Significance
A unique avian-origin H7N9 influenza virus caused 134 human infections with 44 deaths. The host factors contributing to moderate vs. severe disease are not clear. Here, we show that H7N9 severity was associated with a higher level of cytokines/chemokines. We demonstrate that the cytokines in the infected lung were 100- to 1,000-fold higher than those in the plasma. Furthermore, we found that the IFN-induced transmembrane protein-3 (IFITM3) C/C genotype was associated with severe clinical outcome, as reflected by reduced time in seeking medical aid; more rapid progression to acute respiratory distress syndrome; and higher viral load, cytokine/chemokine levels, and mortality rate. Overall, our data suggest that the IFITM3 genotype is a primary driver of the observed differences in clinical outcome after H7N9 infection.
Keywords: avian influenza, clinical outcome
Abstract
A unique avian-origin A/H7N9 influenza virus has so far caused 134 cases with 44 deaths. Probing the host factors contributing to disease severity, we found that lower levels of plasma inflammatory cytokines on hospital admission correlated with faster recovery in 18 patients with A/H7N9 influenza virus, whereas high concentrations of (in particular) IL-6, IL-8, and macrophage inflammatory protein-1β were predictive of a less favorable or fatal outcome. Analysis of bronchoalveolar lavage samples showed up to 1,000-fold greater cytokine/chemokine levels relative to plasma. Furthermore, patients with the rs12252-C/C IFN-induced transmembrane protein-3 (IFITM3) genotype had more rapid disease progression and were less likely to survive. Compared with patients with the rs12252-T/T or rs12252-T/C genotype of IFITM3, patients with the C/C genotype had a shorter time from disease onset to the time point when they sought medical aid (hospital admission or antiviral therapy) and a shorter interval to development of the acute respiratory distress syndrome stage (reflected by shorter intervals between clinical onset and methylprednisolone treatments and higher rates of mechanical ventilator use), as well as experiencing elevated/prolonged lung virus titers and cytokine production and higher mortality. The present analysis provides reported data on the H7N9 influenza-induced “cytokine storm” at the site of infection in humans and identifies the rs12252-C genotype that compromises IFITM3 function as a primary genetic correlate of severe H7N9 pneumonia. Together with rs12252 sequencing, early monitoring of plasma cytokines is thus of prognostic value for the treatment and management of severe influenza pneumonia.
In March 2013, an influenza outbreak caused by a unique avian-origin H7N9 influenza A virus emerged in the Yangtze River Delta on China’s eastern seaboard (1–5). Along with other groups, we characterized this human pathogen and established that the 2013-H7N9 HA is distinct from that of any previously identified H7 influenza A virus (2). To date, the H7N9 virus has caused 134 confirmed cases in nine provinces. Most patients were hospitalized with severe symptoms, particularly pneumonia (97.3%) and acute respiratory distress syndrome (ARDS; 71.2%), leading to high rates (75%) of intensive care unit admissions and mechanical ventilation (66%), and >30% mortality (6, 7).
Our understanding of the relative contributions of viral and host factors to severe influenza, including these H7N9 cases, is far from complete. Multivariate analysis has shown that coexisting medical conditions are the only identified independent risk factors to date for moderate to severe H7N9-associated ARDS (6). It also seems that elevated concentrations of inflammatory mediators in plasma (hypercytokinemia) are indicative of poor disease outcomes in patients with H7N9 infections (5, 8). For example, Chen et al. (5) observed substantially higher plasma cytokine levels in a fatal case of H7N9 virus influenza compared with those in a patient who survived. Zhou et al. (8) then found that the circulating cytokines in H7N9-infected patients could reach concentrations comparable to those characteristic of H5N1 pneumonia. Even so, although suggestive that hypercytokinemia may predict, or reflect, severe disease following H7N9 infection, such observations provide little insight into mechanisms. Furthermore, the question of whether plasma cytokine levels are a direct reflection of what is happening in the infected lung has not been explored previously for any human influenza pneumonia.
As with many influenza cases, it is far from clear why some (but not all) H7N9-infected individuals develop very severe symptoms. What is known is that the IFN-induced transmembrane protein-3 (IFITM3) can restrict influenza virus replication by preventing endocytosed virus particles from entering the host cytoplasm (9, 10). As a consequence, relative to the WT IFITM3+/+ controls, genetically disrupted IFITM3−/− mice are extremely vulnerable to infection with even low pathogenic influenza viruses (11). In humans, homozygosity for the SNP rs12252-C (C/C in contrast to the WT T/T or F1 T/C) is associated with truncation of the N-terminal 21 amino acids of the IFITM3 protein, which, in turn, affects IFITM3 localization (12). This association of the C/C phenotype with severe disease emerged from the analysis of hospitalized patients in China (13) and Europe (11) during the 2009 H1N1 pandemic. However, because the pandemic H1N1 (pH1N1) virus generally caused a mild infection, there was no a priori reason to think that C/C individuals might be even more compromised by the highly pathogenic H7N9 strain.
