Significance
The orphan nuclear receptor small heterodimer partner (SHP) serves as a central regulator of bile acid and cholesterol homeostasis via its transcriptional repression activity. Yet little is known about SHP structure and its mechanism of corepressor recruitment. In this paper, we present the crystal structure of SHP in complex with the transcriptional repressor E1A-like inhibitor of differentiation. Our structural and biochemical studies reveal an unexpected cofactor-binding site on SHP, representing a mode of binding that differs from the conventional understanding of how nuclear receptors recruit transcription cofactors. Disruption of this binding site affects SHP repressor function. Furthermore, the SHP crystal structure provides a rational template for drug design to treat metabolic diseases arising from bile acid and cholesterol imbalances.
Abstract
Small heterodimer partner (SHP) is an orphan nuclear receptor that functions as a transcriptional repressor to regulate bile acid and cholesterol homeostasis. Although the precise mechanism whereby SHP represses transcription is not known, E1A-like inhibitor of differentiation (EID1) was isolated as a SHP-interacting protein and implicated in SHP repression. Here we present the crystal structure of SHP in complex with EID1, which reveals an unexpected EID1-binding site on SHP. Unlike the classical cofactor-binding site near the C-terminal helix H12, the EID1-binding site is located at the N terminus of the receptor, where EID1 mimics helix H1 of the nuclear receptor ligand-binding domain. The residues composing the SHP–EID1 interface are highly conserved. Their mutation diminishes SHP–EID1 interactions and affects SHP repressor activity. Together, these results provide important structural insights into SHP cofactor recruitment and repressor function and reveal a conserved protein interface that is likely to have broad implications for transcriptional repression by orphan nuclear receptors.
Nuclear receptors compose a family of ligand-regulated transcription factors that govern a wide array of cellular activities, including cell proliferation, differentiation, metabolism, and death (1). The nuclear receptor family also contains many orphan members for which no ligand is known (1). Although some of these orphan nuclear receptors function as transcriptional activators, others serve primarily as transcriptional repressors (2). The molecular basis for ligand-regulated transcription by nuclear receptors has been intensely investigated over the past three decades, and structural studies have revealed that ligand-activated receptors use the C-terminal activation function-2 (AF-2) helix (also called helix H12) of the ligand-binding domain (LBD) to recruit coactivators that contain LXXLL motifs (3–9). Antagonist binding destabilizes the AF-2 helix from the canonical LBD fold, thus opening up an extended groove for interactions with the longer LXXXLXXX (L/I) motifs present in nuclear receptor corepressors such as NCoR and SMRT (10). Although these studies have provided a clear molecular basis for how ligands regulate AF-2 helix-dependent transcriptional activity, the molecular basis for ligand-independent repression by orphan nuclear receptors remains poorly understood.
Small heterodimer partner (SHP) is an orphan nuclear receptor that functions primarily as a transcriptional repressor (11). It is regarded as an atypical nuclear receptor because of its lack of a DNA-binding domain (DBD) (12). Instead of binding directly to target genes, SHP heterodimerizes with many DNA-bound nuclear receptors, such as LRH-1 and HNF4α, and inhibits their transcriptional activity (13–15). A two-step mechanism has been proposed for SHP-mediated repression (16, 17). In the first step, SHP uses an internal LXXLL motif to bind to the AF-2 site of its target nuclear receptors, the same site that is required for the binding of coactivators (18). The LXXLL motif of SHP has a relatively high binding affinity for a number of nuclear receptors (18), and SHP binding to the AF-2 site of these nuclear receptors causes the release of coactivators, thus decreasing gene activation. In the second step, SHP actively recruits corepressors to inhibit gene transcription. Identification of SHP-interacting corepressors has been a subject of intense study in recent years (19–25). As a result, a number of proteins have been isolated that bind to SHP in in vitro or in vivo assays, one of which is E1A-like inhibitor of differentiation (EID1) (24).
EID1 was first cloned as an interacting protein for the retinoblastoma protein, which regulates the cell cycle and differentiation (26, 27). EID1 inhibited skeletal muscle cell differentiation by repressing muscle-specific gene expression. This was due to its ability to bind to p300/CBP coactivators and inhibit their histone acetyltransferase (HAT) activity (27). Later, EID1 was found to also interact with SHP (24). Because of its intrinsic transcriptional repression activity, EID1 has been suggested to be a SHP corepressor in several studies (24, 28–30).
Although the detailed molecular basis underlying SHP’s inhibitory function is still under intense investigation, the physiological roles of SHP in regulating bile acid and cholesterol homeostasis in liver are well characterized (13, 14, 31, 32). SHP-mediated repression is a key component of a feedback loop that represses the expression of genes that encode several key hydroxylase enzymes involved in bile acid biosynthesis, such as CYP7A1 and CYP8B1, as well as SHP itself (13, 14). Accordingly, CYP7A1 expression and bile acid synthesis are increased in SHP knockout mice (31, 32). Interestingly, the repressor activity of SHP is itself tightly regulated. SHP inhibition of CYP7A1 in liver is regulated by fibroblast growth factor 15 (FGF15), which is a hormone secreted from the small intestine (33). SHP-mediated repression is abolished in mice lacking FGF15, leading to abnormally high levels of CYP7A1 expression and fecal bile acid excretion (33). A number of retinoid-like compounds have been shown to bind to SHP and enhance its repression of CYP7A1 and CYP8B1 in liver cells (34–36). These findings underscore the importance of understanding the functional and structural basis for SHP’s inhibitory function, which could serve as a drug target for treating metabolic diseases arising from bile acid and cholesterol imbalances.
In this paper, we present a 2.8-Å resolution crystal structure of SHP in complex with a peptide derived from EID1. Unexpectedly, the EID1 peptide is bound to a interface on SHP and mimics the canonical helix H1 of nuclear receptor LBDs. Mutations in this interface affected SHP–EID1 interactions and decreased SHP repressor activity. Furthermore, the SHP–EID1 interface is highly conserved across species, suggesting that their interaction is biologically relevant. Together, our results reveal an interface that regulates the SHP–EID1 interaction and point to a potential molecular mechanism by which SHP recruits corepressors to exert its inhibitory function.
Results
EID1 Peptide Facilitates SHP Crystallization and Diffraction.
