chromatin-associated Notch transcription complexes. Gamma-secretase-inhibitor treatment prevents NOTCH1 nuclear localization, but most genes with NOTCH1-binding sites are insensitive to gamma-secretase inhibitors. Here, we demonstrate that fewer than 10% of NOTCH1-binding sites show dynamic changes in NOTCH1 occupancy when T-lymphoblastic leukemia cells are toggled between the Notch-on and -off states with gamma-secretase inhibiters. Dynamic NOTCH1 sites are functional, being highly associated with Notch target genes, are located mainly in distal enhancers, and frequently overlap with RUNX1 binding. In line with the latter association, we show that expression of IL7R, a gene with key roles in normal T-cell development and in T-lymphoblastic leukemia, is coordinately regulated by Runx factors and dynamic NOTCH1 binding to distal enhancers. Like IL7R, most Notch target genes and associated dynamic NOTCH1-binding sites cooccupy chromatin domains defined by constitutive binding of CCCTC binding factor, which appears to restrict the regulatory potential of dynamic NOTCH1 sites. More remarkably, the majority of dynamic NOTCH1 sites lie in superenhancers, distal elements with exceptionally broad and high levels of H3K27ac. Changes in Notch occupancy produces dynamic alterations in H3K27ac levels across the entire breadth of superenhancers and in the promoters of Notch target genes. These findings link regulation of superenhancer function to NOTCH1, a master regulatory factor and potent oncoprotein in the context of immature T cells, and delineate a generally applicable roadmap for identifying functional Notch sites in cellular genomes.
Notch signaling has a critical developmental role in metazoan animals, and its dysregulation underlies several human developmental disorders and certain cancers such as T-lymphoblastic leukemia (T-LL), in which NOTCH1 gain-of-function mutations occur in ∼60% of cases (1). Physiological and pathological Notch receptor activities are largely mediated by a canonical signaling pathway, through which Notch directly regulates the expression of downstream target genes (for recent review, see ref. 2). Normally, Notch receptors are activated by binding of ligands of the delta–serrate–lag2 family to the Notch ectodomain, an event that renders the juxtamembrane region of Notch susceptible to successive cleavages by ADAM metalloproteases and γ-secretase. The latter cleavage releases the Notch intracellular domain (NICD) from the membrane, allowing it to translocate to the nucleus and form a Notch transcription complex (NTC) with the DNA-binding protein RBPJ and coactivators of the mastermind-like (MAML) family. MAML interacts with the histone acetyltransferase p300 and is essential for activation of transcription (3, 4), and other work suggests that the NOTCH1 NICD also interacts with the CH3 domain of p300 (5). Recent studies have shown that p300 acetylates H3K18 and H3K27 (6), and that the H3K27ac mark is characteristic of active enhancers and correlates with transcription activation (6–9).
In cancers such as T-LL, gain-of-function mutations in NOTCH1 cause excessive Notch activation and exaggerated expression of oncogenic target genes. To further elucidate how NOTCH1 regulates the transcriptomes of T-LL cells, we recently used chromatin immunoprecipitation (ChIP)-Seq to identify RBPJ–NOTCH1-binding sites genomewide in Notch-“addicted” murine and human T-LL cell lines (10). Unexpectedly, we observed that although RBPJ–NOTCH1-binding sites were mainly located within gene promoters (defined as sites <2 kb from a transcriptional start site; TSS), most of these genes did not respond to perturbations of Notch signaling. Conversely, the majority of direct NOTCH1 target genes lacked NOTCH1 binding to their promoters, suggesting that transcriptional response to Notch in T-LL cells is largely mediated through long-range enhancers. Thus, these studies left unresolved both the identity and characteristics of the subset of RBPJ–NOTCH1-binding sites that regulate transcription.
To address this question, we reasoned that functional RBPJ–NOTCH1 genomic-binding sites would be marked by dynamic changes in NOTCH1 occupancy following perturbations of Notch signaling. Here, we use this approach to identify and characterize functional NOTCH1-binding sites genomewide in T-LL cells. Notably, functional sites constitute only a minor subset of NOTCH1-binding sites and are mainly located in distal enhancers, many of which appear to correspond to recently described “superenhancers” (11–14).
Results
Overlap of NOTCH1–RBPJ and ETS1, GABPA, and RUNX1 Binding to the Genomes of Human T-LL Cells.
We noted previously that DNA sequences within 500 base pairs of NOTCH1–RBPJ-binding sites in human T-LL cell lines were highly enriched in binding motifs for ETS (E26-transformation specific) factors, RUNX factors, and the zinc finger transcription factor ZNF143 (10). Like NOTCH1, ETS and RUNX factors are required for normal T-cell development (15–18), suggesting that these factors coordinately regulate the transcriptomes of T-cell progenitors. To confirm the associations suggested by motif analysis, we performed steady-state ChIP-Seq for RUNX1 and two ETS family members, ETS1 and GABPA, in the Notch-addicted human T-LL cell line CUTLL1 (19). We analyzed these data as well as prior steady-state ChIP-Seq data for RBPJ, NOTCH1, ZNF143, and the chromatin marks H3K4me1, H3K4me3, and H3K27me3 (summarized in Table S1). As shown in Fig. S1 A–C, NOTCH1- and RBPJ-binding sites are highly associated with ZNF143-, ETS1-, GABPA-, and RUNX1-binding sites and are mainly located in promoters. NOTCH1–RBPJ-binding sites are also associated with “activating” chromatin marks (H3K4me1 and H3K4me3) and devoid of repressive H3K27me3 marks (Fig. S1A), consistent with the role of NOTCH1–RBPJ complexes as transcriptional activators. An illustrative example of the overlap among these factors and histone marks can be seen in the chromatin landscapes near NOTCH3, a direct NOTCH1 target gene (Fig. S1D).
