Abstract
Dendritic cells (DCs) are the most commonly studied source of the cytokine interleukin-15 (IL-15). Using an IL-15 reporter transgenic mouse, we have recently shown previously unappreciated differences in the levels of IL-15 expressed by subsets of conventional DCs (CD8+ and CD8-). Here we show that IL-15 promoter activity was differentially regulated in subsets of hematopoietically derived cells with IL-15 expression largely limited to myeloid lineages. In contrast, mature cells of the lymphoid lineages expressed little to no IL-15 activity. Surprisingly, we discovered that hematopoietic stem cells (Lin-Sca-1+c-kit+; LSKs) expressed high levels of IL-15 suggesting that IL-15 expression was extinguished during lymphoid development. In the case of T cells, this downregulation was Notch-dependent and occurred in a step-wise pattern coincident with increasing maturation and commitment to a T cell fate. Finally, we further demonstrate that IL-15 expression was also controlled throughout DC development, with key regulatory activity of IL-15 production occurring at the pre-DC branch point leading to the generation of both IL-15+ CD8+ and IL-15-/low CD8-DC subsets. Thus, IL-15 expression is coordinated with cellular fate in myeloid versus lymphoid immune cells.
Introduction
Interleukin-15 (IL-15) is a common γ chain (γc [CD132]) cytokine that functions through a unique mechanism of delivery termed transpresentation. IL-15 has a very high affinity (1.4 × 10-11 M) for its private receptor, IL-15Rα (1). The two proteins bind within the cell and are shuttled to the surface as a complex (2). IL-15 is then presented in trans to responding cells, such as NK cells, CD8 memory T cells, and intraepithelial lymphocytes (IELs)2, that express the other two shared chains of the receptor, γc and IL-2/15Rβ [CD122] (3) The generation of both IL-15 and IL15Rα knockout (KO) mice has greatly facilitated our ability to study the dramatic downstream effects that exist in the absence of IL-15 signaling (4,5). Importantly, transferring NK cells or CD8 T cells from IL-15Rα KO mice to wild-type (WT) recipients demonstrated that IL-15-dependent cells need not express the IL-15Rα chain to mediate the downstream effects of IL-15 signaling (6-8). For this reason, transpresentation is largely accepted as the primary mechanism of IL-15 delivery. Whether cis-presentation of IL-15 occurs in vivo is currently unclear yet remains feasible (9). First, CD8 T cells are known to upregulate IL-15Rα expression following activation (10), and second, impaired development and activation of IL-15-dependent cells is observed in transgenic mice engineered to express a chimeric cytokine receptor composed of the external domain of IL-15Rα fused to the intracellular portion of IL-2Rα, suggesting that cell intrinsic signaling through IL-15Rα is required for the generation of optimal immune responses (11).
Many elegant studies have addressed the cell-cell interactions that drive IL-15-dependent responses, including the cellular sources of IL-15 in vivo [reviewed in 12]. Early studies used the generation of bone marrow (BM) chimeras to distinguish IL-15 derived from hematopoietic (radio-sensitive) versus non-hematopoietic (radio-resistant) sources (6,8,13). In total, these studies showed that there is variation in the requirements for IL-15 from individual sources of IL-15 by individual populations of IL-15-dependent cells. For example, peripheral NK cells and memory CD8 T cells are partially restored when IL-15 production is limited to hematopoietically-derived cells while IL-15 transpresentation by radio-resistant intestinal epithelial cells is necessary and sufficient for the generation and maintenance of CD8αα+ IELs (14). Consistent with their association intracellularly, these studies have also shown that the same cell must express both IL-15 and the IL-15Rα to mediate activity (15,16). Recent approaches have used transgenic expression of IL-15Rα limited to certain subsets of cells and conditional deletion of IL-15Rα using Cre-lox technology to more closely examine the significance of IL-15 from select in vivo sources (17-21). These studies have implicated a role for DCs in transpresenting IL-15 to many subsets of immune cells, yet also suggest that other cellular sources of IL-15 likely work in conjunction with DCs to maintain overall peripheral homeostasis.
The overt lymphoproliferative disease observed when IL-15 is produced in excess is evidence that IL-15 expression must be tightly regulated (22). However, IL-15 itself has proven much more difficult to study. This is likely the combined result of overall low levels of IL-15 in the steady state and possibly poor detection reagents, which in some cases may be further hindered when IL-15 is bound to IL-15Rα. To overcome these hurdles in IL-15 detection, we generated a BAC transgenic mouse strain in which emerald GFP is produced under control of the IL-15 promoter and all upstream regulatory elements (EmGFP/IL-15). We employed this tool to identify previously unappreciated differences in the respective levels of IL-15 produced by individual subsets of conventional DCs during homeostasis and successfully measure changes in IL-15 expression in DCs following virus infection (23). Here we use EmGFP/IL-15 reporter mice to further identify novel subsets of cells capable of producing IL-15 and examine developmental pathways that regulate IL-15 expression in immune cells, which largely result in limiting IL-15 production to cells of the myeloid lineage.
Materials and Methods
Mice
C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and the NCI-Charles River (Fredericksburg, MD), and IL-15-/- mice were purchased from Taconic (Germantown, NY). EmGFP/IL-15 reporter transgenics were bred and maintained at the UConn Health Center. All mice were used between 6 and 20 weeks of age and housed according to the guidelines at the UConn Health Center and MD Anderson Cancer Center. All experiments were performed with the approval of the respective Institutional Animal Care and Usage Committees.
Tissue isolation
Spleen and thymus were harvested in complete media and crushed through a 70um filter. BM was isolated by rapid centrifugation directly into complete media. Peritoneal exudate cells were collected by injecting ∼10ml PBS into the peritoneal cavity and collecting the solution after 5 minutes. Red blood cell lysis was performed by incubating single cell suspensions in tris ammonium chloride for 5 minutes at 37°.
Flow cytometry and cell sorting
All antibodies and Live/Dead® viability dye were purchased from BD Biosciences, Biolegend, and eBioscience. Fc block (anti-CD16/32; clone 2.4G2) was purchased from Bio X Cell (West Lebanon, NH). IL-15 was detected with rabbit anti-IL-15 biotin (Peprotech, Rocky Hill, NJ) followed by streptavidin-APC (Jackson ImmunoResearch Labratories, West Grove, PA). Background staining was determined by examining analogous populations from IL-15-/- mice.
