Abstract
STUDY QUESTION
What is the burden of X chromosome mosaicism in the occurrence of spontaneous menarche (SM) in Turner syndrome (TS)?
SUMMARY ANSWER
SM was significantly associated with X chromosome mosaicism in the TS patients; a mosaicism with around 10% euploid cell line may predict spontaneous pubertal development when determined by molecular-cytogenetic techniques on uncultivated tissues.
WHAT IS KNOWN ALREADY
Spontaneous puberty can be observed in a minority of patients with TS, more frequently, but not exclusively, in those with a high level of 46,XX/45,X mosaicism at standard karyotype. The genetic mechanisms contributing to ovarian function in TS patients are still not determined. However, submicroscopic X-linked and autosomal copy number variations (CNVs) have recently emerged as an important genetic risk category for premature ovarian insufficiency and may be involved in modulating the TS ovarian phenotype.
STUDY DESIGN, SIZE, DURATION
A group of 40 patients with a diagnosis of TS at conventional karyotyping participated in the study; 6 patients had SM and 34 patients had primary amenorrhoea (PA). All clinical data and the patients’ DNA samples were collected over the years at a single paediatric clinic.
PARTICIPANTS/MATERIALS, SETTING, METHODS
The patients' samples were used to perform both genetic (Copy Number Assay) and molecular-cytogenetic (array-CGH and iFISH, interphase-FISH) analyses in order to evaluate the X chromosome mosaicism rate and to detect possible rare CNVs of genes with a known or predicted role in female fertility.
MAIN RESULTS AND THE ROLE OF CHANCE
All TS patients showed variable percentages of the 46,XX lineage, but these percentages were higher in the SM group (P < 0.01). A mosaicism around 10% for the euploid cell line may predict spontaneous pubertal development when determined by molecular-cytogenetic techniques performed in uncultivated tissues. A few CNVs involving autosomal and X-linked ovary-related loci were identified by array-CGH analysis and confirmed by real-time quantitative PCR, including a BMP15 gene duplication at Xp11.22, a deletion interrupting the PAPPA gene at 9q33.1, and an intragenic duplication involving the PDE8A gene at 15q25.3.
LIMITATIONS, REASONS FOR CAUTION
This is a pilot study on a relatively small sample size and confirmation in larger TS cohorts may be required. The ovarian tissue could not be studied in any patients and in a subgroup of patients, the mosaicism was estimated in tissues of different embryonic origin.
WIDER IMPLICATIONS OF THE FINDINGS
The combined determination of X chromosome mosaicism by molecular and molecular-cytogenetic techniques may become useful for the prediction of SM in TS. The detection of CNVs in both X-linked and autosomal ovary-related genes further suggests gene dosage as a relevant mechanism contributing to the ovarian phenotype of TS patients. These CNVs may pinpoint novel candidates relevant to female fertility and generate further insights into the mechanisms contributing to ovarian function.
STUDY FUNDING/COMPETING INTEREST(S)
This study was funded by Telethon Foundation (grant no: GGP09126 to L.P.), the Italian Ministry of the University and Research (grant number: 2006065999 to P.F.) and a Ministry of Health grant ‘Ricerca Corrente’ to IRCCS Istituto Auxologico Italiano (grant number: 08C704-2006). The authors have no conflict of interest to declare.
Keywords: Turner syndrome, ovary, BMP15, X chromosome, copy number variations
Introduction
Turner syndrome (TS) represents one of the most common chromosomal anomalies, with a prevalence of about 1:2500-1:3000 live female births (Sybert and McCauley, 2004). Affected individuals have a variable spectrum of physical and functional alterations likely due to a wide variety of karyotype abnormalities, ranging from 45,X monosomy (50% of cases) to various forms of 45,X/46,XX mosaicism with or without structural defects of the X chromosome in the euploid cell line (Saenger, 1996; Sybert and McCauley, 2004). Among the clinical features of TS, short stature and ovarian defects are present in almost all cases. Ovarian dysfunction in TS women is likely caused by rapid oocyte-loss in the early stages of the meiotic prophase after the 18th week of fetal life, later resulting in ovarian dysgenesis and streak ovaries and partial or complete absence of pubertal development. Consequently, infertility is one of the main problems for TS women (Reynaud et al., 2004). Studies of the X chromosome breakpoints have revealed the existence of two distinct loci on the Xq (Xq13-q21 and Xq23-q27) and one region on the Xp22.1-p11.2 that are significantly associated with the ovarian phenotype (Toniolo, 2006; Persani et al., 2009). Different mechanisms to explain the variable phenotypic manifestations of TS include the haploinsufficiency of X-linked genes that escape X chromosome inactivation, such as SHOX for short stature (Zinn and Ross, 1998; Sybert and McCauley, 2004), and meiotic-chromosome pairing errors (Ogata and Matsuo, 1995). Nevertheless, spontaneous puberty has been observed in 15–20% of 45,X patients (Pasquino et al., 1997; Gracia Bouthelier et al., 2004; Martín Campagne and Roa Llamazares, 2008) and in 32% of mosaic patients, inversely correlating with the severity of chromosomal anomalies at conventional karyotype (Pasquino et al., 1997). Consistently, subjects with 45,X frequently have serum follicle-stimulating hormone (FSH) levels already in the post-menopausal range during infancy, whereas FSH levels are generally normal in patients of the same age with mosaic Turner syndrome (Fechner et al., 2006; Aso et al., 2010). Furthermore, a biochemical parameter of ovarian reserve, the antimüllerian hormone is generally low in infants with 45,X or structural abnormalities of the second X, but similar to controls in mosaic Turner patients (Kallio et al., 2012).
The mechanism supporting pubertal development in a small subset of patients with 45,X is presently unexplained, but low percentage mosaicism, undetected at standard karyotyping, might be a possible explanation. Additionally, submicroscopic copy number variations (CNVs) have recently emerged as an important genetic risk category for premature ovarian insufficiency (POI), both in cases of primary and secondary amenorrhoea. Indeed, in a few cohorts of 46,XX patients diagnosed with POI and analysed by means of high throughput techniques, such as array comparative genomic hybridization (aCGH) and single-nucleotide polymorphism (SNP) array, CNVs affecting several X-linked and autosomal loci with a possible role in female fertility have been detected (Aboura et al., 2009; Dudding et al., 2010; Ledig et al., 2010; Quilter et al., 2010; Liao et al., 2011; McGuire et al., 2011). Very recently, the SNP array technology has been applied to genotype a large cohort of TS patients, demonstrating the equivalence of this method to metaphase cytogenetics for the diagnosis of TS (Prakash et al., 2013). However, the potential involvement of CNVs in the modulation of the TS ovarian phenotype has not been investigated.
In this pilot study, we performed new generation genetic (Copy Number Assay) and molecular-cytogenetic (aCGH and iFISH) investigations in a cohort of TS patients in order to better clarify the synergistic burden of X chromosome mosaicism and rare X-linked and autosomal CNVs on the observed ovarian phenotypes of these patients.
Materials and Methods
Ethics statement
The Institutional Ethic Committees of the IRCCS San Matteo Hospital and IRCCS Istituto Auxologico Italiano approved the study. All participants gave their written informed consent to the study and to the sampling of their tissues.
Patients
Forty patients with TS were recruited at the Paediatric Department of the University of Pavia, in San Matteo Hospital. All the patients had been diagnosed with TS at birth or during the early infancy on the basis of the conventional karyotyping of at least 30 metaphases from cultured peripheral blood lymphocytes.
The patients were classified according to the presence of the spontaneous menarche (SM) and to the results of the conventional karyotyping (Table I). Six of the 40 patients (15%) experienced SM with FSH and LH values at menarche within the normal range. The clinical data of SM patients are summarized in Table II (ovarian phenotype) and Supplementary data, Table SI (extra-ovarian phenotype). The mean age of the SM patients was 18.8 ± 6.7 years (median: 16.5 years). SM occurred at 11.9–14.4 years (mean: 13.1 ± 1.0 years; median: 13 years). One older patient with SM (SM1) underwent secondary amenorrhoea at 18.6 years and was then put on hormone replacement therapy (Table II). The only other TS feature shown by SM1 was short stature (Supplementary data, Table SI). The other 34 patients (>12 years of age; median 18.3 years) had hormone levels in the post-menopausal range on at least two independent determinations (FSH >30 U/l, data not shown) and were classified as primary amenorrhoea (PA).
