Abstract
Chronic thromboembolic pulmonary hypertension (CTEPH) is one of the main causes of severe pulmonary hypertension. However, despite treatment (pulmonary endarterectomy), in approximately 15–20% of patients, pulmonary vascular resistance and pulmonary arterial pressure continue to increase. To date, little is known about the changes that occur in gene expression in CTEPH. The identification of genes associated with CTEPH may provide insight into the pathogenesis of CTEPH and may aid in diagnosis and treatment. In this study, we analyzed the gene expresion profiles of pulmonary artery endothelial cells from 5 patients with CTEPH and 5 healthy controls using oligonucleotide microarrays. Bioinformatics analyses using the Gene Ontology (GO) and KEGG databases were carried out to identify the genes and pathways specifically associated with CTEPH. Signal transduction networks were established to identify the core genes regulating the progression of CTEPH. A number of genes were found to be differentially expressed in the pulmonary artery endothelial cells from patients with CTEPH. In total, 412 GO terms and 113 pathways were found to be associated with our list of genes. All differential gene interactions in the Signal-Net network were analyzed. JAK3, GNA15, MAPK13, ARRB2 and F2R were the most significantly altered. Bioinformatics analysis may help gather and analyze large amounts of data in microarrays by means of rigorous experimental planning, scientific statistical analysis and the collection of complete data. In this study, a novel differential gene expression pattern was constructed. However, further studies are required to identify novel targets for the diagnosis and treatment of CTEPH.
Keywords: chronic thromboembolic pulmonary hypertension, gene, microarray, Gene Ontology, signaling pathway
Introduction
Chronic thromboembolic pulmonary hypertension (CTEPH) is one of the main causes of severe pulmonary hypertension. CTEPH is characterized by the presence of unresolved thromboemboli associated with fibrous stenosis in the proximal pulmonary arteries. Diagnosis is usually made in the advanced stages of the disease when pulmonary vascular resistance (PVR) has increased by 5- to 10-fold. This increase in PVR resistance results in pulmonary hypertension and progressive right heart failure. It may be caused by a single or recurrent pulmonary embolism and/or the local formation of thrombi. The proximal location of pulmonary artery obliteration is the main feature observed in patients with CTEPH that differs from pulmonary arterial hypertension (PAH) (1).
Depending on the localization and extent of proximal thrombotic material, a pulmonary endarterectomy (PEA) may be necessary (2). Approximately half of the pulmonary blood flow dynamics can return to normal levels following PEA. However, in approximately 15–20% of patients, PVR and pulmonary arterial pressure (PAP) continue to rise, thus increasing the mortality rate by up to 4–5% (3). It has been suggested that the reason for the development of the persistent occlusion of the pulmonary artery is a misguided thrombus resolution triggered by infection (4), inflammation (5), autoimmunity, malignancy (6) and/or endothelial dysfunction due to a high presence of phospholipid antibodies and lupus anticoagulants (7,8) rather than prothrombotic factors.
A number of factors, such as C-reactive protein (CRP) (9), endothelin-1 and von Willebrand factor (10) may participate in the pathophysiology of pulmonary hypertension. However, there are hundreds of implicated genomic loci with heterogeneous functions. As a result, there is difficulty in understanding the mechanisms by which this diverse genetic susceptibility translates to a common clinical phenotype. The advent of genome-wide technologies, such as gene expression microarray, has made it possible to achieve a comprehensive view of the alterations in gene expression occurring in CTEPH. In the present study, we used bioinformatics to analyze the differences in gene expression between CTEPH and normal tissue. The different Gene Ontology (GO) terms and pathways revealed the most important mechanisms and candidate genes involved in the development of CTEPH; our data may aid in the development of more individualized treatment regimens according to the genetic characteristics of individual patients.
Patients and methods
Patients
Five consecutive patients with CTEPH were enrolled in this study from the Beijing Chao-Yang Hospital, Capital Medical University, Beijing, China between June 2012 and February 2013. The study was approved by the relevant ethics commitee and all patients provided written consent to participate in this study. All patients were examined using lung ventilation and perfusion scans, right-heart catheterization and pulmonary angiography to confirm the diagnosis. Patients with CTEPH were defined as those having a mean pulmonary arterial pressure (mPAP) of ≥25 mmHg with normal wedge pressure (≤12 mmHg) who had dyspnea on exertion during a period of >6 months on effective anticoagulation. In addition, lung perfusion scans were performed to demonstrate a segmental or larger defect concomitant with a normal ventilation scan (11,12). Finally, chronic thromboembolic findings were confirmed on pulmonary angiography (13). Five healthy controls (donors for lung transplants) were also included.
Microarray analysis
For Affymetrix microarray profiling, total RNA of CTEPH and normal tissues was isolated using TRIzol reagent (Invitrogen, Burlington, ON, Canada) and purified using the RNeasy Mini kit (Qiagen, Hilden, Germany), including a DNase digestion treatment. RNA concentrations were determined by absorbance (A) at 260 nm and quality control standards were A260/A280 = 1.8–2.1, using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
cDNA of pulmonary artery endothelial cells from patients with CTEPH or the normal controls was hybridized to Human Gene 2.0 ST GeneChip® arrays (Affymetrix, Inc., Santa Clara, CA, USA) according to the manufacturer’s instructions. Affymetrix® Expression Console Software (version 1.2.1) was used for microarray analysis. Raw data (CEL files) were normalized at the transcript level using the robust multi-average method (RMA workflow). The median summarization of transcript expressions was calculated. Gene-level data was then filtered to include only those probe sets that are in the ‘core’ metaprobe list, which represent RefSeq genes.
Analysis of differential gene expression
The RVM t-test was applied to filter the differentially expressed genes for the control and experimental group, as the RVM t-test can raise degrees of freedom effectively in the cases of small samples. After the analysis of differentially expressed genes and false discovery rate (FDR) analysis, we selected the differentially expressed genes according to the P-value threshold. A value of P<0.05 was considered to indicate a statistically significant difference, as previously described (14–16). The data of differentially expressed genes were subjected to unsupervised hierarchical clustering (Cluster 3.0) and TreeView analysis (Stanford University, Stanford, CA, USA).
GO analysis
GO analysis was applied to determine the main functions of the differential expression genes according to the GO database, which is the key functional classification of NCBI, which organizes genes into hierarchical categories and identifies the gene regulatory network on the basis of biological process and molecular function (17,18).
Specifically, a two-sided Fisher’s exact test and χ2 test were used to classify the GO categories, and the FDR (19) was calculated to correct the P-value; the smaller the FDR, the smaller the error in judging the P-value. The FDR was defined as:
where Nk refers to the number of Fisher’s test P-values less than χ2 test P-values. We computed P-values for the GO terms of all the differentially expressed genes. Enrichment analysis provides a measure of the significance of the function: as the enrichment increases, the corresponding function is more specific, which helps us to find those GO terms with a more concrete functional description in the experiment. Within the significance category, the enrichment Re was given by Re = (nf/n)/(Nf/N), where nf is the number of flagged genes within the particular category, n is the total number of genes within the same category, Nf is the number of flagged genes in the entire microarray, and N is the total number of genes in the microarray, as previously described (20).