Dynamic Notch-Binding Sites Are Mainly Located in Distal Enhancers and Are Enriched for RUNX1 Binding.
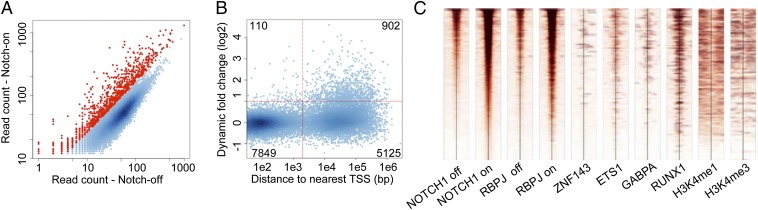
To test the idea that the sensitivity of individual genes to Notch stems from dynamic association of Notch–RBPJ complexes with genomic regulatory elements, we used a method originally described by our group (20) and since exploited by others (21) that relies on reversible inhibition of Notch signaling by gamma-secretase inhibitors (GSI). T-LL cells treated with GSI become depleted of NICD1 and accumulate a pool of membrane-tethered NOTCH1 polypeptides that have undergone ADAM–metalloprotease cleavage. Upon washout of GSI, this pool of partially processed receptors is rapidly cleaved by gamma-secretase, allowing for precisely timed NOTCH1 activation. To study loading of NTCs on regulatory elements in T-LL genomes, we performed ChIP-Seq for RBPJ and NOTCH1 (i) after treatment with GSI for 72 h and (ii) 4 h after GSI washout. Using a linear model to estimate fold-change and a false discovery rate (FDR) of 0.05, we identified 1,012 peaks with significantly increased NOTCH1 occupancy following GSI washout (Fig. 1A and Fig. S2A), hereafter referred to as “dynamic NOTCH1 sites,” which constitute fewer than 10% of NOTCH1 binding sites identified in the “Notch-on” steady-state condition. We used cutoffs based on the bimodal distribution of all 13,986 NOTCH1 peaks (Fig. S2B) to define dynamic and nondynamic sites as being either proximal promoter binding sites (<2 kb from a TSS) or distal enhancer binding sites (>2 kb from a TSS). It is interesting to note that although 57% of the 13,986 NOTCH1 binding sites are located in proximal promoter regions, roughly 90% of the 1,012 dynamic binding sites lie outside of promoters within putative distal enhancers (Fig. 1B and Fig. S2C).
Fig. 1.
NOTCH1 activation reveals dynamic NOTCH1–RBPJ-binding sites. (A) Scatterplot of NOTCH1 ChIP-seq read counts. Each dot represents a NOTCH1 peak identified in CUTLL1 T-LL cells following GSI washout. ChIP-seq reads within 600 bp of a peak summit were counted in the Notch-on and Notch-off states. Red dots indicate dynamic sites. (B) Classes of NOTCH1-binding sites defined by genomic location and dynamism. Each dot is a NOTCH1 peak plotted according to its distance to the nearest gene’s transcriptional start site (TSS) and its signal-fold change from the Notch-off to the Notch-on states. The red vertical line separates proximal promoters and distal enhancers, and the red horizontal line corresponds to a signal-fold-change threshold with an FDR < 0.05. Inset numbers correspond to peaks found in each quadrant. (C) Heat map of dynamic NOTCH1 sites, ranked by ChIP-Seq signal intensity, and associated transcription factor and histone mark signals across a 1-kb window centered on NOTCH1-binding summits.
We then used the same strategy to investigate RBPJ loading onto dynamic NOTCH1 sites. We noted a strong association between NOTCH1 and RBPJ binding to dynamic sites (Fig. 1C), an observation consistent with prior data in fly (22) and mammalian cells (10), suggesting that Notch enhances RBPJ binding to DNA. In contrast to steady-state associations, we found that chromatin proximal to dynamic NOTCH1 sites is enriched for strong RUNX1 binding (55% overlap), and for pervasive (56% overlap), but weaker, ETS1 binding, compared with ZNF143 and GABPA binding (Fig. 1C). This association is in line with motif enrichment analysis performed on genomic sequences within 600 base pairs of the summit of dynamic NOTCH1-binding sites, which revealed that the most highly enriched motifs after RBPJ are those for RUNX factors, followed by E-box–binding factors (Fig. S2D). In line with their predominant nonpromoter location, dynamic NOTCH1 sites are also strongly associated with H3K4me1 marks, which have a significantly broader distribution around dynamic NOTCH1 sites than around nondynamic, nonpromoter NOTCH1 sites (Fig. S2E).