Table I.
Distribution of the cohort of Turner syndrome patients on the basis of conventional karyotype and pubertal development.
| Conventional karyotype | TS full cohort | Complete spontaneous puberty |
|---|---|---|
| n (%) | n (%) | |
| X-monosomy | 31 (77.5) | 2 (33) |
| X-mosaicism with structural abnormalities of the second X | 3 (7.5) | 0 (0) |
| X-mosaicism without structural abnormalities of the second X | 5 (12.5) | 4 (67) |
| Structural abnormalities of the second X | 1 (2.5) | 0 (0) |
| Total | 40 (100) | 6 (100) |
Table II.
Detailed hormone dosages at menarche and next follow-up in the six patients with spontaneous pubertal development.
| Patient | Age (years; months) | Standard karyotype | Menarche (years; months) | Secondary amenorrhoea (years; months) | FSH (UI/l) | LH (UI/l) | E2 (pg/ml) | FSH (UI/l) | LH (UI/l) | E2 (pg/ml) |
|---|---|---|---|---|---|---|---|---|---|---|
| At menarche |
At last follow-up | |||||||||
| SM1a | 32; 8 | 45,X | 14; 5 | 18; 2 | 7.4 | 1.4 | 14 | 132b | 40b | 16b |
| SM2 | 18; 3 | 45,X | 12; 11 | – | 1.7 | 4.7 | 234 | 4.3 | 2.9 | 84.6 |
| SM3 | 15; 9 | 45,X/46,XX | 12; 4 | – | 4.9 | 3.2 | 76.3 | 5.8 | 2.7 | 114 |
| SM4 | 19; 9 | 45,X/46,XX | 14; 1 | – | 6.1 | 3.8 | 55.4 | 7.1 | 1.1 | 64.2 |
| SM5 | 15; 2 | 45,X/46,XX | 13; 1 | – | 4.4 | 4.8 | 61.1 | 3.2 | 6.1 | 80 |
| SM6 | 15; 1 | 45,X/46,XX | 11; 11 | – | 9.8 | 3.7 | 30 | 6 | 2.9 | 24.8 |
aAfter secondary amenorrhoea, SM1 was put on hormone replacement therapy.
bBefore replacement therapy.
E2, 17 β-estradiol; FSH, follicle-stimulating hormone; LH, luteinizing hormone.
Array-CGH analysis
All cases underwent aCGH analysis. Genomic DNA (gDNA) was extracted from the whole blood using the GenEluteTM Blood Genomic DNA kit (Sigma-Aldrich, St. Louis, MO, USA) in accordance with the manufacturer's instructions. Pooled DNA from the peripheral blood of 10 healthy donors (Promega, Southampton, UK), sex-mismatched to the samples, was used as a reference. The genome scan was performed using the Human Genome CGH Microarray Kit 244 K (Agilent Technologies, Santa Clara, CA, USA) which consists of ∼236 000 60-mer oligonucleotide probes covering the entire genome at an average spatial resolution of ∼30 kb. Briefly, 3 µg of DNA from the test and the normal reference samples were processed following the manufacturer's protocol. Dye emission was captured by means of a dual-laser scanner. The images were extracted using the Agilent Feature Extraction 9.1 software, and analysed using the DNA Analytics 4.0 software.
A log ratio plot between the test and reference gDNA was assigned so that any aberrations in the copy number of the test DNA at a particular locus were identified as deviations from a modal value of 0 by the ADM-2 algorithm. CNV classification was performed according to the Database of Genomic Variants (http://projects.tcag.ca/variation/): a CNV was defined as ‘rare’ or ‘common’ if never or previously reported in control subjects, respectively.
The X chromosome mosaic condition was detected qualitatively evaluating, through a mosaicism artificial scale, the vertical right shift of the log ratio profile from the expected value of 0 (Ratio (R): X chromosome copy number of test DNA/X chromosome copy number of reference DNA = 1) towards +1 (R = 2) or +1.58 (R = 3), the latter due to the isochromosome Xq which involves the presence of three copies of the same chromosome region rather than two. The artificial mosaicism scale was created by mixing the blood samples of one normal diploid male with one normal diploid female, as previously described (Ballif et al., 2006). The DNA of the artificial mosaicism representing 75, 50, 30 and 10% of 46,XX cell line was used in aCGH experiments, and the obtained log ratio profile shifts were compared with the aCGH results of all TS patients by overlapping every TS X chromosome array profile to the artificial scale, thus giving an estimation of the patient mosaicism level (Supplementary data, Fig. S1). The mosaicism detection limit of this technique was therefore set at 10%.
FISH analysis
The RPCI-11 BAC clone CTD-2004N6, covering the entire BMP15 gene at Xp11.22, was selected according to the UCSC Genome Browser (http://genome.cse.ucsc.edu, hg19, February 2009) and provided by Invitrogen Ltd (Paisley, UK). The probe was labelled by nick translation with Cy3 (Amersham Biosciences, Chalfont St. Giles, UK) and its physical position was verified on a few female control metaphases derived from peripheral blood lymphocytes.
Multi-colour iFISH analysis was performed in a subset of 13 TS patients (6 SM + 7 PA). For each patient, 300 nuclei derived from PHA-stimulated peripheral blood lymphocytes cultivated for 72 h were analysed by using the CEPX, CEPY, CEP18-Aneuvysion kit (Abbott-Vysis, Chicago, IL, USA), according to the manufacturer's instructions. In addition, a few metaphases were analysed to check the structure of the second X chromosome in the euploid cell line. Furthermore, in patient SM1 dual-colour FISH analysis was performed by using the BAC probe specific for BMP15 in combination with the commercially available centromeric specific probe CEP6 (locus D6Z1) (Abbott-Vysis). In both cases, the combined hybridizations allowed the G1 and G2 cells to be distinguished. In addition, in patient PA4 the presence of a possible mosaic subtelomeric Xp deletion was verified by performing multi-colour FISH analysis using the Vysis ToTelVysion Multi-Color FISH Probe Kit-Mixture 1, which includes the probes TelVysion 1p, TelVysion 1q, TelVysion Xp/Yp and CEP X (Abbott-Vysis). The FISH protocols of Lichter et al. (1990) and Lichter and Cremer (1992) were followed, with minor modifications.
For iFISH analysis on buccal mucosal cells, the subjects were asked to rinse their mouth twice with drinking water. The surface cells were then scraped from the buccal mucosa of both right and left cheek of each individual by using a cytobrush. The cells were removed from the cytobrush by agitation in ice-cold 70% ethanol. After the transport to the laboratory, the cell suspension was centrifuged at 1200 rpm for 10 min and the supernatant was discarded. The pellet was resuspended in fresh 70% ethanol and dropped across a clean slide. The slides were fixed in fixative (1:1 methanol:acetic acid) for 10 min and then air dried. The slides were checked with a phase microscope for the presence of cells with adequate morphology. Multi-colour iFISH analysis was performed on 300 buccal cells as described above, and the FISH protocol of Reddy and Mak (2001) was followed, with minor modifications.
Real-time quantitative PCR
For copy number quantification using real-time quantitative PCR, gDNAs previously extracted from whole blood were analysed for 13 TS patients of the cohort (6 SM and 7 PA). gDNAs were also extracted from buccal epithelium of a subgroup of patients (SM1, SM2, SM3 and PA4) and from vaginal epithelium of SM1, using Oragene DNA Self-Collection Kit (DNA Genotek, Kanata, Ontario, Canada). As controls, gDNAs from 46,XX females, who did not bear CNVs in the regions of interest, and from a 46,XY male were obtained. gDNAs were then used as target templates for the TaqMan Copy Number Assay (Life Technologies, Carlsbad, CA, USA), according to the manufacturer's instructions. The target TaqMan Copy Number mixes were chosen to detect: exon 8 (Hs01858886_cn) of the GYG2 gene (MIM *300198, at Xp22.3); exon 1 (Hs01299108_cn) and exon 2 (HS00957878_cn) of the BMP15 gene (MIM *300247, at Xp11.22); exon 10 (Hs05613398_cn) of the SMARCA1 gene (MIM *300012, at Xq26.1); exon 2 (Hs02373345_cn) and exon 7 (Hs01648393_cn) of the PAPPA gene (MIM *176385, at 9q33.1); intron 2 (Hs03902365) and exon 21 (Hs03005502_cn) of the PDE8A gene (MIM *602972, at 15q25.3); and exon 9 (Hs02909158_cn) of the PRKX gene (MIM *300083, at Xp22.33). The RNase P probe targeting the Ribonuclease P RNA component H1 gene (RPPH1, MIM *608513) on chromosome 14q11.2 was chosen as reference probe. Reactions were run on a 7900HT Real-Time PCR instrument (Life Technologies), SDS Software v2.3, performing the Absolute Quantitation method. Four replicates for each sample were analysed to generate reliable copy number calls. After amplification, data files containing the sample replicate CT values for each reporter dye were imported into the CopyCaller software analysis tool (Life Technologies) to calculate sample copy number values by relative quantification, using the comparative CT (ΔΔCT) method. The method measured the CT difference (ΔCT) between target and reference sequences and then compared the ΔCT values of each sample to the ΔCT of calibrator samples (normal 46,XX females), known to have two copies of the target sequence. Each assay was repeated at least twice.