GO-map
GO-map analysis is the interaction network of the significant GO terms of the differentially expressed genes, and was carried out to integrate the associations between these GO terms by outlining the interactions of related GO terms and summarizing the functional interactions of the differentially expressed genes in diseases (18,20).
Pathway analysis
Pathway analysis was used to identify the common pathways associated with the differentially expressed genes according to the KEGG, BioCarta and Reatome databases. We used the Fisher’s exact test and χ2 test to identify the significant pathways, and the threshold of significance was defined by a P-value and FDR. The enrichment Re was calculated with the equation described above, as previously described (21–23).
Path-Net
Path-Net is the interaction network of the most common pathways associated with the differentially expressed genes, and was built according to the interaction among pathways of the KEGG database to determine the interactions among the significant pathways directly and systemically. It identified the common pathways associated with the differentially expressed genes, as well as the mechanisms behind the activation of a certain pathway (22).
Signal-Net analysis
Using java that allows users to build and analyze molecular networks, network maps were constructed. For instance, if there is confirmative evidence that two genes interact with each other, an interaction edge is assigned between the two genes. The considered evidence is the source of the interaction database from KEGG. Networks are stored and presented as graphs, where nodes are mainly genes (protein, compound, etc.) and edges represent relation types between the nodes, e.g., activation or phosphorylation. The graph nature of Networks peaked our interest to investigate them with powerful tools implemented in R.
In order to investigate the global network, we computationally identified the most important nodes. To this end, we determined the connectivity (also known as the degree) defined as the sum of connection strengths with the other network genes:
In gene networks, the connectivity measures how well a gene correlates with all other network genes. For a gene in the network, the number of source genes of a gene is called the indegree of the gene and the number of target genes of a gene is its outdegree. The character of genes is described by betweenness centrality measures reflecting the importance of a node in a graph relative to other nodes. For a graph G:(V,E) with n vertices, the relative betweenness centrality C’B(v) is defined by:
where σst is the number of shortest paths from s to t, and σst(v) is the number of shortest paths from s to t that pass through a vertex v (24–28).
Data analysis
Numerical data are presented as the means ± standard deviation (SD). Differences between means were analyzed using the Student’s t-test. All statistical analyses were performed using SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA).
Results
Clinical characteristics of the two sample groups
The characteristics of the 5 consecutive patients with CTEPH (3 male, 2 female) enrolled in the study are presented in Table I. In addition, tissue from healthy volunteers was obtained from donors of lung transplants and matched to the patients with CTEPH. All patients with CTEPH did not have lung cancer and underwent anti-vitamin K treatment using warfarin. The median D-dimer level of the patients with CTEPH was 0.499 μg/ml (range, 0–1.32 μg/ml).
Table I.
Clinical characteristics of the patients in this study.
| Sample | No. | Age (years) mean ± SD | Gender (M/F) | mPAP (mmHg) median (range) | PVR (dyne·s·cm−5) | 6 WMT |
|---|---|---|---|---|---|---|
| Healthy controls | 5 | 35.3±10.6 | 2/3 | - | - | - |
| CTEPH patients | 5 | 38.2±14.7 | 2/3 | 55 (33–78) | 1075.4 | 454.7 |
mPAP, mean pulmonary arterial pressure; PVR, pulmonary vascular resistance; CTEPH, chronic thromboembolic pulmonary hypertension; 6 WMT, 6-min walk test.
CTEPH-related differential gene expression profiles
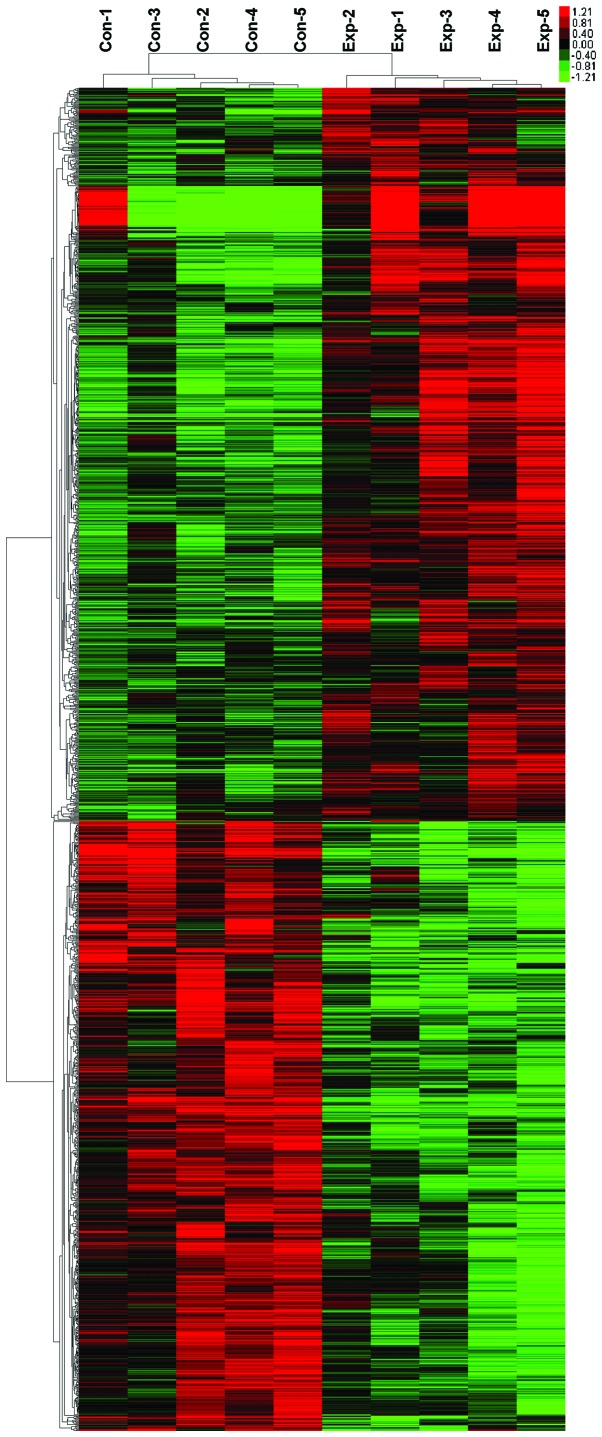
Genome-wide transcriptional profiling of tumors has demonstrated that extensive gene expression occurs after the formation of CTEPH. To investigate the possible changes in gene expression, microarray analysis was used to analyze the gene expression profiles in the patients with CTEPH and normal tissue groups and 1,614 genes with statistically significant changes in expression were identified. Of these, 880 genes were upregulated in the CTEPH samples and 734 were downregulated. Ten genes that were the most significantly upregulated or downregulated according to their P-values are listed (Tables II and III). Hierarchical clustering revealed systematic variations in the expression of genes between the two groups (Fig. 1). The results demonstrated that these differential probes could clearly separate the two groups from the whole samples.
