Abstract
Background & Aims
Photodynamic therapy (PDT) has been shown to be effective in the treatment of high-grade dysplasia (HGD)/mucosal carcinoma in Barrett’s esophagus (BE). Substantial proportions of patients do not respond to PDT or progress to carcinoma despite PDT. The role of biomarkers in predicting response to PDT is unknown. We aimed to determine if biomarkers known to be associated with neoplasia in BE can predict loss of dysplasia in patients treated with ablative therapy for HGD/intramucosal cancer.
Methods
Patients with BE and HGD/intramucosal cancer were studied prospectively from 2002 to 2006. Biomarkers were assessed using fluorescence in situ hybridization performed on cytology specimens, for region-specific and centromeric probes. Patients were treated with PDT using cylindric diffusing fibers (wavelength, 630 nm; energy, 200 J/cm fiber). Univariate and multiple variable logistic regression was performed to determine predictors of response to PDT.
Results
A total of 126 consecutive patients (71 who underwent PDT and 55 patients who did not undergo PDT and were under surveillance, to adjust for the natural history of HGD), were included in this study. Fifty (40%) patients were responders (no dysplasia or carcinoma) at 3 months after PDT. On multiple variable analysis, P16 allelic loss (odds ratio [OR], 0.32; 95% confidence interval [CI], 0.10 – 0.96) predicted decreased response to PDT. BE segment length (OR, 0.71; 95% CI, 0.59 – 0.85), and performance of PDT (OR, 7.17; 95% CI, 2.50 –20.53) were other independent predictors of loss of dysplasia.
Conclusions
p16 loss detected by fluorescence in situ hybridization can help predict loss of dysplasia in patients with BE and HGD/mucosal cancer. Biomarkers may help in the selection of appropriate therapy for patients and improve treatment outcomes.
Barrett’s esophagus (BE) predisposes to esophageal adenocarcinoma, a cancer with one of the fastest increasing incidence rates over the past decade and a highly lethal malignancy once symptoms develop.1,2 High-grade dysplasia (HGD) has been thought to be a marker of progression to carcinoma as well as occult carcinoma.3– 8 Over the past decade, endoscopic therapy has been emerging as an alternative to esophagectomy because of the significant mortality and morbidity associated with esophagectomy.9,10 Different endoscopic ablation techniques with variable success rates have been reported for the treatment of HGD.11–16
Photodynamic therapy (PDT) uses the combination of a photosensitizer and light. Exposure to light of a specific wavelength leads to activation of the photosensitizer followed by creation of singlet oxygen, which causes tissue injury and is thought to be mediated by apoptosis, vascular injury, and the induction of immune and inflammatory responses.17 A randomized multicenter trial compared PDT with surveillance and treatment with omeprazole in patients with HGD. After a 24-month follow-up period, complete ablation of HGD was noted in 77% of patients vs 39% in the omeprazole group.18 These results also have been extended to a 5-year follow-up evaluation.19 We have reported that long-term outcomes (overall mortality and cancer-free survival) are comparable between patients treated with esophagectomy and PDT.20
A number of genetic alterations have been described in BE. These include loss of cell-cycle checkpoint genes such as p16 and p53. Loss or inactivation of these genes by allelic loss (deletions or loss of heterozygosity), point mutations, or promoter hypermethylation (for p16) have been found in a substantial number of patients with BE.21,22 Other genetic alterations involving gains/amplifications of proto-oncogenes (and growth factors/growth factor receptors) also have been described.23,24 We recently showed that a fluorescence in situ hybridization (FISH) assay using a 4-probe combination set consisting of probes to the 8q24, 9p21, 17q11.2, and 20q13.2 loci is able to distinguish between HGD/carcinoma and lesser grades of dysplasia with reasonable sensitivity and specificity (80%).25
Although endoscopic therapies have been successful in ablating HGD and early cancers in a large proportion of patients, it is evident from clinical trials that substantial proportions of patients either do not respond (23%26) or progress to carcinoma (13%26) after treatment with PDT. Limited information is available on whether biomarkers can help predict response to treatment with newer ablative techniques such as PDT.27,28 Identification of biomarkers that predict response to PDT could improve risk stratification in patients with HGD/early cancer and unnecessary toxicity. Patients with favorable biomarker status could be treated endoscopically, whereas an unfavorable profile would identify patients with probable poor response to PDT: this subgroup of patients could be treated with either surgery or other forms of endoscopic therapy.
We hypothesized that assessment of biomarkers can help predict response to ablative treatment in HGD/mucosal cancer in BE: with patients without biomarkers predictive of progression to neoplasia being more likely to respond to ablative therapy than those with biomarkers. The primary aim of this study was to determine if genetic alterations detected by FISH are able to predict loss of dysplasia in patients with HGD/mucosal cancer.
Materials and Methods
Patients with HGD and/or mucosal cancer (defined as carcinoma confined to the mucosa, without invasion of the submucosa) seen in the Barrett’s Esophagus Unit at St Mary’s Hospital in Rochester, Minnesota, between 2002 and 2006 were included in this study.
PDT Group
Inclusion criteria were as follows: (1) presence of HGD/intramucosal carcinoma on biopsy specimens, (2) assessment of biomarkers using FISH before PDT, and (3) treatment with PDT and availability of biopsy results at the first endoscopic surveillance visit (3 months) after PDT. Exclusion criteria included the following: (1) evidence of submucosal invasion of carcinoma on endoscopic mucosal resection (EMR) pathology (these patients were referred to surgery) and (2) patients who were unwilling or unable to consent to the study. Clinical, demographic, and endoscopic data were extracted from a prospectively maintained database, including length of Barrett’s segment, performance of EMR before PDT (including number of EMRs), number of PDT treatments per application, results of post-PDT biopsies (classified as carcinoma, HGD, low-grade dysplasia, nondysplastic BE29), and biomarkers obtained using FISH. HGD was classified into diffuse and focal high-grade dysplasia using previously published criteria.30
Control Group
Patients who elected not to undergo PDT and remain under surveillance (which included endoscopy with surveillance biopsies and/or EMR every 3 months) were included in the control group to adjust for the possible loss of dysplasia as a reflection of the natural history or biology of HGD. The initial assessment and follow-up evaluation were similar to that of the PDT cohort. Cytology specimens were collected and processed for FISH assessment of biomarkers in the same manner as the PDT group. Demographic, clinical, and biomarker data were collected prospectively in this cohort as well.
All patients underwent 4-quadrant biopsies for every centimeter of the involved esophagus. All patients had their diagnosis of HGD or mucosal cancer confirmed by 2 experienced gastrointestinal pathologists using standard criteria.29 Baseline assessments also included endoscopic ultrasound and EMR for any mucosal abnormalities. Computerized tomography scans of the chest and upper abdomen were obtained in all patients.