IL7R Is Coregulated by NOTCH1 and RUNX1 Through 3′ Enhancer Elements.
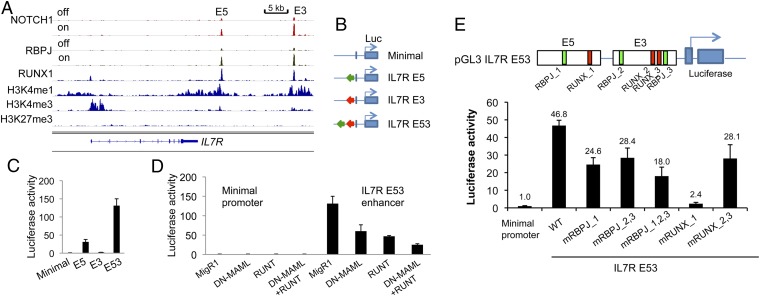
Enrichment of RUNX1 binding near NOTCH–RBPJ dynamic sites suggested that RUNX1 and NOTCH1 might coregulate the expression of certain genes. We studied the functional importance of RUNX1 in human T-LL cells by knocking down RUNX1 and by expressing a dominant negative Runx factor, RUNT. Like GSI and dominant negative MAML1 (DN-MAML, a specific inhibitor of NTCs), knockdown of RUNX1 and expression of RUNT both inhibit T-LL cell growth (Fig. S3 A–C). One candidate gene for coregulation by Notch and Runx is IL7R, a direct Notch target gene that promotes the development of thymocytes and the proliferation of T-LL cells (23, 24). Inspection of chromatin landscapes revealed two dynamic NOTCH1–RBPJ-binding sites located within two distinct regions 3′ of IL7R that contain high levels of the enhancer mark H3K4me1 (Fig. 2A). These same regions also bind RUNX1, raising the possibility that IL7R might be coregulated by Notch and Runx through these putative 3′ enhancers. In support of this idea, GSI, DN-MAML, and RUNT down-regulated expression of IL7R mRNA (Fig. S3D) and IL7R protein levels (Fig. S3E). We also observed that knockdown of RUNX1 decreased IL7R mRNA expression (Fig. S3F), suggesting that the effects of RUNT are mediated, at least in part, through inhibition of RUNX1.
Fig. 2.
Identification and functional characterization of the IL7R 3′ enhancers. (A) Chromatin landscapes around the IL7R locus in human T-LL cells shows the presence of a pair of 3′ enhancers, E5 and E3, each containing a dynamic NOTCH1–RBPJ site and a RUNX1 site. (B) Diagram showing IL7R enhancer reporter constructs. Regions spanning E5, E3, or both (E53) NOTCH1–RBPJ binding sites were cloned into the pGL3-TATA box plasmid. (C) IL7R enhancer elements are active in CUTLL1 T-LL cells. Here and elsewhere, luciferase assays were carried out in triplicate and normalized to the luciferase activity generated by the empty pGL3-TATA box plasmid. (D) IL7R-enhancer activity in CUTLL1 T-LL cells depends on Notch and Runx factors. Notch and Runx factor activity was inhibited by transfection of plasmids encoding DN-MAML and Runt, respectively. (E) Effects of RBPJ and Runx motif mutations on IL7R E53-enhancer reporter gene activity in CUTLL1 cells. The cartoon shows the IL7R E53-enhancer construct and associated RBPJ and Runx motifs. The effects of mutations involving these sites, alone and in combination, are shown in the IL7R E53-enhancer reporter gene assay below. Error bars in C–E represent 1 sd from the mean of data points obtained in triplicate.
We then studied the function of the elements 3′ of IL7R that contain the RBPJ/NOTCH1 and RUNX1 binding sites. We cloned two regions (E5 and E3) encompassing the two dynamic NOTCH1 sites and the adjacent RUNX1 binding sites (Fig. 2B) into a luciferase reporter plasmid containing a TATA box. When introduced into T-LL cells, the IL7R E5 and E3 elements both stimulate transcription individually, and when juxtaposed these two elements stimulate transcription synergistically (Fig. 2C). Like the endogenous IL7R gene, the IL7R reporter gene is down-regulated in T-LL cells by DN-MAML and RUNT (Fig. 2D). Finally, we evaluated the contributions of individual RBPJ- and RUNX1-binding motifs to the transcriptional stimulatory activity of the IL7R enhancer elements in T-LL cells. Mutations in each of these sites decreased firefly luciferase expression, with mutation of the RUNX1 motif in the E5 element being particularly deleterious (Fig. 2E). We conclude that expression of IL7R in T-LL cells is coregulated by binding of RBPJ–NOTCH1 and RUNX1 to a pair of 3′ enhancer elements.
Action of Notch on Target Genes Is Constrained by CCCTC-Binding Factor Domains and Predicted by Dynamic Regulatory Potential.