What we show here is that the IFITM3 rs12252-C/C genotype (vs. C/T or T/T) tends to be predictive of severe H7N9-induced disease. In addition, early and persisting high cytokine/chemokine levels in plasma [especially IL-6, IL-8, and macrophage inflammatory protein (MIP)-1β] are associated with poor clinical outcomes, although whether this is causative or simply a consequence of greater and more prolonged virus replication is not altogether clear. Also apparent from the analysis of bronchoalveolar lavage (BAL) samples is that at least some of those high cytokine/chemokine concentrations in blood are likely a “spillover” from local production in the infected lung. This study thus suggests further approaches that might be used to probe mechanisms, while providing useful predictive measures of outcome that should be of help in managing severe influenza.
Results
High Levels of Inflammatory Cytokines in Plasma Are Predictive of Severe Clinical Outcomes.
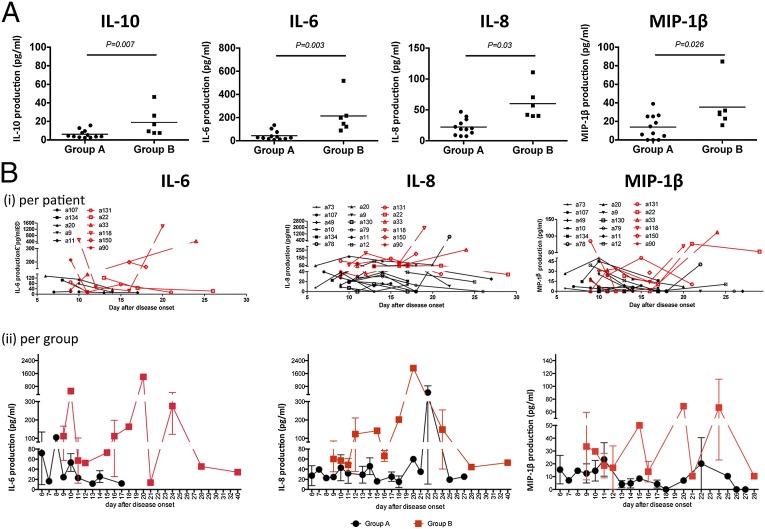
Following admission (ranging from day 6 to day 15 after onset of symptoms), we quantified inflammatory cytokines in the plasma of confirmed H7N9 cases and correlated the findings from these individual kinetic analyses with subsequent clinical outcomes (Table 1). Most consistently elevated were IL-6 and IL-8, whereas IL-10, MIP-1β, and IFN-γ were increased at intermediate levels. Although IL-1β, TNF-α, and MIP-1α were found in several patients, they were detected only at minimal concentrations. Comparing cytokine production in recovered patients discharged from our center by 35 d after onset of illness (group A) with data from those who died (group B), we found significantly higher levels (P < 0.01) of IL-6 and IL-8 in the latter, more clinically compromised group (Fig. 1 A and B and Table S1). All six patients in group B had plasma concentrations >80 pg/mL and ≥40 pg/mL for IL-6 and IL-8, respectively (10-fold higher than normal), a profile found for only 1 (8.3%) of 12 patients in group A. It thus seems that very high blood levels of IL-6 and IL-8 are indicative of high risk of mortality from severe H7N9 influenza disease. Furthermore, IL-10 and MIP-1β were also elevated in group B (Fig. 1 A and B and Table S1), although the differences between groups A and B were less dramatic than for IL-6 and IL-8. Early and continued blood monitoring for these key inflammatory cytokines may thus be helpful for making appropriate decisions regarding case management.
Table 1.