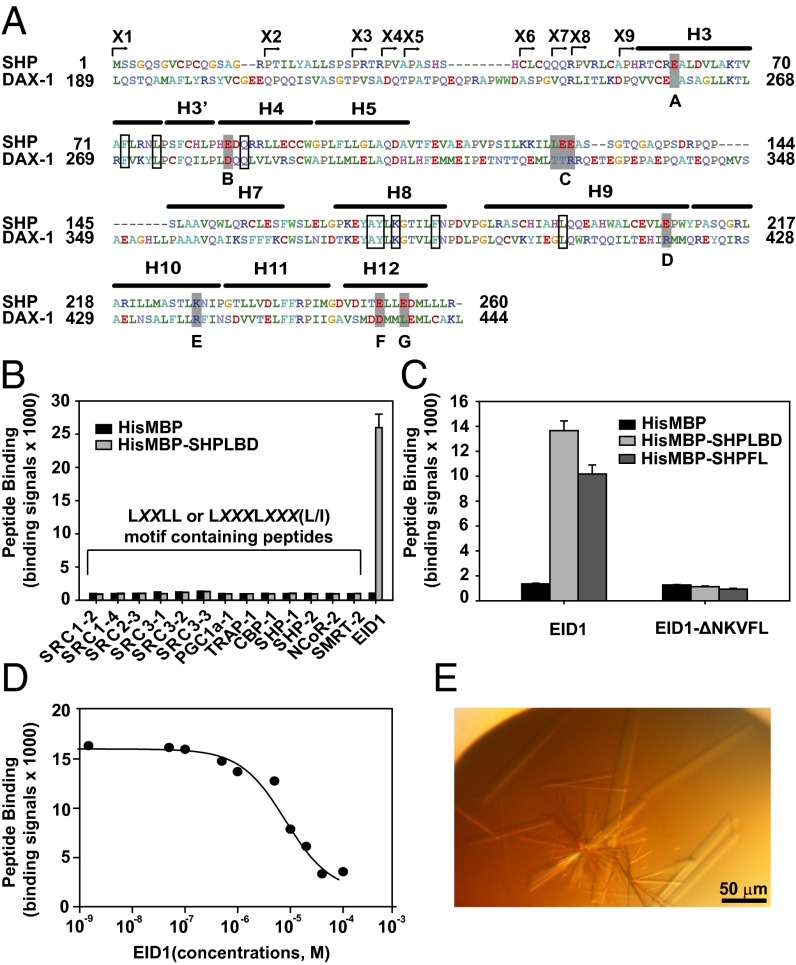
SHP has been a difficult protein for structural studies due to its inherent insolubility when purified. Although initial strategies to express and purify SHP (including insect cell expression, mutations, coexpression with SHP cofactors, and GST fusion) were unsuccessful, ultimately we succeeded using a maltose binding protein (MBP) fusion strategy, which we and others have used extensively to improve protein solubility and crystallization (37, 38). Because the N-terminal region of SHP is not conserved across species (Fig. S1), we generated a series of mouse MBP-SHP proteins that contain SHP N-terminal truncations (named X1–X9 in Fig. 1A and Fig. S1). Interestingly, when SHP putative helices H1 and H2 were removed, MBP-SHP fusion proteins (X6–X9) became highly soluble, with a yield of 10–15 mg per liter of culture, which made crystallization possible (HisMBP-SHPLBD = X6 as an example in Fig. S2A).
Fig. 1.
EID1 peptide facilitates SHP crystallization and diffraction. (A) Amino acid sequence alignment of mouse SHP (NP_035980) and DAX-1 (NP_031456). A series of MBP-SHP N-terminal truncation constructs were made (designated by arrowheads and named X1–X9). Constructs X6–X9 were soluble and tested in crystallization trials. X9 yielded the initial crystals that were further improved by surface mutagenesis. Based on the DAX-1 structure (PDB ID code 3F5C), seven sites (marked A–G) predicted to be on the SHP surface were mutated to the corresponding amino acids in DAX-1 (highlighted in gray) individually or in combination. MBP-SHPBCE (the combination of sites B, C, and E) yielded crystals for final data collection. Residues that mediate the SHP–EID1 interaction (see below) are also conserved in DAX-1 and boxed. Numbers refer to the amino acid positions in the proteins. The canonical LBD helices H1 and H2 are absent in SHP and DAX-1. (B) SHP does not interact with previously described nuclear receptor cofactor peptides. Binding of biotinylated LXXLL or LXXXLXXX (L/I) motif-containing peptides to the SHP LBD (amino acids 46–260) fused to HisMBP was determined in an AlphaScreen assay. The EID1 peptide (C) was used as a positive control. Error bars = SD (n = 3). (C) SHP binds a biotinylated EID1 peptide (biotin-YSGAMHRVSAALEEANKVFLRT) derived from the EID1 middle domain (amino acids 54–120). The binding is lost for a peptide that lacks the NKVFL sequence. SHPLBD (amino acids 46–260) and SHPFL (amino acids 1–260) fused to HisMBP were used in the AlphaScreen assay. Error bars = SD (n = 3). (D) Competitive binding of the EID1 interaction peptide to SHPLBD in the AlphaScreen assay. Biotinylated EID1 peptide (200 nM) from C was incubated with the SHPLBD (amino acids 46–260) HisMBP fusion protein and increasing concentrations of nonbiotinylated EID1 peptide (IC50 = 5–10 µM). Error bars = SD (n = 3). (E) Representative picture of MBP-SHPBCE/EID1 crystals that diffracted up to 2.8 Å.
One of the MBP-SHP constructs (X9, Fig. 1A and Fig. S1) yielded needle-like crystals in initial crystallization trials but not its longer counterparts (X6–X8). Extensive optimization failed to improve their quality. In an effort to overcome this issue, we replaced several surface residues of SHP with the corresponding residues of DAX-1, a closely related orphan receptor for which the crystal structure has been determined (39). This is the same general strategy that we used previously to crystallize the nematode nuclear receptor DAF-12 (8). Based on the sequence alignment of SHP and DAX-1 (PDB ID code 3F5C; sequence identity to SHP = 36%) (39), we mutated, either singly or in combination, a number of residues with flexible side chains that were predicted to be solvent-accessible to the corresponding DAX-1 residues that have less flexible side chains (Fig. 1A). We then tested these mutant SHP proteins in crystallization and functional assays (Figs. S2B and S3). One mutant protein with amino acid changes at positions B, C, and E (Fig. 1A), named MBP-SHPBCE, yielded bigger crystals that diffracted to 6–7 Å (Fig. S2B). These mutations did not affect the ability of SHP to repress LRH-1 activation or EID1 (Fig. S3). Repeated attempts to improve the diffraction limits of these crystals failed.