In vitro culture
For IL-15 detection, 107 whole splenocytes or BM cells were cultured overnight with or without LPS (1ug/ml; Sigma-Aldrich, St. Louis, MO) or IFN-α (300U/ml; PBL Interferon Source and kindly provided by Willem Overwijk [MD Anderson]). For in vitro culture with OP9 stromal cells, experiments were performed as previously described (24). Briefly, ∼4-5 × 104 sorted LSK cells (CD3-CD19-CD11b-CD11c-Gr-1-NK1.1-Ter119-Sca-1+CD117+) were seeded on confluent OP9 or OP9-DL1 stromal cells in OP9 media (MEM Alpha, 20% FBS, pen/strep) supplemented with 5ng/ml Flt-3L and 1ng/ml IL-7 (Peprotech, Rocky Hill, NJ). Cells were replenished with new media and cytokines on d5. To generate BM-derived DCs (BMDCs), 8 × 106 whole BM cells were plated at 2 × 106 cells/ml in 6-well plates with media (IMDM, 10% FBS, 2ME, NEAA, sodium pyruvate, pen/strep) supplemented with 100 ng/ml Flt3L (Celldex Therapeutics, Needham, MA) for 9 days.
Adoptive transfer
∼1-5 × 104purified pre-DCs (CD3-CD19-B220-Ter119-NK1.1-CD11c+MHCIInegCD135+CD172a-) isolated from the BM were transferred to congenic recipients, and the spleens of recipient mice were harvested after 6 days, as previously described (25).
Statistics
Statistical analysis was performed using Prism software (GraphPad, San Diego, CA) using a Student's t-test or ANOVA where appropriate.
Results
IL-15 is expressed at the highest levels in myeloid lineages
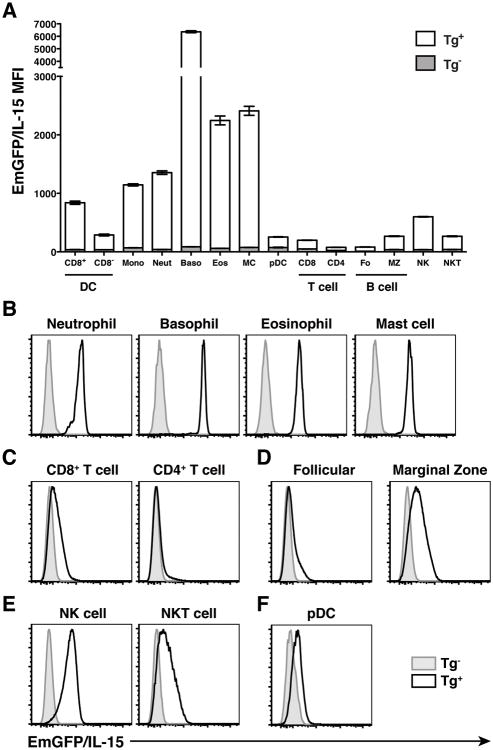
To facilitate the detection of IL-15 in vivo, our lab has recently generated a transgenic BAC reporter mouse in which emerald GFP is expressed under the control of the promoter and upstream regulatory elements of the IL-15 gene locus (23). Our analysis of these mice confirmed previously published studies that show IL-15 production by DCs and monocytes (26,27). However, the ability to use flow cytometry and EmGFP expression as a surrogate for IL-15 promoter activity has revealed previously unappreciated differences in the relative levels of EmGFP/IL-15 expression in individual subsets of cells, particularly CD8+ and CD8- DCs (23 and Fig 1A), and these results were recently corroborated using a second transgenic IL-15 reporter line (28). In order to further identify potentially novel cellular sources of IL-15, we performed a large-scale analysis of both innate and adaptive immune cell subsets in transgenicmice (Tg+) and transgene negative (Tg-) littermate controls using the mean florescence intensity (MFI) of EmGFP as an indicator of IL-15 transcription (Fig 1A). The phenotypic definitions used to identify each population of immune cells by flow cytometry are shown in Supplementary Table 1. We found that EmGFP/IL-15 was highly expressed in splenic myeloid cells including neutrophils, basophils, and eosinophils and mast cells isolated from the peritoneal cavity (Fig 1A and 1B). Alternatively, we found a lack of EmGFP/IL-15 expression in lymphoid lineages, particularly CD4+ T cells and follicular B cells (Fig 1C and 1D). Interestingly, NK and NKT cells along with marginal zone B cells, which are described as more “innate-like” subsets of the immune system, maintained low but detectable levels of EmGFP/IL-15 expression (Fig 1D and 1E). However, unlike their conventional DC counterparts, plasmacytoid DCs expressed EmGFP/IL-15 at very low levels (Fig 1F). Thus, we observed an apparent bifurcation regarding IL-15 promoter activity that was largely associated with myeloid versus lymphoid cell fate.
Figure 1. IL-15 is broadly expressed in myeloid cells.
(A) Individual subsets of immune cells were identified in the spleens of EmGFP reporter transgenic (Tg+) and transgene negative (Tg-) littermate controls. The gating strategy for each subset is described in Supplemental Table 1. Data is presented graphically as the MFI of EmGFP/IL-15 in each subset of immune cells (mean ± SEM). Representative histograms depicting relative levels of EmGFP/IL-15 expression in each population are shown in B-F. Data is representative of at least 3 experiments. Tg- (gray histograms), Tg+ (black line)
Initial attempts to measure cell-surface IL-15 expression using a flow cytometry-based detection method have been largely unsuccessful. However, two reports have detected IL-15 protein in BM-derived DCs and Gr-1+ polymorphonuclear cells (29,30) and using the same reagents we have also detected surface IL-15 on subsets of DCs (17). Given the success of this particular antibody in detecting transpresented IL-15, we sought to measure IL-15 protein directly ex vivo on the populations of granulocytes that exhibited high levels of IL-15 transcription based on EmGFP expression. Unfortunately, we were unable to detect IL-15 expression directly ex vivo on any of the cell populations identified in Figure 1 isolated from the spleen or BM (Supplemental Figure 1A). However, we were also unable to detect IL-15 directly ex vivo on CD11c+ splenocytes (Supplemental Figure 1B). In our previous studies, IL-15 was only detectable when the cells were subjected to an overnight in vitro culture or exposed to TLR ligands (17). Therefore, we decided to take a similar approach to detect IL-15 protein on granulocytic cells. When we cultured splenocytes overnight in media alone, we detected low levels of IL-15 on neutrophils and eosinophils, using CD11chi DCs and IL-15-/- mice as a control for IL-15 staining (Figure 2A). Interestingly, when we performed similar overnight cultures with BM cells, we were also able to detect surface IL-15 on neutrophils, eosinophils, and macrophages (Figure 2B). To further explore the potential of these novel sources of IL-15 to produce protein, we stimulated cells from the spleen and BM with LPS or IFN-α to determine if these known inducers of IL-15 could augment IL-15 expression in the above populations. Surprisingly, all subsets were able to upregulate IL-15 expression to some degree compared to both unstimulated controls and stimulated IL-15 KO cells (Figure 2A and 2B). Thus, in vitro detection of IL-15 in neutrophils and eosinophils correlated with the findings from our reporter mice indicating that these granulocytic populations have the capacity to produce IL-15 protein.
Figure 2. Detection of IL-15 protein from granulocytic myeloid cells.