Cytology Specimen Acquisition and Slide Preparation
Cytology specimen acquisition and slide preparation were performed as previously described25 using a standard cytology brush (Hobbs Medical Inc, Stafford Springs, CT). Cytology specimens for FISH were obtained during the endoscopy immediately preceding PDT.
FISH
Esophageal brushing cells were harvested, fixed, and placed on a slide as previously described.25 The following fluorescently labeled DNA probes then were hybridized to the specimens: 8q24.12-q24.13 (C-MYC), 9p21 (p16), 17p13.1 (p53), 17q11.2-q12 (HER-2), and 20q13.2 (Abbott Molecular Inc, Des Plaines, IL). The slide then was washed and stained with the nuclear counter-stain 4′,6-diamidino-2-phenylindole (DAPI I; Abbott Molecular Inc). Fluorescence microscopy, with unique band filters specific for each of the probe fluorophores, was used to analyze and record all observed signal patterns for 100 (50 minimum) consecutive, noninflammatory, nonsquamous cells. Enumeration was performed without knowledge of the patient’s clinical or histologic diagnosis. Receiver operator characteristic (ROC) curves were used to determine optimal cut-off values for the probes.9,8,17,20 Based on these ROC curves, a specimen was considered positive for p16 loss if 11% or more of cells showed hemizygous 9p21 loss, 6% or more of cells showed homozygous 9p21 loss, 11% or more of cells showed a mixture of hemizygous and homozygous 9p21 loss, or 5% or more of cells showed gains of 8q24, 17q11.2, or 20q13. Based on the average percentage of P53 loss ± 3 SD observed in a normal value study (unpublished data), specimens were considered positive for p53 loss if 14% or more of cells showed 17p13.1 loss. Multiple gains was defined as gains of 2 or more of the following loci, 8q24, 17q11.2, or 20q13, using previously defined thresholds.
PDT
Porfimer sodium (Photofrin; Axcan Pharma, Mont-Saint-Hilaire, Quebec, Canada) was used as a photosensitizer, at a dose of 2 mg/kg. Photofrin was administered intravenously 48 hours before photoradiation. Photoradiation was performed using a bare cylindric diffusing fiber. The cylindric diffusing fibers were either 2.5-or 5.0-cm long (Fibers Direct, Andover, MA). The cylindric diffusing fiber was passed through the accessory channel of the endoscope and placed in the center of the esophageal lumen. The light was delivered from a laser (Lambda Plus; Coherent, Palo Alto, CA; or Diomed; Diomed Inc, Andover, MA) producing 630 nm light with an adjusted power output of 400 mW/cm fiber, delivering a total energy of 200 J/cm fiber energy to the mucosa.
EMR
Focal endoscopically visible lesions underwent EMR for diagnostic purposes to determine histology and exclude carcinoma. EMR was performed as previously described.31 EMR was performed using a commercially available EMR cap (EMR-001; Olympus America Inc, Center Valley, PA). Initially, 5–10 mL of diluted epinephrine (1:200,000) solution was injected into the submucosa underneath the lesion. Subsequently, the lesion was suctioned into the cap after positioning of a crescent snare. The snare then was closed with application of cautery current removing the tissue. PDT was delayed a minimum of 4 weeks if an EMR was performed to allow healing of the EMR site(s).
Follow-Up Evaluation
All patients were placed on twice-a-day proton pump inhibitor therapy at initial evaluation and continued proton pump inhibitor therapy at this dose for at least 2 years after PDT. Patients were educated carefully regarding PDT and its complications, especially dysphagia and photosensitivity by the physicians, nurse practitioner, and clinical coordinators. Follow-up evaluation included endoscopic surveillance with biopsies and EMR if indicated. Four-quadrant biopsy specimens were taken every 1–2 cm across the entire segment that had Barrett’s. Any visible lesions were biopsied separately or, if suspicious, were removed by EMR. Surveillance was performed every 3 months for 2 years, then every 6 months for 1–2 years if HGD was eliminated. If HGD persisted, patients were followed up at 3-month intervals. If low-grade dysplasia was present, then patients were followed up every 6 months. If only nondysplastic Barrett’s mucosa or normal squamous mucosa was present at 2 years, patients were followed up annually.
The primary end point for the study was defined as the absence of carcinoma or any dysplasia in biopsy specimens taken at the 3-month follow-up visit after PDT.
Sample Size Estimation/Statistical Methods
We estimated there was approximately 80% power to detect univariate associations between an individual biomarker and 3-month response rates corresponding to differences of roughly 17% to 19% in the rates for presence vs absence of any particular biomarker. A logistic regression model to discriminate between (3-month) responders vs nonresponders using a combination of the biomarkers could consider several biomarkers simultaneously assuming roughly 50–55 responders at 3 months. We estimated that a sample size of 100 patients would provide a sufficient number of patients with a response to support this statistical analysis, using both univariate and multiple logistic regression methods.
Continuous variables were summarized as means (± SD) or medians (interquartile range) as warranted. The associations of baseline characteristics with response outcome were assessed for continuous variables using the 2-sample t test or the Wilcoxon rank-sum test depending on the distribution of the baseline variable. The univariate associations of response with baseline categoric data were assessed using the chi-square test (or the Fisher exact test when necessary because of small individual cell frequencies). The univariate associations with 3-month response were summarized as odds ratios (ORs) with 95% confidence intervals (95% CIs) from the coefficients in univariate logistic regression models. Individual ORs for each of the biomarkers adjusted for clinically relevant variables (age, sex, length of the Barrett’s segment, PDT, and EMR) also were computed from corresponding logistic regression models. In addition, a model with the clinical variables, P16 loss, P53 loss, and multiple gains, was examined. Given the limited number of patients with response (N = 50), additional multiple variable models (clinical variables plus subsets of biomarkers) were explored informally to identify potential combinations of biomarkers that might provide a useful prediction model. ROC curves were constructed using the predicted probabilities for several models.
A P value of less than .05 was considered statistically significant and the P values for the univariate statistical tests were not corrected for multiple testing because the potential predictive usefulness of each biomarker was of interest. We also decided a priori to include a variable combining multiple gains and/or gains at the 8q24/17q13.1/20q13 loci in the analysis because these markers share a common pathogenesis.
Associations of BE length with biomarker category were assessed using the Wilcoxon rank-sum test and the association of clinical and biomarker variables with having undergone PDT was assessed using the Wilcoxon rank-sum test, the chi-square test, or the Fisher exact test. Data management and statistical analysis were performed using SAS software (SAS Institute Inc, Cary, NC).