Table II.
Most significantly upregulated genes.
| Gene symbol | P-value | Geom mean of intensities in CTEPH group | Geom mean of intensities in healthy controls | Fold-change |
|---|---|---|---|---|
| PTGS2 | 1.25E-05 | 155.56 | 38.18 | 4.07 |
| TBX15 | 2.35E-05 | 95.07 | 23.66 | 4.02 |
| FMO3 | 3.00E-05 | 120.33 | 16.06 | 7.49 |
| LRRC32 | 3.03E-05 | 445.65 | 151.99 | 2.93 |
| FAM100B | 3.87E-05 | 181.47 | 92.94 | 1.95 |
| NEIL3 | 4.62E-05 | 16.92 | 10 | 1.69 |
| LOC100507286 | 8.50E-05 | 33.52 | 10.1 | 3.32 |
| LY75 | 9.46E-05 | 166.81 | 70.72 | 2.36 |
| ITIH3 | 1.27E-04 | 79.24 | 15.24 | 5.2 |
| TRPV2 | 1.29E-04 | 45.02 | 24.8 | 1.82 |
Table III.
Most significantly downregulated genes.
| Gene symbol | P-value | Geom mean of intensities in CTEPH group | Geom mean of intensities in healthy controls | Fold-change |
|---|---|---|---|---|
| CHRDL1 | 5.00E-07 | 11.14 | 344.38 | 0.032 |
| FREM1 | 1.00E-06 | 13.58 | 69.85 | 0.19 |
| BNC2 | 1.20E-06 | 33.6 | 92.91 | 0.36 |
| ACADL | 1.30E-06 | 13.98 | 72.87 | 0.19 |
| UNC13C | 3.10E-06 | 10 | 27.03 | 0.37 |
| NCAM1 | 3.20E-06 | 17.13 | 75.75 | 0.23 |
| PRUNE2 | 3.30E-06 | 221.46 | 706.72 | 0.31 |
| KLHDC5 | 4.50E-06 | 25.88 | 57.87 | 0.45 |
| FERMT2 | 7.40E-06 | 222.13 | 409.8 | 0.54 |
| FAM198A | 7.60E-06 | 24.03 | 48.54 | 0.5 |
Figure 1.
Unsupervised classification of chronic thromboembolic pulmonary hypertension (CTEPH) and healthy control samples based on gene expression profiling. Classification of 10 pulmonary artery endothelial cell samples using the differentially expressed 2098-probe sets. Expression data are depicted as a data matrix where each row represents a gene and each column represents a sample. Expression levels are depicted according to the color scale shown at the top. Red and green indicate expression levels above and below the median, respectively. The magnitude of deviation from the median is represented by the color saturation.
GO analysis of differentially expressed genes in CTEPH and GO map
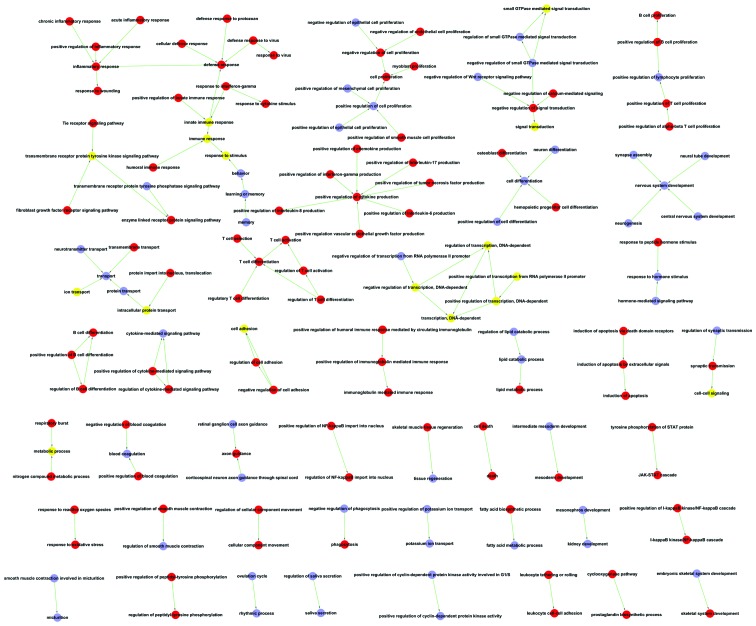
Significant progress in data mining has provided a wide range of bioinformatics analysis options. For example, GO, which has been proven to be extremely useful for the mining of functional and biological significance from very large datasets (17,18), can produce a controlled vocabulary used for dynamic maintenance and interoperability between genome databases. GO analysis of the differentially expressed genes in the two groups was performed. A total of 235 GO items associated with upregulated genes and 177 GO items associated with downregulated genes in the two groups were obtained (Table IV). For the GO items listed in the table, a GO map was constructed to further define the results of GO analysis (Fig. 2). In the GO map, items regarding defense response were the most common between the two groups, suggesting that the formation of thrombi was mainly caused by tissue response to various stimuli, such as inflammation and immune response. Furthermore, items regarding cell proliferation, signal transduction and cytokine production were also very common (high enrichment score). All these items indicated that apart from the traditional knowledge of the role of thrombosis in the development of CTEPH, other mechanisms, such as signaling pathway and cytokines may also play an important role in its pathogenesis.
Table IV.
GO items and enrichment scores.