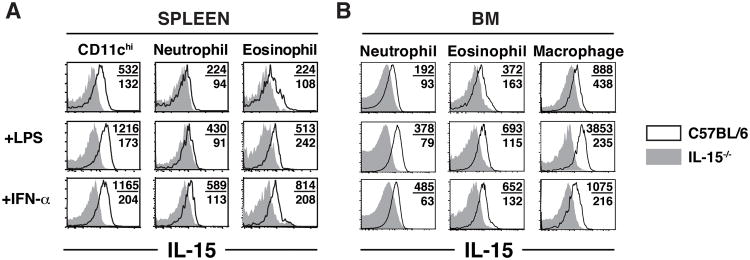
(A,B) 107 cells from the spleen (A) or BM (B) of WT (black lines) and IL-15-/- mice (gray filled histograms) were incubated O/N in the absence or presence of 1ug/ml LPS or 300U/ml IFN-α. Numbers indicate the MFI of IL-15 staining for WT (top) and IL-15-/- samples (bottom). Plots are representative of 2-3 experiments.
Hematopoietic progenitors express high levels of IL-15
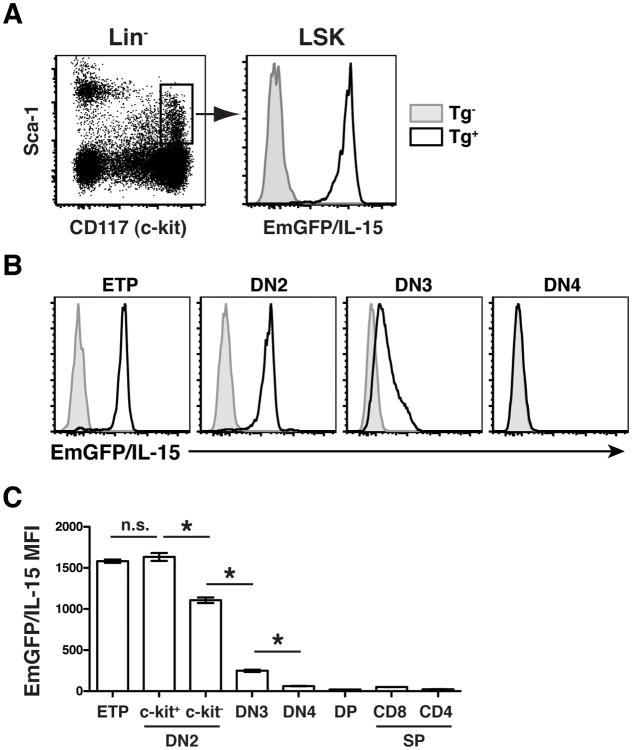
All immune cells are derived from a small population of hematopoietic stem cells (HSCs) maintained in the adult BM. Because we observed such dramatic variation in the levels of EmGFP/IL-15 expressed by mature cells of the hematopoietic system, we asked to what degree EmGFP/IL-15 was expressed by hematopoietic progenitors in the BM of transgenic reporter mice. We hypothesized that HSCs would lack IL-15 expression and that IL-15 would be upregulated in myeloid cells, thus rendering them capable of transpresenting IL-15 and delivering a survival signal to responding NK and T cells. Surprisingly, LSK cells (Lineage-Sca-1+c-kit+), which constitute a heterogeneous population of both long-term and short-term HSCs, expressed uniformly high levels of EmGFP/IL-15 (Figure 3A). Thus, contrary to our hypothesis, this result suggested that alternative mechanisms selectively limited IL-15 expression in lymphoid lineages.
Figure 3. Murine HSCs express high levels of EmGFP/IL-15.
(A) HSCs were identified in the BM of reporter mice and littermate controls as Lin-Sca-1+c-kit+ (LSK). Data are representative of at least 2 independent experiments. Lin- = CD3-CD19-CD11b-CD11c-Gr-1-NK1.1-Ter119-. (B-C) Thymocytes from reporter mice and littermate controls were examined for expression of EmGFP/IL-15. Double negative (DN) thymocyte populations were defined as follows: CD3-CD4-CD8-; ETP, c-kit+CD25-; DN2, CD44highCD25+, both c-kit+ and c-kit-; DN3, CD44-CD25+; DN4, CD44-CD25-. Double positive (DP) cells were CD3-CD4+CD8+, and single positive (SP) cells were either CD3+CD4-CD8+ or CD3+CD4+CD8-. The MFI of EmGFP/IL-15 is shown graphically for each population in B. Tg- (gray histograms), Tg+ (black line).
Since T cells largely lacked EmGFP/IL-15 expression and the pathways that drive T cell commitment are well known, we examined the regulation of IL-15 during T cell development in the thymus. We harvested thymocytes from Tg+ reporters and littermate controls and determined the MFI of EmGFP/IL-15 expression in the CD3-CD4-CD8- double negative (DN), CD3-CD4+CD8+ double positive, and CD3+ single positive CD4+ and CD8+ subsets. Like the LSK cells, early thymic progenitors (ETPs), the c-kit+ fraction of the CD44highCD25- DN1 subset (31), expressed high levels of EmGFP/IL-15 (Fig 3B and 3C). Such elevated levels were maintained within the c-kit+ fraction of CD25-expressing DN2 cells as well. The downregulation of EmGFP/IL-15 began as DN2 cells lost c-kit expression (Figure 3C), and EmGFP/IL-15 expression was significantly reduced at the transition between DN2 and DN3 cells (Fig 3B and 3C). By the DN4 stage, EmGFP/IL-15 expression was completely lost in committed T cells, and it remained absent from all downstream subsets including double positive and single positive cells (Fig 3B and 3C). Thus, these data suggested that the signaling pathways controlling the progression of DN2 to DN3 cells could be responsible for initiating the downregulation of IL-15.