Results
A total of 126 unselected patients (71 who underwent PDT and 55 patients who did not undergo PDT, labeled as controls) were included in this study. The median age of the patients was 68.4 years (interquartile range, 60.5–75 y). A total of 113 patients (90%) were men. A total of 104 (83%) had HGD and the remainder had mucosal cancer at study entry. Of the 104 HGD patients, 81 (80%) had diffuse HGD and the remainder had focal HGD. The mean BE segment length was 5.2 cm (SD, 3.4 cm; range, 1–16 cm). A total of 109 (86%) patients had EMR performed before PDT. Overall, 50 (40%) patients did not have evidence of any dysplasia or carcinoma on surveillance biopsies taken at 3 months after PDT or at the first 3-month surveillance in controls (38 of 71 patients who underwent PDT and 12 of 55 controls). Among these, 28 patients had no BE and 22 had nondysplastic BE. Among nonresponders, 25 had low-grade dysplasia, 48 had HGD, and 3 had carcinoma. The mean follow-up period of patients after PDT was 13.8 months (SD, 8.9 mo).
No significant association of PDT therapy with age, sex, BE segment length, and having undergone EMR was detected (Table 1). The distribution of biomarkers in those patients without PDT also was comparable with that in patients who underwent PDT except for P53 allelic loss, which was more prevalent in the control group (Table 1).
Table 1.
Baseline Characteristics in PDT and Control Groups
| Variable | PDT group (N = 71) | Control group (N = 55) | P value |
|---|---|---|---|
| Mean age, y (SD) | 67.4 (10.3) | 67.5 (11.3) | .86a |
| Male sex, N (%) | 65 (91) | 48 (87) | .43 |
| Mean BE segment length, cm (SD) | 5.5 (3.3) | 4.9 (3.4) | .20# |
| EMR before PDT, N (%) | 59 (83) | 50 (91) | .20 |
| P16 allelic loss, N (%) | 25 (35) | 15 (27) | .34 |
| P53 allelic loss, N (%)b | 14 (21) | 20 (38) | .04 |
| C-MYC gain, N (%) | 33 (47) | 25 (45) | .85 |
| HER2-NEU gain, N (%) | 16 (23) | 17 (31) | .33 |
| 20q13.2 gain, N (%) | 24 (34) | 21 (38) | .65 |
| Multiple gains, N (%) | 25 (35) | 22 (40) | .62 |
Obtained using the Kruskal–Wallis test; remaining P values obtained using the chi-square test.
Data available for 120 patients.
The distribution of genetic alterations detected by FISH in the entire cohort of patients is shown in Table 2. No association of biomarker status with baseline HGD/mucosal cancer status was detected (Table 2).
Table 2.
Distribution of Genetic Alterations Detected by FISH in Patients With HGD and Mucosal Cancer
| Biomarker | HGD, N (%) (N = 104) | Mucosal cancer, N (%) (N = 22) | P valuea |
|---|---|---|---|
| Overall FISH positivity | 70 (68) | 16 (73) | .58 |
| P16 loss | 35 (32) | 6 (27) | .26 |
| P53 lossb | 28 (28) | 6 (30) | .85 |
| C-MYC (8q24) gain | 43 (46) | 12 (54) | .74 |
| HER2-NEU (17q) gain | 27 (27) | 6 (27) | .42 |
| 20q13 gain | 34 (33) | 11 (50) | .15 |
| Multiple gains | 36 (35) | 11 (50) | .19 |
| Multiple gains/any gains | 50 (51) | 12 (55) | .25 |
Obtained using the chi-square test or the Fischer exact test as appropriate.
Data available for 120 patients (100 patients with HGD and 20 with mucosal cancer).
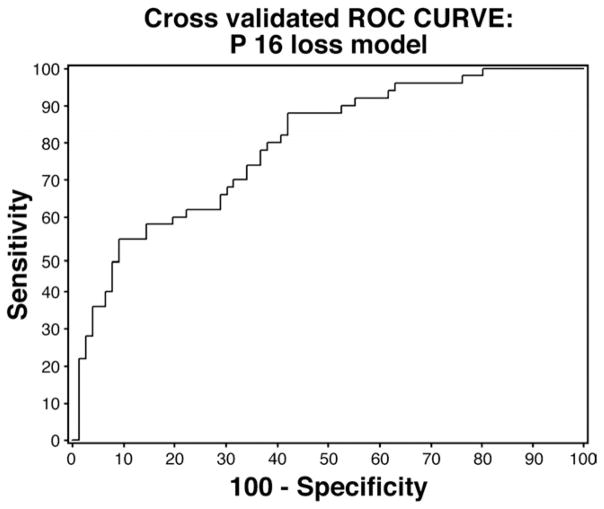
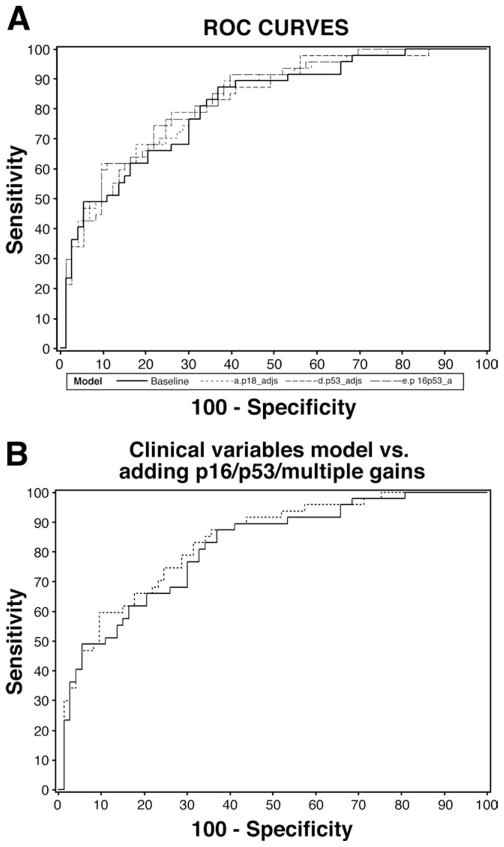
On univariate analysis (Table 3), increasing BE segment length, P16 allelic loss, and P53 allelic loss predicted lack of response. Performance of PDT increased the odds of response on univariate analysis. Age, sex, EMR, and other biomarkers were not significant predictors on univariate analysis. ORs for individual biomarkers adjusted for clinical variables (age, sex, BE segment length, PDT, and EMR) are presented in Table 4. P16 allelic loss was a significant predictor of lack of response, with those with P16 loss having a 75% reduction in the odds for response (loss of dysplasia at 3 months after ablative therapy) relative to those without P16 loss. A model with P16 loss, P53 loss, and multiple gains adjusting for clinical variables also was examined and the results are shown in Table 5. P16 allelic loss was a significant predictor of lack of response after adjusting for all clinical variables and P53 loss as well as multiple gains. Additional analyses did not reveal any interaction between P16 and P53 loss (ie, the odds for response in those with P16 loss did not significantly depend on P53 status). ROC curves were constructed for several models. The primary analyses indicated a model for P16 loss (adjusted for clinical variables) was the only model with a statistically significant biomarker and thus a cross-validated (leave-one-out method) model was used to generate the ROC curve for this model (Figure 1). Additional ROC curves for other models: (1) P53 loss, (2) P16 loss, (3) both P16 and P53 loss (Figure 2A), and (4) P16 loss, P53 loss, and multiple gains (Figure 2B), in each case adjusting for clinical variables, also are presented. A multivariable model with all clinical variables and biomarkers showed increasing BE segment length (OR, 0.69; 95% CI, 0.56 – 0.84) and PDT (OR, 7.09; 95% CI, 2.38–21.15) to be significant predictors of loss of dysplasia, with P16 loss being borderline significant (OR, 0.34; 95% CI, 0.11–1.05). However, this model with 12 variables may be overfit given the limited number of patients (N = 50) with response and the potential overlap among individual biomarkers.