Inclusion of short peptides containing coactivator LXXLL motifs or corepressor LXXXLXXX(L/I) motifs has facilitated the crystallization and diffraction of a number of nuclear receptor LBDs (4, 8, 18). We thus examined the interaction of HisMBP-SHPLBD (X6, Fig. S2A) with a number of LXXLL- or LXXXLXXX(L/I)-containing peptides (refer to sequences in Table S1), but none of them interacted with SHP (Fig. 1B). It was reported previously that EID1, a conserved transcriptional repressor that inhibits p300/CBP coactivators, was able to interact with SHP via its middle domain (amino acids 54–120) (24). Accordingly, a number of biotinylated peptides that cover the mouse EID1 middle region were designed and tested in SHP-binding assays (Fig. 1 C and D). One EID1 peptide encompassing the sequence RVSAALEEANKVFL bound to SHP (IC50 ∼5–10 µM in a homologous competition assay), whereas the same peptide in which the NKVFL sequence was deleted (EID1-ΔNKVF) did not (Fig. 1C). We also tested the HisMBP fused full length SHP protein (SHPFL). The binding results in Fig. 1C indicated that the deleted putative SHP helices H1 and H2 did not affect EID1 binding by comparing SHPLBD and SHPFL. Identification of the EID1 peptide is key to SHP crystallization because, when the peptide was included during crystallization, diffraction of MBP-SHPBCE (refer to above and Fig. 1A) crystals was improved to 2.8 Å, which allowed us to determine its structure (Fig. 1E).
Crystal Structure of the SHP–EID1 Complex.
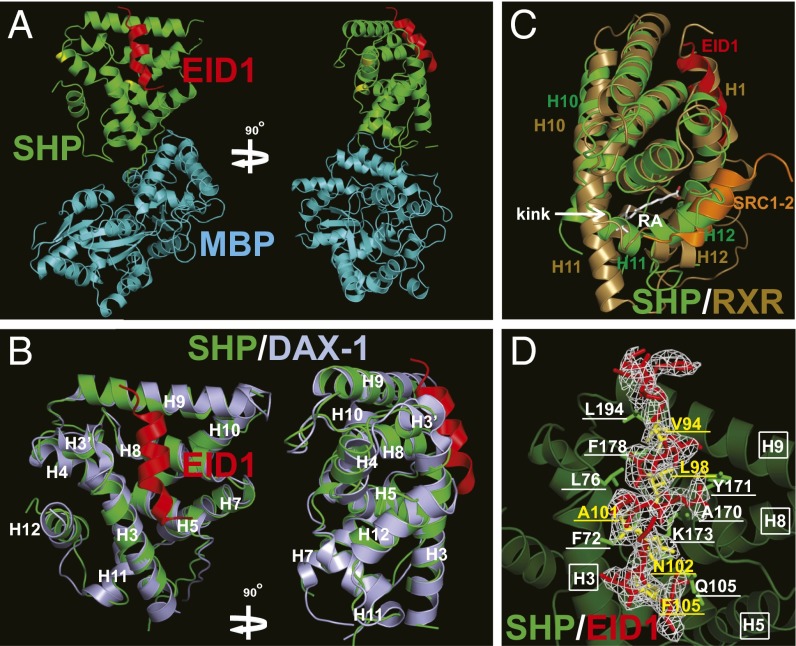
The SHP–EID1 structure was solved by molecular replacement using MBP as a model (see statistics in Table 1), which revealed that each noncrystallographic asymmetric unit contained one complex (Fig. 2A). The overall architecture of SHP is similar to that of DAX-1 (Fig. 2B). SHP helix H12 occupies its own AF-2 site by mimicking coactivator LXXLL motifs (Fig. 2C). A clear kink is observed between helices H10 and H11, which results in the collapse of helix H11 into the space that corresponds to the ligand-binding pocket of ligand-regulated receptors (compare Fig. 2 B and C), leaving no room for ligand binding. Thus, the SHP structure is in an auto-repressed and ligand-free conformation, resembling those of several other orphan nuclear receptors, including DAX-1, COUP-TFII, TR4, and PNR (Fig. 2B and Fig. S4) (38–41).
Table 1.
Data collection and refinement statistics for MBP-SHP/EID1
| Data collection | |
| Space group | P212121 |
| Resolution, Å | 30–2.8 |
| Cell parameters a, b, c, Å | 56.39, 105.15, 136.28 |
| β, ° | 90 |
| Total reflections | 205,691 |
| Unique reflections | 20,723 |
| Rsym | 0.176 (0.754) |
| I/σ | 12.8 (2.2) |
| Completeness, % | 100 |
| Redundancy | 9.9 (8.2) |
| Structure determination and refinement | |
| Resolution, Å | 30–2.8 |
| No. of reflections | 17,181 |
| No. of residues | 605 |
| No. of solvent molecules | 49 |
| No. of non-H atoms | 4,493 |
| Rwork, % | 19.46 |
| Rfree, % | 24.12 |
| rmsd bonds, Å | 0.009 |
| rmsd angles, ° | 1.07 |
| Average B factor, Å2 | 15.1 |
Values for the highest-resolution shell (2.9–2.8Å) are given in parentheses.
Fig. 2.
Structural analysis of SHP/EID1. (A) A ribbon model of MBP (cyan)-SHP (green) in complex with EID1 (red). The surface mutations are highlighted in yellow. (B) Superposition of SHP (green) onto DAX-1 (light blue, PDB ID code 3F5C). EID1 is shown in red. Helices H3–H12 are labeled in white. (C) Superposition of the SHP (green) and ligand-bound RXR (gold, PDB ID code 1FM6) structures. Compared with RXR helices H10 and H11, there is a kink between SHP helices H10 and H11, which causes the space hindrance that occludes the ligand-binding pocket of SHP. RA, cis-retinoic acid in white, is the ligand for RXR. EID1 (red) overlaps with RXR helix H1. The RXR-interacting peptide SRC1-2 (orange) overlaps with SHP helix H12. (D) Representative Fo-Fc electron density omit map contoured at 1.0 σ for EID1 (red). The EID1 amino acids involved in SHP binding are highlighted in yellow and underlined. EID1-binding amino acids in SHP are labeled in white and underlined; they are contributed by helices H3, H5, H8, and H9, which are labeled in white and boxed.