Notch signaling mediates IL-15 downregulation during thymopoiesis
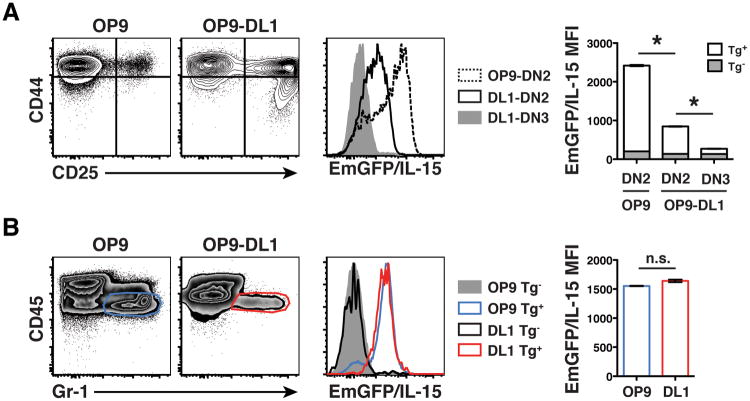
The role of Notch in driving T cell development has been well studied (32), and it is thought that Notch signaling not only initiates the necessary program for T cell development but also turns off any residual expression of myeloid-specific factors (33). To test if Notch signaling was sufficient to drive IL-15 downregulation, we cultured purified LSK cells in vitro with OP9 stromal cells expressing the Notch ligand delta-like 1 (DL1). We sorted EmGFP+ LSK cells from Tg+ mice and EmGFP- LSKs from Tg-littermate controls and cultured them in the presence of Flt3L and IL-7 for 9 days (24). At that time, a large population of CD25+ DN2 and DN3 cells could easily be identified. Similar to our in vivo analysis, we found that the DN2 to DN3 transition driven by Notch signaling included a significant reduction in the MFI of EmGFP/IL-15 (Figure 4A). Interestingly, CD25+ DN2 cells grown on OP9-DL1 cells had significantly reduced EmGFP/IL-15 expression compared to the small number of CD25+ DN2 cells that emerge when LSKs are cultured on control OP9 stromal cells. Thus, in OP9-DL1 cultures, the abundance of Notch ligands may have initiated some degree of EmGFP/IL-15 downregulation in DN2 cells that preceded the complete silencing of IL-15 expression and additional phenotypic changes associated with the DN3 stage. However, the presence of Notch ligands did not universally downregulate EmGFP/IL-15 expression because the small population of Gr-1+ cells that escaped Notch-driven differentiation signals and developed on the OP9-DL1 stromal cells expressed identical levels of EmGFP/IL-15 as Gr-1+ cells that emerged after co-culture with control OP9 cells lacking DL1 expression (Figure 4B). This finding suggested that even in the presence of Notch signals, acquisition of a myeloid fate includes the maintained expression of IL-15.
Figure 4. Notch-driven regulation of IL-15 gene expression during thymopoiesis.
(A-B) LSK cells were sorted from Tg+ and Tg- mice and cultured with OP9 or OP9-DL1 stromal cells in the presence of Flt3L and IL-7 for 9 days, as previously described.24 In A, contour plots (left) indicate the level of CD44 and CD25 expression on the population of Gr-1- cells. The relative level of EmGFP/IL-15 in CD25+ DN2 (CD44high) or DN3 (CD44low) cells grown on either OP9 or OP9-DL1 cells is shown in the histogram (center) and depicted graphically (right). * p < 0.05 using ANOVA followed by a Tukey post test. In B, cells were harvested and analyzed for expression of CD45 and Gr-1 to identify hematopoietically-derived granulocytes (density plots, left). The relative level of EmGFP/IL-15 expression in each population is shown in the histogram (center) and graphically (right). n.s.; not significant using Student's t-test. Results are representative of 2 individual experiments.
Coordinated regulation of IL-15 throughout DC development
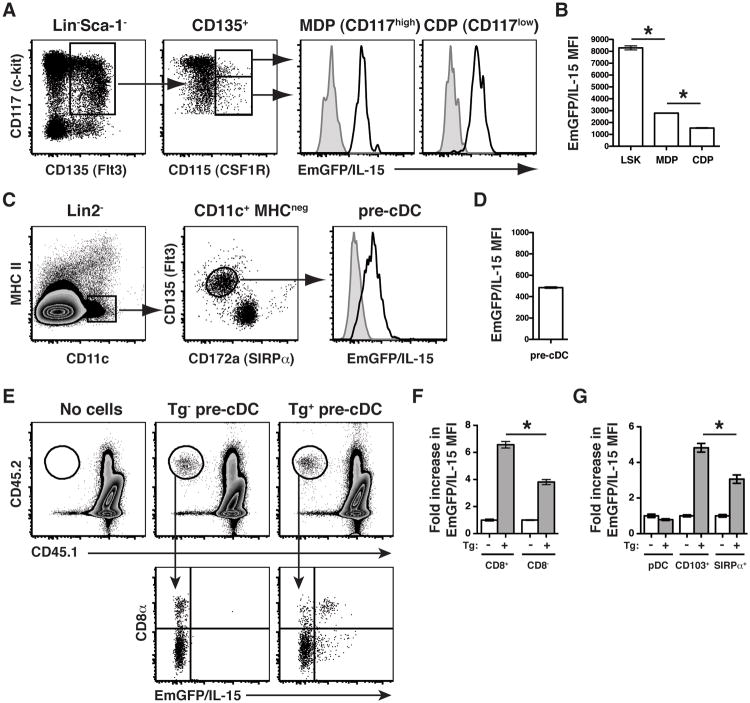
Thus far, our results provided new evidence that IL-15 expression was controlled as immune cells progress towards a mature lymphoid fate. Considering the unique differences in EmGFP/IL-15 production between CD8+ and CD8- DCs, we hypothesized that IL-15 would also be regulated throughout DC development, rather than being controlled exclusively by extrinsic inflammatory mediators. Therefore, we tracked EmGFP/IL-15 expression in BM precursor cells downstream of LSKs but upstream of DC development. Both macrophage-DC progenitors and common DC progenitors expressed high levels of EmGFP/IL-15, albeit at significantly reduced levels compared to the LSK cells (Figure 5A and 5B). Thus, there was a progressive downregulation of EmGFP/IL-15 through each stage of DC development suggesting continuous regulatory activity at the IL-15 locus. Pre-DCs, the final precursor population prior to commitment to either the CD8+ or CD8- DC lineage (25), expressed low but uniform levels of EmGFP/IL-15 (Figure 5C and 5D).
Figure 5. IL-15 expression is tightly regulated throughout DC development.
BM cells were harvested from Tg+ mice (black line) and Tg- littermate controls (gray histograms) to determine EmGFP/IL-15 expression by (A-B) Lin-Sca-1-CD135+macrophage-DC progenitors (MDP), common DC progenitors (CDP), and (C-D) Lin2-CD11c+MHCIInegCD135+CD172a- pre-DC cells. Gating and histograms of EmGFP/IL-15 expression are shown in A&C and depicted graphically in B&D. The level of EmGFP/IL-15 expressed by LSK cells (see Figure 3) is shown for statistical comparison in B. Lin2- = CD3-CD19-B220-Ter119-NK1.1- (E-F) pre-DCs were purified by FACS from Tg+ and Tg- controls and transferred to naïve CD45.1 recipients between 3-4 weeks of age. After 6 days, the spleens of recipient mice were harvested to determine the phenotype of donor-derived cells. Spleens were first stained with anti-CD45.2-APC followed by incubation with anti-APC microbeads to enrich for the donor-derived population by MACS prior to flow cytometric analysis. One mouse that received no donor cells is shown as a control. Data in E is concatenated from one of two independent experiments with 3-4 recipient mice per group per experiment. The data in F is pooled from the two independent experiments. (G) BM cells from Tg+ and Tg- mice were cultured in vitro with Flt3L, and after 9 days, cells were harvested and analyzed by flow cytometry. pDCs were gated as B220+CD11cint, and cDC populations (either CD103+ or SIRPα+) were first gated as B220-CD11c+. The data is presented as the fold increase in MFI of EmGFP over the average MFI of EmGFP from Tg- controls for each population (F and G). * p < 0.05
Since we observed homogeneous expression of IL-15 in the pre-DC precursor population, our results suggested that the overall level of IL-15 expression by a given DC subset was linked to the final developmental program for that particular subset. Therefore, to determine whether EmGFP/IL-15+ pre-DCs were progenitors of both EmGFP/IL-15+ CD8+ DCs and EmGFP/IL-15-/low CD8- DCs, sorted pre-DCs from the BM of Tg+ mice and Tg-controls were transferred to naïve congenic recipients. After 6 days, EmGFP/IL-15 expression by donor-derived DCs was measured. As previously shown (25), pre-DCs gave rise to both DC subsets with preferential generation of CD8- DCs. Nevertheless, we detected two populations of DCs (CD8+ and CD8-) expressing significantly different levels of EmGFP/IL-15 (Figure 5E and 5F) derived from a population of pre-DCs expressing uniform levels of EmGFP/IL-15 (Figure 5C). Thus, while little is known with regard to the precise transcriptional elements that regulate the development of CD8+ versus CD8- DCs, these results indicated that regulation at the IL-15 locus occurred during this cell fate decision.