Table 3.
Predictors of Absence of Dysplasia or Adenocarcinoma at 3-Month Follow-Up Evaluation on Univariate Analyses
| Variable (N = data available on) | Groups | Proportion of responders | OR (95% CI) |
|---|---|---|---|
| Age, y (n = 126) | ≤67 y | 40% (23/57) | 1.00 (0.97–1.04) |
| >67 y | 39% (27/69) | ||
| Male sex (n = 126) | Yes | 39% (44/113) | 1.34 (0.42–4.26) |
| No | 46% (6/13) | ||
| EMR before PDT (n = 126) | Yes | 37% (40/109) | 0.41 (0.14–1.15) |
| No | 59% (10/17) | ||
| BE segment length (n = 126) | ≤5 cm | 50% (40/79) | 0.77 (0.67–0.88) |
| >5 cm | 21% (10/47) | ||
| PDT (n = 126) | Yes | 54% (38/71) | 4.13 (1.87–9.11) |
| No | 22% (12/55) | ||
| P16 loss (n = 126) | Yes | 15% (6/41) | 0.32 (0.14–0.75) |
| No | 50% (36/72) | ||
| P53 loss (n = 120) | Yes | 21% (7/34) | 0.30 (0.12–0.76) |
| No | 47% (40/86) | ||
| C-MYC (8q24) gain (n = 125) | Yes | 41% (20/49) | 0.91 (0.44–1.86) |
| No | 39% (30/77) | ||
| HER2-NEU (17q) gain (n = 124) | Yes | 28% (5/18) | 0.50 (0.21–1.20) |
| No | 42% (45/108) | ||
| 20q13 gain (n = 125) | Yes | 33% (9/27) | 0.68 (0.32–1.45) |
| No | 41% (41/99) | ||
| Multiple gains (n = 125) | Yes | 33% (16/49) | 0.70 (0.33–1.49) |
| No | 44% (34/77) | ||
| Multiple gains or any gains (n = 125) | Yes | 33% (25/75) | 0.84 (0.41–1.72) |
| No | 50% (25/50) |
Table 4.
Biomarker Predictors of Absence of Dysplasia and Adenocarcinoma at 3-Month Follow-Up Evaluation
| Biomarker | OR (95% CI) |
|---|---|
| P16 loss | 0.25 (0.09–0.75) |
| P53 loss | 0.45 (0.16–1.32) |
| C-MYC gain | 1.07 (0.44–2.56) |
| HER2-NEU gain | 0.60 (0.03–44.00) |
| 20 q gain | 0.50 (0.01–37.49) |
| Multiple gains | 0.15 (0.01–3.72) |
OR for each biomarker adjusted for the following clinical variables: age, sex, Barrett’s segment length, PDT, and EMR.
Table 5.
Predictors of Absence of Dysplasia and Adenocarcinoma at 3-Month Follow-Up Evaluation on Multivariable Analysis (N = 120)
| Variable | OR (95% CI) |
|---|---|
| Age | 1.02 (0.97–1.07) |
| Male sex | 1.67 (0.37–7.60) |
| EMR before PDT | 0.30 (0.08–1.18) |
| BE segment length | 0.71 (0.59–0.85) |
| PDT | 7.17 (2.50–20.53) |
| P16 loss | 0.32 (0.10–0.96) |
| P 53 loss | 0.53 (0.14–1.97) |
| Multiple gains | 1.19 (0.37–3.79) |
Area under curve, 0.83 (SE, 0.03).
Figure 1.
ROC curve for cross-validated model (see Materials and Methods section) with P16 loss (adjusted for clinical variables: age, sex, Barrett’s segment length, PDT, and EMR, N = 126). Area under the curve, 0.79 (SE, 0.03).
Figure 2.
(A) ROC curves for models with clinical variables (age, sex, Barrett’s segment length, PDT, and EMR), P16 loss, P53 loss, both P16 and P53 loss each adjusted for clinical variables. —, Model with clinical variables: AUC, 0.83 (SE, 0.03); - - -, model with clinical variables and P16 loss: AUC, 0.83 (SE, 0.03); – – –, model with clinical variables and P53 loss: AUC, 0.82 (SE, 0.03); — — —, model with clinical variables and P16 and P53 loss: AUC, 0.83 (SE, 0.03). (B) ROC curve for model with P16 loss and P53 loss and multiple gains, adjusting for clinical variables. AUC, 0.83 (SE, 0.03). AUC, area under the curve.
Discussion
In this large prospective study, we assessed genetic alterations detected by FISH as predictive factors for response to PDT in patients with BE and HGD/intramucosal cancer. We found that P16 loss was an independent predictor of a lack of response (defined as the absence of dysplasia on biopsy specimens taken at the first 3-month follow-up evaluation), after adjusting for other clinical predictors of response (BE segment length and performance of EMR). This was a large prospective study that assessed the utility of biomarkers assessed by FISH for predicting response to ablative therapy in human subjects being treated for esophageal neoplasia.