| GO name | Enrichment score |
|---|---|
| T cell selection | 34.56136364 |
| T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell | 34.56136364 |
| Cell surface pattern recognition receptor signaling pathway | 34.56136364 |
| Dicarboxylic acid transport | 34.56136364 |
| Cyclooxygenase pathway | 34.56136364 |
| Positive regulation of mast cell activation | 34.56136364 |
| Regulatory T cell differentiation | 34.56136364 |
| Positive regulation of interleukin-10 biosynthetic process | 34.56136364 |
| Positive regulation of interleukin-4 biosynthetic process | 34.56136364 |
| RNA destabilization | 34.56136364 |
| Negative regulation of calcium-mediated signaling | 34.56136364 |
| Regulation of cytokine-mediated signaling pathway | 23.04090909 |
| Positive regulation of immunoglobulin mediated immune response | 23.04090909 |
| Protein deglycosylation | 23.04090909 |
| Detection of biotic stimulus | 23.04090909 |
| Regulation of smooth muscle cell migration | 23.04090909 |
| Viral envelope fusion with host membrane | 23.04090909 |
| Lipoxygenase pathway | 23.04090909 |
| Response to vitamin B3 | 23.04090909 |
| Regulation of NF-kappaB import into nucleus | 23.04090909 |
| Positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process | 23.04090909 |
| Regulation of cellular component movement | 23.04090909 |
| Adhesion to symbiont | 23.04090909 |
| Nucleotide-binding oligomerization domain containing 1 signaling pathway | 23.04090909 |
| Connective tissue replacement involved in inflammatory response wound healing | 20.73681818 |
| Response to peptidoglycan | 20.73681818 |
| Regulation of B cell differentiation | 20.73681818 |
| Regulation of T cell activation | 20.73681818 |
| Nucleotide-binding oligomerization domain containing 2 signaling pathway | 20.73681818 |
| Positive regulation of alpha-beta T cell proliferation | 18.85165289 |
| Chronic inflammatory response | 17.28068182 |
| Regulation of T cell differentiation | 17.28068182 |
| Platelet activating factor biosynthetic process | 17.28068182 |
| Tyrosine phosphorylation of STAT protein | 17.28068182 |
| Pyridine nucleotide biosynthetic process | 17.28068182 |
| Regulation of cholesterol transport | 17.28068182 |
| Negative regulation of collagen biosynthetic process | 17.28068182 |
| Very-low-density lipoprotein particle clearance | 17.28068182 |
| Positive regulation of interferon-alpha biosynthetic process | 17.28068182 |
| Negative regulation of follicle-stimulating hormone secretion | 17.28068182 |
| Antigen processing and presentation of exogenous peptide antigen via MHC class II | 14.81201299 |
| Positive regulation of interleukin-2 biosynthetic process | 14.23114973 |
| Negative regulation of signal transduction | 14.13873967 |
| Chronological cell aging | 13.82454545 |
| Membrane raft polarization | 13.82454545 |
| Response to molecule of fungal origin | 13.82454545 |
| Leukotriene production involved in inflammatory response | 13.82454545 |
| Macrophage derived foam cell differentiation | 13.82454545 |
| Membrane to membrane docking | 13.82454545 |
| Negative regulation of granulocyte differentiation | 13.82454545 |
| Tie receptor signaling pathway | 13.82454545 |
| Positive regulation of hair follicle development | 13.82454545 |
| Positive regulation of chemokine production | 12.96051136 |
| Positive regulation of macrophage chemotaxis | 12.96051136 |
| Lymphocyte chemotaxis | 12.96051136 |
| Negative thymic T cell selection | 11.52045455 |
| Negative regulation of blood vessel endothelial cell migration | 11.52045455 |
| Positive thymic T cell selection | 11.52045455 |
| Positive regulation of humoral immune response mediated by circulating immunoglobulin | 11.52045455 |
| Positive regulation of cytokine-mediated signaling pathway | 11.52045455 |
| Positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | 11.52045455 |
| Negative regulation of lipid storage | 11.52045455 |
| Death | 11.52045455 |
| Cellular response to nutrient | 11.52045455 |
| Positive regulation of actin filament bundle assembly | 11.52045455 |
| Very-low-density lipoprotein particle assembly | 11.52045455 |
| Positive regulation of T-helper 1 cell differentiation | 11.52045455 |
| Myoblast proliferation | 11.52045455 |
| Negative regulation of focal adhesion assembly | 11.52045455 |
| T-helper 1 type immune response | 10.63426573 |
| Enzyme linked receptor protein signaling pathway | 10.36840909 |
| Negative regulation of plasminogen activation | 10.36840909 |
| Leukocyte tethering or rolling | 10.36840909 |
| Cell recognition | 9.874675325 |
| Positive regulation of necrotic cell death | 9.874675325 |
| Positive regulation of T cell receptor signaling pathway | 9.874675325 |
| Angiogenesis involved in wound healing | 9.874675325 |
| Placenta blood vessel development | 9.874675325 |
| Prostaglandin biosynthetic process | 9.600378788 |
| Immunoglobulin mediated immune response | 9.600378788 |
| Defense response to protozoan | 9.425826446 |
| Positive regulation of cytokine production | 9.216363636 |
| Leukocyte cell-cell adhesion | 9.095095694 |
| RNA catabolic process | 9.095095694 |
| I-kappaB kinase/NF-kappaB cascade | 8.84127907 |
| Cellular defense response | 8.640340909 |
| Microglial cell activation involved in immune response | 8.640340909 |
| Initiation of viral infection | 8.640340909 |
| Positive regulation of interferon-gamma biosynthetic process | 8.640340909 |
| Respiratory burst | 8.640340909 |
| Negative regulation of T cell-mediated cytotoxicity | 8.640340909 |
| N-glycan processing | 8.640340909 |
| Chylomicron remnant clearance | 8.640340909 |
| Regulation of mast cell degranulation | 8.640340909 |
| Positive regulation of interleukin-8 biosynthetic process | 8.640340909 |
| Negative regulation of nitric-oxide synthase activity | 8.640340909 |
| Maternal process involved in parturition | 8.640340909 |
| Platelet dense granule organization | 8.640340909 |
| Branching involved in embryonic placenta morphogenesis | 8.640340909 |
| Positive regulation of calcium-mediated signaling | 8.064318182 |
| Humoral immune response | 8.023173701 |
| Chemotaxis | 8.018236364 |
| Response to reactive oxygen species | 7.975699301 |
| Defense response | 7.68030303 |
| Positive regulation of T cell proliferation | 7.513339921 |
| Negative regulation of interleukin-12 production | 7.406006494 |
| Skeletal muscle tissue regeneration | 7.406006494 |
| Positive regulation of innate immune response | 7.406006494 |
| Lymph node development | 7.406006494 |
| Cellular response to lipoteichoic acid | 7.406006494 |
| Positive regulation of interleukin-1 beta secretion | 7.276076555 |
| Response to exogenous dsRNA | 6.912272727 |
| Decidualization | 6.912272727 |
| Regulation of peptidyl-tyrosine phosphorylation | 6.912272727 |
| Germinal center formation | 6.912272727 |
| Natural killer cell activation | 6.912272727 |
| Positive regulation of interleukin-17 production | 6.912272727 |
| Defense response to Gram-positive bacterium | 6.646416084 |
| Hemopoietic progenitor cell differentiation | 6.583116883 |
| JAK-STAT cascade | 6.538636364 |
| Acute inflammatory response | 6.480255682 |
| Cellular copper ion homeostasis | 6.480255682 |
| Positive regulation of B cell differentiation | 6.480255682 |
| Response to gamma radiation | 6.283884298 |
| Neutrophil chemotaxis | 6.283884298 |
| T cell activation | 6.232377049 |
| Metabolic process | 6.099064171 |
| T cell differentiation | 6.099064171 |
| Sprouting angiogenesis | 6.099064171 |
| Positive regulation of tumor necrosis factor biosynthetic process | 6.099064171 |
| Positive regulation of survival gene product expression | 6.099064171 |