Just as the OP9-DL1 system can be used to mimic T cell development in vitro, culturing BM cells in vitro with Flt3L can be used to generate a diverse mixture of conventional and plasmacytoid DCs. Importantly, unlike BMDCs generated by culture with GM-CSF, the CD103+ and CD172α+ (SIRPα+) subsets of cDCs produced in Flt3L-containing cultures more accurately reflect the CD8+ and CD8- subsets observed in vivo (34). We hypothesized that if the level of IL-15 expressed by individual subsets of DCs was linked to their cellular fate, the Flt3L-derived BMDCs should exhibit a similar pattern of EmGFP/IL-15 expression between CD103+ and CD172α+ DCs as did CD8+ and CD8- DCs, respectively. Indeed, the in vitro generation of BMDCs with Flt3L exactly mimicked the pattern of IL-15 expression by DC subsets in vivo (Figure 5G). pDCs lacked expression of EmGFP/IL-15, and CD103+ DCs expressed significantly higher levels of EmGFP/IL-15 than CD172α+ DCs when compared to the average MFI of EmGFP/IL-15 in BMDCs grown from Tg- controls (Figure 5G). Thus, our findings suggested that Flt3 signaling initiates a developmental program that includes the regulation of IL-15 expression in developing DCs.
Discussion
IL-15 is an essential cytokine required for the development and maintenance of several populations of immune cells. However, the unfortunate combination of low expression levels and, perhaps, poor detection reagents has made studying certain elements of IL-15 biology extremely difficult. By using a transgenic IL-15 reporter mouse, we showed that IL-15 expression can be detected in numerous subsets of hematopoietically-derived immune cells. Our findings indicated that granulocytic cells such as neutrophils, eosinophils, and basophils have the potential to make IL-15 in vivo. However, it is important to note that proven reagents for detecting IL-15 protein following in vitro culture failed to detect any measurable levels of IL-15 on any myeloid cells, including DCs, directly ex vivo. Thus, while the IL-15 reporter communicates transcriptional rather than translational activity, it nevertheless remains an excellent tool for dissecting the regulation of IL-15 expression in vivo.
Studies have shown that certain subsets of IL-15-dependent cells require IL-15 to be transpresented by a specific subset of cells. For example, limiting IL-15 transpresentation to CD11c+ cells was largely sufficient to restore memory CD8 T cells in the periphery but failed to reconstitute the IEL compartment of the intestine (17). Alternatively, IELs require IL-15 to be delivered by intestinal epithelial cells (18,21). Because many of the progenitor and granulocytic populations that we have here-in shown to express IL-15 are present at extremely low frequencies, it will likely be difficult to examine their individual contributions in maintaining IL-15-dependent homeostasis in vivo. In addition, it is highly likely that some degree of compensatory mechanisms exists when manipulation of IL-15/IL-15Rα expression occurs. For example, when IL-15Rα is conditionally deleted from both CD11c+ and LysM+ cells, the defect in NK cell homeostasis is not exacerbated above deletion in either subset alone, suggesting that CD11c+ cells and LysM+ cells provide similar functions in the absence of the other (21).
The BM has been suggested as a site of long-term memory CD8 and CD4 T cell maintenance (35-38), and this is perhaps the result of IL-15 reservoirs present in that site. Snell et al. identified a subset of memory CD8 T cells in the BM that expressed high levels of GITR (glucocorticoid-induced TNF receptor) but were absent from secondary lymphoid tissues in the periphery and also from the BM of IL-15-deficient mice (39). They further showed that memory CD8T cells upregulated GITR in response to IL-15 signaling in vitro and upregulated GITR upon migration into the BM suggesting that IL-15 signals were delivered specifically within the BM. Similarly, studies in humans have demonstrated the close proximity of CD8 and CD4 memory T cells to IL-15-producing cells in the BM (40). While others have shown that IL-15 can be expressed by BM stromal cells (26,41), ours is the first report describing IL-15 expression by hematopoietic progenitor cells residing in the BM. Interestingly, HSCs have been shown to reside in a specific BM niche using a CXCL12/CXCR4-dependent mechanism (42), and memory T cell behavior in the BM can also be influenced by CXCL12 (36). Furthermore, IL-15 signaling can induce CXCR4 expression on in vitro-generated CD4 memory T cells which perhaps results in the juxtaposition of memory T cells with IL-15-producing hematopoietic progentiors (43). Regardless, evidence clearly suggests that some IL-15 is derived from hematopoietic sources in the BM, enough to significantly increase the presence of NK cells in the BM of IL-15 KO mice reconstituted with WT BM (14). However, it is interesting to note that deletion of IL-15Rα from DCs had no effect on the presence of memory CD8 T cells specifically within the BM (21). This may not be surprising considering the relatively low numbers of DCs present in this location and the expression of IL-15 by a number of other cell types that likely act in conjunction with stromal cells to drive IL-15-dependent responses in the BM.