The most unexpected feature of the SHP–EID1 structure is the binding mode of EID1. Instead of binding to the canonical AF-2 site of SHP, EID1 is bound to SHP by mimicking helix H1 of the nuclear receptor LBD, which is illustrated by superposition of SHP to the ligand-bound RXR LBD (Fig. 2C). We termed the SHP EID1-binding pocket as the helix H1 pocket to distinguish it from the classic AF-2/H12 pocket. The SHP/EID1-binding site consists of residues from SHP helices H3, H5, H8, and H9 that form a hydrophobic patch to adopt the EID1 helix (Fig. 2D and Fig. S5A). The SHP residues that interact with EID1 include six hydrophobic residues (F72, L76, A170, Y171, F178, and L194) and two hydrophilic residues (Q105 and K173), which form hydrogen bonds with EID1. The EID1 residues that interact with SHP include four hydrophobic residues (V94, L98, A101, and F105) and one hydrophilic residue (N102), which hydrogen-bond with SHP (Fig. 2D and Fig. S5A).The SHP and EID1 residues that mediate the SHP–EID1 interaction are conserved across species (Figs. S1 and S5B), suggesting that their interaction is biologically relevant. Interestingly, although the SHP residues are also conserved in DAX-1 (Fig. 1A), and although SHP shares a similar 3D structure with DAX-1, EID1 did not bind to DAX-1 (Fig. S6A), indicating that the SHP–EID1 interface is receptor-specific.
Analysis of the SHP–EID1 Interaction.
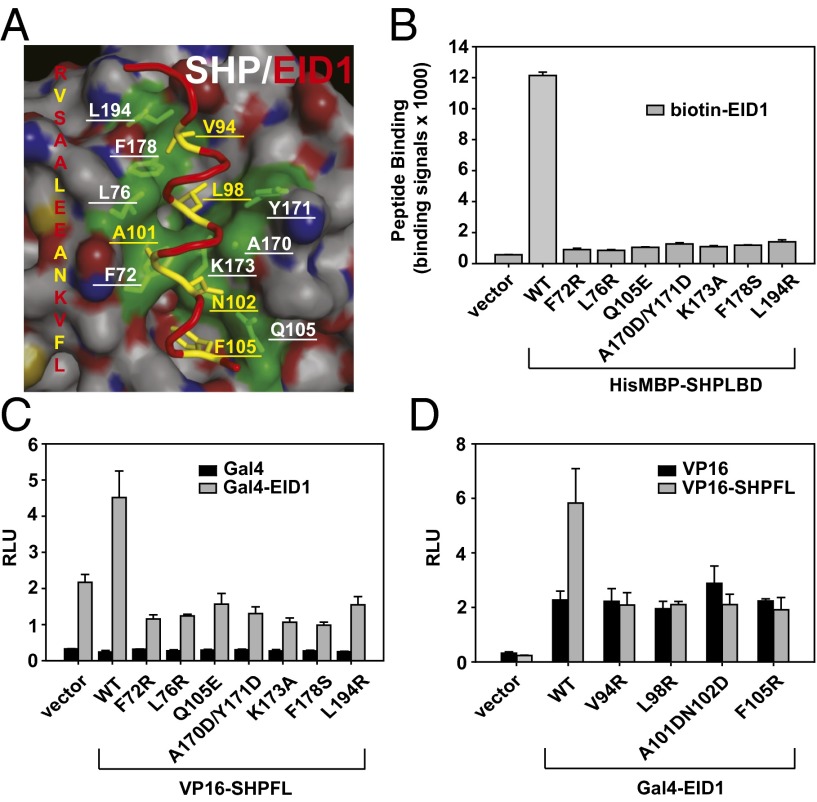
To determine the role of SHP interface residues in EID1 binding, we mutated individual EID1-contacting residues in SHP (Fig. 3A) and measured the effects on SHP–EID1 interactions in AlphaScreen binding assays. As shown in Fig. S6B, most of these mutated proteins (except for SHP F178S) retained their ability to interact with LRH-1, indicating that they retained their functional folds. In contrast, all mutations severely affected SHP binding to the EID1 peptide, consistent with a critical role for these residues in EID1 recognition (Fig. 3B). The results were further confirmed in mammalian two-hybrid assays (Fig. 3 C and D). Mutations of EID1-contacting residues in the full-length SHP (Fig. 3C) or SHP-contacting residues in EID1 (Fig. 3D) abolished SHP–EID1 interactions.
Fig. 3.
Analysis of the SHP–EID1 interaction. (A) Close-up presentation of the SHP–EID1 interface. SHP amino acids involved in the interaction are shown by a green transparent surface and labeled in white. EID1 amino acids involved in the interaction are marked in yellow and underlined. (B) In vitro binding assays to examine the interaction of SHP mutant proteins with EID1 peptide. SHPLBD (amino acids 46–260) was fused to HisMBP and used in assays. Error bars = SD (n = 3). (C) Mammalian two-hybrid assays to analyze the interaction of SHP mutant proteins with EID1. SHPFL (amino acids 1–260) wild-type (WT) and mutant proteins were expressed as VP16 activation domain (VP16) fusion proteins and mouse EID1 (amino acids 54–120) as Gal4-DNA–binding domain (Gal4) fusion proteins. RLU, relative light unit, which is normalized with Renilla luciferase as transfection control. Error bars = SD (n = 3). (D) Mammalian two-hybrid assays to examine the interaction of EID1 mutants with SHPFL. Error bars = SD (n = 3).
Functional Analysis of the SHP–EID1 Interface.