Demographics, clinical information, and cytokine production of 18 H7N9 virus-infected patients
| Group | Patient no. | Sex | Age, y | Underlying medical disorders | Days from disease onset to |
Initial viral load | 1–2 d after admission, pg/mL |
CRP**, mg/L | Clinical outcome (discharged after days of onset or death) | |||||
| Cytokines/chemokines | ||||||||||||||
| Initiation of oseltamivir | First methylprednisolone treatment | Admission | IL-10* | IL-6** | IL-8** | MIP-1β* | ||||||||
| Group A (recovery outcome) | a73 | M | 53 | None | 5 | 5 | 6 | 2.82 | 2.4 | 9.4 | 7.6 | 4.4 | 24.4 | 14 |
| a107 | M | 47 | None | 5 | 5 | 7 | 3.97 | 4.5 | 16.3 | 39.4 | 7.1 | 43.8 | 17 | |
| a49 | M | 68 | Hypertension II | 6 | No | 8 | 3.11 | 3.6 | 29 | 13.1 | 18.5 | 78.9 | 18 | |
| a10 | M | 65 | Hypertension arthritis | 4 | No | 8 | 2.54 | 1.8 | 13.8 | 9.2 | 0 | 10.8 | 18 | |
| a134 | F | 74 | Hypertension III, coronary heart disease (NYHA IV), diabetes II, cholecystitis | 8 | No | 8 | 3.71 | 3.8 | 105.2 | 22.6 | 14.6 | 51.2 | 21 | |
| a78 | M | 74 | None | 8 | 12 | 10 | <2 | 5.4 | 39.5 | 19 | 5.9 | 24.8 | 22 | |
| a20 | F | 81 | Arrhythmia | 5 | 6 | 6 | 3.49 | 15.7 | 134.6 | 47 | 26.6 | 131 | 23 | |
| a9 | M | 67 | None | 5 | 4 | 9 | NA | 7.6 | 76 | 20.3 | 25.4 | 45.1 | 23 | |
| a130 | M | 67 | Diabetes II | 11 | 9 | 9 | NA | 2.6 | 23.7 | 7.5 | 0 | 84 | 27 | |
| a79 | M | 78 | Hypertension II | 11 | 15 | 10 | 2.36 | 2.9 | 25.9 | 28.1 | 25.4 | 74.5 | 31 | |
| a11 | F | 75 | Hypertension II, coronary heart disease (NYHA IV), diabetes | 8 | 9 | 9 | 3.05 | 12.8 | 17.4 | 18.6 | 0 | 106 | 33 | |
| a12 | M | 62 | Hypertension | 9 | 5 | 8 | 2.15 | 9.6 | 34.7 | 34.6 | 39 | 34.7 | 35 | |
| Average | 67.7 | 7 | 7.7 | 8 | 3.02 | 6.1 | 43.8 | 22.3 | 13.9 | 59.1 | ||||
| Group B (fatal outcome) | a131 | F | 79 | Arrhythmia, coronary heart disease (NYHA IV), chronic bronchitis, primary biliary cirrhosis | 6 | 6 | 8 | 4.75 | 16.9 | 88.6 | 110.7 | 84.6 | 82.5 | Death |
| a22 | M | 58 | Hypertension | 9 | 8 | 12 | <2 | 9.3 | 122 | 40.4 | 23 | 80.5 | ||
| a33 | M | 56 | None | 3 | 7 | 8 | 26.2 | 146.9 | 42 | 31.7 | 115 | |||
| a118 | M | 88 | Hypertension III, coronary heart disease (NYHA IV), diabetes II, Chronic bronchitis | 7 | 8 | 9 | 5.67 | 46.3 | 516.9 | 57.1 | 29.7 | 196 | ||
| a150 | M | 80 | None | 7 | 7 | 15 | <2 | 7.6 | 198 | 70.4 | 27.8 | 128 | ||
| a90 | M | 74 | Coronary heart disease (NYHA III) | 6 | 6 | 8 | 3.32 | 7.6 | 215.7 | 40 | 15.9 | 93.8 | ||
| Average | 68.5 | 6.3 | 7 | 10 | 4.58 | 19 | 214.7 | 60.1 | 35.5 | 120.1 | ||||
All patients in group A recovered within 40 d after the onset of illness, and all patients in group B died. CRP, C-reactive protein; F, female; M, male; NA, not available; NYHA, New York Heart Association.
P < 0.05 between group A and group B; **P < 0.01 between group A and group B.
Fig. 1.
Cytokines and chemokines associated with severe H7N9 influenza infection in plasma and BAL fluid. (A) Elevated levels of IL-10, IL-6, IL-8, and MIP-1β in the patients’ plasma correlate with fatal clinical outcomes, as specified in Table 1. Cytokine concentrations were assessed within 1–2 d after hospital admission; patients who recovered and were discharged within 35 d were assigned to group A (survival group), and those died from the infection comprise group B. (B) Kinetics of IL-6, IL-8, and MIP-1β levels in plasma during the course of H7N9 infection, represented for (i) individual patients and (ii) group A vs. group B, as outlined in Table S1. Cytokine and chemokines were measured with cytometric bead array (CBA) kits.
No bacterial infections during the early hypercytokinemia were identified by clinical laboratory tests, possibly due to the fact that the patients were receiving intensive antibiotic treatment before their diagnosis with H7N9 virus. However, with the persistence of inflammation, bacteria infection occurred in patients who showed a huge blimp of inflammatory cytokines in their plasma (Fig. 1B).
Greatly Elevated Levels of Inflammatory Cytokines in the Pneumonic Lung.