Our studies show that steady-state IL-15 expression is linked to a myeloid cell fate. There are numerous transcriptional networks at play in the myeloid versus lymphoid cell fate decision, and it is highly unlikely that a single transcription factor will be identified that regulates all instances of IL-15 expression in hematopoietic cells. However, one potential candidate is the ets family member PU.1 (encoded by Sfpi1), which is essential for myeloid cell development, including macrophages and DCs (44-48). Also important for B cell development, gradations of PU.1 expression in progenitor cells drive the generation of B lymphocytes versus macrophages (49). Alternatively, concurrent with commitment to the T cell lineage is a loss of expression of myeloid-specific transcription factors (33,50). In fact, constitutive expression of PU.1 is sufficient to block T cell development at the DN to double positive transition (51). However, recent studies have shown that ETPs maintain a significant level of PU.1 expression and retain myeloid potential, while DN3 cells, which are committed to T cell development, lack PU.1 expression (33). We observed a similar pattern of IL-15 expression in developing T cells where IL-15 was expressed at high levels by ETPs, but Notch signaling led to the downregulation of IL-15 in DN3 and DN4 cells, perhaps by over-riding PU.1-initiated lineage commitment (52-54). Thus, silencing of IL-15 in the lymphoid lineages occurred as progenitors were directed towards a lymphoid fate. Since preliminary studies have suggested that PU.1 can bind to a site in the first intron of the IL-15 gene locus (EV Rothenberg, personal communication), the possibility exists that PU.1 can modulate IL-15 expression in vivo. Interestingly, retroviral transduction of DN3 or DN4 cells with PU.1 enforces a myeloid cell fate when cultured on OP9 stromal cells (50). Whether or not this transcriptional reprogramming would include the upregulation of IL-15 could reveal a direct role for PU.1 in driving IL-15 transcription.
We further found that IL-15 expression was continuously modulated during myelopoiesis, particularly in the development of DC subsets. The Flt3-Flt3L signaling axis plays a dominant role in controlling DC development (55). This conclusion has been established in vivo (56,57) but is also apparent using Flt3L (as opposed to GM-CSF) to generate DCs from BM progenitors in vitro (34). Considering these two commonly used approaches for generating BMDCs, it is interesting to note that previous work shows that CD8+ memory T cells are better maintained by Flt3L-derived DCs than those grown in GM-CSF (58), which could suggest a preferential role for Flt3 signaling in driving optimal IL-15 expression by terminally differentiated DCs. Flt3-Flt3L is also critical for the generation of pDCs, which we have shown lack IL-15 expression. pDCs are unique from their conventional counterparts because they share similarities with developing lymphocytes, including the expression of RAG genes (59). Since we have shown that IL-15 expression is turned off in lymphoid lineages during their development, perhaps the transcriptional network that is responsible for communicating lymphoid-esque characteristics onto this population of innate cells could also be responsible for IL-15 downregulation during their development. Finally, which signals and transcription factors collaborate to control the differentiation of pre-DCs into CD8+ versus CD8- cDCs is currently unknown. However our data, both in vivo and in vitro, indicated that CD8+ DCs express higher levels of IL-15. Id2, IRF8, and Batf3 have all been shown to be required for CD8+ DC development but whether or not these specific transcription factors act at the IL-15 locus remains to be examined. The differential expression of IL-15 by cDC subsets has consequences in vivo as both CD8+ DCs and IL-15Rα were required to maintain a unique population of CD122+ antigen-inexperienced CD8 T cells that exhibit a memory phenotype (28). Importantly, our previous studies have demonstrated that both subsets of cDCs have the capacity to upregulate IL-15 expression following virus-induced inflammation in an IFNaR-dependent fashion suggesting the downregulation of IL-15 by immature CD8- DCs does not involve permanent modifications to the gene locus (23).
In conclusion, the use of an IL-15 reporter transgenic system has facilitated the identification of immune cells with the potential to produce IL-15 in vivo. We have shown a large-scale bifurcation in the two lineages of hematopoietically-derived cells where-by IL-15 expression is primarily limited to myeloid cells and downmodulated during lymphoid cell development. Surprisingly, IL-15 was expressed at high levels by HSCs in the BM, and IL-15 regulation occurred during both thymopoiesis and DC development. In total, our findings suggested that molecular mechanisms responsible for the development of cells in both myeloid and lymphoid lineages were also able to exert regulatory control at the IL-15 gene locus.
Supplementary Material
Acknowledgments
The authors thank Quynh-Mai Pham and the UConn Health Center Flow Cytometry Core for technical assistance and Juan Carlos Zuniga-Pflücker for OP9 and OP9-DL1 stromal cells.
Footnotes
S.L.C is supported by a postdoctoral fellowship (PF-11-152-01-LIB) from The American Cancer Society, and S.W.S was supported by an NIH predoctoral training grant in Cancer Immunology CA009598. Additional support provided by NIH grants 1RC1HL100569 (NIHLB) (to H.L.A.), AI 070910 (to K.S.S.), and R01 AI76457 and U01 AI 095544 (to L.L.).
Abbreviations used in this article: BM, bone marrow; BMDC, bone marrow-derived dendritic cell; DC, dendritic cell; DL1, delta-like 1; DN, double negative; EmGFP, emerald GFP; ETP, early thymic progenitor; HSC, hematopoietic stem cell; IEL, intraepithelial lymphocyte; KO, knockout; LSK, Lin-Sca-1+c-kit+; MFI, mean fluorescence intensity; Tg+ or Tg-, transgene positive or negative; WT, wild-type
References
- 1.Giri JG, Kumaki S, Ahdieh M, Friend DJ, Loomis A, Shanebeck K, DuBose R, Cosman D, Park LS, Anderson DM. Identification and cloning of a novel IL-15 binding protein that is structurally related to the alpha chain of the IL-2 receptor. EMBO J. 1995;15:3654–3633. doi: 10.1002/j.1460-2075.1995.tb00035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duitman EH, Orinska Z, Bulanova E, Paus R, Bulfone-Paus S. How a cytokine is chaperoned through the secretory pathway by complexing with its own receptor: lessons from interleukin-15 (IL-15)/IL-15 receptor alpha. Mol Cell Biol. 2008;28:4851–4861. doi: 10.1128/MCB.02178-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubois S, Mariner J, Waldmann TA, Tagaya Y. IL-15Ralpha recycles and presents IL-15 In trans to neighboring cells. Immunity. 2002;17:537–547. doi: 10.1016/s1074-7613(02)00429-6. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy MK, Glaccum M, Brown SN, Butz EA, Viney JL, Embers M, Matsuki N, Charrier K, Sedger L, Willis CR, Brasel K, Morrissey PJ, Stocking K, Schuh JCL, Joyce S, Peschon J. Reversible defects in natural killer and memory CD8 T cell lineages in Interleukin-15-deficient mice. J Exp Med. 2000;191:771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lodolce JP, Boone DL, Chai S, Swain RE, Dassopoulos T, Trettin S, Ma A. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity. 1998;9:669–676. doi: 10.1016/s1074-7613(00)80664-0. [DOI] [PubMed] [Google Scholar]
