Abstract
Bacillus pumilus is characterized by a higher oxidative stress resistance than other comparable industrially relevant Bacilli such as B. subtilis or B. licheniformis. In this study the response of B. pumilus to oxidative stress was investigated during a treatment with high concentrations of hydrogen peroxide at the proteome, transcriptome and metabolome level. Genes/proteins belonging to regulons, which are known to have important functions in the oxidative stress response of other organisms, were found to be upregulated, such as the Fur, Spx, SOS or CtsR regulon. Strikingly, parts of the fundamental PerR regulon responding to peroxide stress in B. subtilis are not encoded in the B. pumilus genome. Thus, B. pumilus misses the catalase KatA, the DNA-protection protein MrgA or the alkyl hydroperoxide reductase AhpCF. Data of this study suggests that the catalase KatX2 takes over the function of the missing KatA in the oxidative stress response of B. pumilus. The genome-wide expression analysis revealed an induction of bacillithiol (Cys-GlcN-malate, BSH) relevant genes. An analysis of the intracellular metabolites detected high intracellular levels of this protective metabolite, which indicates the importance of bacillithiol in the peroxide stress resistance of B. pumilus.
Introduction
Bacillus pumilus is a Gram-positive, rod-shaped and endospore-forming bacterium closely related to the industrially relevant bacteria Bacillus subtilis and Bacillus licheniformis. B. pumilus represents a potential alternative host for the industrial production of enzymes. For the evaluation and optimization of fermentation processes with this organism a comprehensive knowledge on its physiology and stress adaptation is required.
During fermentation processes a variety of stresses (e.g. salt, heat and oxidative stress) can impair the fitness of the production host and the quality of the fermentation product [1]–[3]. B. pumilus strains are highly resistant against UV radiation and hydrogen peroxide, which may explain the finding of viable spores of B. pumilus in hostile environments such as the interior of the Sonoran desert basalt and spacecrafts [4], [5]. This natural potential and resistances of B. pumilus could be a major benefit for the improvement of industrial production strains, since oxidative stress can occur in all phases of fermentation processes [1]–[3].
Reactive oxygen species (ROS) such as superoxide (O2 · −), hydrogen peroxide (H2O2) and hydroxyl radical (OH·) are successive one-electron-reduction products of molecular oxygen and therefore occur in all aerobically living organisms [3], [6], [7]. Increased ROS production that exceeds the cell defense capacity leads to oxidative stress in the cell and to the oxidation of nucleic acids, proteins and lipids [2], [3], [8]–[10].
In B. subtilis, the cellular defense against oxidative stress is ensured by the detoxification of harmful agents, protection of macromolecules and the repair or removal of damaged molecules. The oxidative stress response of this organism is regulated by specific transcriptional regulators, such as PerR, SigB, LexA/RecA, Spx and OhrR, as previously described in detail [11]–[13]. The oxidative stress response of B. pumilus differs significantly from the response in B. subtilis, as major oxidative stress genes of B. subtilis are missing in the genome of B. pumilus, such as the catalase KatA or alkyl hydroperoxide reductase AhpCF. For some of these genes no homologs could be found in the B. pumilus genome. This leads to the questions, which genes compensate the missing genes and are thus responsible for the oxidative stress resistance of B. pumilus. In this study we used a combination of proteomics, transcriptomics and metabolomics to investigate the individual peroxide stress response of B. pumilus.
Materials and Methods
2.1 Strain, Media, Growth and Cell Sampling
Bacillus pumilus Jo2 (DSM 14395) was used for all experiments described in this study. Cells were grown aerobically at 37°C and 180 rpm in minimal medium containing 15 mM (NH4)2SO4, 8 mM MgSO4×7 H2O, 27 mM KCl, 7 mM Na-citrate×2 H2O, 50 mM Tris-HCl (pH 7.5) supplemented with 1.8 mM KH2PO4, 2 mM CaCl2, 1 µM FeSO4×7 H2O, 10 µM MnSO4×4 H2O, 4.5 mM glutamate, 0.2% w/v glucose and 0.04 µM biotin. Exponentially growing cells at an OD500 nm of 0.6 were exposed to a final concentration of 2 mM hydrogen peroxide. Proteome samples were taken from unstressed cultures before and 10 as well as 30 minutes after exposure to hydrogen peroxide. Samples were pulse-labeled with L-[35S]-methionine for 5 min, as described by Hoi et al. [14]. Samples for preparative gels were prepared from unlabeled cells 30 and 60 min after exposure to H2O2 [14]. Preparative gels were used only for spot identification via mass spectrometry.
Samples for RNA extraction were taken before (control) and 3 and 8 min after addition of H2O2. Cell samples for RNA extraction were mixed with 0.5 volumes of ice-cold killing buffer (20 mM Tris-HCl pH 7.5, 5 mM MgCl, 20 mM NaN3), and immediately harvested at 10000×g for 5 min at 4°C.
2.2. Scanning Electron Microscopy
For the scanning electron microscopy, the cells were separated from the culture medium by filtration through a 0.2 µm pore size polycarbonate filter. The filter were placed in fixation solution (1% glutaraldehyde, 4% paraformaldehyde, 50 mM NaN3 in 5 mM HEPES [pH 7.4]) for 1 h at room temperature and 4°C overnight. After fixation, the samples were treated with 2% tannic acid for 1 h, 1% osmium tetroxide for 2 h, 1% thiocarbohydrazide for 30 min, 1% osmium tetroxide overnight, and 2% uranyl acetate for 30 min with washing steps in between. The samples were dehydrated in a graded series of aqueous ethanol solutions (10–100%) and then critical point-dried. Finally, filter were mounted on aluminum stubs, sputtered with gold/palladium and examined in a scanning electron microscope EVO LS10 (Carl Zeiss microscopy GmbH, Oberkochen, Germany).
2.3 Transmission Electron Microscopy
Cells were fixed in 1% glutaraldehyde, 4% paraformaldehyde, 50 mM NaN3 in 5 mM HEPES for 1 h at room temperature and then at 4°C overnight. Subsequent to embedding the cells in low gelling agarose, cells were postfixed in 2% osmium tetroxide for 2 h at 4°C. After dehydration in graded series of ethanol (20–100%) for 10 min each step with 0.5% uranyl acetate in 70% ethanol for 30 min (at 4°C) in between, the material was embedded in Epon. Sections were cut on an ultramicrotome (Reichert Ultracut, Leica UK Ltd, Milton Keynes, UK), stained with uranyl acetate and lead citrate and analyzed with a transmission electron microscope LEO 906 (Carl Zeiss microscopy GmbH, Oberkochen, Germany).
2.4 2D-Gel Electrophoresis
Cytosolic protein extracts were loaded onto IPG-strips in the pH-range 4–7 (GE Healthcare Bio-Sciences AB, Finland) using 100 µg protein for labeled samples and 500 µg for preparative gels. 2D-PAGE was performed as described by Büttner et al. [15]. Autoradiography of radioactively labeled gels was performed as previously described [14]. Preparative gels were stained with Coomassie Brilliant Blue as described by Voigt et al. [16]. Proteins were excised from preparative gels, digested and the peptide solution spotted onto MALDI targets using the Ettan Spot Handling Workstation (GE Healthcare, UK). Identification was performed using MALDI-TOF-MS/MS (Proteome Analyzer 5800 MDS Sciex, USA) and an in-house B. pumilus Jo2 (DSM 14395) database as described by Wolf et al. [17]. Protein quantification was done with the Delta2D proteome software (Decodon, Germany).