| Induction of positive chemotaxis | 6.099064171 |
| Response to progesterone stimulus | 6.048238636 |
| Negative regulation of endothelial cell proliferation | 5.958855799 |
| Response to bacterium | 5.760227273 |
| Response to interferon-gamma | 5.760227273 |
| Induction of apoptosis via death domain receptors | 5.760227273 |
| Negative regulation of blood coagulation | 5.760227273 |
| Positive regulation of smooth muscle contraction | 5.760227273 |
| Inflammatory response | 5.541909621 |
| Negative regulation of growth of symbiont in host | 5.529818182 |
| Oligosaccharide metabolic process | 5.457057416 |
| Cellular component movement | 5.368755516 |
| Negative regulation of cell adhesion | 5.236570248 |
| Mesoderm development | 5.236570248 |
| Copper ion transport | 5.184204545 |
| Positive regulation of blood coagulation | 5.184204545 |
| Negative regulation of phosphorylation | 5.184204545 |
| Positive regulation of NF-kappaB import into nucleus | 5.184204545 |
| Positive regulation of blood vessel endothelial cell migration | 5.184204545 |
| Nitrogen compound metabolic process | 4.937337662 |
| Positive regulation vascular endothelial growth factor production | 4.937337662 |
| Positive regulation of interleukin-8 production | 4.937337662 |
| Response to cytokine stimulus | 4.883670949 |
| Positive regulation of interleukin-6 production | 4.838590909 |
| Response to inorganic substance | 4.767084639 |
| Response to vitamin D | 4.767084639 |
| Amino acid transport | 4.712913223 |
| B cell proliferation | 4.712913223 |
| Positive regulation of erythrocyte differentiation | 4.712913223 |
| Response to interleukin-1 | 4.670454545 |
| Positive regulation of interferon-gamma production | 4.564708405 |
| Lipopolysaccharide-mediated signaling pathway | 4.430944056 |
| Positive regulation of B cell proliferation | 4.412088975 |
| Positive regulation of smooth muscle cell proliferation | 4.388744589 |
| Negative regulation of T cell proliferation | 4.320170455 |
| Response to virus | 4.291934046 |
| Ion transport | 4.203409091 |
| Protein import into nucleus, translocation | 4.189256198 |
| Negative regulation of NF-kappaB transcription factor activity | 4.10050077 |
| B cell differentiation | 4.066042781 |
| Heterophilic cell-cell adhesion | 4.066042781 |
| Response to mechanical stimulus | 3.879336735 |
| Immune response | 3.769560495 |
| Positive regulation of tumor necrosis factor production | 3.75666996 |
| Cholesterol efflux | 3.736363636 |
| B cell receptor signaling pathway | 3.736363636 |
| Induction of apoptosis | 3.697923681 |
| Phagocytosis | 3.456136364 |
| Positive regulation of nitric oxide biosynthetic process | 3.388368984 |
| Positive regulation of NF-kappaB transcription factor activity | 3.352968114 |
| Regulation of cell adhesion | 3.344648094 |
| Rho protein signal transduction | 3.323208042 |
| Positive regulation of inflammatory response | 3.323208042 |
| Anti-apoptosis | 3.318806442 |
| Response to stimulus | 3.291558442 |
| Response to peptide hormone stimulus | 3.200126263 |
| Protein complex assembly | 3.19027972 |
| Cellular calcium ion homeostasis | 3.174002783 |
| Elevation of cytosolic calcium ion concentration | 3.141942149 |
| Positive regulation of angiogenesis | 3.110522727 |
| Response to wounding | 3.049532086 |
| Cell-cell signaling | 3.034012838 |
| Induction of apoptosis by extracellular signals | 3.005335968 |
| Defense response to virus | 3.005335968 |
| Response to lipopolysaccharide | 2.948362775 |
| Positive regulation of peptidyl-tyrosine phosphorylation | 2.94139265 |
| Protein homooligomerization | 2.928929122 |
| Cellular amino acid metabolic process | 2.840660025 |
| Positive regulation of I-kappaB kinase/NF-kappaB cascade | 2.757555609 |
| Cell adhesion | 2.737533753 |
| Fatty acid biosynthetic process | 2.728528708 |
| Interspecies interaction between organisms | 2.696985024 |
| Osteoblast differentiation | 2.69309327 |
| Transmembrane receptor protein tyrosine kinase signaling pathway | 2.546626794 |
| Signal transduction | 2.529953417 |
| Activation of MAPK activity | 2.449230494 |
| Innate immune response | 2.367216687 |
| Skeletal system development | 2.319554606 |
| Response to oxidative stress | 2.273773923 |
| Response to hypoxia | 2.15335599 |
| Cell proliferation | 2.122933884 |
| Cell death | 2.057224026 |
| Multicellular organismal development | 1.901434245 |
| Negative regulation of cell proliferation | 1.840545992 |
| Lipid metabolic process | 1.78765674 |
| Positive regulation of transcription from RNA polymerase II promoter | 0.643002114 |
| Transmembrane transport | 0.599113562 |
| Small GTPase-mediated signal transduction | 0.433824648 |
| Regulation of transcription, DNA-dependent | 0.407975625 |
| Intracellular protein transport | 0.399553337 |
| Axon guidance | 0.360014205 |
| Translation | 0.261432403 |
| DNA repair | 0.23247554 |
| Antigen processing and presentation of peptide antigen via MHC class I | 0.174552342 |
| Positive regulation of transcription, DNA-dependent | 0.17004361 |
| Protein folding | 0.164970709 |
| Protein ubiquitination | 0.154291802 |
| mRNA processing | 0.152926388 |
| Xenobiotic metabolic process | 0.12042287 |
| Synaptic transmission | 0.116172651 |
| Negative regulation of transcription, DNA-dependent | 0.112945633 |
| Mitotic cell cycle | 0.104100493 |
| Fibroblast growth factor receptor signaling pathway | 0.101950925 |
| Transcription, DNA-dependent | 0.016632033 |
| Transcription, DNA-dependent | −0.039880623 |
| Translation | −0.062686789 |
| Blood coagulation | −0.076875635 |
| Intracellular protein transport | −0.079838087 |
| DNA repair | −0.092905756 |
| Regulation of small GTPase-mediated signal transduction | −0.160604524 |
| Viral reproduction | −0.162175997 |
| Innate immune response | −0.227046396 |
| Mitotic cell cycle | −0.249614261 |
| Positive regulation of transcription, DNA-dependent | −0.254833747 |
| Cytokine-mediated signaling pathway | −0.264766564 |
| Negative regulation of transcription, DNA-dependent | −0.270823316 |
| Immune response | −0.355455602 |
| Positive regulation of transcription from RNA polymerase II promoter | −0.385450859 |
| Negative regulation of transcription from RNA polymerase II promoter | −0.389854531 |
| Small GTPase-mediated signal transduction | −0.404535246 |
| Protein transport | −0.408638731 |
| Proteolysis | −0.541854957 |
| Regulation of transcription, DNA-dependent | −0.628876401 |
| Transport | −1.651693671 |
| Signal transduction | −1.661658778 |
| Positive regulation of cell proliferation | −1.826982685 |
| Response to ethanol | −2.273925035 |
| Regulation of cell shape | −2.419618529 |
| Cell differentiation | −2.426430518 |
| Kidney development | −2.458065857 |
| Cell-cell signaling | −2.530440751 |
| Homophilic cell adhesion | −2.617008461 |
| Muscle organ development | −2.736337463 |
| Neuron differentiation | −2.825179589 |
| Chromatin modification | −2.900517711 |
| Lipid catabolic process | −2.95971195 |
| Regulation of cell growth | −2.986376022 |
| Sodium ion transport | −2.99537113 |
| Calcium ion transport | −3.107697548 |
| Inner ear morphogenesis | −3.139088432 |
| Fatty acid metabolic process | −3.1873821 |
| Response to stimulus | −3.288568834 |
| Palate development | −3.333928404 |
| Neuron projection morphogenesis | −3.341610266 |
| Potassium ion transport | −3.452997275 |
| Transmembrane receptor protein tyrosine kinase signaling pathway | −3.489344615 |
| Cellular response to insulin stimulus | −3.489344615 |
| Memory | −3.511522653 |
| Response to hormone stimulus | −3.511522653 |
| Nervous system development | −3.540414928 |