The specific chromosomal loci examined in this study (listed in Table 1) were chosen based on a previous study from our institution. We found that a probe set containing FISH probes to these loci distinguished adenocarcinoma and HGD from lesser grades of dysplasia with reasonable sensitivity and specificity.25 In addition, we selected these probes for their reliable performance characteristics (quality of hybridization), as well as the observation that gain/amplification at proto-oncogene loci may be a more specific indicator of neoplasia than chromosomal losses (because normal cells may show artifactual losses owing to signal overlap or incomplete hybridization).25,32
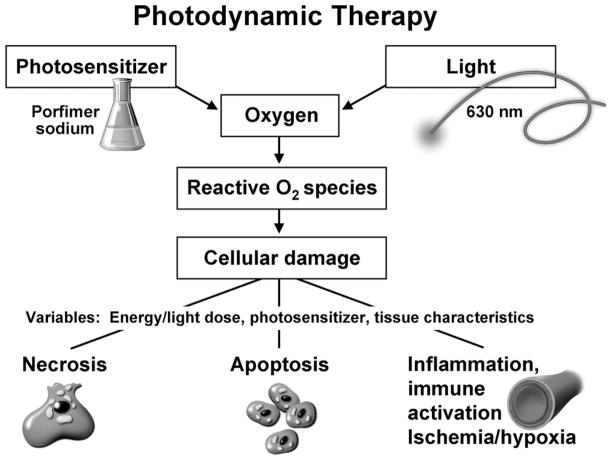
PDT induces tissue damage by the interaction of light, oxygen, and a photosensitizer (Figure 3).33–36 Few investigators have studied the influence of biomarkers on treatment outcomes in esophageal neoplasia. Foultier et al27 assessed the influence of cell DNA content (measured by flow cytometry) on outcomes in patients with early esophageal cancer treated by PDT. Aneuploidy was associated with poor response, with only 5 of 15 patients with aneuploidy achieving complete remission, compared with 12 of 17 patients without aneuploidy. It is unclear if the investigators adjusted for other factors influencing response. We previously reported in abstract form28 that nonresponders to PDT had a higher prevalence of P53 mutations (5 of 9) than responders (0 of 10) (P < .01). A similar difference was seen with P16 promoter hypermethylation without statistical significance. Our previous report was limited by its retrospective nature, small sample size, and the absence of adjustment for other response predictors. The current study addresses these shortcomings by its prospective nature, adequate sample size, and adjustment for other clinical predictors of response. Clinical studies in patients with hematologic, breast, and colon cancers also have suggested that the loss of P53 function is a predictor of poor treatment response to chemotherapy and radiation.37–39
Figure 3.
Mechanism of action of PDT. Reactive oxygen species produced by PDT after the interaction of light, photosensitizer, and oxygen mediate cellular damage by multiple mechanisms. The exact proportion of damage caused by each mechanism is dependent on the energy dose delivered (high doses causing more necrosis, lower doses cause apoptosis and ischemia, in addition to necrosis)56 as well as the photosensitizer used (sensitizers accumulating in the mitochondria and lysosomes are more likely to initiate apoptosis, whereas those accumulating in the cell membranes cause more necrosis).33
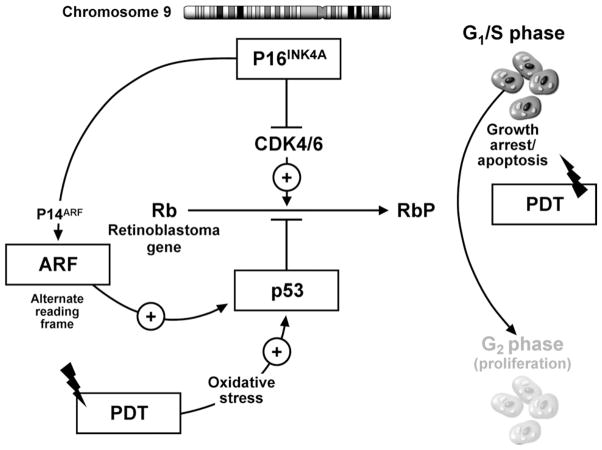
A number of genetic alterations have been described in BE. These include inactivation of cell-cycle checkpoint (P16 and P53) genes by multiple mechanisms (allelic loss, mutation and promoter hypermethylation in the case of P16)21,22,28,40 – 44 as well as gains/amplification at different proto-oncogene loci.24,45– 47 P16 and P53 genes regulate the cell cycle. p16INK4A (p16) is encoded by the INK4a/CDKN2A gene located on chromosome 9p21. p16 is a cyclin-dependent kinase (CDK) inhibitor that blocks the activity of CDK4 and CDK6 (Figure 4). P53 is well established as a key gene that monitors the cell for damage by triggering apoptosis when significant injury has occurred. Cells acquiring P53 lesions appear to be less susceptible to spontaneous and radiation/chemotherapy-induced apoptosis.48,49 Links between the p16 and p53 pathways are present and mediated by p14ARF (ARF, alternate reading frame; Figure 4). Therefore, loss of the INK4a gene potentially may disrupt 2 cell control pathways, one through p16INK4A/CDK4/6/pRB and the other through p14ARF/MDM2/p53.50
Figure 4.
Putative model of the influence of cell-cycle check point genes on PDT-induced cellular apoptosis. The p16 locus on chromosome 9 can transcribe 2 proteins: (1) P16INK4 protein, which inhibits CDK4 and CDK 6, leading to the inhibition of phosphorylation of the retinoblastoma gene product (Rb), causing inhibition of cell-cycle progression and growth arrest, and (2) p14ARF, which inhibits the degradation of p53 protein by MDM2, thereby potentiating the P53-mediated inhibition of cell-cycle progression and causing cell-cycle arrest. PDT causes oxidative stress, which can activate cellular apoptosis mechanisms by P53-dependent and -independent mechanisms. In the presence of intact p16 (p14 ARF) and p53 function, PDT can induce cell injury by apoptosis. Loss of p16 and p53 function allows the progression of cells to the G2 phase of the cell cycle, leading to cell proliferation. This may provide cells with a survival advantage, leading to decreased response to PDT.
The decreased response to PDT in patients with P16 loss may be owing to resistance of cells with these biomarkers to the effects of PDT. It is known that one of the mechanisms of tissue damage by PDT is the induction of apoptosis.33 Inactivation or loss of the P16 locus also may be associated with the loss of the ARF-mediated regulation of the P53 locus (Figure 4). In addition, loss of the P16 locus may lead to loss of P53-dependent apoptosis initiation. Hence, cells with loss of P16 function may be more resistant to PDT-induced apoptosis, allowing them to survive PDT. Fisher et al51 found that cells with mutated P53 were significantly less sensitive to PDT than those with intact P53 function. They also52 subsequently found that cells with inactive P53 function showed decreased apoptosis after exposure to PDT. There are no prior reports assessing the influence of P16 loss on PDT response. Unlike the earlier-described studies, our current study is a prospective in vivo study of this phenomenon in human beings with HGD and BE. In vitro cell models lack the ability to account for ischemia induced by PDT (a major mechanism of action of PDT). This is especially relevant in terms of altered mitochondrial function in a hypoxic milieu, given that mitochondria are crucial organelles in the initiation of apoptosis by PDT. The possibility of cells with genetic alterations having an alternate mode of cell repair that allows them to overcome damage inflicted by PDT also has been suggested.53 In the current study, P53 loss detected by FISH was a predictor of lack of response at 3 months on univariate analysis. However, P53 allelic loss did not prove to be a significant predictor on multivariable analysis.