2.5 Microarray Experiment
Total RNA of B. pumilus was prepared by the acid phenol method [18] with the modifications described elsewhere [19]. The isolated RNA was treated with DNase (RNase-free DNase Set, Quiagen, Germany) and subsequently concentrated and cleaned (RNA cleanup and concentration Kit, Norgen Biotek, Canada). Quantity of RNA was determined on a microscale spectrophotometer (Nanodrop ND-1000, Peqlab Biotechnologie GmbH, Germany) and RNA integrity was analyzed using a capillary electrophoresis system (Bioanalyzer 2100, Agilent Technologies, USA). Synthesis and purification of fluorescently labeled cDNA was carried out according to Schroeter et al. [20] with minor modifications described below. After the labeling and clean-up step [20], 600 ng of respective Cy3- and Cy5 -labeled cDNA were admixed (ad. 44 µl), denaturated and mixed with 11 µl pre-warmed blocking agent and 60 µl hybridization buffer (both Gene expression hybridization kit, Agilent Technologies, USA). 100 µl of the emerging cDNA mixture, respectively, were used for any hybridization. Custom-made B. pumilus Jo2 4×44 K gene expression microarrays were obtained from Agilent Technologies (https://earray.chem.agilent.com/earray/), containing 60-mer Oligonucleotide probes (SurePrint technology, Agilent Technologies). Probe design was performed on the chromosome sequence of B. pumilus Jo2 (Sequence Intellectual Property of Henkel KGaA). In addition to the annotated open reading frames (ORFs), ORFs were predicted using (i) Glimmer 3.0 [21], (ii) ZCURVE [22], (iii) Genemark HMM [23], and (iv) Prodigal [24]. Predicted ORFs were added to the design provided that: (i) they were non-overlapping with existing ORFs; or (ii) they were in the reverse complementary strand of existing ORFs. On the annotated and predicted ORFs, up to 5 probes were designed. Altogether, a total of 41377 probes were designed by means of OligoWiz 2.1.3 [25] using default parameters for prokaryotic long-mers. The arrays were hybridized and washed according to the manufacturer’s instructions (Two-Color Microarray-Based Gene Expression Analysis Protocol, Agilent Technologies, USA), followed by a last wash step with acetonitrile (Carl Roth GmbH+Co. KG, Germany) for 30 sec. Microarrays were scanned using the Agilent scanner Type G2565CA with high resolution upgrade G2539A and the software Scan Control 8.4.1 (Agilent Technologies, USA). Data were extracted from scanned images using Agilent's Feature Extraction Software (version 10.5.1.1; Agilent Technologies, USA) using default settings. A common reference type of design was employed, and data from three biological replicate hybridizations for each point in time were used for data analysis. Spot signals were normalized using Lowess as described earlier [26]. Next, for each ORF a signal was determined by taking the median signal of the up to 5 probes per ORF. Differential regulation was determined from the biological triplicate measurements by false-discovery rate (FDR) from the Cyber-T p-values [27] by means of multiple testing correction [26]. Differential regulation was defined as a 2-fold or higher differential expression with a FDR cut-off value of 0.05 or lower.
2.6 Metabolomic Analysis of Thiols as their Monobromobimane-derivatives
Cells were grown in minimal medium as described above and exponentially grown cells from 10 ml culture medium were harvested before oxidative stress, 10, 30 and 60 min after addition of hydrogen peroxide. The isolation of LMW-thiols for HPLC analysis was performed as described previously [28]. In brief, after centrifugation the cells were washed with 50 mM Tris–HCl (pH 8.0) and resuspended in 50% acetonitrile containing 20 mM Tris–HCl (pH 8.0), 1 mM penicillamine as internal standard and 2 mM monobromobimane (mBBr). Control samples were resuspended without penicillamine and 5 mM N-ethylmaleimide (NEM) was used prior to addition of mBBr. Thiols were extracted at 60°C and directly labeled with mBBr. Labeling reaction was stopped with aqueous methane sulfonic acid in a final concentration of 5 mM. BSmB (monobromobimane-derivative of BSH) standards were synthesized as described previously [7], [29]. For detection and quantification of LMW-thiols, ion pairing HPLC was performed as described before [30]. For absolute quantification the ratio peak area thiol/peak area internal standard was used and an eight-point calibration between 10 nM and 2000 nM was generated.
2.7 Prediction of the PerR Consensus Sequence
Prediction of the PerR consensus sequence was done with the PRODORIC® database (http://prodoric.tu-bs.de/vfp/index2.php) release 8.9 [31] using the consensus sequence as described by Fuangthong et al. [32].
Results and Discussion
3.1 Effects of H2O2 on Growth and Cell Morphology
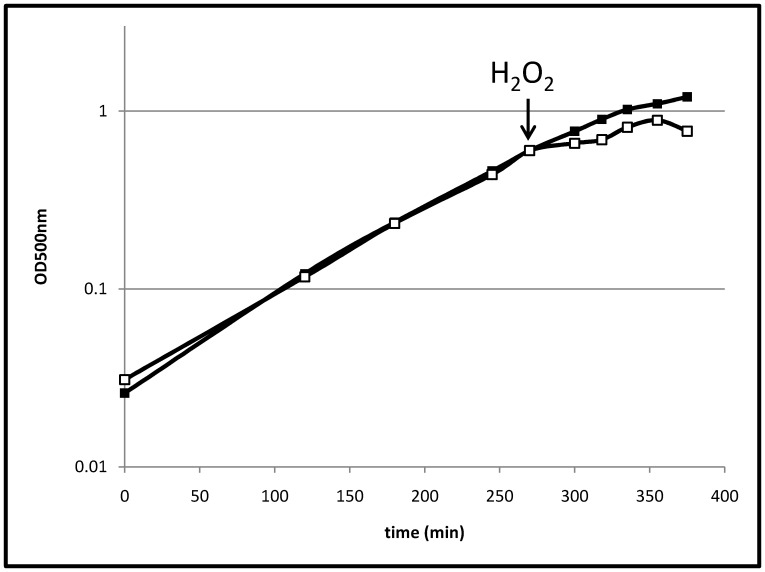
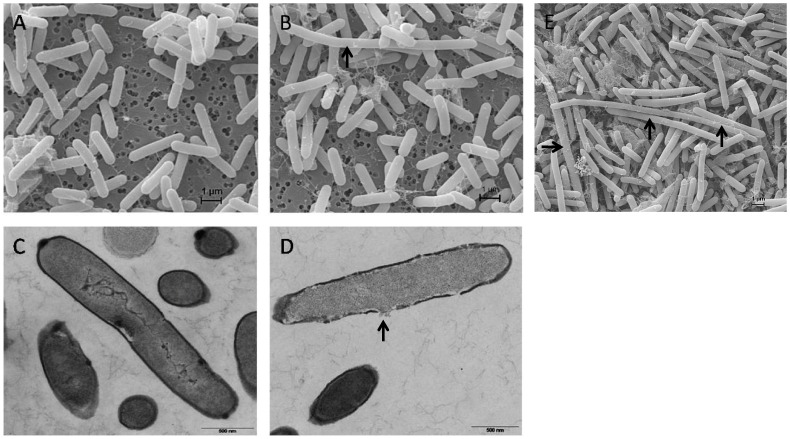
Exponentially growing B. pumilus cells were treated with 2 mM hydrogen peroxide. Thus, the concentration of H2O2 that was used to trigger the stress in this study was about 40-fold higher than those used for comparable analyses with B. subtilis or B. licheniformis [13], [20]. The highest peroxide concentrations allowing growth for B. subtilis and B. licheniformis were 4 and 1 mM, respectively (Table S1). B. pumilus is still able to grow with 20 mM hydrogen peroxide. This indicates a striking resistance of B. pumilus against peroxide stress. Compared to unstressed cells, growth was significantly impaired for a short time (approximately 15 min) after the H2O2 treatment (Figure 1). However, after that time, cells continued to grow for about one hour. An electron microscopy analysis indicated that after exposure to H2O2 most of the cells are morphologically intact, but some of the cells exhibited major damage of their envelope (Figure 2D). Furthermore, scanning electron microscopy revealed some atypically long cells (up to approximately 10–20% two hours after H2O2 treatment, Figure 2B, 2E) indicating an impact of hydrogen peroxide on processes involved in cell division.
Figure 1. Growth of B. pumilus.
Growth of B. pumilus under control conditions (filled squares) and stressed with 2 mM H2O2 at OD500 nm 0.6 (empty squares).
Figure 2. Electron microscopy micrographs.
Scanning (A,B,E) and transmission (C,D) electron microscopy micrographs of B. pumilus cells under control conditions (A,C), 30 min (B,D) and 120 min after treatment with 2 mM H2O2 (E).
3.2 Global Expression Profile
All values presented for up- and downregulation of genes or proteins are fold change values. The analysis of the soluble intracellular proteome of B. pumilus revealed 54 significantly upregulated and 111 downregulated proteins 10 min after H2O2 treatment (with a threshold of 2-fold, Table 1, Table S2, Figure 3). For the visualization of the fast and early response on proteome level, a labeling with 35S-methionine was necessary. 30 minutes after initiating the stress, 73 proteins were up- and 59 proteins downregulated (Table 1, Table S2 and S3, Figure 4). Transcriptome analysis revealed an at least 2-fold increased transcription of 181 genes three minutes after treatment with H2O2; 76 of them were more than 3-fold upregulated. Eight minutes after treatment, the transcription of 558 genes appeared at least 2-fold increased (307 genes with an at least 3-fold increased transcription). Three minutes after the stress, 266 genes were transcribed with an at least 3-fold lower rate than under control conditions, for 296 genes this decreased transcription rate was shown eight minutes after treatment. To indicate quality of the transcriptome results, raw data for individual probes for selected genes (which were not found to be induced in the proteome analysis) are presented in Table S4. These data show similar basal values and changes following addition of hydrogen peroxide for all five probes corresponding to a gene.