Automated sequencing
The entire coding sequence and intron–exon junctions of the BMP15 gene (GenBank numbers AF082349.1 and AF082350.1) were analysed in patient SM1 by denaturing high-performance liquid chromatography (DHPLC) of PCR-amplified fragments on the WAVE apparatus automatic instrument (Transgenomics, Omaha, NE, USA), as previously described (Rossetti et al., 2009).
Statistical analysis
Fisher's exact probability test was used to determine whether the prevalence of mosaic karyotypes was significantly different between groups of patients with SM or PA as well as whether the prevalence of the identified autosomal ‘common’ CNVs was significantly different between TS patients with PA and a control group of 55 healthy women, who presented a normal spontaneous pubertal development and have successfully reproduced. A significance test assigns a P-value equal to or smaller than 5%.
Results
High-resolution a-CGH analysis detects rare copy number variants in X-linked and autosomal genes relevant to ovarian function
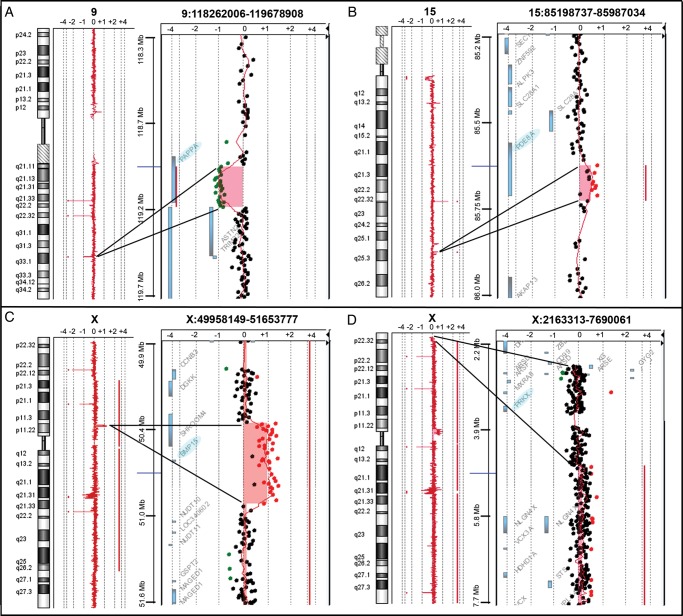
Through high-resolution aCGH analysis on 40 TS patients, one X-linked and 16 autosomic heterozygous CNVs were identified (Table III).All of these CNVs affect genes with a known or a possible role in female fertility. Of the 17 CNVs, 3 were rare and involved the PAPPA, PDE8A and BMP15 genes (Fig. 1A–C). In addition, in patient PA4, a rare mosaic deletion of at least 2 Mb (chrX:2700316-4739185, hg19) at Xp22.33-p22.32 was detected (Fig. 1D), thus revealing the presence of a structural abnormality of the second X chromosome not previously identified by conventional cytogenetic analysis. In order to investigate a possible association between the lack of pubertal development in TS patients and the identified 14 autosomal ‘common’ CNVs, we performed a case–control genetic association study comparing the frequency of the common CNVs in the PA group (34 individuals) with that found in a group of 55 healthy women with normal puberty. None of these CNVs were significantly enriched in PA TS in comparison with the control population (data not shown).
Table III.
Detailed list of the identified CNVs affecting genes potentially related to female fertility.
| CNVa | Physical positionb | Chromosomal band | Size (kb) | Genes included in the CNVc | Patient ID |
|---|---|---|---|---|---|
| Rare | |||||
| Loss | chr9:118971743-119187538 | 9q33.1 | 216 | PAPPA, ASTN2 | PA5 |
| Gain | chr15:85591615-85666309 | 15q25.3 | 75 | PDE8Ad | PA4 |
| Gain | chrX:50400649-50955142 | Xp11.22 | 554 | BMP15, SHROOM4 | SM1 |
| Common | |||||
| Gain | chr3:100347505-100419624 | 3q12.2 | 72 | GPR128e | SM3 |
| Gain | chr3:100380047-100419624 | 3q12.2 | 40 | GPR128e | PA7 |
| Gain | chr5:69711984-70314582 | 5q13.2 | 603 | SERF1A, SMN2, NAIPe | PA26 |
| Gain | chr5:69705562-70587018 | 5q13.2 | 881 | SERF1A, SMN2, NAIP, GTF2H2 | PA37 |
| Loss | chr5:69705562-70309855 | 5q13.2 | 604 | SERF1A, SMN2, NAIP | PA40 |
| Loss | chr6:266079-307998 | 6p25.3 | 42 | DUSP | SM3 |
| Loss | chr6:259528-293493 | 6p25.3 | 34 | DUSP | PA17 |
| Loss | chr6:283968-375949 | 6p25.3 | 92 | DUSP | PA31 |
| Gain | chr6:259528-317679 | 6p25.3 | 58 | DUSPe | PA33 |
| Gain | chr6:259528-293493 | 6p25.3 | 34 | DUSPe | PA34 |
| Loss | chr8:15952011-16015454 | 8p22 | 63 | MSR1 | PA41 |
| Loss | chr22:39359112-39385485 | 22q13.1 | 26 | APOBEC3A, APOBEC3B | SM5 |
| Loss | chr22:39359112-39385485 | 22q13.1 | 26 | APOBEC3A, APOBEC3B | PA18 |
| Loss | chr22:39359112-39385485 | 22q13.1 | 26 | APOBEC3A, APOBEC3B | PA38 |
aCopy number variations classification has been performed according to the Database of Genomic Variants (http://projects.tcag.ca/variation/).
bAccording to the genome assembly hg19 (UCSC Genome Browser, release February 2009, http://genome.cse.ucsc.edu, hg19).
cGenes likely to be implicated in female fertility are indicated in bold.
dIntragenic duplication likely perturbing gene expression.
ePartial gene duplications involving either 5’ or 3’ end must be further molecularly characterized to clarify the actual perturbation of gene expression.
Figure 1.
Detection of copy number variations. High-resolution aCGH analysis 244 K Agilent identified in patient PA5 a deletion of at least 216 kb at 9q33.1 (chr9:118971743-119187538, hg19), which interrupts the PAPPA gene coding region (A), in patient PA4 a duplication of at least 75 kb at 15q25.3 (chr15:85591615-85666309, hg19), which includes part of the PDE8A gene coding region (B), and in patient SM1 a duplication of at least 554 kb at Xp11.22 (chrX:50400649-50955142, hg19,), which contains the entire BMP15 gene (C). In addition, a mosaic deletion of at least 2 Mb at Xp22.33-p22.32 (chrX:2700316-4739185, hg19), which includes the entire PRKX gene, was detected in patient PA4 (D). The most important genes involved in the identified microrearrangements are highlighted in light blue.