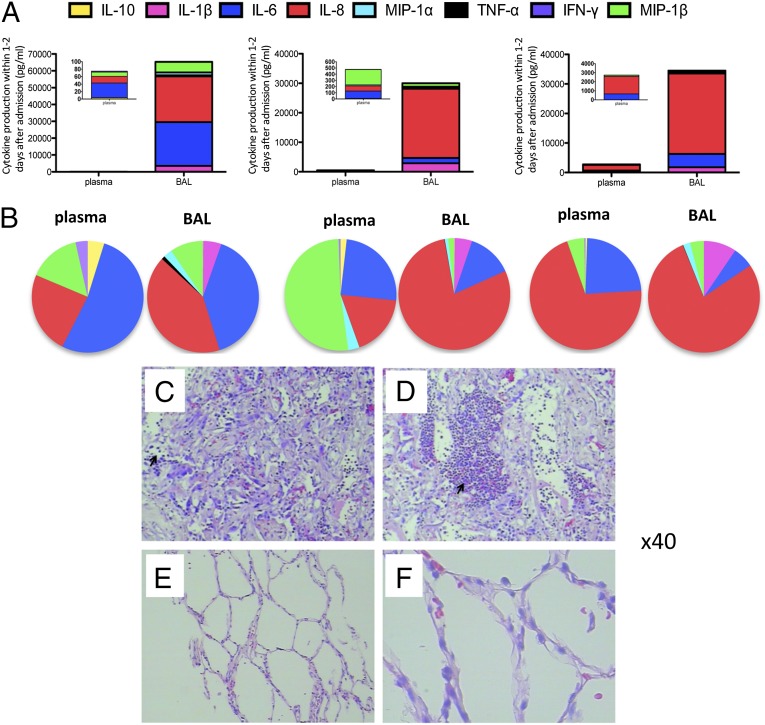
As part of the clinical assessment, we collected BAL fluids from three fatal cases. These were sampled at day 20 (a118), day 28 (a33), and day 33 (a22) after onset of illness. Plasma samples were collected at the same time points. For these individuals, levels of IL-1β, IL-6, IL-8, IL-10, TNF-α, IFN-γ, MIP-1α, and MIP-1β were substantially elevated for contemporary plasma and BAL samples, with the BAL concentrations being 100-fold (MIP-1α and MIP-1β) to 1,000-fold (IL-1β, IL-6, and IL-8) higher and TNF-α being only detected in the BAL samples (Fig. 2A and Table S2). Because we infused and collected ∼5 mL of BAL from the right lower lobe, it is likely that cytokines were diluted during the lung washing process. However, given that the plasma volume for an adult human being is ∼2.7–3.0 L, it looks likely that those mediators that are at very high relative levels in the BAL are produced locally in the infected lung and then spilled over into the blood. This interpretation fits with the fact that in two of three patients, the proportions (rather than absolute amounts) of the different cytokines present in the BAL and plasma were very similar.
Fig. 2.
Cytokine levels and lung histology in fatal cases. (A) Massively increased levels of cytokines and chemokines at the site of infection (BAL) compared with plasma, measured for three individual patients (a33, a22, and a118), as outlined in Table S2. (B) Comparison of cytokine patterns between plasma and BAL within individuals. (C–F) Diffused alveolar damage with thickening of the alveolar interstitium with extensive lymphocyte infiltration and hyaline membrane formation was observed in the lung of two H7N9-infected patients (a118 and a33). (C and D) Arrows point to cellular infiltration into the alveoli. (E and F) Normal lung samples from H7N9-negative subjects are shown, with no inflammation observed. Paraffin-embedded sections were stained with H&E. (Magnification: C–F, 40×.)
The huge differential in concentration between BAL and plasma was not observed for IFN-γ and, to some extent, IL-10, which could reflect dysfunctional antiviral immunity in patients hospitalized with severe H7N9 infection. The fact that IFN-γ is at equivalent concentrations in the BAL and plasma for two of these three cases could also indicate that IFN-γ is either being effectively consumed/used in the infected lung or that it is largely produced in the lymphoid tissue. Overall, these findings suggest that monitoring the plasma early for inflammatory cytokines/chemokines is likely to be of prognostic value. Furthermore, these data present the direct comparison of cytokine/chemokine levels in BAL and plasma in severe human influenza pneumonia.
The severity of lung pathology was confirmed histologically for two fatal cases. The profile of diffuse alveolar damage with extensive lymphocyte infiltration and hyaline membrane formation observed for these patients is consistent with the idea that secreted inflammatory mediators are involved in lesion pathogenesis (Fig. 2 C–F). Thickening of the alveolar interstitium, together with extensive inflammatory cell infiltration partitioned by small, flattened air spaces, was observed in one patient. Taken together, all these data are supportive of the idea that greatly elevated cytokine/chemokine levels are associated with the severe lung damage and airway compromise that would be expected for severe influenza pneumonia.