Table 1. Selected induced genes and proteins.
| ORF ID | gene | transcriptome | proteome | Regulon in other Bacilli | |||
| 3 min | 8 min | 10 min | 30 min | ||||
| BPJ13600 | zinc-transporting ATPase ZosA | zosA | 12.74 | 28.72 | perR | ||
| BPJ25410 | glutamyl-tRNA reductase HemA | hemA | 3.44 | 3.99 | perR | ||
| BPJ25390 | porphobilinogen deaminase HemC | hemC | 2.68 | 3.90 | perR | ||
| BPJ25370 | delta-aminolevulinic acid dehydratase HemB | hemB | 2.52 | 3.72 | perR | ||
| BPJ25400 | putative cytochrome C biogenesis protein HemX | hemX | 2.86 | 4.25 | perR | ||
| BPJ25380 | uroporphyrinogen III synthase HemD | hemD2 | 2.68 | 4.23 | perR | ||
| BPJ25360 | glutamate-1-semialdehyde 2,1-aminomutase HemL | hemL | 2.75 | 3.56 | perR | ||
| BPJ21690 | Fur family ferric uptake regulation protein Fur | fur | 1.92 | 3.62 | perR | ||
| BPJ11620 | transcriptional regulator Spx | spxA | 4.14 | 3.31 | perR/spx/sigB | ||
| BPJ11610 | putative N-acetyltransferase YjbC | yjbC | 2.41 | 4.41 | perR/spx/sigB/sigM/sigW/sigX | ||
| BPJ09760 | catalase KatX2 | katX2 | 6.96 | 10.69 | 15.18 | 21.09 | sigB/sigF |
| BPJ34450 | putative ABC transporter permease YwjA | ywjA | 1.57 | 4.47 | fur | ||
| BPJ30810 | hydroxamate siderophore ABC transporter ATP-bindingprotein FhuC | fhuC1 | 1.51 | 2.46 | fur | ||
| BPJ30830 | hydroxamate siderophore ABC transporter permeaseFhuB | fhuB1 | 1.52 | 4.01 | fur | ||
| BPJ30820 | hydroxamate siderophore ABC transporter permeaseFhuG | fhuG1 | 1.53 | 3.20 | fur | ||
| BPJ08440 | ABC transport system permease | bpj08440 | 4.11 | 7.49 | fur | ||
| BPJ08430 | putative iron complex transport system substratebinding protein | bpj08430 | 4.54 | 7.43 | fur | ||
| BPJ08420 | putative HTH-type transcriptional regulator | bpj08420 | 3.58 | 5.59 | fur | ||
| BPJ08580 | putative nitroreductase YfhC | yfhC | 2.67 | 5.00 | 1.10 | fur | |
| BPJ08410 | ferredoxin–NADP reductase 2 | bpj08410 | 3.90 | 3.83 | fur | ||
| BPJ37570 | AraC family transcriptional regulator/putativeFeuA-like substrate-binding domain ybbB | ybbB | 4.93 | 12.84 | fur | ||
| BPJ37580 | iron complex ABC transporter substrate-binding proteinFeuA | feuA | 3.21 | 10.04 | fur, btr, citB | ||
| BPJ37590 | putative bacillibactin esterase YbbA | ybbA | 5.24 | 18.42 | fur/btr/citB | ||
| BPJ07970 | C56 family peptidase YfkM | yfkM | 2.94 | 7.61 | 7.39 | 3.09 | fur/sigB |
| RBPU30260 | FeS cluster assembly protein SufB | sufB | 1.87 | 2.10 | 1.73 | Fe/S cluster biogenesis | |
| RBPU30280 | cysteine desulfurase SufS | sufS | −1.77 | 2.69 | Fe/S cluster biogenesis | ||
| RBPU30290 | FeS cluster assembly permease SufD | sufD | 1.73 | Fe/S cluster biogenesis | |||
| RBPU30300 | FeS cluster assembly ATPase SufC | sufC | 2.52 | 2.13 | Fe/S cluster biogenesis | ||
| BPJ11040 | diaminobutyrate–2-oxoglutarate aminotransferase RhbA | rhbA | −1.11 | 11.10 | siderophore synthesis | ||
| BPJ11080 | rhizobactin siderophore biosynthesis protein RhbE | rhbE | −1.18 | 5.28 | siderophore synthesis | ||
| BPJ11090 | rhizobactin siderophore biosynthesis protein RhbF | rhbF | −1.72 | 3.02 | siderophore synthesis | ||
| BPJ35800 | iron complex ABC transporter ATP-binding protein FhuC | fhuC2 | 3.88 | 7.85 | iron uptake | ||
| BPJ35810 | iron complex ABC transporter permease FhuB | fhuB2 | 3.32 | 7.15 | iron uptake | ||
| BPJ35770 | putative iron complex ABC transporter permease FhuG | fhuG2 | 2.31 | 4.39 | iron uptake | ||
| BPJ35780 | putative iron complex ABC transporter substrate-binding protein FhuD | fhuD | 2.72 | 5.57 | iron uptake | ||
| BPJ35830 | putative iron transport-associated protein/putative siderophore | bpj35830 | 3.65 | 5.84 | iron uptake | ||
| BPJ35840 | putative heme uptake protein IsdC | bpj35840 | 4.91 | 7.62 | iron uptake | ||
| BPJ35850 | putative iron transport-associated protein | bpj35850 | 3.89 | 6.47 | iron uptake | ||
| BPJ28430 | DinB-like domain-containing protein YuaE | yuaE | 2.25 | 2.87 | spx | ||
| BPJ31980 | thioredoxin-disulfide reductase TrxB | trxB | 3.97 | 3.93 | 3.59 | spx | |
| BPJ29110 | putative NADH-dependent butanol dehydrogenase YugJ | yugJ | 2.32 | 1.08 | 4.60 | spx | |
| BPJ19830 | methionine sulfoxide reductase MsrA | msrA | 1.46 | 2.24 | spx | ||
| BPJ19820 | peptide-methionine sulfoxide reductase MsrB | msrB | 1.48 | 2.27 | spx | ||
| BPJ25870 | thioredoxin TrxA | trxA | 1.40 | 2.58 | spx/ctsR/sigB | ||
| BPJ35200 | NADPH-dependent nitro/flavin reductase NfrA | nfrA | 2.50 | 2.47 | 5.21 | spx/sigD/spo0A | |
| BPJ24450 | cystathionine gamma-lyase MccB | mccB | −1.58 | 7.58 | spx/cymR | ||
| BPJ17710 | putative cell division suppressor protein YneA | yneA | 2.24 | 44.25 | lexA/SOS | ||
| BPJ10180 | 3′-5′ exoribonuclease YhaM | yhaM | 0.71 | 2.81 | lexA/SOS | ||
| BPJ21860 | DNA polymerase 4 | polY1 | 10.68 | lexA/SOS | |||
| BPJ32300 | excinuclease ABC subunit B | uvrB | 7.22 | 2.52 | 4.29 | lexA/SOS | |
| BPJ32290 | excinuclease ABC subunit A | uvrA | 1.49 | 6.75 | lexA/SOS | ||
| BPJ25860 | excinuclease ABC subunit UvrC | uvrC | 3.65 | lexA/SOS | |||
| BPJ17700 | repressor LexA | lexA | 1.55 | 5.66 | lexA/SOS | ||
| BPJ17730 | DUF896 family protein YnzC | ynzC | 0.65 | 8.85 | lexA/SOS | ||
| BPJ12460 | phage-like PBSX protein XkdA | xkdA | 3.10 | 17.84 | lexA/SOS | ||
| BPJ17720 | resolvase-like protein YneB | yneB | 1.38 | 17.03 | lexA/SOS | ||
| BPJ10160 | putative exonuclease YhaO | yhaO | 8.76 | lexA/SOS | |||