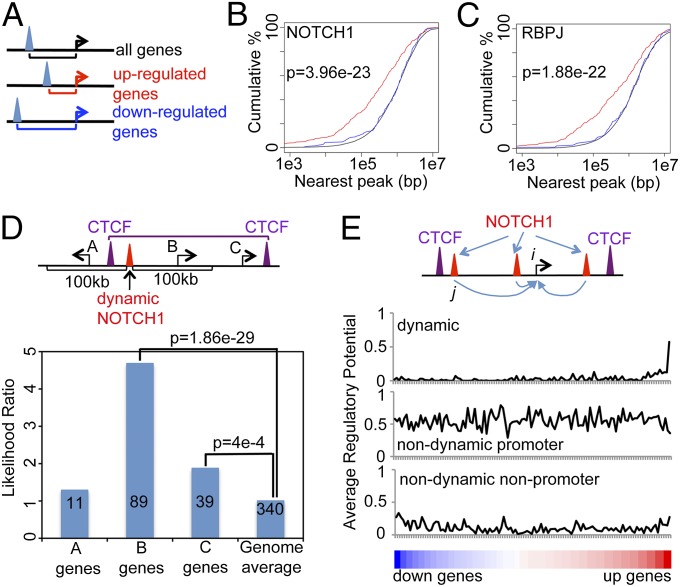
We next investigated the genomic relationships between dynamic NOTCH1/RBPJ sites and NOTCH1 target genes more broadly. We used previously published gene-expression profiling data (10) to identify 340 genes that were significantly up-regulated and 187 genes that were significantly down-regulated (FDR < 0.05) after GSI washout. We found that dynamic NOTCH1–RBPJ sites are much more likely to be near the TSSs of up-regulated genes than down-regulated genes, which by contrast are no more likely to be near dynamic NOTCH1–RBPJ sites than any randomly selected gene (Fig. 3 A–C). These relationships are consistent with the current view that NOTCH1–RBPJ complexes only act as transcriptional activators.
Fig. 3.
Target gene activation through dynamic NOTCH1-binding sites. (A–C) Dynamic NOTCH1–RBPJ-binding sites are preferentially located near genes that are up-regulated by Notch. The distance from the nearest binding site to the TSS of each gene was recorded (A) and cumulative distributions of 340 up-regulated genes (red), 187 down-regulated genes (blue), and the genomic background (black) were plotted for dynamic NOTCH1 (B) and dynamic RBPJ sites (C). P values were calculated by the Kolmogorov–Smirnov test. (D) Constitutive CTCF-binding sites define domains that restrict NOTCH1 regulation of nearby genes. Genes near dynamic NOTCH1 sites were classified into three categories: A, within 100 kb but in a different CTCF domain; B, within 100 kb and in the same CTCF domain; C, more than 100 kb away and in the same CTCF domain. The ratio between the likelihood of finding an activated gene in each category and the likelihood of finding an activated gene randomly are shown in the bar plot; numbers in each bar correspond to the activated genes in each category. P values were calculated by Fisher exact test. (E) Relationships between regulatory potentials and Notch-dependent changes in gene expression. The schematic shows how regulatory potential is calculated (see Materials and Methods for details). The lower panels show relationships between regulatory potentials of NOTCH1-binding sites and differential expression of genes (see Supporting Information for details).
In mammalian genomes, functional chromatin domains are hypothesized to be delineated by constitutive CCCTC-binding factor (CTCF) sites (25–27). We partitioned the genome using constitutive CTCF-binding sites derived from Encyclopedia of DNA elements (ENCODE) data and asked if CTCF domains restrict the regulatory activity of dynamic NOTCH1 sites. We grouped genes into three categories based on the position of their TSSs relative to (i) constitutive CTCF-binding sites and (ii) the dynamic NOTCH1 sites. We found that genes located in the same CTCF domain as one or more dynamic NOTCH1-binding sites are much more likely to be activated by NOTCH1 than genes located in a different CTCF domain, regardless of how close they are to the nearest dynamic NOTCH1 site (Fig. 3D).
In most instances, dynamic NOTCH1-associated CTCF domains contained one NOTCH1 target gene and one to five dynamic NOTCH1 sites (summarized in Fig. S4A). Exceptions to this general rule were observed, however. The most notable of these is found in the GTPase of the immunity-associated protein family (GIMAP family) gene cluster, which lies within a CTCF domain on chromosome 7q36 and encodes a family of GTPases implicated in regulation of lymphocyte development, survival, and homeostasis (28). Three dynamic NOTCH1–RBPJ sites lie within this cluster, and five flanking GIMAP genes (GIMAP2, GIMAP1, GIMAP5, GIMAP6, and GIMAP7) score as NOTCH1 target genes (Fig. S4B).