| Cell adhesion | −3.575097603 |
| Embryonic digit morphogenesis | −3.710683639 |
| Muscle contraction | −3.732970027 |
| Positive regulation of glucose import | −3.9462826 |
| Learning or memory | −4.062349736 |
| Synapse assembly | −4.14359673 |
| Odontogenesis | −4.14359673 |
| Multicellular organismal development | −4.157006428 |
| Positive regulation of canonical Wnt receptor signaling pathway | −4.213827183 |
| Behavior | −4.2498428 |
| Regulation of heart contraction | −4.361680769 |
| Response to morphine | −4.361680769 |
| Neurogenesis | −4.479564033 |
| Negative regulation of insulin secretion | −4.603996367 |
| Negative regulation of epithelial cell proliferation | −4.73553912 |
| Branching morphogenesis of a tube | −4.73553912 |
| Embryonic skeletal system development | −4.93285325 |
| Positive regulation of mesenchymal cell proliferation | −4.972316076 |
| Tissue regeneration | −5.022541491 |
| Negative regulation of Wnt receptor signaling pathway | −5.179495913 |
| Central nervous system development | −5.217862549 |
| Neurotransmitter transport | −5.404691387 |
| Retinal ganglion cell axon guidance | −5.404691387 |
| Positive regulation of blood pressure | −5.404691387 |
| Calcium-dependent cell-cell adhesion | −5.452100961 |
| Activation of phospholipase C activity by G-protein coupled receptor protein signaling Pathway coupled to IP3 second messenger | −5.599455041 |
| Middle ear morphogenesis | −5.715305835 |
| Eye development | −6.542521153 |
| Positive regulation of epithelial cell proliferation | −6.605733918 |
| Positive regulation of cell differentiation | −6.683220533 |
| Mammary gland development | −6.90599455 |
| Regulation of smooth muscle contraction | −6.90599455 |
| Positive regulation of tyrosine phosphorylation of Stat5 protein | −6.90599455 |
| Hemopoietic stem cell proliferation | −6.90599455 |
| Neural tube development | −7.206255183 |
| Calcium ion transport into cytosol | −7.312229524 |
| Mesonephros development | −7.769243869 |
| Peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan | −7.769243869 |
| Prostate gland growth | −7.769243869 |
| Ion transport | −8.119209809 |
| Regulation of synaptic transmission | −8.28719346 |
| Positive regulation of insulin-like growth factor receptor signaling pathway | −8.28719346 |
| Positive regulation of cyclin-dependent protein kinase activity | −8.28719346 |
| Vagina development | −8.28719346 |
| Cardiac muscle tissue development | −8.879135851 |
| Transmembrane receptor protein tyrosine phosphatase signaling pathway | −9.207992734 |
| Choline metabolic process | −9.207992734 |
| Negative regulation of epinephrine secretion | −9.207992734 |
| Protein insertion into membrane | −9.207992734 |
| Type II pneumocyte differentiation | −9.207992734 |
| Activation of protein kinase B activity | −9.562146301 |
| Metabolic process | −9.749639365 |
| Regulation of respiratory gaseous exchange by neurological system process | −10.35899183 |
| Creatine metabolic process | −10.35899183 |
| Retinal metabolic process | −10.35899183 |
| Peptide biosynthetic process | −10.35899183 |
| Growth hormone receptor signaling pathway | −10.35899183 |
| Hormone-mediated signaling pathway | −11.30071836 |
| Rhythmic process | −11.50999092 |
| Cardiac left ventricle morphogenesis | −11.8388478 |
| Positive regulation of cyclin-dependent protein kinase activity involved in G1/S | −11.8388478 |
| Positive regulation of lymphocyte proliferation | −11.8388478 |
| Ovulation cycle | −13.8119891 |
| Negative regulation of norepinephrine secretion | −13.8119891 |
| Taurine metabolic process | −13.8119891 |
| Negative regulation of actin filament polymerization | −13.8119891 |
| Positive regulation of potassium ion transport | −13.8119891 |
| Urinary bladder development | −13.8119891 |
| Morphogenesis of an epithelial fold | −13.8119891 |
| Ciliary neurotrophic factor-mediated signaling pathway | −13.8119891 |
| Aromatic compound catabolic process | −16.57438692 |
| Negative regulation of phagocytosis | −16.57438692 |
| Tertiary branching involved in mammary gland duct morphogenesis | −16.57438692 |
| Cellular response to heparin | −16.57438692 |
| Regulation of vasodilation | −17.7582717 |
| Nucleoside triphosphate catabolic process | −20.71798365 |
| Saliva secretion | −20.71798365 |
| Female genitalia morphogenesis | −20.71798365 |
| Smooth muscle contraction involved in micturition | −20.71798365 |
| Regulation of prostatic bud formation | −20.71798365 |
| Pericardium morphogenesis | −27.6239782 |
| Lateral sprouting involved in mammary gland duct morphogenesis | −27.6239782 |
| Regulation of protein metabolic process | −41.4359673 |
| Negative regulation of the force of heart contraction involved in baroreceptor response to Increased systemic arterial blood pressure | −41.4359673 |
| Renin secretion into blood stream | −41.4359673 |
| Regulation of thyroid hormone mediated signaling pathway | −41.4359673 |
| Negative regulation of leukocyte chemotaxis | −41.4359673 |
| Pyruvate transport | −41.4359673 |
| Nucleoside diphosphate catabolic process | −41.4359673 |
| Glycolate metabolic process | −41.4359673 |
| Negative regulation of lamellipodium assembly | −41.4359673 |
| Muscle hypertrophy | −41.4359673 |
| Myotube cell development | −41.4359673 |
| Mevalonate transport | −41.4359673 |
| Male somatic sex determination | −41.4359673 |
| Spinal cord patterning | −41.4359673 |
| Orbitofrontal cortex development | −41.4359673 |
| Cell-cell adhesion involved in neuronal-glial interactions involved in cerebral cortex radial glia guided migration | −41.4359673 |
| Corticospinal neuron axon guidance through spinal cord | −41.4359673 |
| Neural plate mediolateral regionalization | −41.4359673 |
| Cellular potassium ion homeostasis | −41.4359673 |
| Regulation of mismatch repair | −41.4359673 |
| Positive regulation of phospholipase A2 activity | −41.4359673 |
| Retinol transport | −41.4359673 |
| Response to luteinizing hormone stimulus | −41.4359673 |
| Positive regulation of locomotion | −41.4359673 |
| Sequestering of neurotransmitter | −41.4359673 |
| Carnitine catabolic process | −41.4359673 |
| Homocysteine catabolic process | −41.4359673 |
| Regulation of adenylate cyclase activity | −41.4359673 |
| Phosphatidic acid metabolic process | −41.4359673 |
| Regulation of saliva secretion | −41.4359673 |
| Paraxial mesoderm structural organization | −41.4359673 |
| Intermediate mesoderm development | −41.4359673 |
| Urothelial cell proliferation | −41.4359673 |
| Induction of negative chemotaxis | −41.4359673 |
| Regulation of lipid catabolic process | −41.4359673 |
| Negative regulation of small GTPase-mediated signal transduction | −41.4359673 |
| Micturition | −41.4359673 |
| Activation of prostate induction by androgen receptor signaling pathway | −41.4359673 |
| Neural plate pattern specification | −41.4359673 |
| Dermatome development | −41.4359673 |
| Negative regulation of activation-induced cell death of T cells | −41.4359673 |
| Cellular response to chemical stimulus | −41.4359673 |
| Negative regulation of smooth muscle cell chemotaxis | −41.4359673 |
| Negative regulation of mononuclear cell migration | −41.4359673 |
| Pattern specification involved in metanephros development | −41.4359673 |
| Negative regulation of neutrophil chemotaxis | −41.4359673 |
| Metanephric cap mesenchymal cell proliferation involved in metanephros development | −41.4359673 |
| Positive regulation of non-canonical Wnt receptor signaling pathway | −41.4359673 |
| Positive regulation of Wnt receptor signaling pathway involved in dorsal/ventral axis specification | −41.4359673 |
Figure 2.