- 6.Koka R, Burkett PR, Chien M, Chai S, Chan F, Lodolce JP, Boone DL, Ma A. Interleukin (IL)-15R[alpha]-deficient natural killer cells survive in normal but not IL-15R[alpha]-deficient mice. J Exp Med. 2003;197:977–984. doi: 10.1084/jem.20021836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lodolce JP, Burkett PR, Boone DL, Chien M, Ma A. T cell-independent interleukin 15Ralpha signals are required for bystander proliferation. J Exp Med. 2001;194:1187–1194. doi: 10.1084/jem.194.8.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burkett PR, Koka R, Chien M, Chai S, Chan F, Ma A, Boone DL. IL-15Ralpha expression on CD8+ T cells is dispensable for T cell memory. Proc Natl Acad Sci U S A. 2003;100:4724–4729. doi: 10.1073/pnas.0737048100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olsen SK, Ota N, Kishishita S, Kukimoto-Niino M, Murayama K, Uchiyama H, Toyama M, Terada T, Shirouzu M, Kanagawa O, Yokoyama S. Crystal Structure of the interleukin-15.interleukin-15 receptor alpha complex: insights into trans and cis presentation. J Biol Chem. 2007;282:37191–37204. doi: 10.1074/jbc.M706150200. [DOI] [PubMed] [Google Scholar]
- 10.Schluns KS, Williams K, Ma A, Zheng XX, Lefrançois L. Cutting Edge: Requirement for IL-15 in the generation of primary and memory antigen-specific CD8 T cells. J Immunol. 2002;168:4827–4831. doi: 10.4049/jimmunol.168.10.4827. [DOI] [PubMed] [Google Scholar]
- 11.Wu Z, Xue HH, Bernard J, Zeng R, Issakov D, Bollenbacher-Reilley J, Belyakov IM, Oh S, Berzofsky JA, Leonard WJ. The IL-15 receptor {alpha} chain cytoplasmic domain is critical for normal IL-15Ralpha function but is not required for trans-presentation. Blood. 2008;112:4411–4419. doi: 10.1182/blood-2007-03-080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castillo EF, Schluns KS. Regulating the immune system via IL-15 transpresentation. Cytokine. 2012;59:479–490. doi: 10.1016/j.cyto.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schluns KS, Klonowski KD, Lefrançois L. Trans-regulation of memory CD8 T cell prolferation by IL-15Rα+ bone marrow-derived cells. Blood. 2004;103:988–994. doi: 10.1182/blood-2003-08-2814. [DOI] [PubMed] [Google Scholar]
- 14.Schluns KS, Nowak EC, Cabrera-Hernandez A, Puddington L, Lefrancois L, Aguila HL. Distinct cell types control lymphoid subset development by means of IL-15 and IL-15 receptor alpha expression. Proc Natl Acad Sci U S A. 2004;101:5616–5621. doi: 10.1073/pnas.0307442101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandau MM, Schluns KS, Lefrancois L, Jameson SC. Cutting edge: transpresentation of IL-15 by bone marrow-derived cells necessitates expression of IL-15 and IL-15R alpha by the same cells. J Immunol. 2004;173:6537–6541. doi: 10.4049/jimmunol.173.11.6537. [DOI] [PubMed] [Google Scholar]
- 16.Burkett PR, Koka R, Chien M, Chai S, Boone DL, Ma A. Coordinate expression and trans presentation of interleukin (IL)-15Ralpha and IL-15 supports natural killer cell and memory CD8+ T cell homeostasis. J Exp Med. 2004;200:825–834. doi: 10.1084/jem.20041389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stonier SW, Ma LJ, Castillo EF, Schluns KS. Dendritic cells drive memory CD8 T-cell homeostasis via IL-15 transpresentation. Blood. 2008;112:4546–4554. doi: 10.1182/blood-2008-05-156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma LJ, Acero LF, Zal T, Schluns KS. Trans-presentation of IL-15 by intestinal epithelial cells drives development of CD8alphaalpha IELs. J Immunol. 2009;183:1044–1054. doi: 10.4049/jimmunol.0900420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castillo EF, Stonier SW, Frasca L, Schluns KS. Dendritic cells support the in vivo development and maintenance of NK cells via IL-15 trans-presentation. J Immunol. 2009;183:4948–4956. doi: 10.4049/jimmunol.0900719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castillo EF, Acero LF, Stonier SW, Zhou D, Schluns KS. Thymic and peripheral microenvironments differentially mediate development and maturation of iNKT cells by IL-15 transpresentation. Blood. 2010;116:2494–2503. doi: 10.1182/blood-2010-03-277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mortier E, Advincula R, Kim L, Chmura S, Barrera J, Reizis B, Malynn BA, Ma A. Macrophage- and Dendritic-Cell-Derived Interleukin-15 Receptor Alpha Supports Homeostasis of Distinct CD8(+) T Cell Subsets. Immunity. 2009;31:811–822. doi: 10.1016/j.immuni.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 22.Fehniger TA, Suzuki K, Ponnappan A, VanDeusen JB, Cooper MA, Florea SM, Freud AG, Robinson ML, Durbin J, Caligiuri MA. Fatal leukemia in interleukin 15 transgenic mice follows early expansions in natural killer and memory phenotype CD8+ T cells. J Exp Med. 2001;193:219–231. doi: 10.1084/jem.193.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colpitts SL, Stoklasek TA, Plumlee CR, Obar JJ, Guo C, Lefrancois L. Cutting edge: the role of IFN-alpha receptor and MyD88 signaling in induction of IL-15 expression in vivo. J Immunol. 2012;188:2483–2487. doi: 10.4049/jimmunol.1103609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmes R, Zuniga-Pflucker JC. The OP9-DL1 system: generation of T-lymphocytes from embryonic or hematopoietic stem cells in vitro. Cold Spring Harb Protoc. 2009 doi: 10.1101/pdb.prot5156. [DOI] [PubMed] [Google Scholar]
- 25.Liu K, Victora GD, Schwickert TA, Guermonprez P, Meredith MM, Yao K, Chu FF, Randolph GJ, Rudensky AY, Nussenzweig M. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324:392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grabstein KH, Eisenman J, Shanebeck K, Rauch C, Srinivasan S, Fung V, Beers C, Richardson J, Schoenborn MA, Ahdieh M. Cloning of a T cell growth factor that interacts with the beta chain of the interleukin-2 receptor. Science. 1994;264:965–968. doi: 10.1126/science.8178155. [DOI] [PubMed] [Google Scholar]
- 27.Mattei F, Schiavoni G, Belardelli F, Tough DF. IL-15 is expressed by dendritic cells in response to type I IFN, double- stranded RNA, or lipopolysaccharide and promotes dendritic cell activation. J Immunol. 2001;167:1179–1187. doi: 10.4049/jimmunol.167.3.1179. [DOI] [PubMed] [Google Scholar]