Other factors that may impact NOTCH1 regulation include the number, dynamism, and spacing of NOTCH1-binding sites relative to any gene’s TSS. To test the effect of these variables, we calculated the “regulatory potential” for each differentially expressed gene using a distance-weighted metric (29) that takes into account the number and spacing of NOTCH1-binding sites (Fig. 3E, Top; see Materials and Methods for details). This metric was calculated for dynamic sites, nondynamic promoter sites, and nondynamic nonpromoter sites (Fig. 3E). We found that the genes that are up-regulated upon NOTCH activation are highly enriched for those with high dynamic NOTCH1 regulatory potentials. Of note, although thousands of genes have nondynamic NOTCH1-binding sites in their promoter regions that yield a high average regulatory potential, there is no correlation between the calculated regulatory potential of these genes and actual changes in gene expression following perturbation of Notch signaling.
These results suggest that the calculated dynamic regulatory potential can help to identify direct Notch target genes, particularly those that are under the control of distal enhancer elements. In line with this idea, 87 genes with high dynamic regulatory potential were up-regulated following GSI washout (Table S2). This list includes many previously identified putative direct NOTCH1 target genes, most of which appear to be regulated in part or in whole by distal-enhancer elements.
Dynamic Notch Binding Activates Target Genes Through Interactions with H3K27ac-Marked Superenhancers.
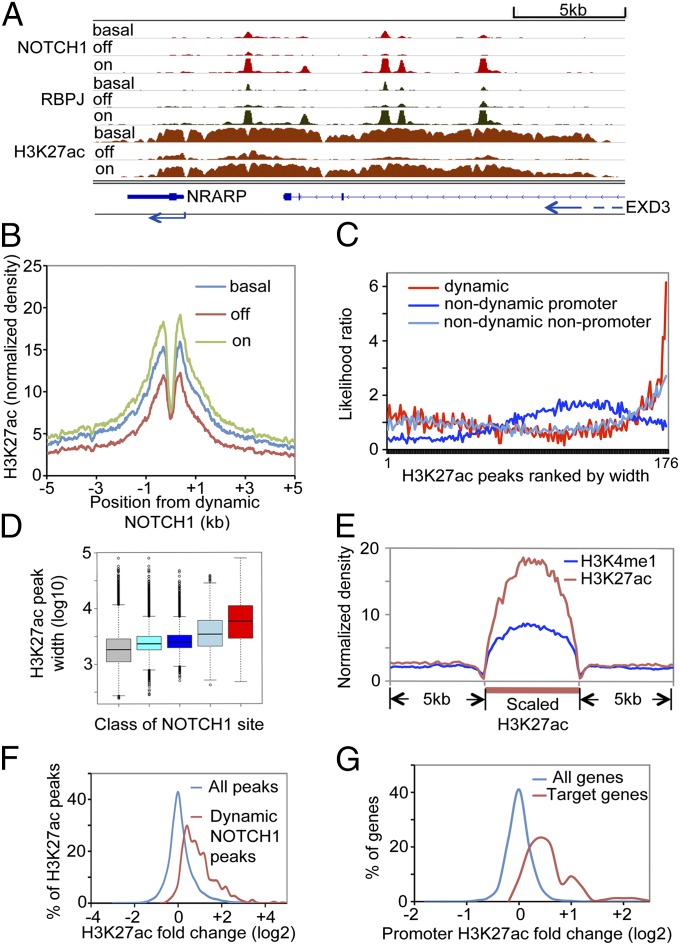
We further investigated how dynamic NOTCH1 sites induce transcriptional activation by studying histone H3K27 acetylation (H3K27ac), an active enhancer mark, under steady-state, Notch-off, and Notch-on conditions using the GSI washout strategy. We noted that the H3K27ac landscapes around dynamic NOTCH1 sites near Notch target genes are remarkably sensitive to alterations in Notch signaling. For example, five dynamic NOTCH1 sites located 5′ of the Notch-regulated ankyrin repeat protein (NRARP) gene body lie within a 20-kb H3K27ac peak that shows striking Notch-dependent changes across its full breadth (Fig. 4A). Similarly, the three dynamic NOTCH1 sites located 5′ of the HES5 gene body are found within a >10-kb H3K27ac peak that also shows large Notch-dependent changes (Fig. S5A) that are accompanied by marked changes in HES5 expression (Fig. S5B). The wide breadth of H3K27ac peaks associated with these dynamic NOTCH1 sites earmarks these regions as superenhancers, recently described genomic elements linked to the regulation of genes that are involved in oncogenesis and differentiation (11, 12, 14).
Fig. 4.