| BPJ16880 | recombinase RecA | recA | 1.63 | 7.22 | 4.94 | 9.58 | lexA/SOS/comK |
| BPJ35170 | minor extracellular serine protease Vpr | vpr | 1.58 | 2.23 | lexA/SOS/phoP | ||
| BPJ21470 | hypothetical protein YpuD | ypuD | 1.93 | 7.12 | lexA/SOS/sigB/sigM | ||
| BPJ10170 | putative ATPase YhaN | yhaN | 8.73 | lexA/SOS | |||
| BPJ13450 | ATP-dependent Clp protease ATP-binding subunit ClpE | clpE | 2.78 | 45.41 | ctsR | ||
| BPJ25460 | ATP-dependent protease ATP-binding subunit ClpX | clpX | 2.67 | ctsR | |||
| BPJ00800 | DNA repair protein RadA | radA | 10.02 | ctsR/sigB | |||
| BPJ00760 | transcriptional regulator CtsR | ctsR | 9.40 | ctsR/sigB | |||
| BPJ00770 | transcriptional regulator McsA | mcsA | 10.26 | ctsR/sigB | |||
| BPJ31850 | ATP-dependent Clp protease proteolytic subunit ClpP | clpP | 1.79 | 4.26 | 8.74 | 1.73 | ctsR/sigB |
| BPJ00780 | putative ATP:guanido phosphotransferase McsB | mcsB | 1.43 | 8.87 | ctsR/sigB/sigF | ||
| BPJ00790 | ATP-dependent Clp protease ClpC | clpC | 6.44 | ctsR/sigB/sigF | |||
| BPJ00810 | DNA integrity scanning protein DisA | disA | 5.15 | ctsR/sigB/sigM | |||
| BPJ15470 | adenylyl-sulfate kinase CysC | cysC | 23.93 | 1.60 | cymR | ||
| BPJ15480 | uroporphyrin-3 C-methyltransferase CysG | cysG | 1.49 | 10.53 | cymR | ||
| BPJ15460 | sulfate adenylyltransferase Sat | sat | 1.37 | 13.07 | cymR | ||
| BPJ20800 | tryptophan synthase alpha subunit TrpA | trpA | 2.16 | 13.51 | TRAP | ||
| BPJ20810 | tryptophan synthase beta subunit TrpB | trpB | 1.13 | 13.99 | TRAP | ||
| BPJ20820 | N-(5′-phosphoribosyl)anthranilate isomerase TrpF | trpF | 1.59 | 2.09 | TRAP | ||
| BPJ20830 | indole-3-glycerol-phosphate synthase TrpC | trpC | 2.36 | 1.49 | 10.53 | TRAP | |
| BPJ20840 | anthranilate phosphoribosyltransferase TrpD | trpD | 1.44 | 2.72 | 1.37 | 13.07 | TRAP |
| BPJ20850 | anthranilate synthase component 1 | trpE | 2.61 | TRAP | |||
| BPJ12980 | transcriptional regulator OhrR | ohrR | 2.54 | ohrR | |||
| BPJ12970 | peroxiredoxin OhrA | ohrA | 10.66 | 9.88 | 6.29 | 1.30 | ohrR |
| BPJ12990 | peroxiredoxin OhrB | ohrB | 2.10 | sigB/ohrR | |||
| BPJ19510 | putative bacillithiol biosynthesis deacetylase YojG | yojG | 3.31 | bacillithiol-related | |||
| BPJ20020 | DUF1094 family protein YphP | yphP | 1.78 | 2.22 | bacillithiol-related | ||
| BPJ21140 | putative thioredoxin reductase YpdA | ypdA | 0.67 | 2.58 | bacillithiol-related | ||
| BPJ22220 | DUF1094 family protein YqiW | yqiW | 2.58 | bacillithiol-related | |||
| BPJ31300 | glycine betaine/carnitine/choline ABC transporter permease OpuCD | opuCD | 3.03 | glycine betaine transport | |||
| BPJ31310 | glycine betaine/carnitine/choline ABC transporter substrate-binding protein OpuCC | opuCC | 0.88 | 2.62 | glycine betaine transport | ||
| BPJ31320 | glycine betaine/carnitine/choline ABC transporter permease OpuCB | opuCB | 0.92 | 2.74 | glycine betaine transport | ||
| BPJ31330 | glycine betaine/carnitine/choline ABC transporter ATP-binding protein OpuCA | opuCA | 0.94 | 2.46 | glycine betaine transport | ||
| BPJ02950 | glycine betaine ABC transporter ATP-binding protein OpuAA | opuAA | 2.76 | 10.75 | glycine betaine transport | ||
| BPJ02960 | glycine betaine ABC transporter membrane protein | opuAB | 2.41 | 10.19 | glycine betaine transport | ||
| BPJ02970 | glycine betaine ABC transporter substrate-binding protein | opuAC | 2.37 | 7.48 | glycine betaine transport | ||
| BPJ29360 | Na+/H+ antiporter subunit MrpA | mrpA | 4.57 | sodium transport | |||
| BPJ29370 | Na+/H+ antiporter subunit MrpB | mrpB | 4.77 | sodium transport | |||
| BPJ29380 | Na+/H+ antiporter subunit MrpC | mrpC | 3.60 | sodium transport | |||
| BPJ29390 | Na+/H+ antiporter subunit MrpD | mrpD | 1.48 | 3.87 | sodium transport | ||
| BPJ29400 | Na+/H+ antiporter subunit MrpE | mrpE | 1.51 | 3.20 | sodium transport | ||
| BPJ29410 | Na+/H+ antiporter subunit MrpF | mrpF | 1.72 | 3.38 | sodium transport | ||
| BPJ29420 | Na+/H+ antiporter subunit MrpG | mrpG | 1.96 | 2.60 | sodium transport | ||
Selected genes and proteins that are induced in H2O2 treated B. pumilus cells.
Genes and proteins are listed, which could be assigned to putative regulons known from other Bacilli. Complete lists of upregulated as well as downregulated genes/proteins is given in supporting information Tables S2 and S3. For transcriptome, selected genes are shown for 3 and 8 minutes after stress compared to the control conditions (0 min). For a complete list of induced and repressed genes see Table S3. Differential regulation was determined from the biological triplicate measurements by false-discovery rate (FDR) from the Cyber-T p-values [27] by means of multiple testing correction [26]. Differential regulation was defined as a two-fold or higher differential expression with a FDR cut-off value of 0.05 or lower. Protein quantification was performed by the Delta 2D software (Decodon) from 3 biological replicates with a FDR cut-off value of 0.05 or lower.
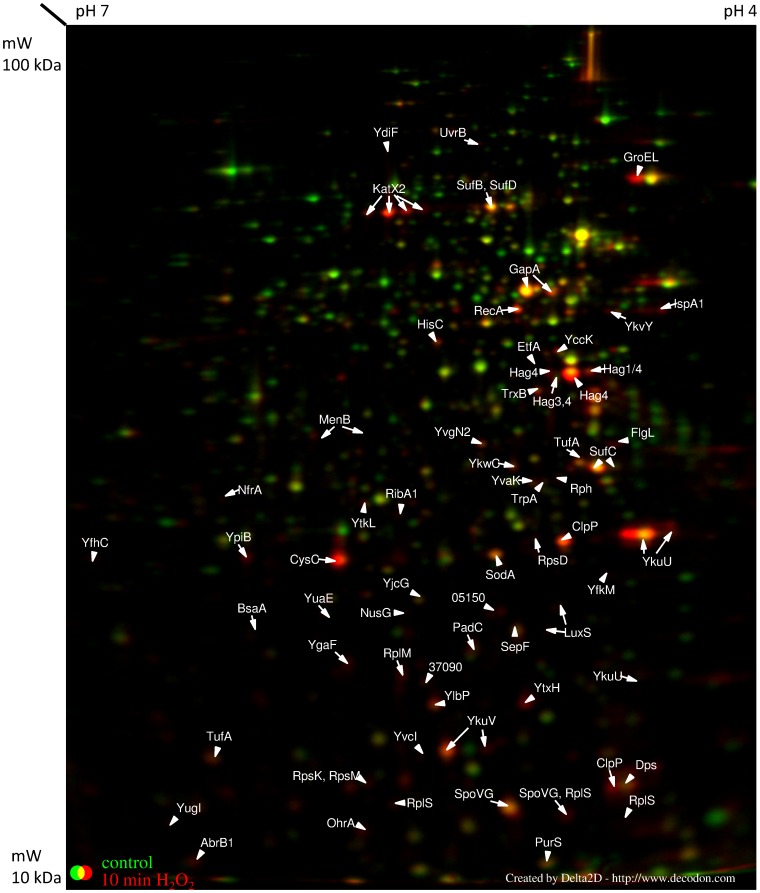
Figure 3. Cytosolic proteome 10 min after H2O2 treatment.
The cytosolic proteome of B. pumilus cells 10 min after H2O2 treatment. Cell samples were labeled with L–[35S]-methionine during the exponential growth phase (OD500 nm 0.6), and 10 min after H2O2 addition. Proteins were separated in a pH gradient 4 (right) –7 (left).
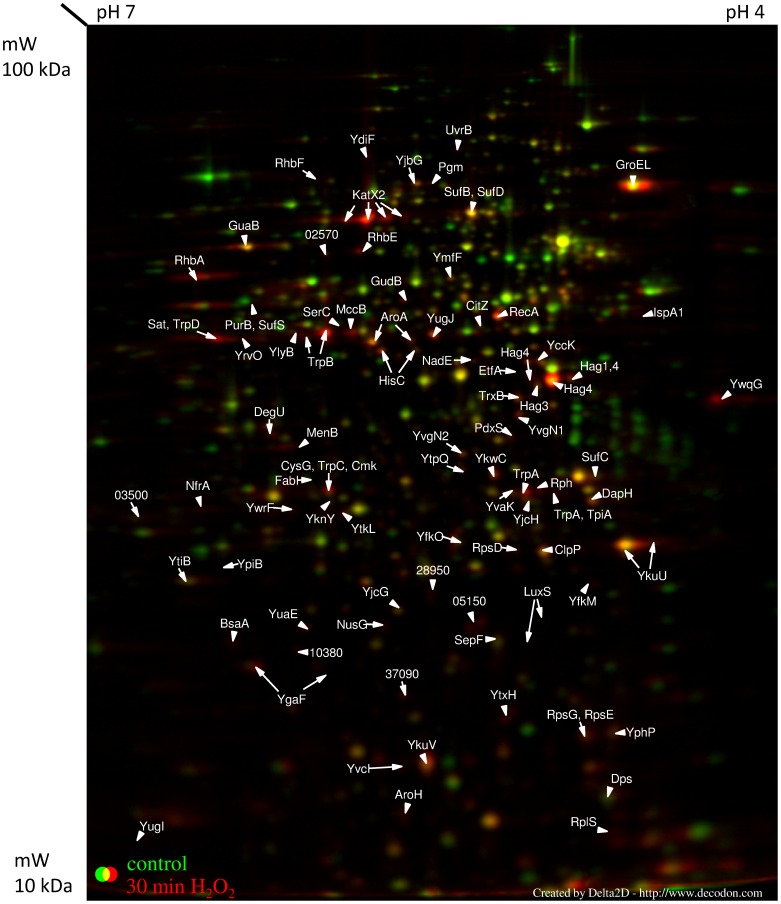
Figure 4. Cytosolic proteome 30 min after H2O2 treatment.