Association of the rs-12252-C Variant with Disease Severity.
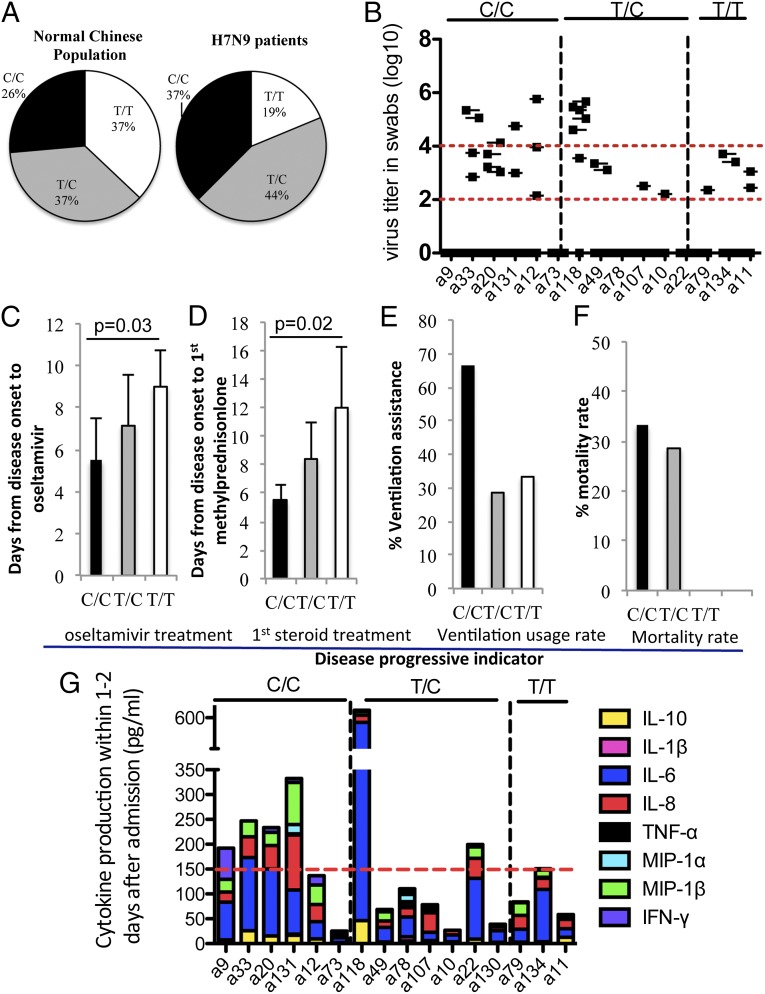
Given that the SNP rs12252-C variant was associated with clinical compromise and elevated plasma cytokine levels for the generally mild 2009 pH1N1 virus (11, 13), it seemed appropriate to probe this correlation for our 16 hospitalized H7N9 patients. Sequence analysis (Fig. 3A and Table S3) of peripheral blood mononuclear cell DNA showed that 6 (37.5%) of 16 patients had the C/C genotype, 7 (43.75%) of 16 patients had the T/C genotype, and 3 (18.75%) of 16 patients had the T/T WT genotype. The proportions of C/C and T/C individuals in our H7N9-hospitalized patients were thus selectively increased relative to prevalence rates of 26.5% (C/C), 36.5% (T/C), and 37% (T/T) in the normal (data obtained from the 1,000 genome project) healthy Chinese population (Fig. 3A). Further examination of the clinical records showed a trend in the time from disease onset to hospital admission within distinct IFITM3 groups, with the C/C cohort reporting a mean of 6 d before seeking hospital admission, compared with 7.7 d for the T/C patients and 8.3 d for the T/T patients. Evidence of more rapid disease progression (Fig. 3D and Table S3) was also reflected in clinical decisions, leading to significantly reduced times to first dosing with methylprednisolone (average of 5.5 d for C/C patients and average of 12 d for T/T patients; P = 0.03), with one of three T/T cases considered not to require this steroid treatment. This pattern was also apparent for the first oseltamivir treatment, with the intervals being 5.5 d for the C/C patients and significantly increased to 9 d for the T/T patients (Fig. 3C; P = 0.02). A similar trend was observed in mortality rates. Fatal outcomes were recorded for 33.3% of the H7N9-infected C/C individuals, 28% of the T/C group, and none of the WT T/T cases (Fig. 3F). Apart from the increased trend in lethality, mechanical ventilation [or extracorporeal membrane oxygenation respiratory assistance (ECMO)] was required in four (66.7%) of six C/C genotype patients, two (28%) of seven T/C genotype patients, and one (33.3%) of three T/T genotype patients (Fig. 3E). In view of the small number of patients with the WT T/T phenotype (n = 3), the reported trends also reflect a bias in IFITM3 genotype distribution.