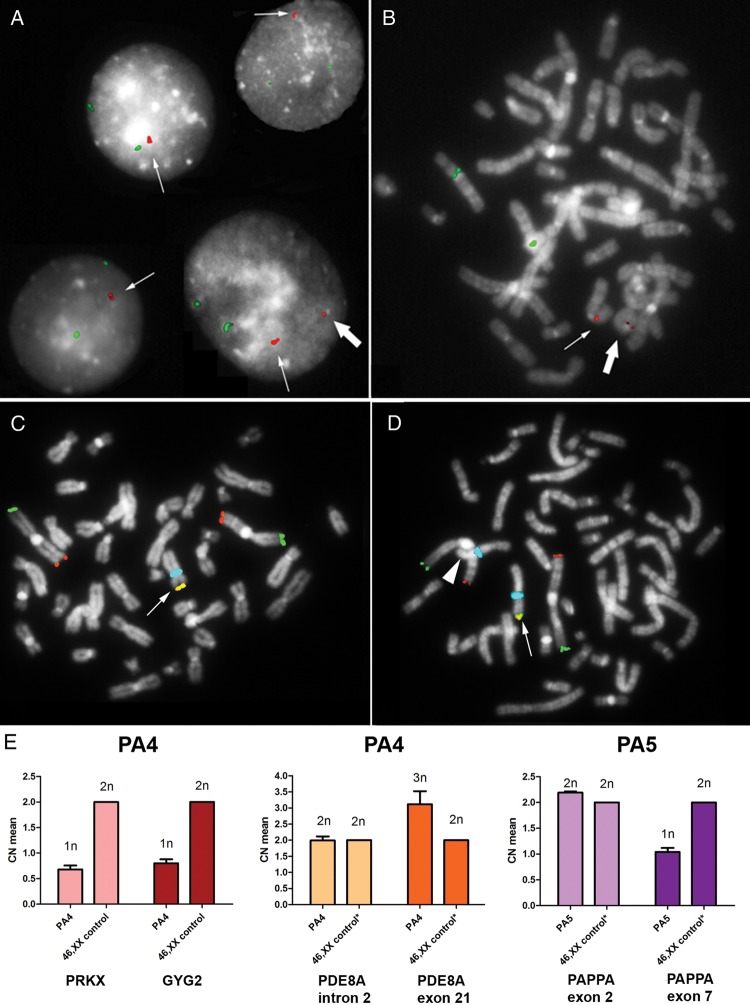
To clarify the structural characteristics of the BMP15 duplication in patient SM1 (Fig. 1C), a specific FISH analysis was performed. A duplicated signal was observed in all the analysed cells (n = 100), thus underlying the presence of a tandem duplication (Fig. 2A). Interestingly, an additional single signal was found in a few nuclei and metaphases (9%), due to the presence of an euploid cell line, which was characterized by a ring(X) chromosome in addition to the conserved one (Fig. 2A and B). A wild-type coding sequence of BMP15 was detected in SM1.
Figure 2.
Validation and structural characterization of the identified CNVs. (A) iFISH analysis performed on 100 nuclei from patient SM1 characterized the rearrangement involving BMP15 earlier identified by array-CGH analysis as a tandem duplication (thin arrow). In the 9% of the cells an additional smaller signal was identified (big arrow) which represents the low-level mosaic euploid cell line carrying a ring(X) chromosome as seen in the patient's metaphases (B). The BAC probe CTD-2004N6, specific for BMP15, is shown in red and the CEP6 probe (locus D6Z1) is in green. (C and D) Multi-colour FISH analysis on metaphases from patient PA4 identified both the 45,X (C) and the 46,XX (D) cell lineages, and confirmed the presence of a heterozygous Xp terminal deletion in the euploid cell lineage (D), as demonstrated by the loss of the expected subtelomeric Xp signal (arrowhead) compared with the conserved X chromosome (arrow). The TelVysion 1p probe is shown in green, the TelVysion 1q is in red, the TelVysion Xp/Yp is in yellow (red + green merged) and CEPX (locus DXZ1) is in blue. (E) Real-time quantitative PCR on gDNAs from patients PA4 and PA5. PA4's copy number (CN) profile for both the PRKX and GYG2 genes, located at Xp22.32, showed only one copy of both genes (predicted CN = 1n), and confirmed the heterozygous duplication of part of the PDE8A gene, located at 15q25.3, by comparing the regions within (dark orange bar, 3n) and outside the duplication (light orange bar, 2n). The CN profile of patient PN5 identified the heterozygous deletion interrupting the PAPPA gene, located at 9q33.1, by comparing the region not affected (light purple bar, 2n) with the deleted one (dark purple bar, 1n). All samples were also compared with 46,XX control (calibrator) gDNA. *The CN profile of the interested regions has been previously tested by array-CGH analysis. Calculated mean CNs are shown on the left-axis, while predicted CNs are displayed on the top of each bar. SD, standard deviation, is represented by the error bars (CN means of the calibrator have been calculated as 2n by the analysis software and therefore no SD values are shown).
The presence of a terminal deletion in the euploid cell line of patient PA4 (Fig. 1D) was confirmed by multi-colour FISH analysis, using a probe specific for the chromosome Xp subtelomeric region (Fig. 2C and D), and by real-time quantitative PCR, using specific Taqman probes for the GYG2 and PRKX genes (Fig. 2E), which revealed one copy of both genes compared with a 46,XX control. Similarly, we could also confirm the rare duplication of part of the PDE8A gene in PA4 (Figs 1B and 2E), and the rare deletion interrupting the PAPPA gene in PA5 (Figs 1A and 2E).
The mosaicism level influences the occurrence of SM in TS
On the basis of the conventional cytogenetic results, the frequency of mosaicism in our cohort was significantly higher in the group of patients with pubertal development than in those with PA (67 versus 12%, P < 0.01) (Table I). The X chromosome mosaic condition was confirmed by the aCGH analysis on peripheral blood cells using the mosaicism artificial scale (Supplementary data, Fig. S1, Table IV). We demonstrated that all SM patients, including SM1 and SM2, earlier reported as 45,X, showed an appreciable shift of the X chromosome log ratio profile compared with only 4 out of 34 PA patients (Table IV). All SM patients except SM1 were found to be carriers of 45,X/46,XX mosaics ≥10% (Table IV). Of note, in SM1 the profile shift was limited to the region Xp22.1–Xq26.2, thus clarifying the structure of the mosaic ring(X) chromosome, which was previously identified by iFISH analysis (Figs 1C, 2B and Supplementary data, Fig. S1, Table IV). Otherwise, the four cytogenetically ascertained mosaic patients of the PA group showed mosaicism levels >10%, but invariably contained abnormalities of the second X chromosome (Table IV).
Table IV.
Detailed molecular and molecular-cytogenetic results.
| Patient | quantitative real-time PCR |
aCGH | iFISHb |
Conventional karyotypingc | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
BMP15 at Xp11.22 |
GYG2 at Xp22.33 |
SMARCA1 at Xq25 |
X chromosome profile shift, % of mosaicism |
CEPX at Xp11.1-q11 |
|||||||||
| pb | bm | vm | pb | bm | vm | pb | bm | vm | pb | pblc | bm | pblc | |
| SM | |||||||||||||
| SM1a | 2n | 2n | 2n | n | n | n | n | n | n | +, <10% r(X)(p22.1q26.2) | ss[91]/ds[9] | NP* | 45,X |
| SM2 | n | n | NP* | n | n | NP* | n | n | NP* | +, 10% | ss[93.2]/ds[6.8] | ss[90.7]/ds[9.3] | 45,X |
| SM3 | n | n | NP* | n | n | NP* | n | n | NP* | +, 10% | ss[82]/ds[18] | ss[80]/ds[20] | 45,X/46,XX |
| SM4 | 2n | NP* | NP* | 2n | NP* | NP* | 2n | NP* | NP* | +, 30% | ss[26]/ds[74] | NP* | 45,X/46,XX |
| SM5 | 2n | NP* | NP* | 2n | NP* | NP* | 2n | NP* | NP* | +, 75% | ss[19]/ds[81] | NP* | 45,X/46,XX |
| SM6 | 2n | NP* | NP* | 2n | NP* | NP* | 2n | NP* | NP* | +, 75% | ss[19]/ds[81] | NP* | 45,X/46,XX |
| PA | |||||||||||||
| PA1 | n | NP* | NP* | n | NP* | NP* | n | NP* | NP* | Absent | ss[99]/ds[1] | NP* | 45,X |
| PA2 | n | NP* | NP* | n | NP* | NP* | n | NP* | NP* | Absent | ss[99.4]/ds[0.6] | NP* | 45,X |
| PA3 | n | NP* | NP* | n | NP* | NP* | n | NP* | NP* | Absent | ss[99]/ds[1] | NP* | 45,X |
| PA4 | n | 2n | NP* | n | n | NP* | n | 2n | NP* | +, 10-30% del(X)(p22.33→p22.32) | ss[63.7]/ds[36.3] | ss[36]/ds[64] | 45,X/46,XX |
| PA5 | n | NP* | NP* | n | NP* | NP* | 3n | NP* | NP* | − (Xp) + (Xq), % NA | ss[15]/ds[85] | NP* | 45,X/46,X,i(Xq) |
| PA6 | n | NP* | NP* | n | NP* | NP* | 3n | NP* | NP* | − (Xp) + (Xq), % NA | ss[23]/ds[77] | NP* | 45,X/46,X,i(Xq) |
| PA7 | n | NP* | NP* | n | NP* | NP* | n | NP* | NP* | − (Xp) + (Xq), % NA | ss[85]/ds[15] | NP | 45,X/46,X,i(Xq) |
| PA8-PA40 | NP | NP | NP | NP | NP | NP | NP | NP | NP | Absent | NP | NP | 45,X |
aThe patient has a BMP15 wild-type sequence.