GO map of significant differentially expressed genes: red circles represent the GO categories of upregulated genes, lavender circles represent the GO categories of downregulated genes and yellow circles represent the GO categories associated with both upregulated and downregulated genes.
Pathway analysis of differentially expressed genes in CTEPH and Path-Net
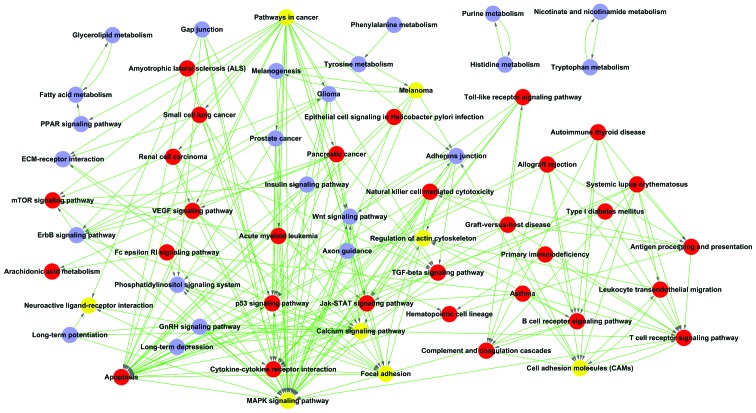
To determine the involvment of signal transduction pathways in CTEPH, related pathways were analyzed according to the functions and interactions of the differentially expressed genes. By using pathway analysis with the threshold of significance defined on the basis of P<0.05, a great number of significant pathways was found (Tables V and VI). The high enriched pathways targeted by overexpressed genes were involved in cytokine-cytokine receptor interaction, leishmaniasis and cell adhesion molecules (CAMs). By contrast, significant pathways corresponding to underexpressed genes appeared to be responsible for focal adhesion, neuroactive ligand-receptor interaction and arrhythmogenic right ventricular cardiomyopathy (ARVC). However, for the pathways listed which seemed to not be relevant to CTEPH, Path-Net was used to analyze the different pathways to identify the most important ones (Fig. 3). In Path-Net, the MAPK signaling pathway and apoptosis had the largest enrichment degree, which suggests that they are not the most significant variation pathways but participate in pathway regulation. Identifying these pathways may help us regulate the related pathways and control the development of CTEPH.
Table V.
Upregulated significant pathways.
| Pathway name | -LgP |
|---|---|
| Cytokine-cytokine receptor interaction | 23.42004685 |
| Leishmaniasis | 21.90107349 |
| Cell adhesion molecules (CAMs) | 20.65682392 |
| Chagas disease | 15.37890961 |
| T cell receptor signaling pathway | 13.94901928 |
| Hematopoietic cell lineage | 13.74068504 |
| Natural killer cell mediated cytotoxicity | 13.17649459 |
| Chemokine signaling pathway | 13.14867922 |
| Primary immunodeficiency | 13.11509338 |
| Phagosome | 12.87858647 |
| Antigen processing and presentation | 12.1066584 |
| Intestinal immune network for IgA production | 12.08192561 |
| Malaria | 11.93017145 |
| Toll-like receptor signaling pathway | 11.56038144 |
| Allograft rejection | 11.39679734 |
| Graft-versus-host disease | 11.05163889 |
| B cell receptor signaling pathway | 10.95069165 |
| Type I diabetes mellitus | 10.72782064 |
| Viral myocarditis | 10.43654793 |
| NOD-like receptor signaling pathway | 9.858981253 |
| Asthma | 8.781008205 |
| Jak-STAT signaling pathway | 8.780882537 |
| Leukocyte transendothelial migration | 8.69910665 |
| Amoebiasis | 8.385599375 |
| Systemic lupus erythematosus | 8.176130358 |
| Autoimmune thyroid disease | 8.170383587 |
| Fc gamma R-mediated phagocytosis | 7.492623136 |
| Fc epsilon RI signaling pathway | 6.80371883 |
| Complement and coagulation cascades | 6.801648207 |
| Apoptosis | 5.705278343 |
| Lysosome | 5.334893435 |
| Pathways in cancer | 4.192720575 |
| Regulation of actin cytoskeleton | 4.003028031 |
| p53 signaling pathway | 3.535108478 |
| Cytosolic DNA-sensing pathway | 3.092429362 |
| Neuroactive ligand-receptor interaction | 3.078524076 |
| Calcium signaling pathway | 2.822547274 |
| Epithelial cell signaling in Helicobacter pylori infection | 2.818702804 |
| MAPK signaling pathway | 2.752907643 |
| RIG-I-like receptor signaling pathway | 2.693488553 |
| Neurotrophin signaling pathway | 2.625676671 |
| Pantothenate and CoA biosynthesis | 2.610488424 |
| Acute myeloid leukemia | 2.603797719 |
| Arachidonic acid metabolism | 2.514704889 |
| Focal adhesion | 2.36365754 |
| TGF-beta signaling pathway | 2.223171679 |
| Prion diseases | 2.198269973 |
| Olfactory transduction | 2.139616901 |
| mTOR signaling pathway | 2.121221511 |
| Amyotrophic lateral sclerosis (ALS) | 2.079595295 |
| Endocytosis | 1.906955325 |
| VEGF signaling pathway | 1.890777366 |
| Aldosterone-regulated sodium reabsorption | 1.851879925 |
| Shigellosis | 1.780192486 |
| Glycosaminoglycan biosynthesis - keratan sulfate | 1.771193081 |
| Small cell lung cancer | 1.633206157 |
| Other glycan degradation | 1.615330588 |
| Salivary secretion | 1.530885213 |
| Renal cell carcinoma | 1.501166586 |
| Pancreatic cancer | 1.501166586 |
| Melanoma | 1.473244623 |
| Ether lipid metabolism | 1.40495844 |
-LgP, negative logarithm of the P-value.