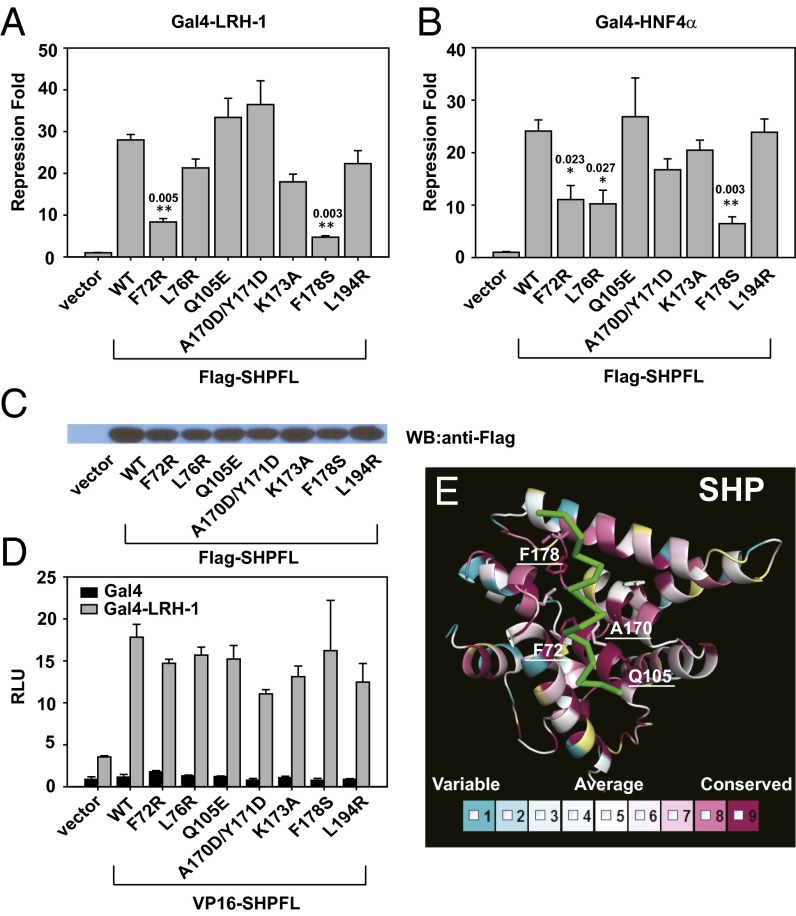
Next, we investigated the importance of the integrity of the SHP–EID1 interface in SHP repression. SHP inhibits LRH-1 and HNF4α transcriptional activity (13–15). Hence we used Gal4-LRH-1 and Gal4-HNF4α to study repressor activity of SHP wild type (WT) and mutants. Mutations of F72, F178, and L76 decreased SHP function significantly on LRH-1 and HNF4α (Fig. 4 A and B). These mutations did not change SHP expression level or its ability to bind the receptor (Fig. 4 C and D), suggesting that the mutations impaired SHP’s ability to recruit corepressors. Finally, we examined the conservation of the SHP–EID1 interface by including SHP sequences from additional species that are available in the protein databank (32 species in total; refer to Table S2). As illustrated in the protein conservation map (Fig. 4E), the amino acids that form the EID1-binding pocket are highly conserved, particularly F72, Q105, A170, and F178.
Fig. 4.
Functional analysis of the SHP–EID1 interface. (A) Gal4-LRH-1 was used in reporter assays to examine repressor activity of SHP mutants. Repression fold was calculated by comparing RLU with SHP to RLU without SHP. The results were then analyzed using Student’s independent-sample t test. The statistical significance level was set to be P < 0.05 (one asterisk) or P < 0.01 (two asterisks). The numbers above asterisks indicate P values. Error bars = SD (n = 3). (B) Gal4-HNF4α was used in reporter assays to examine repressor activity of SHP mutants. Error bars = SD (n = 3). (C) Western blot was performed to check expression level of SHP mutants in cells. The same amount of cell lysates was loaded. (D) Mammalian two-hybrid assays to examine the interaction of LRH-1 with SHPFL WT and mutants. Error bars = SD (n = 3). (E) SHP conservation heat map compiled from 32 species (Table S2) using the program ConSurf (http://consurftest.tau.ac.il/). The heat map color code bar (Lower) displays nine equally spaced bins of relative sequence conservation. Residues shown in white (bin 5) represent the average level of conservation across the SHP LBD, whereas bin 1 (blue) represents the residues with the lowest relative conservation and bin 9 (purple) represents the residues with the highest relative conservation. The EID1 helix is shown in green and the SHP residues surrounding EID are shown in stick presentation.
Discussion
SHP is an atypical nuclear receptor in that it lacks a DBD but is able to interact directly with other nuclear receptors and to repress their transcriptional activity. These characteristics make SHP an intriguing target for structural studies (11). However, determining the structure of SHP has proven difficult due to solubility issues. In this study, we overcame these difficulties by using MBP fusion and domain-mapping strategies, which allowed us to obtain large amounts of soluble SHP protein for crystallization and biochemical analysis. Through a combination of N-terminal deletions, surface mutations, and inclusion of an EID1 peptide, we were able to crystallize the SHP–EID1 complex and solve its structure. The structure reveals an unexpected SHP–EID1 interface, where EID1 adopts a helical structure that mimics helix H1 in the canonical LBD fold. Mutations in the SHP–EID1 interface disrupt EID1 binding to SHP. Together, these results provide crucial mechanistic insights into SHP-mediated cofactor recruitment and repression (24, 28, 30).
SHP functions predominantly as a transcriptional repressor as exemplified by its physiological role in interacting with LRH-1 and HNF4α to inhibit CYP7A1 expression and bile acid synthesis. A number of proteins have been isolated as SHP corepressors, including histone deacetylases (SIRT1, HDAC1, and HDAC3) (25, 28) and EID1, which inhibits p300/CBP HAT activity (24, 26, 27). However, the mechanism whereby these corepressors interact with SHP has been poorly understood. An important clue to unraveling this mechanism comes from our sequence analysis and crystallization experiments indicating that SHP does not have canonical H1 and H2 helices. First, the N-terminal amino acids corresponding to H1 and H2 are not conserved across species and contain many prolines that disfavor the formation of helices. Second, repeated attempts to crystallize full-length SHP-containing putative H1 and H2 sequences have failed. Third, our cocrystal structure of SHP and the EID1 peptide reveals that SHP lacks helix H1 of the canonical nuclear receptor LBD fold. Instead, the EID1 peptide mimics helix H1 to become an integral part of the SHP LBD fold. While the physiological role of EID1 in SHP repression is still under investigation, the integrity of the EID1–SHP interface appears critical to SHP repressor activity based on our studies.