bThe euploid cell line was generally estimated by FISH at a higher rate than the expected rate on the basis of the aCGH profile probably due to a selection bias in favour of the euploid cell line during cell culture.
cAt least 30 metaphases were analysed.
bm, buccal mucosa; ds, double signal; n, copy number; NA, not available due to the lack of an artificial mosaicism scale ad hoc; NP, not performed; NP*, not performed due to the lack of proper samples; PA, primary amenorrhoea; pb, peripheral blood; pblc, peripheral blood lymphocytes cultures; SM, spontaneous menarche; ss, single signal; vm, vaginal mucosa; +, present; −, absent.
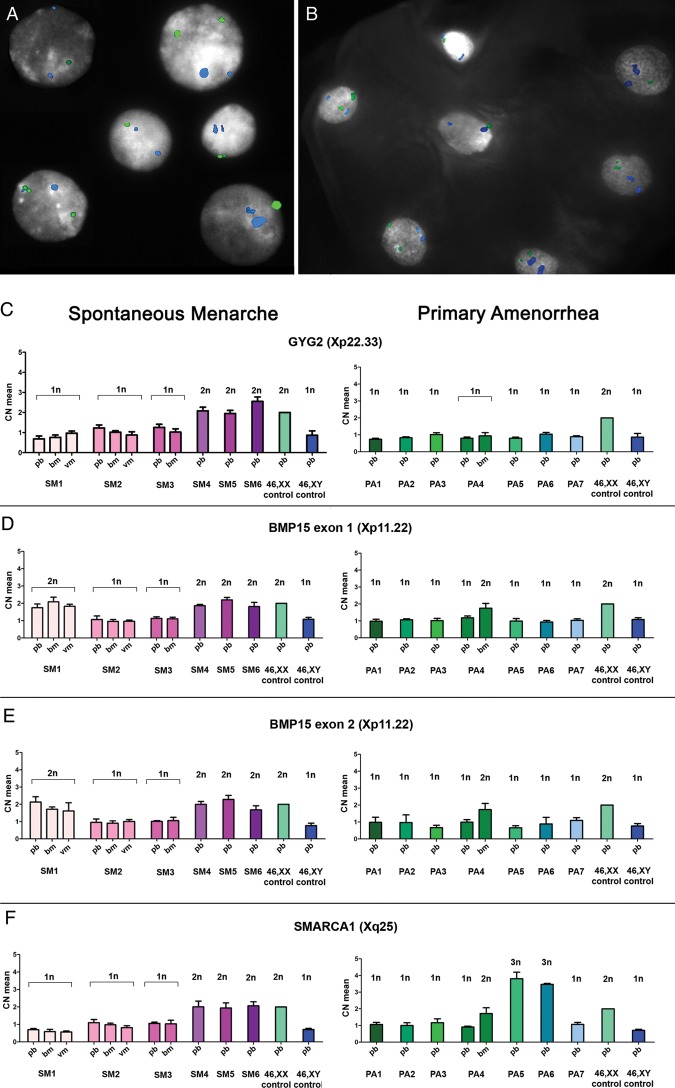
The mosaicism rates were then evaluated by multi-colour iFISH analysis and real-time quantitative PCR on peripheral blood. These two approaches were performed on a subgroup of 13 patients, SM1-6 and PA1-7, including the only 4 patients with PA who were mosaics at aCGH analysis (PA4-7) and 3 representative cases with absent mosaicism signals (Table IV). The iFISH analysis revealed a mosaic condition in PHA-stimulated cultures of all SM and PA patients, based on the presence of one or two signals specific for the X chromosome centromeric α-sat DNA (locus DxZ1) (Fig. 3A), even if at a very low degree (≤1%) in the three 45,X patients PA with absent X chromosome shift at aCGH (Table IV). The same approach was carried out on the buccal mucosa of SM2, SM3 and PA4. The mosaicism rate found in peripheral blood was confirmed in buccal mucosa cells of SM2 and SM3, whereas a double amount of the euploid cell line was identified in PA4 (Table IV, Fig. 3B).
Figure 3.
X chromosome mosaicism evaluation. In patient PA4, the euploid cell line mosaic condition was established by iFISH analysis both in peripheral blood lymphocyte culture (A) and in buccal mucosa (B) where a double amount of 46,XX cells was identified. The CEPX probe (locus DXZ1) is shown in green and the CEP18 (locus D18Z1) is in blue. (C–F) Mosaicism levels in SM patients (pink bars on the left) and PA patients (green-light blue bars on the right) were evaluated by real-time quantitative PCR in peripheral blood (pb), buccal (bm) and vaginal (vm) mucosa, depending on tissues availability. SM1, harbouring the BMP15 duplication, SM4, SM5 and SM6 all showed two copies of BMP15 (D and E). Two copies of GYG2 and SMARCA1 genes were identified in SM4, SM5 and SM6 (C and F) who have mosaicism levels ≥30% (sensitivity cut-off of the method), whereas SM1, SM2 and SM3 showed only one copy of GYG2 and SMARCA1 (mosaicism level <10%) (C and F). SM1's copy number profile was confirmed also in bm and vm. PA1-7 showed one copy of BMP15, GYG2 and SMARCA1 genes (C and F), except for PA5 and PA6 who presented with more than two copies of SMARCA1 (3n) because of the isochromosome(Xq) in >30% of the euploid cell line (F). In PA4's bm, real-time quantitative PCR confirmed the increased copy number mosaic levels for BMP15 and SMARCA1 (2n) and the mosaic deletion at Xp22.33-p22.32 including GYG2. All samples were also compared with 46,XX (calibrator) and 46,XY control gDNAs. Calculated mean CNs are shown on the left-axis, while predicted CNs are displayed on the top of each bar. SD, standard deviation, is represented by the error bars (CN means of controls have been calculated 2n by the analysis software and therefore no SD values are shown).
A mean of two copies of GYG2, BMP15 and SMARCA1 genes, which are located at chromosome Xp22.3, Xp11.22 and Xq25, respectively, were identified by real-time quantitative PCR in the only the three patients with a mosaicism level ≥30% (SM4-6), thus establishing a sensitivity threshold of around 30% for the mosaicism detection by means of this technique (Table IV, Fig. 3C–F). As expected, two copies of BMP15 were also detected in SM1 (Table IV, Fig. 3D and E). All the seven PA patients showed one copy of the three analysed genes, except for PA5 and PA6 who presented with more than two copies of SMARCA1 because of a duplication of the only Xq arm in >30% of the cells (Table IV, Fig. 3C–F). For SM1-3 and PA4, this analysis could be further extended to another tissue of different embryonic origin, such as buccal epithelium or vaginal epithelium, confirming the same blood copy number profile for BMP15, GYG2 and SMARCA1, except for PA4 who showed an increment only in BMP15 and SMARCA1 copy numbers (Table IV, Fig. 3C–F).