Fig. 3.
Rs-12252-C association with disease severity in patients hospitalized with a H7N9 influenza virus. Due to the T→C nucleotide mutation at the SNP12252 position, human IFITM3 can have three homoforms (C/C with N-terminal 21 amino acids truncated IFITM3, T/T with WT IFITM3, and F1 T/C). (A) Comparison of allele frequencies in the normal Chinese population (from the 1,000 genomes project) and in patients with H7N9 virus from the SHAPHC. Also shown are association of the rs-12252-C group with higher influenza viral loads (quantified by real-time RT-PCR on days 8–13 after disease onset) (B), shorter time (days) from disease onset to oseltamivir treatment (C), shorter time to first methylprednisolone treatment (D), greater use of extracorporeal membrane oxygenation or mechanical respiratory ventilation (E), increased mortality rates (F), and elevated cytokines/chemokines in plasma (G). In B, dashed lines depict viral titres (log 10) of 2 and 4. In G, dashed line refers to the highest cytokines/chemokines level found for the T/T genotype.
Reduced Capacity of rs-12252-C/C Genotype Patients to Control H7N9 Virus Replication.
Our kinetic analysis indicated that the H7N9 influenza virus replicates actively between days 8 and 13 after clinical onset. Analysis of peak virus titers (log10 copies per milliliter, determined by real-time PCR for cDNA copies from throat swabs in viral transport medium) showed that the C/C genotype patients were less able to control H7N9 virus production, with four of six showing titers >104, whereas that was true for only one of five T/C genotype patients and none of the T/T genotype group (Fig. 3B). Considering that the H7N9 influenza A virus is thought to replicate 10- to 100-fold more in the human lung than in the trachea (8), the difference between the mean value for C/C (103.9) and T/T (103.0) genotypes in the trachea may be even more striking for the deeper regions of the respiratory tract. All patients were treated with oseltamivir, so it seems reasonable to assume that this was not a confounding variable.
Increased Serum Levels of IL-6, IL-8, and MIP-1β in the C/C Genotype Patients.
Plasma levels of IL-6, IL-8, and MIP-1β were generally higher in the C/C group vs. the T/C and T/T groups (Fig. 3G), as suggested from the earlier pH1N1 study. Our data showed that cytokine/chemokine levels were greatly elevated in the C/C genotype patients compared with the T/C and T/T genotype patients (Fig. 3G). Although all the T/T cohort had total cytokine levels <150 pg/mL, four of six of the C/C cases had total cytokine levels >150 pg/mL (Fig. 3G). Overall, our study provides an association of the IFITM3 C/C genotype with disease severity and higher cytokine and chemokine levels in response to H7N9 influenza virus infection.
Discussion
The newly identified H7N9 influenza A virus has caused a number of fatalities (2–5, 7). Although infected children usually suffer only a mild disease course (8), elderly subjects can be much more severely affected (2–5), indicating that there is little (if any) cross-protection from earlier influenza episodes and that the decline in primary adaptive immunity with age is a determining factor. Similar to what has been observed for the H5N1 pathogen (14), our findings indicate that high levels of plasma inflammatory cytokines/chemokines early in the course of the H7N9 infection are correlated with severe clinical outcomes. As reported previously for H7N9 infection (5, 8), IL-6 and IL-8 are of particular interest, with the concentrations being even higher than those recorded earlier for H5N1-infected patients (8). In addition to those earlier findings, the patient pool size and kinetic intensity of the present sampling/analysis provide unique insights into the correlation between hypercytokinemia and disease severity in patients with H7N9 influenza virus infection.
Whether these greatly elevated cytokine/chemokine levels offer an explanation for why the H7N9 virus causes such severe disease, or are simply a correlate of inflammation and pathology, cannot be determined from this type of analysis, but this kinetic study does suggest an approach, based on early monitoring of plasma cytokines, that could inform clinical decision making. Why these particular cytokines are prominent, as distinct from TNF-α, which shares a key signaling pathway with IL-6 and IL-8, is not clear, although we have, of course, no information on relative rates of consumption, utilization, and/or degradation. The analysis also suggests, but by no means proves, that much of the plasma cytokine/chemokine pool is derived by diffusion from the inflamed and infected respiratory tract. The concentrations of inflammatory cytokines in the BAL were as much as 103-fold higher than those found in plasma. It is likely of value to regard the plasma as a “window” for IL-6 and IL-8 levels in the damaged lung, an insight that has only been possible because we had the opportunity to analyze several BAL samples from severely compromised individuals. Further studies are needed to compare viral titers and cytokine levels in patients with milder influenza infections or severe seasonal influenza disease.