It is of interest to note that not all of the mutations in SHP that affected EID1’s interaction (Fig. 3 C and D) resulted in loss of repression (Fig. 4 A and B). A likely explanation for this difference is that SHP recruits other corepressors through the helix H12 site that act independently of EID1. Indeed, previous studies have shown SHP helix H12 is crucial for its repressor activity (24), and EID1 is known to form a multiple-component complex with other corepressors to regulate SHP’s inhibitory activity (20, 22, 23). For this reason, mutation of the SHP–EID1 interface residues F72, L76, and F178 might be expected to compromise both EID1 binding and repression activity, whereas mutations of other SHP–EID1 interface residues may still retain SHP repressor activity because they permit the assembly of other non-EID1–dependent corepressors through the H12 helix. Another possible explanation is that SHP may recruit a different set of corepressors through the same interface that requires F72, L76, and F178 as key residues mediating these interactions. Validation of these potential interactions will require future experimentation.
The molecular basis for SHP–EID1 interactions is very different from that for interactions between traditional ligand-regulated receptors and corepressors such as NCoR and SMRT. In the case of ligand-regulated receptors, antagonist binding displaces the C-terminal helix H12 from the canonical LBD fold, thus opening the AF-2 pocket for the binding of the LXXXLXXX (L/I) motifs present in NCoR and SMRT (10). In the case of SHP, its AF-2 site is occupied by its own helix H12. Instead, SHP uses a helix H1 pocket to accommodate EID1, which lacks the classical corepressor LXXXLXXX (L/I) motif. Several other orphan nuclear receptors such as DAX-1, TR4, TLX, and PNR function primarily as transcription repressors to regulate diverse physiological programs (2, 42, 43). These orphan nuclear receptors directly recruit receptor-specific corepressors instead of common corepressors such as SMRT and NCoR (17, 24, 42–45). Although the mechanisms of cofactor recruitment by these orphan nuclear receptors are unclear, the known corepressors for these receptors do not contain LXXLL or LXXXLXXX (L/I) motifs. Based on these observations, we speculate that the H1 helix pocket may be the alternative to the AF-2 site for the recruitment of corepressors by these specialized orphan receptors. Consistent with this notion, repressor orphan nuclear receptors DAX-1, TR4, and PNR also lack an H1 helix (Fig. 2B and Fig. S4 B and C) (38, 39, 41), thus providing an interaction surface for recruitment of their corresponding corepressors. It is worth noting that for ligand-regulated receptors, the isolated H1 helix can be assembled in trans with the rest of the LBD in a ligand-dependent manner, and further, ligand binding is enhanced by the inclusion of helix H1 in the canonical LBD (46).
It is noteworthy that our SHP structure is a ligand-free structure with helix H10 collapsed into the ligand-binding pocket, much like the apo-RXR structure (47, 48). Interestingly, synthetic retinoid-like compounds such as 3-Cl-AHPC have been shown to bind and regulate SHP activity (34–36). We speculate that binding of these pharmacophores might rearrange the kinked helix H10, thus allowing formation of the AF-2 cofactor-binding pocket. Consistent with this hypothesis, 3-Cl-AHPC can promote SHP interaction with LXXLL-containing peptides (Fig. S7A). In contrast, EID1 binding to SHP is independent of the presence of 3-Cl-AHPC (Fig. S7B). This suggests that SHP has two cofactor-binding sites, including one that is ligand-dependent (and thus potentially druggable) via the C-terminal AF-2 site and the other that is ligand-independent via the EID1-binding site near the helix H1 pocket. Thus, our SHP–EID1 structure complex provides an explanation for SHP cofactor recruitment and repressor function, and it reveals a protein interface that regulates nuclear receptor functions.
Materials and Methods
Plasmids and Reagents.
The mutants used for crystallization, AlphaScreen assays, or cotransfection assays were created by site-directed mutagenesis using the QuikChange method (Stratagene) and verified by sequencing.
Protein Preparation and Crystallization.
For biochemical assays, mouse SHP (amino acids 46–260) was cloned into an engineered pETDuet-1 (Novagen) plasmid, in which HisMBP was introduced in front of its original multiple cloning sites. HisMBP-SHPFL (amino acids 1–260; the last 10 cysteines were mutated to serines for solubility) and HisMBP-DAX-1LBD (NP_031456, amino acids 205–472) were cloned in the same way. Biotinylated MBP-LRH1 (NP_995582, amino acids 299–541) was constructed as described elsewhere (49). For crystallization trials, SHP (amino acids 55–260) was cloned into the MBP vector described previously (37). The proteins were expressed in BL21 (DE3) cells, first purified by amylose-resin chromatography (Biolabs) and then followed by size-exclusion chromatography.
The MBP-SHP crystals were grown at 20 °C in sitting drops containing 1.0 μL of the protein solution (10 mg/mL) and 1.0 μL of the well solution containing 0.1 M Tris/Cl, pH 8.5, 25% (wt/vol) PEG1000. The MBP-SHP/EID1 crystals were grown at 20 °C in sitting drops containing 1.0 μL of the protein solution (10 mg/mL) and 1.0 μL of the well solution containing 0.1 M 2-(cyclohexylamino ethanesulfonic acid), pH 9.5, 30% (wt/vol) PEG3000. The molecular ratio of MBP-SHP to the EID1 peptide (YSGAMHRVSAALEEANKVFLRTARAGDALDG) was 1:1.5. In general, crystals appeared within 2 d and grew to the final size in about 1 wk, at which time they were flash-frozen and stored in liquid nitrogen.
Data Collection, Structure Determination, Refinement, Superposition, and Conservation Heat Map.
The diffraction data were collected with a MAR225 CCD detector at the 21-ID beamline at the Advanced Photon Source at Argonne National Laboratory (Argonne, IL). The observed reflections were reduced, merged, and scaled with DENZO and SCALEPACK in the HKL2000 package. The structure was determined with the PHASER program by molecular replacement using the crystal structures of MBP and DAX-1 as the models. Manual model building was carried out with the programs O (50) and COOT (51), and the structure was refined with crystallography NMR software (52) and the CCP4 program refmac5 (53). The conservation heat map was compiled from 32 SHP sequences using the program ConSurf (http://consurftest.tau.ac.il) (54).The data collection and structure determination statistics are summarized in Table 1.