Discussion
In the present work, we have analysed by different molecular and molecular-cytogenetic approaches a series of 40 TS patients, 6 of them with a spontaneous pubertal development. The results of this pilot study suggest that a mosaicism <10% for the euploid cell lineage, as measured by modern molecular-cytogenetic techniques on uncultivated tissues, may predict the lack of future pubertal development. When confirmed on larger cohorts, this approach may become a useful complement to the previously proposed biochemical markers (Fechner et al., 2006; Aso et al., 2010; Kallio et al., 2012). The whole of our data, including those collected on tissues of different embryological origins (peripheral blood, buccal and vaginal mucosa) in a subgroup of TS patients, are consistent with (i) the advanced hypothesis that all TS patients bear some degree of mosaicism (Held et al., 1992; Ranke and Saenger, 2001) and (ii) the positive correlation between the TS mosaic karyotype and the probability of a spontaneous pubertal onset (Pasquino et al., 1997). Indeed, mosaicism-like signals were detected by aCGH in four PA patients, but the additional studies confirmed the presence of mosaicisms with cells harbouring abnormalities of the second X chromosome.
Therefore, aCGH represents a valuable alternative method for TS diagnosis, as recently proposed for SNP array (Prakash et al., 2013). The aCGH could indeed detect structural abnormalities of the second X in the euploid cell lines, which were not identified by standard karyotype, and precisely maps the breakpoints, as seen in SM1 and PA4 (Table IV, Fig. 1C and D). By avoiding the growth bias in favour of the euploid cell line during in vitro culture, aCGH was able to detect the presence of the euploid cell lineages at low mosaic levels (around 10%) in two SM patients, both previously karyotyped as 45,X (Tables I and IV). Of note, the use of combined molecular-cytogenetic approaches in different tissues would be of help also in unveiling cryptic 45,X/46,XY mosaics. Nevertheless, in contrast with the SNP array, a quantitative estimation of X chromosome mosaicism rate cannot be performed directly by analysing the aCGH profile shift and an ad hoc mosaicism scale must be applied (Supplementary data, Fig. S1).
The patients SM1 and PA4 represent two cases apparently in contrast with the ‘10% mosaicism threshold’. The autosomic or X-linked CNVs identified in these cases involve genes related to ovarian function, and may thus contribute to explain the discrepancy between the karyotype and the ovarian phenotype (Table IV). In SM1, the aCGH analysis identified a duplication at Xp11.22 containing the entire sequence of the only BMP15 gene. BMP15 (MIM *300247) is located within the Xp locus associated with premature ovarian failure (POF) and encodes an oocyte-derived growth factor of the TGF-β superfamily, involved in follicular development as a critical regulator of many granulosa cells processes and ovulation rate (McNatty et al., 2004; Persani et al., 2011). Evidence of the fundamental role of this factor in follicular development came from animal models (rodents and sheeps) and humans (Dube et al., 1998; Galloway et al., 2000; Di Pasquale et al., 2004; Hanrahan et al., 2004). Several mutational screenings performed in different cohorts of patients affected with 46,XX Primary Ovarian Insufficiency described the significant association with loss-of-function variations in BMP15 (Persani et al., 2011), and functional analysis of some of the reported variants were consistent with a mechanism of haploinsufficiency (Rossetti et al., 2009; Persani et al., 2010; Wang et al., 2010). More recently, a deletion of about 4 Mb at Xp11.23-p11.22, including the entire BMP15 gene, has been identified in a POF patient by SNP array analysis (McGuire et al., 2011). As a double dose of BMP15 seems to be required for the normal rate of follicular depletion in 46,XX women and to provide an adequate anti-apoptotic effect on granulosa cells (Persani et al., 2010), our data are consistent with the hypothesis that a BMP15 duplication on the conserved X chromosome might have preserved the adequate gene dosage in the developing ovary of SM1, thus leading to the conservation of a certain amount of functional follicles at pubertal age and the ability to compensate for the presence of a cell line with r(X)(p22.1q26.2). Indeed, such a structural abnormality generates the complete monosomy of the Xpter-Xp22.1 and Xq26.2-Xqter regions (Fig. 1C), which would be expected to be less stable compared with a cell line with a non-rearranged chromosome X (i.e. SM2) (Table IV). However, SM1 underwent secondary amenorrhoea after 4 years of regular menses (Table II). It is therefore likely that the haploinsufficiency of possible ovarian determinants located in the monosomic regions contributed to the POF, as well as the haploinsufficiency of the SHOX gene (MIM *312865), located at Xp22.3, which is responsible for the patient's short stature (Supplementary data, Table SI). In the other patients with SM, it is reasonable that a correct dosage of several X-linked candidate factors, including BMP15, likely due to a sufficient amount of the euploid cell line in the ovary, can sustain the ovarian development and the folliculogenesis at least for some years, delaying a complete ovarian insufficiency.
The major role of different loci at Xp rather than at Xq for the ovarian phenotype may be strengthened by the PA5-7 cases who have one copy of BMP15 and Xq trisomy due to an isochromosome (Xq) in the euploid cell line (Table IV). Accordingly, Hagen et al. (2010) described spontaneous puberty in only 1 out of 7 TS patients with a 45,X/46,X,i(Xq) karyotype. Interestingly, PA5 carries a heterozygous partial deletion of the PAPPA gene, which could play an additional deleterious role on ovarian phenotype (Figs 1A and 2E). Indeed, the involvement of PAPPA, the pregnancy-associated plasma protein-A, in modulating ovarian function by control of the bioavailability of ovarian insulin-like growth factor is known (Ohnishi et al., 2005) and, recently, it has been demonstrated that the lack of this functional metalloproteinase compromises mouse ovarian steroidogenesis and female fertility (Nyegaard et al., 2010).
The second remarkable case was PA4, whose molecular-cytogenetic findings revealed a mosaicism level (in both peripheral blood and buccal mucosa) that was similar to that of SM patients (Table IV). Nevertheless, aCGH analysis identified a structural abnormality of an X chromosome in the 46,XX cell lineage (i.e. a rare mosaic deletion at Xp22.33-p22.32), which might have contributed to the ovarian defect (Figs 1D and 2C–E). The deleted region, indeed, includes among others the PRKX gene which encodes an X-linked cAMP-dependent protein kinase expressed in the ovary, with a possible role in renal epithelial morphogenesis and in macrophage and granulocyte maturation (Zimmermann et al., 1999). Although a correlation between this protein and female fertility has never been reported, a microduplication affecting the only PRKX gene has previously been reported in a patient with POF (menarche at age 14 years and menopause at age 37 years) (Dudding et al., 2010). Furthermore, bioinformatic analysis of PRKX predicted functional partners (the progesterone receptor, the androgen receptor and different members of adenylate ciclase family) supports its role in female meiosis as these proteins are known to be involved in the fine regulation of oocyte maturation (Sun et al., 2009; Gleicher et al., 2011; Kim et al., 2011). In addition, PA4 was a carrier of an autosomal microduplication, affecting the PDE8A gene coding region and potentially resulting in PDE8A haploinsufficiency (Figs 1B and 2E). PDE8A encodes a cAMP-phosphodiesterase, an enzyme that catalysis the transformation of cyclic nucleotides into 5′ nucleotides, whose functional presence in the bovine ovarian follicle has recently been demonstrated (Sasseville et al., 2009).
These data would support the idea that defects in autosomal loci may contribute to the ovarian phenotype of TS patients in addition to the haploinsufficiency of X-linked genes (Persani et al., 2009), as previously reported in a few series of patients with POI (Aboura et al., 2009; Ledig et al., 2010; Liao et al., 2011; McGuire et al., 2011).
Considering the autosomal ‘common’ CNVs identified in our cohort (Table III), the two involving the NAIP and DUSP22 genes have recently been proposed as candidates for POF (Aboura et al., 2009). Indeed, NAIP encodes the neuronal apoptosis inhibitory protein and is overexpressed in breast cancer patients with unfavourable clinical features (Choi et al., 2007), whereas DUSP22, the dual specificity phosphatases gene, is involved in the regulation of estrogen receptor α-mediated signalling (Sekine et al., 2007). Here, the association of the ovarian defect with both NAIP and DUSP22 CNVs failed to be statistically significant in the case–control analysis, although a P-value of 0.06 has been assigned to DUSP22 gains. The other relevant CNVs were selected because of their high expression level in the ovary (e.g. the GPR128 gene encoding a G protein-coupled receptor, and the MSR1 gene coding for a Macrophage Scavenger Receptor) or because of a likely involvement in ovarian function (e.g. the cluster of APOBEC genes coding for the apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3) (see the following reviews: Wu et al., 2004; Chiu and Greene, 2008; Wang et al., 2008; Koning et al., 2009; Turner et al., 2011). Although these CNVs are currently considered as benign variants, and the affected genes cannot be considered as susceptibility loci for ovarian failure in TS, we cannot rule out that their presence, individually or in combination with other abnormalities, may have contributed to the premature ovarian failure in the corresponding TS patients.