The respiratory tract represents an independent mucosal compartment, with innate and adaptive immune profiles that can be distinct from those occurring at other organs or sites (15, 16). Because most influenza A viruses multiply mainly in respiratory epithelium, it is logical to think that the predominant innate and adaptive immune responses reflect cellular recruitment to the lung and, as part of the effort both to eliminate the pathogen and to maintain the integrity of the organ, the local production of a spectrum of cytokines and chemokines (17, 18). That interpretation indeed fits with the unique, albeit limited, BAL data. Of the prominent mediators found, IL-8 is known to promote the infiltration of neutrophils (19), whereas MIP-1α and MIP-1β attract monocytes and macrophages (17, 18, 20). In addition, little IL-10 was found in the BAL from three severe cases, which may be reflective of unrestrained inflammation, because the up-regulation of IL-10 is generally thought to counterbalance, and prevent excessive immune activation. Furthermore, the absence of IFN-γ from the BAL focuses attention away from the roles that natural killer cells and T cells, which tend to produce this cytokine in mice infected experimentally with the H1N1 and H3N2 influenza A viruses, may play in this infection. Whether IL-10 and IFN-γ could be of value for treatment might be tested first in animal models.
The continued recruitment of immune cells into the lung correlates with the progressive deterioration of oxygen exchange and exacerbated hypoxia, leading finally to ARDS. Because ARDS is the major cause for death in H5N1, severe acute respiratory syndrome, and H7N9 infection, it is clearly a priority to work out how we might attenuate such extreme functional compromise without delaying virus clearance. We need to know more, and it is possible that expanding a spectrum of anti-inflammatory treatment could be of benefit. Thus, the administration of immunomodulators or giving IFN-α via aerosol or mechanical ventilation might be considered. Because adaptive immunity develops mainly in the secondary lymphoid tissues, any topical application of anti-inflammatories via the respiratory tract may be less deleterious than if larger amounts are given systemically.
The rs12252-C allele is found at an incidence of 25.8% in the Beijing Chinese population, 27% in the southern Chinese population, and 38.2% in the Japanese population (data from the 1,000 genome project). Due to the high risk shown for pH1N1 (11, 13) and the prevalence of rs12252-C in these East Asian cohorts, it was obviously important to study the role of this specific SNP marker in H7N9 influenza infection. As reported above, we found that compromised IFITM3 function is broadly correlated with disease severity, indicating that early identification of this defect may be beneficial for appropriate patient management. What the correlation between high chemokine/cytokine levels and the C/C phenotype further suggests is that we need to develop a better benefit/risk understanding of the role of these inflammatory mediators in respiratory disease. Furthermore, due to the association of rs12252-C with the severity of both low pathogenic pH1N1 (13) and high pathogenic H7N9 (our study) infections, and because of the high frequency of the C/C genotype in the Chinese population, it appears important that the vaccination programs in East Asia recommend that individuals with a C/C genotype be preferentially targeted for routine influenza immunization. However, a thorough assessment of the relative risk of vaccination, as well as its protective efficacy in the C/C group, needs to be determined first.
In summary, our data demonstrate that inflammatory immune responses linked to the C/C IFITM3 genotype during H7N9 infection play an important role in the pathogenesis of the influenza disease. Early screening for the IFITM3 genotype and monitoring of inflammatory cytokines in plasma might help to evaluate the severity of disease, and thereby to apply proper treatment and management to prevent influenza-associated deaths.
Methods
Subjects and Samples.
A total of 18 patients with H7H9 infection confirmed by real-time PCR were admitted to the Shanghai Public Health Clinical Center (SHAPHC). Plasma was collected every 2–3 d, and BAL samples were obtained from three patients. The clinical information included patient demographics plus daily monitoring of treatment, disease progression, and/or recovery. Viral titers were determined by real-time PCR for cDNA copies from throat swabs as log10 copies per milliliter of viral transport medium. Written informed consent was obtained from all participants. The overall study was reviewed and approved by the SHAPHC Ethics Committee.
Cytometric Bead Array Analyses of Cytokines and Chemokines.
Cytokines and chemokines in plasma and BAL fluid were quantified using a human inflammatory cytokine kit or a human soluble protein flex-set system (BD Biosciences). Briefly, 30 μL of plasma or BAL was serially diluted and mixed with various cytokine and chemokine capture beads, followed by PE detection beads, and incubated in the dark at room temperature for 3 h. Following acquisition on a BD FACSAria II (BD Biosciences), data were analyzed by CBA analysis software (BD Biosciences).
Sequencing and Genotyping of rs12252.