Dynamic NOTCH1 sites are preferentially located within H3K27ac-marked superenhancers. (A) Chromatin landscapes near the NOTCH1 target gene NRARP. The flanking EXD3 gene is not expressed in T-LL. (B) Composite profiles of H3K27ac flanking dynamic NOTCH1 sites under steady-state (basal), Notch-off, and Notch-on conditions. (C) Enrichment of dynamic NOTCH1 sites in the broadest H3K27ac peaks. Genomic H3K27ac peaks (n = 35,244) were ranked by peak width and grouped into 176 bins, each containing 200 peaks. H3K27ac peaks associated with dynamic or nondynamic NOTCH1 sites were counted in each bin, and the ratio of (i) the likelihood of NOTCH1 binding to H3K27ac peaks and (ii) the average likelihood NOTCH1 binding to all genomic H3K27ac peaks is plotted for each class of NOTCH1 sites. (D) Distribution of H3K27ac peak width for all H3K27ac peaks (gray), all promoter H3K27ac peaks (cyan), H3K27ac peaks associated with nondynamic promoter NOTCH1 (dark blue), nondynamic nonpromoter NOTCH1 (light blue), and dynamic NOTCH1 (red) is shown. (E) Composite profiles of H3K27ac and H3K4me1 on dynamic NOTCH1 associated H3K27ac peaks (see Supporting Information for details). (F) Fold-change distribution of H3K27ac peaks associated with dynamic NOTCH1 sites versus all H3K27ac peaks. Fold change is the ratio of H3K27ac level in the Notch-on and Notch-off states. (G) Fold-change distribution of promoter H3K27ac levels on NOTCH1 target genes and all genes. H3K27ac level is measured as the normalized read count within 2 kb of transcriptional start sites.
Guided by these local observations, we next assessed the effect of Notch on H3K27ac peaks genomewide. We found that 83% (843 out of 1,012) of dynamic NOTCH1 sites overlap with H3K27ac peaks, and that H3K27ac levels flanking dynamic NOTCH1 sites are reduced by GSI treatment and restored by GSI washout (Fig. 4B and Fig. S5C); by contrast, H3K27ac levels near nondynamic NOTCH1 sites are insensitive to short-term treatment with GSI and subsequent GSI washout (Fig. S5D). Moreover, H3K27ac peaks associated with dynamic NOTCH1 sites are much more likely than nondynamic sites to be located in the widest H3K27ac peaks in the genome (Fig. 4C), and as a result are significantly broader than the genomewide average sizes of all H3K27ac peaks and H3K27ac peaks associated with nondynamic NOTCH1 sites (Fig. 4D). Overall, the median width of H3K27ac peaks associated with dynamic NOTCH1 sites is 5,863 bp, with 56% of peaks being over 5 kb in width, while the median width of all 35,244 H3K27ac peaks in the CUTLL cell genome is 1,830 bp, with only 9% being over 5 kb in width (Fig. S5E). The particularly large size of distal regulatory elements associated with dynamic NOTCH1 sites is also reflected in a broad distribution of the enhancer mark H3K4me1 around this subset of H3K27ac peaks (Fig. 4E), an association that was hinted at in Fig. S2E. Furthermore, changes in H3K27ac levels between Notch-off and Notch-on states are selectively observed near dynamic NOTCH1-binding sites (Fig. 4F), and increased H3K27ac near dynamic sites is accompanied by increased H3K27 acetylation of Notch target gene promoters (Fig. 4G), both of which are consistent with transcriptional regulatory function for the dynamic NOTCH1 sites.
To further characterize Notch-sensitive superenhancers, we performed ChIP-Seq for p300, a factor recruited by Notch transcription complexes (5) that acetylates H3K27 (6), as well as BRD4 and mediator (MED1), factors enriched in superenhancers (11, 12, 14). As anticipated, dynamic NOTCH1 sites were associated with p300 binding (Fig. S5F), and H3K27ac-marked NOTCH1-associated superenhancers were accompanied by broad regions of BRD4 and MED1 binding (Fig. S5G). The Notch-sensitive superenhancers near the Notch target genes NRARP, HES5, DTX1, and IGF1R (Fig. S6) illustrate that p300 tends to bind near dynamic NOTCH1–RBPJ sites, and that BRD4 and MED1 bind more broadly across entire superenhancer regions.
To extend the association between dynamic NOTCH1 binding and superenhancer function to other T-LL lines, we studied how changes in NOTCH1 activity affect the DTX1 superenhancer in the T-LL cell lines KOPT-K1, DND41, and HPBALL, each of which have gain-of-function mutations involving NOTCH1 (1). Similar to CUTLL cells, we observed that depletion of NOTCH1 with GSI results in loss of H3K27ac across the entire breadth of the DTX1 superenhancer in all of these lines (Fig. S7). Taken together, these findings suggest that Notch–superenhancer interactions are of general importance in regulation of gene expression in Notch-addicted T-LL cells.
Discussion
ChIP-Seq has revealed in great detail where transcription factors bind in genomes, but factor-binding sites generally greatly outnumber regulated genes, and many functionally important binding sites lie in distal enhancers rather than promoters. These considerations make identification of binding sites that are directly involved in transcriptional regulation challenging. To overcome this limitation, we used small-molecule Notch-pathway inhibitors to toggle between the Notch-on and Notch-off states, and by doing so identified a relatively small subset of dynamic NOTCH1 sites in T-LL cells that are highly associated with Notch target genes. Our results underscore the importance of using transcription-factor perturbations to identify binding sites that govern dynamic changes in gene expression. Factors that determine the dynamism (or lack thereof) of NOTCH1 sites in T-LL genomes remain to be established. It also remains to be determined whether functional roles different from acute regulation of direct Notch target genes exist for nondynamic NOTCH1 sites.