In conclusion, the present work supports gene dosage as a relevant mechanism contributing to the ovarian phenotype of TS patients. However, definitive confirmation of the reported findings should be obtained by an extensive CNV structural characterization and expression studies of candidate genes in animal models. Further studies on large TS cohorts are needed to elucidate the burden of X-linked dosage-sensitive genes in spontaneous pubertal development. These should evaluate the X chromosome mosaicism rates in tissues of different embryonic origin, including the ovary. The first duplication rearrangement of BMP15 in a 45,X TS patient with SM brings additional evidence for a prominent determinant role of this Xp-linked gene on ovarian folliculogenesis. The detection of CNVs in autosomal ovary-related genes may generate further insights on the fundamental mechanisms contributing to ovarian function.
Supplementary data
Supplementary data are available at http://humrep.oxfordjournals.org/.
Authors’ roles
The authors were responsible for the following aspects of the study. C.Cas.: study design, acquisition, analysis and interpretation of data from array-CGH and FISH analysis; manuscript draft preparation; R.R.: study design, acquisition, analysis and interpretation of data from real-time quantitative PCR, manuscript draft preparation; D.R.: acquisition, analysis and interpretation of data from array-CGH; M.P.R.: chromosomal sample collection and preparation; C.Cac.: sample preparation and clinical data collection; E.B.: acquisition, analysis and interpretation of data from DHPLC analysis; V.C.: patient recruitment and clinical data collection; P.I.: study design and critical revision of the manuscript; D.L.: patient recruitment and critical revision of manuscript; P.F. and L.P.: study design and coordination, data interpretation and critical revision of the manuscript. All authors reviewed and approved the final version of the manuscript.
Funding
This work was partially supported by Telethon Foundation, Italy (grant number: GGP09126 to L.P.), the Italian Ministry of the University and Research (grant number: 2006065999 to P.F.) and a Ministry of Health grant “Ricerca Corrente” to Istituto Auxologico Italiano IRCCS (grant number: 08C704-2006). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Funding to pay the Open Access publication charges for this article was provided by the Foundation Telethon Italia.
Conflict of interest
The authors have none to declare.
Supplementary Material
Acknowledgements
The authors thank the patients and their families for their cooperation.
References
- Aboura A, Dupas C, Tachdjian G, Portnoï MF, Bourcigaux N, Dewailly D, Frydman R, Fauser B, Ronci-Chaix N, Donadille B, et al. Array comparative genomic hybridization profiling analysis reveals deoxyribonucleic acid copy number variations associated with premature ovarian failure. J Clin Endocrinol Metab. 2009;9411:4540–4546. doi: 10.1210/jc.2009-0186. [DOI] [PubMed] [Google Scholar]
- Aso K, Koto S, Higuchi A, Ariyasu D, Izawa M, Miyamoto Igaki J, Hasegawa Y. Serum FSH level below 10 mIU/mL at twelve years old is an index of spontaneous and cyclical menstruation in Turner syndrome. Endocr J. 2010;57:909–913. doi: 10.1507/endocrj.k10e-092. [DOI] [PubMed] [Google Scholar]
- Ballif BC, Rorem EA, Sundin K, Lincicum M, Gaskin S, Coppinger J, Kashork CD, Shaffer LG, Bejjani BA. Detection of low-level mosaicism by array CGH in routine diagnostic specimens. Am J Med Genet A. 2006;140:2757–2767. doi: 10.1002/ajmg.a.31539. [DOI] [PubMed] [Google Scholar]
- Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- Choi J, Hwang YK, Choi YJ, Yoo KE, Kim JH, Nam SJ, Yang JH, Lee SJ, Yoo KH, Sung KW, et al. Neuronal apoptosis inhibitory protein is overexpressed in patients with unfavourable prognostic factors in breast cancer. J Korean Med Sci. 2007;22(Suppl):S17–S23. doi: 10.3346/jkms.2007.22.S.S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pasquale E, Beck-Peccoz P, Persani L. Hypergonadotropic ovarian failure associated with an inherited mutation of human bone morphogenetic protein-15 (BMP15) gene. Am J Hum Genet. 2004;75:106–111. doi: 10.1086/422103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube JL, Wang P, Elvin J, Lyons KM, Celeste AJ, Matzuk MM. The bone morphogenetic protein 15 gene is X-linked and expressed in oocytes. Mol Endocrinol. 1998;12:1809–1817. doi: 10.1210/mend.12.12.0206. [DOI] [PubMed] [Google Scholar]
- Dudding TE, Lawrence O, Winship I, Froyen G, Vandewalle J, Scott R, Shelling AN. Array comparative genomic hybridization for the detection of submicroscopic copy number variations of the X chromosome in women with premature ovarian failure. Hum Reprod. 2010;25:3159–3160. doi: 10.1093/humrep/deq284. [DOI] [PubMed] [Google Scholar]
- Fechner PY, Davenport ML, Qualy RL, Ross JL, Gunther DF, Eugster EA, Huseman C, Zagar AJ, Quigley CA Toddler Turner Study Group. Differences in follicle-stimulating hormone secretion between 45,X monosomy Turner syndrome and 45,X/46,XX mosaicism are evident at an early age. J Clin Endocrinol Metab. 2006;91:4896–4902. doi: 10.1210/jc.2006-1157. [DOI] [PubMed] [Google Scholar]
- Galloway SM, McNatty KP, Cambridge LM, Laitinen MP, Juengel JL, Jokiranta JL, McLaren RJ, Luiro K, Dodds KG, Montgomery GW, et al. Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nat Genet. 2000;25:279–283. doi: 10.1038/77033. [DOI] [PubMed] [Google Scholar]
- Gleicher N, Weghofer A, Barad DH. The role of androgens in follicle maturation and ovulation induction: friend or foe of infertility treatment? Reprod Biol Endocrinol. 2011;9:116. doi: 10.1186/1477-7827-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracia Bouthelier R, Oliver Iguacel A, Gonzalez Casado I, Alcalde de Alvaré A. Optimization of treatment in Turner's syndrome. J Pediatr Endocrinol Metab. 2004;17:427–434. [PubMed] [Google Scholar]
- Hagen CP, Main KM, Kjaergaard S, Juul A. FSH, LH, inhibin B and estradiol levels in Turner syndrome depend on age and karyotype: longitudinal study of 70 Turner girls with or without spontaneous puberty. Hum Reprod. 2010;25:3134–3141. doi: 10.1093/humrep/deq291. [DOI] [PubMed] [Google Scholar]
- Hanrahan JP, Gregan SM, Mulsant P, Mullen M, Davis GH, Powell R, Galloway SM. Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovis aries) Biol Reprod. 2004;70:900–909. doi: 10.1095/biolreprod.103.023093. [DOI] [PubMed] [Google Scholar]
- Held KR, Kerber S, Kaminsky E, Singh S, Goetz P, Seemanova E, Goedde HW. Mosaicism in 45,X Turner syndrome: does survival in early pregnancy depend on the presence of two sex chromosomes? Hum Genet. 1992;88:288–294. doi: 10.1007/BF00197261. [DOI] [PubMed] [Google Scholar]
- Kallio S, Aittomäki K, Piltonen T, Veijola R, Liakka A, Vaskivuo TE, Dunkel L, Tapanainen JS. Anti-Mullerian hormone as a predictor of follicular reserve in ovarian insufficiency: special emphasis on FSH-resistant ovaries. Hum Reprod. 2012;27:854–860. doi: 10.1093/humrep/der473. [DOI] [PubMed] [Google Scholar]
- Kim J, Bagchi IC, Bagchi MK. Control of ovulation in mice by progesterone receptor-regulated gene networks. Mol Hum Reprod. 2011;15:821–828. doi: 10.1093/molehr/gap082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol. 2009;83:9474–9485. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledig S, Röpke A, Wieacker P. Copy number variants in premature ovarian failure and ovarian dysgenesis. Sex Dev. 2010;4:225–232. doi: 10.1159/000314958. [DOI] [PubMed] [Google Scholar]