AlphaScreen Binding Assays.
The binding of the cofactor peptides to SHP was determined by AlphaScreen luminescence proximity assays as described (8, 18, 41). Reaction mixtures consisted of 50 nM HisMBP fusion proteins, 200 nM biotinylated peptides/proteins, 10 μg/mL nickel chelate-coated acceptor beads (PerkinElmer Life Sciences), and 10 μg/mL streptavidin-coated donor beads (PerkinElmer Life Sciences) in a buffer containing 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS), pH 7.4, 50 mM NaF, 50 mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), and 0.1 mg/mL BSA. The peptides used in our studies are listed in Table S1.
Cell Reporter Assay and Statistics Analysis.
AD293 cells were cultured and cotransfected in 24-well plates as reported (41). Mouse SHPFL (amino acids 1–260) WT and mutants were cloned into the p3xFLAG-CMV expression vector (Sigma, E4401). A GSAGSA linker was added in front of the SHP gene. VP16-SHPFL was constructed by fusing mouse SHP (amino acids 1–260) to the VP16 vector. Gal4-EID1 was constructed by fusing mouse EID1 middle domain (amino acids 54–120) to the Gal4 vector. Gal4-LRH-1 and HNF4α were constructed as described elsewhere (15). Statistical analysis was performed using Excel (38). Comparisons were performed using Student’s independent-sample t test (two-tailed distribution and two-sample equal variance). The statistical significance level was set at P < 0.05 or P < 0.01.
Supplementary Material
Acknowledgments
We thank Z. Wawrzak and J. S. Brunzelle for assistance in data collection at the beamlines of sector 21 (Life Sciences Collaborative Access Team), which is funded in part by the Michigan Economic Development Corporation and Michigan Technology Tri-Corridor Grant 085P1000817. Use of the Advanced Photon Source was supported by the Office of Science of the US Department of Energy. This work was supported by the Jay and Betty Van Andel Foundation; Ministry of Science and Technology (China) Grants 2012ZX09301001-005 and 2012CB910403; Amway (China); National Institutes of Health Grants R01 DK071662 (to H.E.X.), R01 DK067158 (to D.J.M. and S.A.K.), and T32 DK007307 (to C.Z.); The Robert A. Welch Foundation (Grants I-1275 to D.J.M. and I-1558 to S.A.K.); and the Howard Hughes Medical Institute (D.J.M.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4NUF).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322827111/-/DCSupplemental.
References
- 1.Mangelsdorf DJ, et al. The nuclear receptor superfamily: The second decade. Cell. 1995;83(6):835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, Dufau ML. Gene silencing by nuclear orphan receptors. Vitam Horm. 2004;68:1–48. doi: 10.1016/S0083-6729(04)68001-0. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, et al. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol Cell. 2005;17(4):491–502. doi: 10.1016/j.molcel.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Krylova IN, et al. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120(3):343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 5.Gampe RT, Jr, et al. Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol Cell. 2000;5(3):545–555. doi: 10.1016/s1097-2765(00)80448-7. [DOI] [PubMed] [Google Scholar]
- 6.Jin L, et al. Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma. Mol Endocrinol. 2010;24(5):923–929. doi: 10.1210/me.2009-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rha GB, Wu G, Shoelson SE, Chi YI. Multiple binding modes between HNF4alpha and the LXXLL motifs of PGC-1alpha lead to full activation. J Biol Chem. 2009;284(50):35165–35176. doi: 10.1074/jbc.M109.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhi X, et al. Structural conservation of ligand binding reveals a bile acid-like signaling pathway in nematodes. J Biol Chem. 2012;287(7):4894–4903. doi: 10.1074/jbc.M111.315242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Lambert MH, Xu HE. Activation of nuclear receptors: A perspective from structural genomics. Structure. 2003;11(7):741–746. doi: 10.1016/s0969-2126(03)00133-3. [DOI] [PubMed] [Google Scholar]
- 10.Xu HE, et al. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature. 2002;415(6873):813–817. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- 11.Båvner A, Sanyal S, Gustafsson JA, Treuter E. Transcriptional corepression by SHP: Molecular mechanisms and physiological consequences. Trends Endocrinol Metab. 2005;16(10):478–488. doi: 10.1016/j.tem.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Seol W, Choi HS, Moore DD. An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science. 1996;272(5266):1336–1339. doi: 10.1126/science.272.5266.1336. [DOI] [PubMed] [Google Scholar]
- 13.Goodwin B, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6(3):517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 14.Lu TT, et al. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6(3):507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 15.Kir S, Zhang Y, Gerard RD, Kliewer SA, Mangelsdorf DJ. Nuclear receptors HNF4α and LRH-1 cooperate in regulating Cyp7a1 in vivo. J Biol Chem. 2012;287(49):41334–41341. doi: 10.1074/jbc.M112.421834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YK, Moore DD. Dual mechanisms for repression of the monomeric orphan receptor liver receptor homologous protein-1 by the orphan small heterodimer partner. J Biol Chem. 2002;277(4):2463–2467. doi: 10.1074/jbc.M105161200. [DOI] [PubMed] [Google Scholar]
- 17.Seol W, Chung M, Moore DD. Novel receptor interaction and repression domains in the orphan receptor SHP. Mol Cell Biol. 1997;17(12):7126–7131. doi: 10.1128/mcb.17.12.7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, et al. Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner. Proc Natl Acad Sci USA. 2005;102(27):9505–9510. doi: 10.1073/pnas.0501204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie YB, et al. SMILE, a new orphan nuclear receptor SHP-interacting protein, regulates SHP-repressed estrogen receptor transactivation. Biochem J. 2008;416(3):463–473. doi: 10.1042/BJ20080782. [DOI] [PubMed] [Google Scholar]