- 28.Sosinowski T, White JT, Cross EW, Haluszczak C, Marrack P, Gapin L, Kedl RM. CD8alpha+ Dendritic Cell Trans Presentation of IL-15 to Naive CD8+ T Cells Produces Antigen-Inexperienced T Cells in the Periphery with Memory Phenotype and Function. J Immunol. 2013;190:1936–1947. doi: 10.4049/jimmunol.1203149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyazaki S, Ishikawa F, Shimizu K, Ubagai T, Edelstein PH, Yamaguchi K. Gr-1high polymorphonuclear leukocytes and NK cells act via IL-15 to clear intracellular Haemophilus influenzae in experimental murine peritonitis and pneumonia. J Immunol. 2007;179:5407–5414. doi: 10.4049/jimmunol.179.8.5407. [DOI] [PubMed] [Google Scholar]
- 30.Mortier E, Woo T, Advincula R, Gozalo S, Ma A. IL-15Ralpha chaperones IL-15 to stable dendritic cell membrane complexes that activate NK cells via trans presentation. J Exp Med. 2008;205:1213–1225. doi: 10.1084/jem.20071913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, Meraz A, Bhandoola A. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 32.Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 33.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 34.Brasel K, De ST, Smith JL, Maliszewski CR. Generation of murine dendritic cells from flt3-ligand-supplemented bone marrow cultures. Blood. 2000;96:3029–3039. [PubMed] [Google Scholar]
- 35.Becker TC, Coley SM, Wherry EJ, Ahmed R. Bone marrow is a preferred site for homeostatic proliferation of memory CD8 T cells. J Immunol. 2005;174:1269–1273. doi: 10.4049/jimmunol.174.3.1269. [DOI] [PubMed] [Google Scholar]
- 36.Mazo IB, Honczarenko M, Leung H, Cavanagh LL, Bonasio R, Weninger W, Engelke K, Xia L, McEver RP, Koni PA, Silberstein LE, von Andrian UH. Bone marrow is a major reservoir and site of recruitment for central memory CD8+ T cells. Immunity. 2005;22:259–270. doi: 10.1016/j.immuni.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 37.Parretta E, Cassese G, Barba P, Santoni A, Guardiola J, Di Rosa F. CD8 cell division maintaining cytotoxic memory occurs predominantly in the bone marrow. J Immunol. 2005;174:7654–7664. doi: 10.4049/jimmunol.174.12.7654. [DOI] [PubMed] [Google Scholar]
- 38.Tokoyoda K, Zehentmeier S, Hegazy AN, Albrecht I, Grun JR, Lohning M, Radbruch A. Professional memory CD4+ T lymphocytes preferentially reside and rest in the bone marrow. Immunity. 2009;30:721–730. doi: 10.1016/j.immuni.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 39.Snell LM, Lin GH, Watts TH. IL-15-dependent upregulation of GITR on CD8 memory phenotype T cells in the bone marrow relative to spleen and lymph node suggests the bone marrow as a site of superior bioavailability of IL-15. J Immunol. 2012;188:5915–5923. doi: 10.4049/jimmunol.1103270. [DOI] [PubMed] [Google Scholar]
- 40.Herndler-Brandstetter D, Landgraf K, Jenewein B, Tzankov A, Brunauer R, Brunner S, Parson W, Kloss F, Gassner R, Lepperdinger G, Grubeck-Loebenstein B. Human bone marrow hosts polyfunctional memory CD4+ and CD8+ T cells with close contact to IL-15-producing cells. J Immunol. 2011;186:6965–6971. doi: 10.4049/jimmunol.1100243. [DOI] [PubMed] [Google Scholar]
- 41.Mrozek E, Anderson P, Caligiuri MA. Role of interleukin-15 in the development of human CD56+ natural killer cells from CD34+ hematopoietic progenitor cells. Blood. 1996;87:2632–2640. [PubMed] [Google Scholar]
- 42.Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 43.Jourdan P, Vendrell JP, Huguet MF, Segondy M, Bousquet J, Pene J, Yssel H. Cytokines and cell surface molecules independently induce CXCR4 expression on CD4+ CCR7+ human memory T cells. J Immunol. 2000;165:716–724. doi: 10.4049/jimmunol.165.2.716. [DOI] [PubMed] [Google Scholar]
- 44.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 45.McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, Maki RA. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 46.Scott EW, Fisher RC, Olson MC, Kehrli EW, Simon MC, Singh H. PU.1 functions in a cell-autonomous manner to control the differentiation of multipotential lymphoid-myeloid progenitors. Immunity. 1997;6:437–447. doi: 10.1016/s1074-7613(00)80287-3. [DOI] [PubMed] [Google Scholar]
- 47.Guerriero A, Langmuir PB, Spain LM, Scott EW. PU.1 is required for myeloid-derived but not lymphoid-derived dendritic cells. Blood. 2000;95:879–885. [PubMed] [Google Scholar]
- 48.Carotta S, Dakic A, D'Amico A, Pang SH, Greig KT, Nutt SL, Wu L. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 2010;32:628–641. doi: 10.1016/j.immuni.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 49.DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 50.Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 51.Anderson MK, Weiss AH, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity. 2002;16:285–296. doi: 10.1016/s1074-7613(02)00277-7. [DOI] [PubMed] [Google Scholar]
- 52.Franco CB, Scripture-Adams DD, Proekt I, Taghon T, Weiss AH, Yui MA, Adams SL, Diamond RA, Rothenberg EV. Notch/Delta signaling constrains reengineering of pro-T cells by PU.1. Proc Natl Acad Sci U S A. 2006;103:11993–11998. doi: 10.1073/pnas.0601188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rothenberg EV, Scripture-Adams DD. Competition and collaboration: GATA-3, PU.1, and Notch signaling in early T-cell fate determination. Semin Immunol. 2008;20:236–246. doi: 10.1016/j.smim.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Del Real MM, Rothenberg EV. Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3. Development. 2013;140:1207–1219. doi: 10.1242/dev.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmid MA, Kingston D, Boddupalli S, Manz MG. Instructive cytokine signals in dendritic cell lineage commitment. Immunol Rev. 2010;234:32–44. doi: 10.1111/j.0105-2896.2009.00877.x. [DOI] [PubMed] [Google Scholar]
- 56.Mckenna HJ, Stocking KL, Miller RE, Brasel K, De ST, Maraskovsky E, Maliszewski CR, Lynch DH, Smith J, Pulendran B, Roux ER, Teepe M, Lyman SD, Peschon JJ. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95:3489–3497. [PubMed] [Google Scholar]
- 57.Waskow C, Liu K, rrasse-Jeze G, Guermonprez P, Ginhoux F, Merad M, Shengelia T, Yao K, Nussenzweig M. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat Immunol. 2008;9:676–683. doi: 10.1038/ni.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frasca L, Stonier SW, Overwijk WW, Schluns KS. Differential mechanisms of memory CD8 T cell maintenance by individual myeloid cell types. J Leukoc Biol. 2010;88:69–78. doi: 10.1189/jlb.1209816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shigematsu H, Reizis B, Iwasaki H, Mizuno S, Hu D, Traver D, Leder P, Sakaguchi N, Akashi K. Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity. 2004;21:43–53. doi: 10.1016/j.immuni.2004.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.