Inactivation of the P16 locus by any mechanism has been shown to enable clonal expansion of cells.21 This also may provide these cells with a proliferation advantage allowing them to colonize wounded epithelium after PDT. The P16 genotype has been shown to correlate with the median BE segment length.21 On multivariable analysis (Table 5), BE segment length was an independent predictor of response, adjusting for P16 loss. We also found a statistically significant positive correlation between P16 loss and length of the Barrett’s segment in our cohort (Wilcoxon rank-sum test, P = .003).
Performance of EMR before PDT was a borderline significant predictor of decreased response on multivariable analysis (OR, 0.30; 95% CI, 0.08–1.18). The median number of EMRs performed was 2 (range, 1–12). EMR was performed for either visible lesions (71%) or in patients with flat dysplasia (no identifiable lesion, with HGD diagnosed on mucosal biopsy alone, 29%). We have previously reported that patients with nodules are more likely to progress to carcinoma (60% of those with nodules progressed to cancer, compared with 23% without nodules; hazard ratio, 2.6 [range, 1.2–5.3]).30 The presence of visible lesions necessitating EMRs is perhaps a surrogate marker of more aggressive biology making these patients less likely to respond to PDT. Focal HGD was associated with a numerically lower (although not statistically significant) proliferation index compared with diffuse HGD in the earlier-described study.
Despite the large sample size and prospective nature of the study, this study had some potential limitations. We defined response to PDT as the absence of dysplasia at 3 months (first follow-up evaluation after PDT). Previous studies have used different criteria for response to PDT: from the absence of HGD on biopsy specimens at any follow-up endoscopy26 to the absence of HGD in 2 successive endoscopies.54 We chose a more stringent definition of response, and picked the first follow-up endoscopy because this was most likely to represent the effects of PDT. At our center, if dysplasia is found at the follow-up endoscopy, other techniques such as EMR are used to remove residual areas of dysplasia, which potentially would confound assessment of response to PDT alone. However, the possibility that the lack of dysplasia on biopsy specimens at 3 months may reflect sampling error cannot be excluded. We also recognized that the absence of dysplasia at 3 months may be a reflection of the biology (or natural history) of dysplasia in patients with markers of progression. We attempted to address this by including a comparable control group of patients who did not undergo PDT in the model; however, PDT remained a strong predictor of response after adjusting for other factors (OR, 7). The proportion of patients with P1621 and P53 loss43,55 in our cohort was somewhat lower than that reported in the literature. Compared with gene analysis, dual-probe FISH (using a locus-specific probe for 17p13.1 and a centromeric probe, CEP 17) had moderate sensitivity (68.4%), but high specificity (95.8%)44 for the detection of loss of heterozygosity. Loss of P53 function occurs by more than one mechanism; dual-probe FISH only identifies the subgroup with a chromosome copy number change. However, FISH may be more sensitive than loss of heterozygosity analysis, because loss of heterozygosity analyses typically require that at least 70% of cells in a sample be tumor to be able to detect loss of heterozygosity.44 FISH also can be performed on cells in metaphase or interphase. This is extremely useful because the majority of cells in any cell population are in inter-phase. FISH also can detect submicroscopic changes that are not apparent on digital image analysis or flow cytometry as well as ascertain the nature of the alteration at a specific locus because of its superior resolution. We also performed FISH on cytology specimens because the use of tissue sections could lead to sectioning artifacts, which could compromise FISH results.
In conclusion, this large prospective study identified p16 allelic loss when detected by FISH as predicting response to PDT in patients with BE and HGD/mucosal cancer. Identification of biomarkers may help in the selection of appropriate therapy for patients and improve treatment outcomes.
Acknowledgments
Supported by National Institutes of Health grants R01 CA111603-01A1, R01CA097048, and R21CA122426-01. A patent has been filed for the fluorescence in situ hybridization probe set discussed in this study. Dr Halling and Ms Shannon Brankley are listed as inventors on the filed patent. Drs Halling and Wang, and Ms Brankley have the potential to receive royalties from the sale of this fluorescence in situ hybridization probe set.
Abbreviations used in this paper
- BE
Barrett’s esophagus
- CDK
cyclin-dependent kinase
- CI
confidence interval
- EMR
endoscopic mucosal resection
- FISH
fluorescence in situ hybridization
- HGD
high-grade dysplasia
- OR
odds ratio
- PDT
photodynamic therapy
- ROC
receiver operator characteristic
References
- 1.Devesa SS, Blot WJ, Fraumeni JF., Jr Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer. 1998;83:2049–2053. [PubMed] [Google Scholar]
- 2.Provenzale D, Schmitt C, Wong JB. Barrett’s esophagus: a new look at surveillance based on emerging estimates of cancer risk. Am J Gastroenterol. 1999;94:2043–2053. doi: 10.1111/j.1572-0241.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- 3.Reid BJ, Levine DS, Longton G, et al. Predictors of progression to cancer in Barrett’s esophagus: baseline histology and flow cytometry identify low- and high-risk patient subsets. Am J Gastroenterol. 2000;95:1669–1676. doi: 10.1111/j.1572-0241.2000.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnell TG, Sontag SJ, Chejfec G, et al. Long-term nonsurgical management of Barrett’s esophagus with high-grade dysplasia. Gastroenterology. 2001;120:1607–1619. doi: 10.1053/gast.2001.25065. [DOI] [PubMed] [Google Scholar]