The cytosolic proteome of B. pumilus cells 30 min after H2O2 treatment. Cell samples were labeled with L–[35S]-methionine during the exponential growth phase (OD500 nm 0.6), and 30 min after H2O2 addition. Proteins were separated in a pH gradient 4 (right) –7 (left).
To compare the physiological changes in H2O2 treated B. pumilus cells with the oxidative stress responses of other organisms, the upregulated genes and proteins were assigned to putative regulons known from related organisms like B. subtilis and B. licheniformis [13], [20]. 139 of the upregulated genes and proteins could be assigned to these putative regulons (Table S2). The thus classified genes and proteins identified in this study are summarized and discussed below.
3.3 PerR Regulon
The PerR regulon is known to be highly induced by oxidative stress caused by hydrogen peroxide and paraquat [13]. As shown previously for B. licheniformis, the B. pumilus genome encodes a PerR regulator protein with a high level of identity (93%) to the PerR-protein known from B. subtilis [20]. Transcription of the perR gene was significantly increased immediately after stress (Table 1). This indicates a regulation mechanism of PerR in H2O2 treated B. pumilus cells that is similar to the de-repression model reported for B. subtilis [33].
In our study genes assigned to a putative PerR regulon, including those encoding the regulator proteins Fur and SpxA as well as the zinc-uptake protein ZosA, the heme biosynthesis complex HemABCD2LX and the general stress protein YjbC were significantly induced at transcriptional level (Table 1).
Strikingly, some of the PerR-regulated genes exhibiting the highest induction in B. subtilis cells subjected to hydrogen peroxide, are absent from the genome of the B. pumilus strain used in our study, as well as from a previously published B. pumilus genome [34]. This applies e.g. for the genes encoding the catalase KatA and the DNA-protection protein MrgA. Furthermore, B. pumilus lacks not only the genes ahpC and ahpF, encoding subunits of the alkyl hydroperoxide reductase, but there are no genes annotated with this function in the genome.
Instead of KatA, a gene annotated as catalase KatX2 (53% sequence similarity to B. subtilis KatX) was significantly induced in B. pumilus cells at transcriptional and translational level (up to 10 and 20-fold, respectively, Table 1). Thereby, KatX2 was one of the proteins with the highest induction rates detected. B. subtilis and B. licheniformis subjected to hydrogen peroxide exhibit a more than 100-fold induction of KatA [13], [20]. KatX2 comprises about 0.38% of the cytoplasmic protein present in the gel before addition of hydrogen peroxide. The values for B. subtilis and B. licheniformis are 0.13% in both strains (personal communication C. Scharf, B. Voigt). After addition of hydrogen peroxide KatX2 comprises about 3.8% of the cytoplasmic protein. This is comparable to the value of 3.6% for B. licheniformis (personal communication B. Voigt) but higher than the value for B. subtilis (1.2%, personal communication C. Scharf). These values indicate that in B. pumilus there is a higher synthesis of KatX2 already in unstressed cells compared to B. subtilis and B. licheniformis KatA explaining the lower induction rate. In B. subtilis, KatX is the major spore catalase and under control of SigB and SigF [35], [36]. We detected a B. subtilis PerR consensus sequence [32] containing 2 mismatches about 90 bases in front of the start codon of KatX2 indicating a possible involvement of PerR in its regulation.
3.4 Fur Regulon and Fe-metabolism
The PerR-regulated fur gene of B. pumilus, shows 95% similarity to the fur gene known from B. subtilis and was induced 3.6-fold after stress [32]. The regulator protein Fur of B. subtilis controls the expression of genes responsible for iron uptake [37]. Immediately after exposure to H2O2, cytosolic iron concentration is considerably reduced to prevent the formation of OH• by the Fenton reaction [13]. Upregulation of the Fur-controlled genes may be a reaction of the cells to optimize iron uptake in order to face the resulting iron limitation. Alternatively it might be that Fur is H2O2 sensitive as it is in E. coli [38].
Nine genes of a putative Fur regulon showed a significantly increased expression in B. pumilus cells after H2O2 treatment, including the ABC transporter system fhuB1C1G1 (Table 1). The fhuC gene was induced by H2O2 in B. subtilis and B. licheniformis, too [13], [20]. Further Fur regulon member genes known to be induced by H2O2 in B. subtilis showing an induction in our study were ykuN, ykuP (flavodoxins) and the hypothetical protein ykuO. With an about 30-fold higher mRNA level 8 minutes after treatment, these were among the highest upregulated genes in this putative regulon. The putative nitroreductase YfhC, also induced in H2O2 stressed B. subtilis cells, was the only member of the putative Fur regulon we observed to be upregulated at translational level.
The gene ywjA, encoding another ABC transporter of yet unknown function, the peptidase encoding gene yfkM and the bacillibactin esterase encoding gene ybbA were upregulated, too. These genes are Fur-regulated in B. subtilis, but they were not upregulated by H2O2 in this organism [13], [39]. In B. subtilis and B. licheniformis, the siderophore biosynthesis complex encoded by dhbACEBF was strongly upregulated by H2O2. In our study, these genes showed no significant changes in their expression level.
Other genes that exhibited higher transcription rates after H2O2 treatment were the iron ABC transporter protein encoding gene feuA and its upstream-located regulator ybbB (renamed btr in B. subtilis) [40]. Unlike B. subtilis, the B. pumilus genome encodes a second Fhu-related iron uptake system. Our study showed an induction of the genes encoding FhuC2-FhuB2-BPJ35820 as well as fhuG2 and fhuD immediately after subjecting the cells to the stress. Two further putative iron transporter systems, bpj35830-bpj35840-bpj35850 and bpj08420-bpj08430-bpj08440, were induced, too. The proteins encoded by the latter genes showed no significant homology to any protein known from related Bacillus species.
Furthermore, the proteomic approach revealed a strong induction of the siderophore synthesis proteins RhbA, RhbE and RhbF, encoded by the rhbABCDEF-operon (Table 1). A rather slight induction at the translational level was shown for the iron/sulfur cluster biogenesis proteins SufB, SufS, SufD and SufC as previously shown for B. licheniformis [20]. The sufU gene was found to be only slightly upregulated at the mRNA level.
3.5 Spx Regulon and Bacillithiol
Another regulator protein assigned to the putative PerR regulon is SpxA, controlling the expression of the Spx regulon in B. subtilis [41], [42]. This gene exhibited an about 4-fold increased transcription rate in H2O2 stressed B. pumilus cells. Some of the genes and proteins attributed to a putative Spx regulon in B. pumilus appeared to have rather moderately increased expression rates or were not induced after H2O2 treatment.
In our study we detected six genes of a putative Spx regulon to be induced following H2O2 treatment (Table 1). The proteins encoded by three of them, nitro/flavinreductase NfrA, putative NADPH-dependent butanol dehydrogenase YugJ and thioredoxin-disulfide reductase TrxB, were induced in H2O2 treated cells, too. Upregulation of msrAB (methionine sulfoxide reductase operon) and trxA (thioredoxin) was detected at transcriptional level only. The proteins TrxA and TrxB are described to act in direct detoxification of hydrogen peroxide [43]–[45]. Cystathionine gamma-lyase MccB and DinB-like domain-containing protein YuaE showed an induction only at proteome level.
The Spx-regulated srf operon, mediating competence and metabolic functions in B. subtilis, is absent in the B. pumilus genome as shown before for B. licheniformis [42], [46], [47].