Roughly 90% of dynamic NOTCH1 sites in T-LL cells lie outside of gene promoters. One factor that is hypothesized to influence pairing of distal regulatory elements with promoters is their spatial relationship to constitutive CTCF-binding sites, which appear to organize the mammalian genome into functional chromatin domains (25–27). Using constitutive CTCF binding data from the ENCODE consortium, we identified a strong bias toward colocalization of dynamic NOTCH1 sites and high-confidence direct Notch target genes within the same CTCF domain. Our data, however, do not exclude the possibility that some genes are regulated by Notch responsive enhancers across CTCF boundaries; indeed, some Notch target genes (such as MYC) lack candidate enhancers, and some putative Notch responsive enhancers fall within CTCF domains without evident NOTCH target genes. This is similar to the current view of the CTCF interactome in embryonic stem cells, in which most CTCF-delineated chromatin domains are created by loops formed between loci on the same chromosome, but a significant minority involves pairs of loci on different chromosomes (26, 30). Studies using methods that provide an unbiased view of three-dimensional chromatin organization will likely be needed to elucidate how Notch regulates certain target genes, such as MYC.
Transcription is regulated by the interplay of transcription factors, chromatin regulators, and core components of the basal transcriptional machinery. A recent study showed that in T-LL cells the chromatin regulators LSD1, PHF8, AF4p12, and BRG1 associate with NTCs on regulatory elements such as the DTX1 and IL7R enhancers, and are required for expression of at least a subset of NOTCH target genes (31). An additional finding of our work is the strong correlation between the occupancy of dynamic NOTCH1-binding sites and levels of H3K27 acetylation of Notch response elements and associated target gene promoters. Both NICD1 and MAML1 bind p300 (3–5), which carries out H3K27 acetylation (6), and in line with this we observe that p300 is recruited to dynamic NOTCH1 sites genomewide. Runx factors also recruit p300 to chromatin, and it is possible that optimal recruitment of p300 to Notch response elements such as the IL7R enhancers requires both Notch and Runx factors. Of note, Yatim et al. identified RUNX1 as a component of the NOTCH1 interactome in T-LL cells (31), raising the possibility that NOTCH1 and RUNX1 might physically contact each other on genomic response elements; however, dynamic NOTCH1 sites and nearby RUNX1 sites do not show any preferred spacing in T-LL genomes, making direct physical interaction unlikely as a general rule. Further work will be needed to define the basis and extent of Notch/Runx transcriptional interplay in T-LL cells and normal hematopoietic progenitors.
Perhaps the most striking observation emerging from this study is the association of functional Notch binding sites with superenhancers. Superenhancers are recently characterized “giant” regulatory switches that appear to have important roles in regulating the expression of genes that control lineage specification during development (14) and of oncogenes in transformed cells (11, 14). Our findings suggest that NOTCH1, an oncoprotein and master regulator of T-lineage specification, functions in large part in T-LL cells through dynamic binding to superenhancers. NOTCH1 unloading and reloading are associated with extensive and dramatic changes in the H3K27ac levels of superenhancers that are spatially associated with robust NOTCH1 target genes. It will be of interest to determine if the interaction of Notch with superenhancers noted in T-LL cells extends to other cellular contexts, and if so, whether superenhancers will prove to have a general role in integrating signaling inputs involving Notch and other pathways.
Materials and Methods
Cell Culture and GSI Washout Studies.
Human T-LL CUTLL1, KOPT-K1, DND41, and HPBALL cells were cultured as previously reported (20). T-LL cells were treated with the GSI compound E (1 μM) for 72 h to establish the Notch-off state, and Notch was then reactivated by washing out GSI, as described (20). Notch-on cells were harvested 4 h after GSI washout.
ChIP and Next-Generation Sequencing.
ChIP-seq was performed as described (10). Antibody information is provided in the SI Materials and Methods. ChIP-seq data have been deposited in the Gene Expression Omnibus (accession number GSE51800). Primer sequences for local ChIP are given in the Supporting Information.
ChIP-Seq Data Analysis.
Uniquely mapped, nonredundant sequence reads aligned to human genome build hg19 were retained (32). Genomic enrichment was identified using MACS 1.4 under a P value threshold of 10−5 (29, 33). Dynamic binding sites were determined using a linear model described in Supporting Information. Heat maps and composite profiles of ChIP-seq enrichment were generated as described (34).
Regulatory Potential Calculation and Data Integration.
For each gene i, the regulatory potential, Pi, of associated NOTCH1-binding sites was calculated by Pi = Σ exp-[Δij/λ], where ∆ij is the distance from the TSS of the gene i to the jth binding site located within the CTCF domain (see schematic in Fig. 3E), and λ is a scale factor determined by empirical fitting (29). T-score and FDR of differentially expressed genes were determined using linear models for microarray data (LIMMA) (35). The rank mean in Table S2 was calculated as the arithmetic mean of the ranks of a gene in the regulatory potential list and in the differential expression t-score list. Additional details are in Supporting Information.
Reporter Constructs.