Genomic DNA was extracted from PMBCs using the QIAamp DNA Blood Mini Kit (Qiagen), and the exon 1 region of IFITM3 containing rs12252 was amplified by PCR. The amplification was performed using the following forward and reverse primers: 5′-GGAAACTGTTGAGAAACCGAA-3′ and 5′-CATACGCACCTTCACGGAGT-3′ (Shanghai Generay Biotech). The PCR products were purified and then sequenced on an Applied Biosystems 3730xl DNA Analyzer (GATC Biotech). SNPs were identified using Chromas (ABI).
Statistical Analyses.
A parametric independent t test was used to calculate P values for descriptive variables based on the assumptions of normal distribution and homogeneity of variance, and a nonparametric Mann–Whitney test was used to calculate P values for variables not meeting the assumptions of normal distribution and homogeneity. All statistical tests were considered significant at P < 0.05, and data were analyzed using SPSS software (IBM; version 17.0) and Prism 5 (GraphPad).
Supplementary Material
Acknowledgments
This work was supported by the National Grand Program on Key Infectious Disease Control (Grants 2012ZX10001-006 and 2013ZX10001-002), China Ministry of Health; the 973 National Key Basic Research Project (Grant 2012CB519005), Ministry of Science and Technology of the People’s Republic of China; and Australian National Health and Medical Research Council (NHMRC) Program Grant APP567122 (to P.C.D.). Z.W. is an NHMRC China-Australia Exchange Fellow, and K.K. is an NHMRC Career Development Fellowship Level 2 Fellow.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321748111/-/DCSupplemental.
References
- 1.Wen Y, Klenk H-D. H7N9 avian influenza virus—Search and re-search. Emerging Microbes Infect. 2013;2:e18. doi: 10.1038/emi.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao R, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368(20):1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 3.Li Q, et al. Preliminary Report: Epidemiology of the Avian Influenza A (H7N9) Outbreak in China. N Engl J Med. 2013 10.1056/NEJMoa1304617. [Google Scholar]
- 4.Yang F, et al. A fatal case caused by novel H7N9 avian influenza A virus in China. Emerging Microbes and Infections. 2013 doi: 10.1038/emi.2013.22. 10.1038/emi.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: Clinical analysis and characterisation of viral genome. Lancet. 2013;381(9881):1916–1925. doi: 10.1016/S0140-6736(13)60903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao HN, et al. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med. 2013;368(24):2277–2285. doi: 10.1056/NEJMoa1305584. [DOI] [PubMed] [Google Scholar]
- 7.Yu H, et al. Human infection with avian influenza A H7N9 virus: An assessment of clinical severity. Lancet. 2013;382(9887):138–145. doi: 10.1016/S0140-6736(13)61207-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou J, et al. Biological features of novel avian influenza A (H7N9) virus. Nature. 2013;499(7459):500–503. doi: 10.1038/nature12379. [DOI] [PubMed] [Google Scholar]
- 9.Brass AL, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139(7):1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feeley EM, et al. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7(10):e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everitt AR, et al. GenISIS Investigators MOSAIC Investigators IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484(7395):519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia R, et al. The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J Virol. 2012;86(24):13697–13707. doi: 10.1128/JVI.01828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang YH, et al. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat Commun. 2013;4:1418. doi: 10.1038/ncomms2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Jong MD, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12(10):1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohlmeier JE, Woodland DL. Immunity to respiratory viruses. Annu Rev Immunol. 2009;27:61–82. doi: 10.1146/annurev.immunol.021908.132625. [DOI] [PubMed] [Google Scholar]
- 16.Tamura S, Kurata T. Defense mechanisms against influenza virus infection in the respiratory tract mucosa. Jpn J Infect Dis. 2004;57(6):236–247. [PubMed] [Google Scholar]
- 17.Maines TR, et al. Pathogenesis of emerging avian influenza viruses in mammals and the host innate immune response. Immunol Rev. 2008;225:68–84. doi: 10.1111/j.1600-065X.2008.00690.x. [DOI] [PubMed] [Google Scholar]
- 18.Fritz RS, et al. Nasal cytokine and chemokine responses in experimental influenza A virus infection: Results of a placebo-controlled trial of intravenous zanamivir treatment. J Infect Dis. 1999;180(3):586–593. doi: 10.1086/314938. [DOI] [PubMed] [Google Scholar]
- 19.Baggiolini M, Walz A, Kunkel SL. Neutrophil-activating peptide-1/interleukin 8, a novel cytokine that activates neutrophils. J Clin Invest. 1989;84(4):1045–1049. doi: 10.1172/JCI114265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Driscoll KE. Macrophage inflammatory proteins: Biology and role in pulmonary inflammation. Exp Lung Res. 1994;20(6):473–490. doi: 10.3109/01902149409031733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.