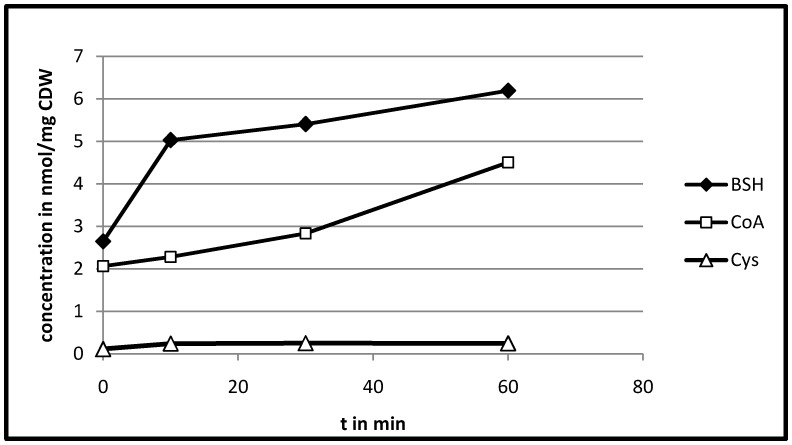
We noticed an increased transcription of ypdA and yqiW as well as an induction of the yphP gene product (Table 1). These genes co-occur with bacillithiol (Cys-GlcN-malate, BSH) synthesis genes [48]. However, only one gene encoding a protein involved in bacillithiol synthesis, yojG was transcribed at a slightly elevated level (Table S2). Bacillithiol is one of the major thiols in B. subtilis and known to be involved in resistance against organic peroxide stress and disulfide stress [7], [49], [50]. For further investigation, we analyzed the cytosolic metabolome of H2O2 treated B. pumilus cells concerning the concentration of thiol compounds. Our analysis revealed a bacillithiol level of 2.6 nmol per mg cell dry weight already under control conditions. Similar BSH concentrations have been detected in B. subtilis (0.6–2.2 nmol per mg) [7], [48], [51]. Ten minutes after H2O2 treatment, the cytosolic concentration of bacillithiol increased to 5 nmol per mg cell dry weight (Figure 5). The increase continued up to a concentration of about 6.2 nmol per mg cell dry weight 60 minutes after stress. Since only one bacillithiol synthesis gene (yojG, renamed bshB2 in B. subtilis) was slightly upregulated, increase of bacillithiol concentration in the cells might be regulated allosterically, for example, by an oxidation of the BSH pool leading to a relief of feedback inhibition. [52], [53].
Figure 5. Concentration of thiol compounds in B. pumilus cells.
Cytosolic concentration of bacillithiol (BSH), CoA and cysteine (Cys) per mg cell dry weight (CDW) during the exponential growth phase (OD500 nm 0.6 at 0 min) and 10, 30 and 60 min after H2O2 treatment.
3.6 SOS Regulon
H2O2 treatment leads to the formation of OH• by Fenton reaction, which exhibits a high DNA-damaging potential. Lowering the concentration of iron in the cells reduces this threat. As a result, B. subtilis and B. licheniformis cells subjected to oxidative stress caused by H2O2, induced the SOS regulon, regulated by the proteins RecA and LexA, responsible for repair of DNA [13], [20], [54], [55].
The proteomic analysis displayed the induction of two proteins, excinuclease subunit UvrB and the recombinase RecA, assigned to a putative SOS regulon in B. pumilus following H2O2 treatment (Table 1). The transcriptomic approach added further 13 upregulated genes belonging to this putative regulon; among them the excinuclease subunits encoding genes uvrA and uvrC. The operon yneABynzC, induced by H2O2 and involved in suppression of cell division in B. subtilis, was also strongly induced in our study [13], [56]. This might be an explanation for the formation of atypically long cells as described above. Showing an about 44-fold increased transcription rate, yneA belongs to the strongest induced genes observed in our study. Furthermore, the putative DNA double-strand break repair cluster yhaONM exhibited a significantly higher transcription rate following H2O2 addition [57].
3.7 CtsR Regulon
The CtsR regulon, mediating repair and/or degradation of misfolded and damaged proteins, was induced by several oxidative stressors in B. subtilis and B. licheniformis [13], [20], [58]. In our study, we detected an upregulation of nine genes assigned to a putative CtsR regulon in B. pumilus indicating a significant impact of H2O2 on protein quality (Table 1). The operon ctsR-mcsAB-clpC was transcribed with significantly higher intensity after the addition of H2O2 as well as the genes clpE, clpX and clpP, encoding members of the proteolytic complex. Only ClpP was observed to be induced at the protein level. Furthermore, the DNA repair protein encoding gene radA and the DNA integrity scanning protein encoding gene disA showed higher transcription rates compared to control conditions.
3.8 SigB Regulon
Besides the induction of the above described putative regulons more or less directly associated to oxidative stress, H2O2 treated cells exhibited an upregulation of 47 genes known to be under control of the general stress sigma factor SigB in B. subtilis (Table 1) [59], [60]. A part of a putative SigB-regulon in B. pumilus detected to be upregulated in our study was the sigB gene itself with its signal cascade genes rsbRSTUVW and rsbX indicating an activation of the putative regulon via the general stress response cascade known from B. subtilis [61].
Another of these putative SigB-dependent genes, encoding the putative universal stress protein NhaX, showed the highest induction rate detected in this study (more than 60-fold). Further strongly upregulated genes are the regulator protein encoding gene mgsR and ydaG (general stress protein), both also detected to be induced in H2O2 stressed B. licheniformis cells [20]. The upregulated genes mgsR and ydaG encode proteins with still unknown functions. Six of the upregulated putative SigB-dependent genes could be also detected to be induced in the proteomic approach. The putative general stress protein YtxH is among the strongest induced proteins (about 14-fold). The putative iron storage/DNA protecting protein Dps, providing peroxide resistance in B. anthracis, was induced in H2O2 treated B. pumilus cells, too [62].
3.9 CymR Regulon
The results of our study showed an upregulation of several proteins belonging to a putative CymR regulon. In B. subtilis, it is described to be involved in regulation of the sulfur metabolism [63]. An induction of genes belonging to this regulon has been shown in cells afflicted with oxidative stress caused by paraquat, but not stress caused by H2O2 [13]. Our proteome study showed a strong induction of three putatively CymR-regulated proteins. The adenylyl-sulfate kinase (CysC) was with an induction of about 24-fold the strongest induced protein. An upregulation of the sulfate adenylyltransferase (Sat) catalyzing sulfate assimilation to 3′-phospho-adenylylsulfate was also detected (Table 1). Further proteins involved in cysteine biosynthesis were not significantly upregulated. The third upregulated protein is the uroporphyrin-3 C-methyltransferase (CysG). This enzyme catalyzes a reaction in a branch in the heme pathway producing precorrin2. An induction of the enzymes that continue the pathway from precorrin2 to siroheme could not be detected.
3.10 Other B. pumilus Upregulated Genes/proteins
The OhrR-regulated peroxiredoxin-encoding gene ohrA is reported to be involved in organic peroxide resistance in B. subtilis [64]. Following H2O2 treatment, there was no induction of this gene observed in B. subtilis and B. licheniformis [13], [20]. In our study, we observed a strongly induced expression of this gene at transcriptional and translational level indicating an involvement of this peroxiredoxin in the H2O2 resistance of B. pumilus (Table 1). Transcription of the other organic peroxide resistance peroxiredoxin (ohrB) as well as their regulator gene ohrR was also slightly induced in hydrogen peroxide treated B. pumilus cells.
H2O2 treatment induced some additional regulator genes. One of them is fadR, encoding a regulator protein mediating fatty acid degradation in B. subtilis [65]. Two genes putatively controlled by FadR, etfAB - encoding the electron transfer flavoprotein alpha and beta subunit, were also induced (Table S2). Another regulator, AbrB1, controlling the expression of genes induced by transition from exponential to stationary growth in B. subtilis [66], was induced at transcriptional and translational level. Similar results, but with significantly higher induction rates in the proteomic approach, were observed for the AbrB1-regulated peroxiredoxin YkuU and thiol-disulfide oxidoreductase YkuV. Furthermore, several putative regulator genes with still unknown targets were observed to be upregulated. Bpj13620, bpj17020 and ydcI showed the highest changes in their expression rates. Genes encoding a sensor kinase and a response regulator forming the two-component system YhcYZ were significantly induced directly after H2O2 treatment. Its function is also unknown.
Several genes and proteins involved in transport processes were detected to be upregulated following H2O2 stress (Table 1, S2). H2O2 treatment caused an upregulation of the sodium uptake system natAB and the mrpABCDEFG cluster. This operon encodes a sodium excretion system that is considered to be the major sodium excretion system in bacteria and acts in pH homeostasis and multiple resistances in B. subtilis [67], [68].
Strikingly, transcription of the glycine betaine uptake system consisting of opuAA-AB-AC and opuCA-CB-CC-CD was observed to be significantly induced after treatment, indicating that H2O2 impacts osmotic homeostasis in B. pumilus cells [69]. Furthermore, it is worth to mention that H2O2 induced expression of a putative TRAP regulon in B. pumilus cells. An upregulation of the tryptophan-synthesis operon trpABFCDE as well as histidinol-phosphate aminotransferase HisC was observed in our analysis. However, neither addition of tryptophan nor addition of glycine betaine before peroxide treatment brought forth better growth or survival of stressed B. pumilus cells (data not shown).