A luciferase reporter plasmid containing a TATA box was assembled by replacing the SV40 promoter region of pGL3 (Promega) with the sequence GATCTCCAGATATATATAGAGGCCGCCAGGGCCTGCGGATCACACAGA. To create IL7R enhancer reporter constructs, 311 (E5) and 200 bp (E3) genomic DNA sequences were amplified by PCR, sequenced, and cloned into the pGL3 TATA box construct individually or in tandem. Mutations in transcription binding site motifs were created by PCR or by QuikChange site-directed mutagenesis kit (Stratagene) according to provided protocols. Primer and genomic DNA sequences of reporter constructs are available on request.
Reporter Gene Assays.
CUTLL1 cells were transfected with empty pGL3 or pGL3 IL7R enhancer reporter plasmids mixed with an internal control Renilla luciferase plasmid, pRL-TK, at a 50:1 ratio using Lipofectamine LTX reagent (Invitrogen). At 48 h posttransfection, cells were lysed and dual luciferase assays were performed as described (36).
Supplementary Material
Acknowledgments
H.W. is supported by a National Institutes of Health (NIH) training grant (T32HL007627). C.Z. is supported by a fellowship from the Leukemia and Lymphoma Society. W.S.P., S.C.B., X.S.L., and J.C.A. are supported by NIH Grant P01 CA119070, and X.S.L. is supported by NIH Grant R01 GM099409.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Database deposition: The sequence reported in this paper has been deposited in the Gene Expression Omnibus, www.ncbi.nlm.nih.gov/geo/ (accession no. GSE51800).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315023111/-/DCSupplemental.
References
- 1.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 2.Kopan R. Notch signaling. Cold Spring Harb Perspect Biol. 2012;4(10) doi: 10.1101/cshperspect.a011213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallberg AE, Pedersen K, Lendahl U, Roeder RG. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol Cell Biol. 2002;22(22):7812–7819. doi: 10.1128/MCB.22.22.7812-7819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fryer CJ, Lamar E, Turbachova I, Kintner C, Jones KA. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 2002;16(11):1397–1411. doi: 10.1101/gad.991602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oswald F, et al. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol Cell Biol. 2001;21(22):7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin Q, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30(2):249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotney J, et al. Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb. Genome Res. 2012;22(6):1069–1080. doi: 10.1101/gr.129817.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci USA. 2011;108(36):14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lovén J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153(2):320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker SC, et al. NISC Comparative Sequencing Program; National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program Authors; NISC Comparative Sequencing Program Authors Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci USA. 2013;110(44):17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eyquem S, Chemin K, Fasseu M, Bories JC. The Ets-1 transcription factor is required for complete pre-T cell receptor function and allelic exclusion at the T cell receptor beta locus. Proc Natl Acad Sci USA. 2004;101(44):15712–15717. doi: 10.1073/pnas.0405546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204(8):1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zamisch M, et al. The transcription factor Ets1 is important for CD4 repression and Runx3 up-regulation during CD8 T cell differentiation in the thymus. J Exp Med. 2009;206(12):2685–2699. doi: 10.1084/jem.20092024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu S, Zhao DM, Jothi R, Xue HH. Critical requirement of GABPalpha for normal T cell development. J Biol Chem. 2010;285(14):10179–10188. doi: 10.1074/jbc.M109.088740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palomero T, et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006;20(7):1279–1287. doi: 10.1038/sj.leu.2404258. [DOI] [PubMed] [Google Scholar]
- 20.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20(15):2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liefke R, et al. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 2010;24(6):590–601. doi: 10.1101/gad.563210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krejcí A, Bray S. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007;21(11):1322–1327. doi: 10.1101/gad.424607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González-García S, García-Peydró M, Alcain J, Toribio ML. Notch1 and IL-7 receptor signalling in early T-cell development and leukaemia. Curr Top Microbiol Immunol. 2012;360:47–73. doi: 10.1007/82_2012_231. [DOI] [PubMed] [Google Scholar]
- 24.González-García S, et al. CSL-MAML-dependent Notch1 signaling controls T lineage-specific IL-7Ralpha gene expression in early human thymopoiesis and leukemia. J Exp Med. 2009;206(4):779–791. doi: 10.1084/jem.20081922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cuddapah S, et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19(1):24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Handoko L, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43(7):630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hawkins RD, et al. Dynamic chromatin states in human ES cells reveal potential regulatory sequences and genes involved in pluripotency. Cell Res. 2011;21(10):1393–1409. doi: 10.1038/cr.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filén S, Lahesmaa R. GIMAP proteins in T-lymphocytes. J Signal Transduct. 2010;2010:268589. doi: 10.1155/2010/268589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Q, et al. A comprehensive view of nuclear receptor cancer cistromes. Cancer Res. 2011;71(22):6940–6947. doi: 10.1158/0008-5472.CAN-11-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Espinoza CA, Ren B. Mapping higher order structure of chromatin domains. Nat Genet. 2011;43(7):615–616. doi: 10.1038/ng.869. [DOI] [PubMed] [Google Scholar]
- 31.Yatim A, et al. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol Cell. 2012;48(3):445–458. doi: 10.1016/j.molcel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25(15):1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article3. [DOI] [PubMed]
- 36.Malecki MJ, et al. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006;26(12):4642–4651. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.