- 5.Weston AP, Sharma P, Topalovski M, et al. Long-term follow-up of Barrett’s high-grade dysplasia. Am J Gastroenterol. 2000;95:1888–1893. doi: 10.1111/j.1572-0241.2000.02234.x. [DOI] [PubMed] [Google Scholar]
- 6.Rice TW, Falk GW, Achkar E, et al. Surgical management of high-grade dysplasia in Barrett’s esophagus. Am J Gastroenterol. 1993;88:1832–1836. [PubMed] [Google Scholar]
- 7.Falk GW, Rice TW, Goldblum JR, et al. Jumbo biopsy forceps protocol still misses unsuspected cancer in Barrett’s esophagus with high-grade dysplasia. Gastrointest Endosc. 1999;49:170–176. doi: 10.1016/s0016-5107(99)70482-7. [DOI] [PubMed] [Google Scholar]
- 8.Cameron AJ, Carpenter HA. Barrett’s esophagus, high-grade dysplasia, and early adenocarcinoma: a pathological study. Am J Gastroenterol. 1997;92:586–591. [PubMed] [Google Scholar]
- 9.Nigro JJ, Hagen JA, DeMeester TR, et al. Prevalence and location of nodal metastases in distal esophageal adenocarcinoma confined to the wall: implications for therapy. J Thorac Cardiovasc Surg. 1999;117:16–25. doi: 10.1016/s0022-5223(99)70464-2. [DOI] [PubMed] [Google Scholar]
- 10.Orringer MB, Marshall B, Stirling MC. Transhiatal esophagectomy for benign and malignant disease. J Thorac Cardiovasc Surg. 1993;105:265–277. [PubMed] [Google Scholar]
- 11.Overholt BF, Panjehpour M, Haydek JM. Photodynamic therapy for Barrett’s esophagus: follow-up in 100 patients. Gastrointest Endosc. 1999;49:1–7. doi: 10.1016/s0016-5107(99)70437-2. [DOI] [PubMed] [Google Scholar]
- 12.Attwood SE, Lewis CJ, Caplin S, et al. Argon beam plasma coagulation as therapy for high-grade dysplasia in Barrett’s esophagus. Clin Gastroenterol Hepatol. 2003;1:258–263. [PubMed] [Google Scholar]
- 13.Kovacs BJ, Chen YK, Lewis TD, et al. Successful reversal of Barrett’s esophagus with multipolar electrocoagulation despite inadequate acid suppression. Gastrointest Endosc. 1999;49:547–553. doi: 10.1016/s0016-5107(99)70380-9. [DOI] [PubMed] [Google Scholar]
- 14.Buttar NS, Wang KK, Lutzke LS, et al. Combined endoscopic mucosal resection and photodynamic therapy for esophageal neoplasia within Barrett’s esophagus. Gastrointest Endosc. 2001;54:682–688. doi: 10.1067/gien.2001.0003. [DOI] [PubMed] [Google Scholar]
- 15.Pacifico RJ, Wang KK, Wongkeesong LM, et al. Combined endoscopic mucosal resection and photodynamic therapy versus esophagectomy for management of early adenocarcinoma in Barrett’s esophagus. Clin Gastroenterol Hepatol. 2003;1:252–257. [PubMed] [Google Scholar]
- 16.Sharma P, Jaffe PE, Bhattacharyya A, et al. Laser and multipolar electrocoagulation ablation of early Barrett’s adenocarcinoma: long-term follow-up. Gastrointest Endosc. 1999;49:442–446. doi: 10.1016/s0016-5107(99)70040-4. [DOI] [PubMed] [Google Scholar]
- 17.Dougherty TJ, Gomer CJ, Henderson BW, et al. Photodynamic therapy. J Natl Cancer Inst. 1998;90:889–905. doi: 10.1093/jnci/90.12.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Overholt BF, Lightdale CJ, Wang KK, et al. International, multi-center, partially blinded, randomized study of the efficacy of photodynamic therapy (PDT) using porfimer sodium (POR) for the ablation of high-grade dysplasia (HGD) in Barrett’s esophagus (BE): results of 24-month follow up (abstr) Gastroenterology. 2003;124:153. [Google Scholar]
- 19.Overholt BF, Wang KK, Burdick JS, et al. Five-year efficacy and safety of photodynamic therapy with Photofrin in Barrett’s high-grade dysplasia. Gastrointest Endosc. 2007;66:460–468. doi: 10.1016/j.gie.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 20.Prasad GA, Wang KK, Buttar NS, et al. Long-term survival following endoscopic and surgical treatment of high-grade dysplasia in Barrett’s esophagus. Gastroenterology. 2007;132:1226–1233. doi: 10.1053/j.gastro.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong DJ, Paulson TG, Prevo LJ, et al. p16(INK4a) lesions are common, early abnormalities that undergo clonal expansion in Barrett’s metaplastic epithelium. Cancer Res. 2001;61:8284–8289. [PubMed] [Google Scholar]
- 22.Prevo LJ, Sanchez CA, Galipeau PC, et al. p53-mutant clones and field effects in Barrett’s esophagus. Cancer Res. 1999;59:4784–4787. [PubMed] [Google Scholar]
- 23.Walch AK, Zitzelsberger HF, Bruch J, et al. Chromosomal imbalances in Barrett’s adenocarcinoma and the metaplasia-dysplasia-carcinoma sequence. Am J Pathol. 2000;156:555–566. doi: 10.1016/S0002-9440(10)64760-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tselepis C, Morris CD, Wakelin D, et al. Upregulation of the oncogene c-myc in Barrett’s adenocarcinoma: induction of c-myc by acidified bile acid in vitro. Gut. 2003;52:174–180. doi: 10.1136/gut.52.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brankley SM, Wang KK, Harwood AR, et al. The development of a fluorescence in situ hybridization assay for the detection of dysplasia and adenocarcinoma in Barrett’s esophagus. J Mol Diagn. 2006;8:260–267. doi: 10.2353/jmoldx.2006.050118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Overholt BF, Lightdale CJ, Wang KK, et al. Photodynamic therapy with porfimer sodium for ablation of high-grade dysplasia in Barrett’s esophagus: international, partially blinded, randomized phase III trial. Gastrointest Endosc. 2005;62:488–498. doi: 10.1016/j.gie.2005.06.047. [DOI] [PubMed] [Google Scholar]
- 27.Foultier MT, Vonarx-Coinsman V, de Brito LX, et al. DNA or cell kinetics flow cytometry analysis of 33 small gastrointestinal cancers treated by photodynamic therapy. Cancer. 1994;73:1595–1607. doi: 10.1002/1097-0142(19940315)73:6<1595::aid-cncr2820730610>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 28.Krishnadath KK, Wang KK, Taniguchi K, et al. p53 mutations in Barrett’s esophagus predict poor response to photodynamic therapy. Gastroenterology. 2001;120:A413. [Google Scholar]
- 29.Montgomery E, Bronner MP, Goldblum JR, et al. Reproducibility of the diagnosis of dysplasia in Barrett esophagus: a reaffirmation. Hum Pathol. 2001;32:368–378. doi: 10.1053/hupa.2001.23510. [DOI] [PubMed] [Google Scholar]
- 30.Buttar NS, Wang KK, Sebo TJ, et al. Extent of high-grade dysplasia in Barrett’s esophagus correlates with risk of adenocarcinoma. Gastroenterology. 2001;120:1630–1639. doi: 10.1053/gast.2001.25111. [DOI] [PubMed] [Google Scholar]