3.11 Downregulated Genes/proteins
As shown for many other organisms, the adaptation mechanism of B. pumilus cells to oxidative stress includes also a downregulation of vegetative cellular functions. Most of the down-regulated genes encode proteins involved in main metabolic pathways. As shown for B. subtilis and B. licheniformis, expression of the purine and pyrimidine synthesis genes was downregulated as well as genes involved in synthesis of arginine (Table S3) [13], [20]. Contrary to B. subtilis and B. licheniformis, a repression of histidine synthesis genes was not observed. Instead, isoleucine and leucine synthesis genes were expressed in lower amounts following H2O2 treatment. This repression might due to the iron sparing response described by Gaballa et al. [70]. Repression of enzymes involved in branched chain amino acid synthesis has been found during iron starvation in B. subtilis [37]. Furthermore, we observed a reduced expression of most of the aminoacyl-tRNA-synthetases, with the exception of tryptophanyl-tRNA-synthetase trpS, which matched the upregulation of the tryptophan operon.
Strikingly, a stringent response, i.e. a downregulation of ribosomal proteins or elongation factors like fusA, tsf or tufA, as described for other organisms (B. subtilis, B. licheniformis, E. coli) could not be detected in B. pumilus [13], [20], [71].
Conclusion
The combination of proteomics and transcriptomics revealed a specific adaptation of B. pumilus cells caused by the oxidative stress trigger H2O2. Although many of the induced genes and proteins could be assigned to well-known oxidative stress regulons like PerR, CtsR and Fur, there are particular mechanisms detectable which seem to be involved in the remarkable oxidative stress resistance of B. pumilus. The concentration of H2O2 that was used to trigger the stress in our study was about 40-fold higher than those used for comparable analysis of B. subtilis or B. licheniformis. Our study could enlighten several points at which the peroxide stress response of B. pumilus cells is different from its Gram-positive relatives. It is suggested that the catalase KatA is replaced by the catalase KatX2. Furthermore, our study revealed an induction of genes that are highly correlated to bacillithiol synthesis indicating an involvement of bacillithiol in the peroxide stress response of B. pumilus. Metabolome analysis demonstrated a basal level of this protective metabolite but also an increase of the cytosolic bacillithiol concentration during peroxide stress. Furthermore, a considerable set of H2O2 induced unique proteins with so far unknown function could be identified in this study. These proteins are worth to address in follow up studies to elucidate their specific role in the oxidative stress adaptation of this organism. Finally, since B. pumilus is an organism of industrial interest, understanding its oxidative stress response and defining marker genes for the analysis of fermentation processes is important to prevent possible negative influences on the process and the product quality.
Supporting Information
Determination of minimal inhibition concentration of hydrogen peroxide.
(XLSX)
Genes and proteins that are upregulated after addition of hydrogen peroxide.
(XLSX)
Genes and proteins that are downregulated after addition of hydrogen peroxide.
(XLSX)
Individual signal on the array for five probes for selected genes.
(XLSX)
Acknowledgments
We thank Annette Meuche for excellent technical assistance and Chris Hamilton (University of East Anglia) for providing the bacillithiol standard.
Funding Statement
This work was financially supported by funds of the Competence Network “Mipro - Microbes for production: A genomics-based approach to engineer novel industrial production strains” (0315594B) by the German Federal Ministry of Education and Research (BMBF, www.bmbf.de) and the project “Ausbau und Profilierung von COAST Fun-Gene” (UG11043, ESF/IV-BM-B35-0003/12) of the Bildungsministerium of Mecklenburg-Vorpommern (www.regierung-mv.de). The transcriptome analysis was financially supported by the Henkel KGaA (www.henkel.de). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Schweder T, Hecker M (2004) Monitoring of stress responses. Adv Biochem Eng Biotechnol 89: 47–71. [DOI] [PubMed] [Google Scholar]
- 2. Stadtman ER, Levine RL (2003) Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids 25: 207–218. [DOI] [PubMed] [Google Scholar]
- 3. Farr SB, Kogoma T (1991) Oxidative stress responses in Escherichia coli and Salmonella typhimurium . Microbiol Rev 55: 561–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benardini JN, Sawyer J, Venkateswaran K, Nicholson WL (2003) Spore UV and acceleration resistance of endolithic Bacillus pumilus and Bacillus subtilis isolates obtained from Sonoran desert basalt: implications for lithopanspermia. Astrobiology 3: 709–717. [DOI] [PubMed] [Google Scholar]
- 5. Kempf MJ, Chen F, Kern R, Venkateswaran K (2005) Recurrent isolation of hydrogen peroxide-resistant spores of Bacillus pumilus from a spacecraft assembly facility. Astrobiology 5: 391–405. [DOI] [PubMed] [Google Scholar]
- 6. Imlay JA, Fridovich I (1991) Assay of metabolic superoxide production in Escherichia coli . J Biol Chem 266: 6957–6965. [PubMed] [Google Scholar]
- 7. Newton GL, Rawat M, La Clair JJ, Jothivasan VK, Budiarto T, et al. (2009) Bacillithiol is an antioxidant thiol produced in Bacilli. Nat Chem Biol 5: 625–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blokhina O, Virolainen E, Fagerstedt KV (2003) Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann Bot 91 Spec No: 179–194. [DOI] [PMC free article] [PubMed]
- 9. Imlay JA (2003) Pathways of oxidative damage. Annu Rev Microbiol 57: 395–418. [DOI] [PubMed] [Google Scholar]
- 10. Aruoma OI, Halliwell B, Gajewski E, Dizdaroglu M (1991) Copper-ion-dependent damage to the bases in DNA in the presence of hydrogen peroxide. Biochem J 273 (Pt 3): 601–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zuber P (2009) Management of oxidative stress in Bacillus . Annu Rev Microbiol 63: 575–597. [DOI] [PubMed] [Google Scholar]
- 12. Imlay JA (2008) Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77: 755–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mostertz J, Scharf C, Hecker M, Homuth G (2004) Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150: 497–512. [DOI] [PubMed] [Google Scholar]
- 14. Hoi le T, Voigt B, Jürgen B, Ehrenreich A, Gottschalk G, et al. (2006) The phosphate-starvation response of Bacillus licheniformis . Proteomics 6: 3582–3601. [DOI] [PubMed] [Google Scholar]
- 15. Büttner K, Bernhardt J, Scharf C, Schmid R, Mäder U, et al. (2001) A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis . Electrophoresis 22: 2908–2935. [DOI] [PubMed] [Google Scholar]
- 16. Voigt B, Schweder T, Becher D, Ehrenreich A, Gottschalk G, et al. (2004) A proteomic view of cell physiology of Bacillus licheniformis . Proteomics 4: 1465–1490. [DOI] [PubMed] [Google Scholar]
- 17. Wolf C, Hochgräfe F, Kusch H, Albrecht D, Hecker M, et al. (2008) Proteomic analysis of antioxidant strategies of Staphylococcus aureus: diverse responses to different oxidants. Proteomics 8: 3139–3153. [DOI] [PubMed] [Google Scholar]
- 18. Völker U, Engelmann S, Maul B, Riethdorf S, Völker A, et al. (1994) Analysis of the induction of general stress proteins of Bacillus subtilis . Microbiology 140 (Pt 4): 741–752. [DOI] [PubMed] [Google Scholar]
- 19. Homuth G, Masuda S, Mogk A, Kobayashi Y, Schumann W (1997) The dnaK operon of Bacillus subtilis is heptacistronic. J Bacteriol 179: 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schroeter R, Voigt B, Jürgen B, Methling K, Pöther DC, et al. (2011) The peroxide stress response of Bacillus licheniformis . Proteomics 11: 2851–2866. [DOI] [PubMed] [Google Scholar]
- 21. Delcher AL, Harmon D, Kasif S, White O, Salzberg SL (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res 27: 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guo FB, Zhang CT (2006) ZCURVE_V: a new self-training system for recognizing protein-coding genes in viral and phage genomes. BMC Bioinformatics 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borodovsky M MR, Besemer J., Lomsadze A. (2003) Current Protocols in Bioinformatics. [DOI] [PubMed]
- 24. Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, et al. (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wernersson R, Juncker AS, Nielsen HB (2007) Probe selection for DNA microarrays using OligoWiz. Nat Protoc 2: 2677–2691. [DOI] [PubMed] [Google Scholar]
- 26. van Hijum SA, de Jong A, Baerends RJ, Karsens HA, Kramer NE, et al. (2005) A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baldi P, Long AD (2001) A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics 17: 509–519. [DOI] [PubMed] [Google Scholar]
- 28. Pöther DC, Liebeke M, Hochgräfe F, Antelmann H, Becher D, et al. (2009) Diamide triggers mainly S Thiolations in the cytoplasmic proteomes of Bacillus subtilis and Staphylococcus aureus . J Bacteriol 191: 7520–7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sharma SV, Jothivasan VK, Newton GL, Upton H, Wakabayashi JI, et al. (2011) Chemical and Chemoenzymatic syntheses of bacillithiol: a unique low-molecular-weight thiol amongst low G+C Gram-positive bacteria. Angew Chem Int Ed Engl 50: 7101–7104. [DOI] [PubMed] [Google Scholar]