- 20.Sanyal S, et al. Involvement of corepressor complex subunit GPS2 in transcriptional pathways governing human bile acid biosynthesis. Proc Natl Acad Sci USA. 2007;104(40):15665–15670. doi: 10.1073/pnas.0706736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boulias K, Talianidis I. Functional role of G9a-induced histone methylation in small heterodimer partner-mediated transcriptional repression. Nucleic Acids Res. 2004;32(20):6096–6103. doi: 10.1093/nar/gkh947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang S, et al. Coordinated recruitment of histone methyltransferase G9a and other chromatin-modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. Mol Cell Biol. 2007;27(4):1407–1424. doi: 10.1128/MCB.00944-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kemper JK, Kim H, Miao J, Bhalla S, Bae Y. Role of an mSin3A-Swi/Snf chromatin remodeling complex in the feedback repression of bile acid biosynthesis by SHP. Mol Cell Biol. 2004;24(17):7707–7719. doi: 10.1128/MCB.24.17.7707-7719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Båvner A, Johansson L, Toresson G, Gustafsson JA, Treuter E. A transcriptional inhibitor targeted by the atypical orphan nuclear receptor SHP. EMBO Rep. 2002;3(5):478–484. doi: 10.1093/embo-reports/kvf087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chanda D, Xie YB, Choi HS. Transcriptional corepressor SHP recruits SIRT1 histone deacetylase to inhibit LRH-1 transactivation. Nucleic Acids Res. 2010;38(14):4607–4619. doi: 10.1093/nar/gkq227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyake S, et al. Cells degrade a novel inhibitor of differentiation with E1A-like properties upon exiting the cell cycle. Mol Cell Biol. 2000;20(23):8889–8902. doi: 10.1128/mcb.20.23.8889-8902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacLellan WR, Xiao G, Abdellatif M, Schneider MD. A novel Rb- and p300-binding protein inhibits transactivation by MyoD. Mol Cell Biol. 2000;20(23):8903–8915. doi: 10.1128/mcb.20.23.8903-8915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou T, et al. Novel polymorphisms of nuclear receptor SHP associated with functional and structural changes. J Biol Chem. 2010;285(32):24871–24881. doi: 10.1074/jbc.M110.133280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macchiarulo A, Rizzo G, Costantino G, Fiorucci S, Pellicciari R. Unveiling hidden features of orphan nuclear receptors: The case of the small heterodimer partner (SHP) J Mol Graph Model. 2006;24(5):362–372. doi: 10.1016/j.jmgm.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 30.Park YY, et al. Differential role of the loop region between helices H6 and H7 within the orphan nuclear receptors small heterodimer partner and DAX-1. Mol Endocrinol. 2004;18(5):1082–1095. doi: 10.1210/me.2003-0339. [DOI] [PubMed] [Google Scholar]
- 31.Kerr TA, et al. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev Cell. 2002;2(6):713–720. doi: 10.1016/s1534-5807(02)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, et al. Redundant pathways for negative feedback regulation of bile acid production. Dev Cell. 2002;2(6):721–731. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 33.Inagaki T, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2(4):217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Farhana L, et al. Adamantyl-substituted retinoid-related molecules bind small heterodimer partner and modulate the Sin3A repressor. Cancer Res. 2007;67(1):318–325. doi: 10.1158/0008-5472.CAN-06-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miao J, et al. Ligand-dependent regulation of the activity of the orphan nuclear receptor, small heterodimer partner (SHP), in the repression of bile acid biosynthetic CYP7A1 and CYP8B1 genes. Mol Endocrinol. 2011;25(7):1159–1169. doi: 10.1210/me.2011-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dawson MI, et al. An adamantyl-substituted retinoid-derived molecule that inhibits cancer cell growth and angiogenesis by inducing apoptosis and binds to small heterodimer partner nuclear receptor: Effects of modifying its carboxylate group on apoptosis, proliferation, and protein-tyrosine phosphatase activity. J Med Chem. 2007;50(11):2622–2639. doi: 10.1021/jm0613323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pioszak AA, Xu HE. Molecular recognition of parathyroid hormone by its G protein-coupled receptor. Proc Natl Acad Sci USA. 2008;105(13):5034–5039. doi: 10.1073/pnas.0801027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan MH, et al. The crystal structure of the orphan nuclear receptor NR2E3/PNR ligand binding domain reveals a dimeric auto-repressed conformation. PLoS ONE. 2013;8(9):e74359. doi: 10.1371/journal.pone.0074359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sablin EP, et al. The structure of corepressor Dax-1 bound to its target nuclear receptor LRH-1. Proc Natl Acad Sci USA. 2008;105(47):18390–18395. doi: 10.1073/pnas.0808936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kruse SW, et al. Identification of COUP-TFII orphan nuclear receptor as a retinoic acid-activated receptor. PLoS Biol. 2008;6(9):e227. doi: 10.1371/journal.pbio.0060227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou XE, et al. The orphan nuclear receptor TR4 is a vitamin A-activated nuclear receptor. J Biol Chem. 2011;286(4):2877–2885. doi: 10.1074/jbc.M110.168740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang CL, Zou Y, Yu RT, Gage FH, Evans RM. Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes Dev. 2006;20(10):1308–1320. doi: 10.1101/gad.1413606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takezawa S, et al. A cell cycle-dependent co-repressor mediates photoreceptor cell-specific nuclear receptor function. EMBO J. 2007;26(3):764–774. doi: 10.1038/sj.emboj.7601548. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Wang L, Rajan H, Pitman JL, McKeown M, Tsai CC. Histone deacetylase-associating Atrophin proteins are nuclear receptor corepressors. Genes Dev. 2006;20(5):525–530. doi: 10.1101/gad.1393506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altincicek B, et al. Interaction of the corepressor Alien with DAX-1 is abrogated by mutations of DAX-1 involved in adrenal hypoplasia congenita. J Biol Chem. 2000;275(11):7662–7667. doi: 10.1074/jbc.275.11.7662. [DOI] [PubMed] [Google Scholar]
- 46.Pissios P, Tzameli I, Kushner P, Moore DD. Dynamic stabilization of nuclear receptor ligand binding domains by hormone or corepressor binding. Mol Cell. 2000;6(2):245–253. doi: 10.1016/s1097-2765(00)00026-5. [DOI] [PubMed] [Google Scholar]
- 47.Gampe RT, Jr, et al. Structural basis for autorepression of retinoid X receptor by tetramer formation and the AF-2 helix. Genes Dev. 2000;14(17):2229–2241. doi: 10.1101/gad.802300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375(6530):377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 49.Melcher K, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462(7273):602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 51. Emsley P, Cowtan K (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1):2126–2132. [DOI] [PubMed]
- 52.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 53.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 1):247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 54. Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N (2010) ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38(Web Server issue):W529–W533. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.