- 31.Prasad GA, Wang KK, Lutzke LS, et al. Frozen section analysis of esophageal endoscopic mucosal resection specimens in the real-time management of Barrett’s esophagus. Clin Gastroenterol Hepatol. 2006;4:173–178. doi: 10.1016/j.cgh.2005.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sokolova IA, Halling KC, Jenkins RB, et al. The development of a multitarget, multicolor fluorescence in situ hybridization assay for the detection of urothelial carcinoma in urine. J Mol Diagn. 2000;2:116–123. doi: 10.1016/S1525-1578(10)60625-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oleinick NL, Morris RL, Belichenko I. The role of apoptosis in response to photodynamic therapy: what, where, why, and how. Photochem Photobiol Sci. 2002;1:1–21. doi: 10.1039/b108586g. [DOI] [PubMed] [Google Scholar]
- 34.Granville DJ, McManus BM, Hunt DW. Photodynamic therapy: shedding light on the biochemical pathways regulating porphyrin-mediated cell death. Histol Histopathol. 2001;16:309–317. doi: 10.14670/HH-16.309. [DOI] [PubMed] [Google Scholar]
- 35.Cecic I, Korbelik M. Mediators of peripheral blood neutrophilia induced by photodynamic therapy of solid tumors. Cancer Lett. 2002;183:43–51. doi: 10.1016/s0304-3835(02)00092-7. [DOI] [PubMed] [Google Scholar]
- 36.Gomer CJ, Luna M, Ferrario A, et al. Cellular targets and molecular responses associated with photodynamic therapy. J Clin Laser Med Surg. 1996;14:315–321. doi: 10.1089/clm.1996.14.315. [DOI] [PubMed] [Google Scholar]
- 37.O’Connor PM, Jackman J, Jondle D, et al. Role of the p53 tumor suppressor gene in cell cycle arrest and radiosensitivity of Burkitt’s lymphoma cell lines. Cancer Res. 1993;53:4776–4780. [PubMed] [Google Scholar]
- 38.Ozbun MA, Butel JS. Tumor suppressor p53 mutations and breast cancer: a critical analysis. Adv Cancer Res. 1995;66:71–141. doi: 10.1016/s0065-230x(08)60252-3. [DOI] [PubMed] [Google Scholar]
- 39.Goh HS, Elnatan J, Low CH, et al. p53 point mutation and survival in colorectal cancer patients: effect of disease dissemination and tumour location. Int J Oncol. 1999;15:491–498. doi: 10.3892/ijo.15.3.491. [DOI] [PubMed] [Google Scholar]
- 40.Galipeau PC, Cowan DS, Sanchez CA, et al. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett’s esophagus. Proc Natl Acad Sci U S A. 1996;93:7081–7084. doi: 10.1073/pnas.93.14.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamelin R, Flejou JF, Muzeau F, et al. TP53 gene mutations and p53 protein immunoreactivity in malignant and premalignant Barrett’s esophagus. Gastroenterology. 1994;107:1012–1018. doi: 10.1016/0016-5085(94)90225-9. [DOI] [PubMed] [Google Scholar]
- 42.Maley CC, Galipeau PC, Li X, et al. Selectively advantageous mutations and hitchhikers in neoplasms: p16 lesions are selected in Barrett’s esophagus. Cancer Res. 2004;64:3414–3427. doi: 10.1158/0008-5472.CAN-03-3249. [DOI] [PubMed] [Google Scholar]
- 43.Neshat K, Sanchez CA, Galipeau PC, et al. p53 mutations in Barrett’s adenocarcinoma and high-grade dysplasia. Gastroenterology. 1994;106:1589–1595. doi: 10.1016/0016-5085(94)90415-4. [DOI] [PubMed] [Google Scholar]
- 44.Wongsurawat VJ, Finley JC, Galipeau PC, et al. Genetic mechanisms of TP53 loss of heterozygosity in Barrett’s esophagus: implications for biomarker validation. Cancer Epidemiol Biomarkers Prev. 2006;15:509–516. doi: 10.1158/1055-9965.EPI-05-0246. [DOI] [PubMed] [Google Scholar]
- 45.Walch A, Specht K, Bink K, et al. Her-2/neu gene amplification, elevated mRNA expression, and protein overexpression in the metaplasia-dysplasia-adenocarcinoma sequence of Barrett’s esophagus. Lab Invest. 2001;81:791–801. doi: 10.1038/labinvest.3780289. [DOI] [PubMed] [Google Scholar]
- 46.Wang KK, Meltzer SJ, WongKeeSong LM, et al. Methylation biomarkers in prediction of response to photodynamic therapy of Barrett’s esophagus. Gastroenterology. 2007;132:A64. [Google Scholar]
- 47.Sarbia M, Arjumand J, Wolter M, et al. Frequent c-myc amplification in high-grade dysplasia and adenocarcinoma in Barrett esophagus. Am J Clin Pathol. 2001;115:835–840. doi: 10.1309/MXXH-25N3-UAL2-G7XX. [DOI] [PubMed] [Google Scholar]
- 48.Kastan MB, Onyekwere O, Sidransky D, et al. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 49.Lowe SW, Bodis S, McClatchey A, et al. p53 status and the efficacy of cancer therapy in vivo. Science. 1994;266:807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 50.Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta. 1998;1378:F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 51.Fisher AM, Rucker N, Wong S, et al. Differential photosensitivity in wild-type and mutant p53 human colon carcinoma cell lines. J Photochem Photobiol B. 1998;42:104–107. doi: 10.1016/s1011-1344(97)00130-9. [DOI] [PubMed] [Google Scholar]
- 52.Fisher AM, Ferrario A, Rucker N, et al. Photodynamic therapy sensitivity is not altered in human tumor cells after abrogation of p53 function. Cancer Res. 1999;59:331–335. [PubMed] [Google Scholar]
- 53.Gomer CJ, Rucker N, Ferrario A, et al. Expression of potentially lethal damage in Chinese hamster cells exposed to hematoporphyrin derivative photodynamic therapy. Cancer Res. 1986;46:3348–3352. [PubMed] [Google Scholar]
- 54.Pech O, Gossner L, May A, et al. Long-term results of photodynamic therapy with 5-aminolevulinic acid for superficial Barrett’s cancer and high-grade intraepithelial neoplasia. Gastrointest Endosc. 2005;62:24–30. doi: 10.1016/s0016-5107(05)00333-0. [DOI] [PubMed] [Google Scholar]
- 55.Wu TT, Watanabe T, Heitmiller R, et al. Genetic alterations in Barrett esophagus and adenocarcinomas of the esophagus and esophagogastric junction region. Am J Pathol. 1998;153:287–294. doi: 10.1016/S0002-9440(10)65570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piette J, Volanti C, Vantieghem A, et al. Cell death and growth arrest in response to photodynamic therapy with membrane-bound photosensitizers. Biochem Pharmacol. 2003;66:1651–1659. doi: 10.1016/s0006-2952(03)00539-2. [DOI] [PubMed] [Google Scholar]