- 30. Pöther DC, Gierok P, Harms M, Mostertz J, Hochgräfe F, et al. (2013) Distribution and infection-related functions of bacillithiol in Staphylococcus aureus . Int J Med Microbiol 303: 114–123. [DOI] [PubMed] [Google Scholar]
- 31. Münch R, Hiller K, Barg H, Heldt D, Linz S, et al. (2003) PRODORIC: prokaryotic database of gene regulation. Nucleic Acids Res 31: 266–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fuangthong M, Herbig AF, Bsat N, Helmann JD (2002) Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J Bacteriol 184: 3276–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee JW, Helmann JD (2006) The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440: 363–367. [DOI] [PubMed] [Google Scholar]
- 34. Gioia J, Yerrapragada S, Qin X, Jiang H, Igboeli OC, et al. (2007) Paradoxical DNA repair and peroxide resistance gene conservation in Bacillus pumilus SAFR-032. PLoS One 2: e928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bagyan I, Casillas-Martinez L, Setlow P (1998) The katX gene, which codes for the catalase in spores of Bacillus subtilis, is a forespore-specific gene controlled by sigmaF, and KatX is essential for hydrogen peroxide resistance of the germinating spore. J Bacteriol 180: 2057–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Petersohn A, Engelmann S, Setlow P, Hecker M (1999) The katX gene of Bacillus subtilis is under dual control of sigmaB and sigmaF. Mol Gen Genet 262: 173–179. [DOI] [PubMed] [Google Scholar]
- 37. Baichoo N, Wang T, Ye R, Helmann JD (2002) Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol Microbiol 45: 1613–1629. [DOI] [PubMed] [Google Scholar]
- 38. Varghese S, Wu A, Park S, Imlay KR, Imlay JA (2007) Submicromolar hydrogen peroxide disrupts the ability of Fur protein to control free-iron levels in Escherichia coli . Mol Microbiol 64: 822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, et al. (1997) The complete genome sequence of the gram-positive bacterium Bacillus subtilis . Nature 390: 249–256. [DOI] [PubMed] [Google Scholar]
- 40. Gaballa A, Helmann JD (2007) Substrate induction of siderophore transport in Bacillus subtilis mediated by a novel one-component regulator. Mol Microbiol 66: 164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Choi SY, Reyes D, Leelakriangsak M, Zuber P (2006) The global regulator Spx functions in the control of organosulfur metabolism in Bacillus subtilis . J Bacteriol 188: 5741–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nakano S, Küster-Schöck E, Grossman AD, Zuber P (2003) Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis . Proc Natl Acad Sci U S A 100: 13603–13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Spector A, Yan GZ, Huang RR, McDermott MJ, Gascoyne PR, et al. (1988) The effect of H2O2 upon thioredoxin-enriched lens epithelial cells. J Biol Chem 263: 4984–4990. [PubMed] [Google Scholar]
- 44. Chae HZ, Chung SJ, Rhee SG (1994) Thioredoxin-dependent peroxide reductase from yeast. J Biol Chem 269: 27670–27678. [PubMed] [Google Scholar]
- 45. Fernando MR, Nanri H, Yoshitake S, Nagata-Kuno K, Minakami S (1992) Thioredoxin regenerates proteins inactivated by oxidative stress in endothelial cells. Eur J Biochem 209: 917–922. [DOI] [PubMed] [Google Scholar]
- 46. Nakano S, Erwin KN, Ralle M, Zuber P (2005) Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol Microbiol 55: 498–510. [DOI] [PubMed] [Google Scholar]
- 47. Veith B, Herzberg C, Steckel S, Feesche J, Maurer KH, et al. (2004) The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. J Mol Microbiol Biotechnol 7: 204–211. [DOI] [PubMed] [Google Scholar]
- 48. Gaballa A, Newton GL, Antelmann H, Parsonage D, Upton H, et al. (2010) Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc Natl Acad Sci U S A 107: 6482–6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee JW, Soonsanga S, Helmann JD (2007) A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci U S A 104: 8743–8748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chi BK, Gronau K, Mäder U, Hessling B, Becher D, et al. (2011) S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol Cell Proteomics 10: M111 009506. [DOI] [PMC free article] [PubMed]
- 51. Chi BK, Roberts AA, Huyen TT, Bäsell K, Becher D, et al. (2013) S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria. Antioxid Redox Signal 18: 1273–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Upton H, Newton GL, Gushiken M, Lo K, Holden D, et al. (2012) Characterization of BshA, bacillithiol glycosyltransferase from Staphylococcus aureus and Bacillus subtilis . FEBS Lett 586: 1004–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gaballa A, Antelmann H, Hamilton CJ, Helmann JD (2013) Regulation of Bacillus subtilis bacillithiol biosynthesis operons by Spx. Microbiology 159: 2025–2035. [DOI] [PubMed] [Google Scholar]
- 54. Miller MC, Resnick JB, Smith BT, Lovett CM Jr (1996) The Bacillus subtilis dinR gene codes for the analogue of Escherichia coli LexA. Purification and characterization of the DinR protein. J Biol Chem 271: 33502–33508. [PubMed] [Google Scholar]
- 55. Love PE, Lyle MJ, Yasbin RE (1985) DNA-damage-inducible (din) loci are transcriptionally activated in competent Bacillus subtilis . Proc Natl Acad Sci U S A 82: 6201–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kawai Y, Moriya S, Ogasawara N (2003) Identification of a protein, YneA, responsible for cell division suppression during the SOS response in Bacillus subtilis . Mol Microbiol 47: 1113–1122. [DOI] [PubMed] [Google Scholar]
- 57. Krishnamurthy M, Tadesse S, Rothmaier K, Graumann PL (2010) A novel SMC-like protein, SbcE (YhaN), is involved in DNA double-strand break repair and competence in Bacillus subtilis . Nucleic Acids Res 38: 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Leichert LI, Scharf C, Hecker M (2003) Global characterization of disulfide stress in Bacillus subtilis . J Bacteriol 185: 1967–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Petersohn A, Brigulla M, Haas S, Hoheisel JD, Völker U, et al. (2001) Global analysis of the general stress response of Bacillus subtilis . J Bacteriol 183: 5617–5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hecker M, Reder A, Fuchs S, Pagels M, Engelmann S (2009) Physiological proteomics and stress/starvation responses in Bacillus subtilis and Staphylococcus aureus . Res Microbiol 160: 245–258. [DOI] [PubMed] [Google Scholar]
- 61. Hecker M, Völker U (2001) General stress response of Bacillus subtilis and other bacteria. Adv Microb Physiol 44: 35–91. [DOI] [PubMed] [Google Scholar]
- 62. Tu WY, Pohl S, Gizynski K, Harwood CR (2012) The iron-binding protein Dps2 confers peroxide stress resistance on Bacillus anthracis . J Bacteriol 194: 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Even S, Burguiere P, Auger S, Soutourina O, Danchin A, et al. (2006) Global control of cysteine metabolism by CymR in Bacillus subtilis . J Bacteriol 188: 2184–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fuangthong M, Atichartpongkul S, Mongkolsuk S, Helmann JD (2001) OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis . J Bacteriol 183: 4134–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Matsuoka H, Hirooka K, Fujita Y (2007) Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation. J Biol Chem 282: 5180–5194. [DOI] [PubMed] [Google Scholar]
- 66. Perego M, Spiegelman GB, Hoch JA (1988) Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis . Mol Microbiol 2: 689–699. [DOI] [PubMed] [Google Scholar]
- 67. Ito M, Guffanti AA, Oudega B, Krulwich TA (1999) mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J Bacteriol 181: 2394–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kajiyama Y, Otagiri M, Sekiguchi J, Kosono S, Kudo T (2007) Complex formation by the mrpABCDEFG gene products, which constitute a principal Na+/H+ antiporter in Bacillus subtilis . J Bacteriol 189: 7511–7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kempf B, Bremer E (1995) OpuA, an osmotically regulated binding protein-dependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis . J Biol Chem 270: 16701–16713. [DOI] [PubMed] [Google Scholar]
- 70. Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song KB, et al. (2008) The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci U S A 105: 11927–11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. VanBogelen RA, Kelley PM, Neidhardt FC (1987) Differential induction of heat shock, SOS, and oxidation stress regulons and accumulation of nucleotides in Escherichia coli . J Bacteriol 169: 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Determination of minimal inhibition concentration of hydrogen peroxide.
(XLSX)
Genes and proteins that are upregulated after addition of hydrogen peroxide.
(XLSX)
Genes and proteins that are downregulated after addition of hydrogen peroxide.
(XLSX)
Individual signal on the array for five probes for selected genes.
(XLSX)