- Liao C, Fu F, Yang X, Sun YM, Li DZ. Analysis of Chinese women with primary ovarian insufficiency by high resolution array-comparative genomic hybridization. Chin Med J. 2011;124:1739–1742. [PubMed] [Google Scholar]
- Lichter P, Cremer T. Chromosome analysis by non-isotopic in situ hybridization. In: Rooney DE, Czepulkowski BH, editors. Human Cytogenetics. A Practical Approach. Oxford: IRL Press at Oxford University Press; 1992. pp. 157–192. [Google Scholar]
- Lichter P, Tang CJ, Call K, Hermanson G, Evans GA, Housman D, Ward DC. High resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- Martín Campagne E, Roa Llamazares C. Spontaneous puberty and menarche in a patient with Turner syndrome and 45X monosomy. An Pediatr. 2008;70:200–202. doi: 10.1016/j.anpedi.2008.09.008. [DOI] [PubMed] [Google Scholar]
- McGuire MM, Bowden W, Engel NJ, Ahn HW, Kovanci E, Rajkovic A. Genomic analysis using high-resolution single-nucleotide polymorphism arrays reveals novel microdeletions associated with premature ovarian failure. Fertil Steril. 2011;95:1595–1600. doi: 10.1016/j.fertnstert.2010.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNatty KP, Moore LG, Hudson NL, Quirke LD, Lawrence SB, Reader K, Hanrahan JP, Smith P, Groome NP, Laitinen M, et al. The oocyte and its role in regulating ovulation rate: a new paradigm in reproductive biology. Reproduction. 2004;128:379–386. doi: 10.1530/rep.1.00280. [DOI] [PubMed] [Google Scholar]
- Nyegaard M, Overgaard MT, Su YQ, Hamilton AE, Kwintkiewicz J, Hsieh M, Nayak NR, Conti M, Conover CA, Giudice LC. Lack of functional pregnancy-associated plasma protein-A (PAPPA) compromises mouse ovarian steroidogenesis and female fertility. Biol Reprod. 2010;82:1129–1138. doi: 10.1095/biolreprod.109.079517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata T, Matsuo N. Turner syndrome and female sex chromosome aberrations: deduction of the principal factors involved in the development of clinical features. Hum Genet. 1995;95:607–629. doi: 10.1007/BF00209476. [DOI] [PubMed] [Google Scholar]
- Ohnishi J, Ohnishi E, Shibuya H, Takahashi T. Functions for proteinases in the ovulatory process. Biochim Biophys Acta. 2005;1751:95–109. doi: 10.1016/j.bbapap.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Pasquino AM, Passeri F, Pucarelli I, Segni M, Municchi G. Spontaneous pubertal development in Turner's syndrome. Italian Study Group for Turner's Syndrome. J Clin Endocrinol Metab. 1997;82:1810–1813. doi: 10.1210/jcem.82.6.3970. [DOI] [PubMed] [Google Scholar]
- Persani L, Rossetti R, Cacciatore C, Bonomi M. Primary ovarian insufficiency: X chromosome defects and autoimmunity. J Autoimmun. 2009;33:35–41. doi: 10.1016/j.jaut.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Persani L, Rossetti R, Cacciatore C. Genes involved in human premature ovarian failure. J Mol Endocrinol. 2010;45:405. doi: 10.1677/JME-10-0070. [DOI] [PubMed] [Google Scholar]
- Persani L, Rossetti R, Cacciatore C, Fabre S. Genetic defects of ovarian TGF-β-like factors and premature ovarian failure. J Endocrinol Invest. 2011;34:244–251. doi: 10.1007/BF03347073. [DOI] [PubMed] [Google Scholar]
- Prakash S, Guo D, Maslen CL, Silberbach M, Milewicz D, Bondy CA GenTAC Investigators. Single-nucleotide polymorphism array genotyping is equivalent to metaphase cytogenetics for diagnosis of Turner syndrome. Genet Med. 2013 doi: 10.1038/gim.2013.77. doi:10.1038/gim.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilter CR, Karcanias AC, Bagga MR, Duncan S, Murray A, Conway GS, Sargent CA, Affara NA. Analysis of X chromosome genomic DNA sequence copy number variation associated with premature ovarian failure (POF) Hum Reprod. 2010;258:2139–2150. doi: 10.1093/humrep/deq158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranke MB, Saenger P. Turner's syndrome. Lancet. 2001;358:309–314. doi: 10.1016/S0140-6736(01)05487-3. [DOI] [PubMed] [Google Scholar]
- Reddy KS, Mak L. Mosaic unbalanced structural abnormalities confirmed using FISH on buccal mucosal cells. Ann Genet. 2001;44:37–40. doi: 10.1016/s0003-3995(00)01032-7. [DOI] [PubMed] [Google Scholar]
- Reynaud K, Cortvrindt R, Verlinde F, De Schepper J, Bourgain C, Smitz J. Number of ovarian follicles in human fetuses with the 45,X karyotype. Fertil Steril. 2004;81:1112–1119. doi: 10.1016/j.fertnstert.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Rossetti R, Di Pasquale E, Marozzi A, Bione S, Toniolo D, Grammatico P, Nelson LM, Beck-Peccoz P, Persani L. BMP15 mutations associated with primary ovarian insufficiency cause a defective production of bioactive protein. Hum Mutat. 2009;30:804–810. doi: 10.1002/humu.20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenger P. Turner's syndrome. N Engl J Med. 1996;335:1749–1754. doi: 10.1056/NEJM199612053352307. [DOI] [PubMed] [Google Scholar]
- Sasseville M, Albuz FK, Côté N, Guillemette C, Gilchrist RB, Richard FJ. Characterization of novel phosphodiesterases in the bovine ovarian follicle. Biol Reprod. 2009;81:415–425. doi: 10.1095/biolreprod.108.074450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y, Ikeda O, Hayakawa Y, Tsuji S, Imoto S, Aoki N, Sugiyama K, Matsuda T. DUSP22/LMW-DSP2 regulates estrogen receptor-alpha-mediated signaling through dephosphorylation of Ser-118. Oncogene. 2007;26:6038–6049. doi: 10.1038/sj.onc.1210426. [DOI] [PubMed] [Google Scholar]
- Sun QY, Miao YL, Schatten H. Towards a new understanding on the regulation of mammalian oocyte meiosis resumption. Cell Cycle. 2009;1:2741–2747. doi: 10.4161/cc.8.17.9471. [DOI] [PubMed] [Google Scholar]
- Sybert VP, McCauley E. Turner's syndrome. N Engl J Med. 2004;351:1227–1238. doi: 10.1056/NEJMra030360. [DOI] [PubMed] [Google Scholar]
- Toniolo D. X-linked premature ovarian failure: a complex disease. Curr Opin Genet Dev. 2006;6:293–300. doi: 10.1016/j.gde.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Turner EC, Hughes J, Wilson H, Clay M, Mylonas KJ. Conditional ablation of macrophages disrupts ovarian vasculature. Reproduction. 2011;141:821–831. doi: 10.1530/REP-10-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Prossnitz ER, Roy SK. G protein-coupled receptor 30 expression is required for estrogen stimulation of primordial follicle formation in the hamster ovary. Endocrinology. 2008;149:4452–4461. doi: 10.1210/en.2008-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Wen Q, Ni F, Zhou S, Wang J, Cao Y, Ma X. Analyses of growth differentiation factor 9 (GDF9) and bone morphogenetic protein 15 (BMP15) mutation in Chinese women with premature ovarian failure. Clin Endocrinol. 2010;72:135–136. doi: 10.1111/j.1365-2265.2009.03613.x. [DOI] [PubMed] [Google Scholar]
- Wu R, Van der Hoek KH, Ryan NK, Norman RJ, Robker RL. Macrophage contributions to ovarian function. Hum Reprod Update. 2004;10:119–133. doi: 10.1093/humupd/dmh011. [DOI] [PubMed] [Google Scholar]
- Zimmermann B, Chiorini JA, Ma Y, Kotin RM, Herberg FW. PrKX is a novel catalytic subunit of the cAMP-dependent protein kinase regulated by the regulatory subunit type I. J Biol Chem. 1999;274:5370–5378. doi: 10.1074/jbc.274.9.5370. [DOI] [PubMed] [Google Scholar]
- Zinn AR, Ross JL. Turner syndrome and haploinsufficiency. Curr Opin Genet Dev. 1998;8:322–327. doi: 10.1016/s0959-437x(98)80089-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.