Table VI.
Pathways associated with significantly downregulated genes.
| Pathway name | -LgP |
|---|---|
| Focal adhesion | 8.1140073 |
| Neuroactive ligand-receptor interaction | 5.712988 |
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 5.5013372 |
| Calcium signaling pathway | 5.445408 |
| Wnt signaling pathway | 5.1186354 |
| Vascular smooth muscle contraction | 5.0207839 |
| Long-term depression | 4.8678343 |
| Aldosterone-regulated sodium reabsorption | 3.9213362 |
| Axon guidance | 3.8465848 |
| Dilated cardiomyopathy | 3.8147938 |
| Hypertrophic cardiomyopathy (HCM) | 3.4512102 |
| ErbB signaling pathway | 3.2950005 |
| Salivary secretion | 3.2203532 |
| Adherens junction | 3.1261703 |
| Pathways in cancer | 3.0889176 |
| Pancreatic secretion | 2.8145968 |
| Glycine, serine and threonine metabolism | 2.7224771 |
| Glioma | 2.7174562 |
| ECM-receptor interaction | 2.6818514 |
| Progesterone-mediated oocyte maturation | 2.6485594 |
| Glycerolipid metabolism | 2.6032586 |
| Long-term potentiation | 2.5263793 |
| Regulation of actin cytoskeleton | 2.3885129 |
| Tyrosine metabolism | 2.3264949 |
| Phosphatidylinositol signaling system | 2.2235491 |
| GnRH signaling pathway | 2.204073 |
| Melanogenesis | 2.204073 |
| Nicotinate and nicotinamide metabolism | 2.1809844 |
| ABC transporters | 2.093767 |
| Olfactory transduction | 2.080365 |
| Histidine metabolism | 2.0047222 |
| Cell adhesion molecules (CAMs) | 1.9933196 |
| Oocyte meiosis | 1.9319734 |
| MAPK signaling pathway | 1.9265796 |
| Gap junction | 1.9104997 |
| Insulin signaling pathway | 1.8902518 |
| PPAR signaling pathway | 1.8327772 |
| Melanoma | 1.8327772 |
| Fc gamma R-mediated phagocytosis | 1.7609805 |
| Phenylalanine metabolism | 1.7586389 |
| Gastric acid secretion | 1.7471867 |
| Arginine and proline metabolism | 1.6184534 |
| Inositol phosphate metabolism | 1.5877798 |
| Circadian rhythm - mammal | 1.5137608 |
| Tryptophan metabolism | 1.5097806 |
| Amoebiasis | 1.4980899 |
| Chemokine signaling pathway | 1.4705506 |
| Purine metabolism | 1.4329084 |
| Prostate cancer | 1.3808613 |
| Fatty acid metabolism | 1.3705322 |
| Valine, leucine and isoleucine degradation | 1.3705322 |
-LgP, negative logarithm of the P-value.
Figure 3.
Path-Net network of significantly differentially expressed genes: red circles represent the pathways associated with upregulated genes, lavender circles represent the pathways associated with downregulated genes and yellow circles represent the pathways associated with both up- and downregulated genes.
Signal transduction networks in CTEPH
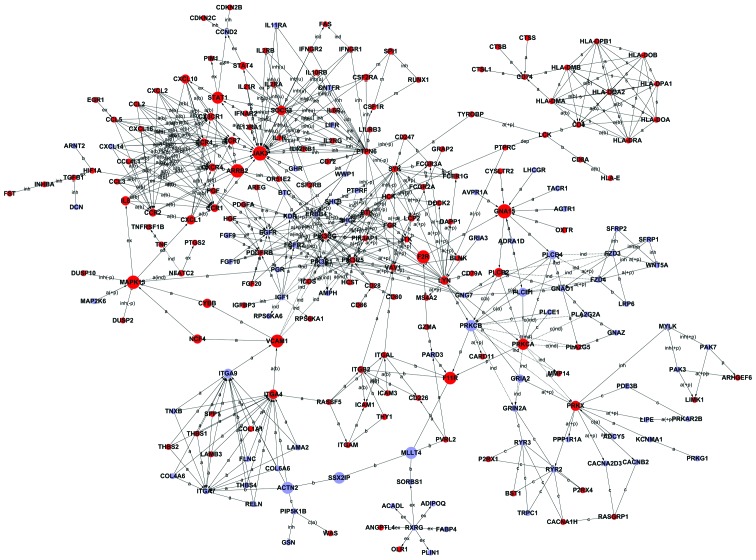
According to the literature and experimental records in the databases, 440 (276+164) genes appearing in previous 113 (62+51) pathways were collected and a diagram of the gene interaction network was drawn up based on these genes (Fig. 4). The total number of genes in the network was 232, and the particular associations between them are listed in Table VII. In the network, cycle nodes represent genes, and the edges between two nodes represent the interactions between genes, which were quantified by betweenness centrality. Betweenness centrality within the network which contains both the direct regulation by degree and the signal transmitting between the genes represents the size of the cycle node. The higher the betweenness centrality, the more common the gene within the network. The clustering coefficient can be used to estimate the complexity of interactions among genes that neighbor the core gene with the exception of core gene participation. The lower the clustering coefficient, the more independent of the core gene is the interaction among genes in the neighborhood of the core gene. Janus kinase 3 (JAK3), guanine nucleotide binding protein (G protein), alpha 15 (Gq class) (GNA15), mitogen-activated protein kinase 13 (MAPK13), arrestin, beta 2 (ARRB2) and coagulation factor II (thrombin) receptor (F2R) were the five main central genes identified by betweenness centrality.
Figure 4.
Signal transduction networks of CTEPH-related genes. Circles represent genes, red circles represent upregulated genes, and blue circles represent downregulated genes. Arrows represent the activation of (a); straight line represents combinations; dotted line represents indirect effects; a, represents activation; ex, represents gene expression; b, represents binding; c, represents compound; ind, represents indirect effects; inh, represents inhibition; u, represents ubiquination, s, represents state change; detailed annotation listed in Table VII.
Table VII.
Characteristics of genes.
| Gene symbol | Description | Betweenness centrality | Degree | Indegree | Outdegree | Style |
|---|---|---|---|---|---|---|
| JAK3 | Janus kinase 3 | 0.038452356 | 31 | 26 | 5 | Up |
| GNA15 | Guanine nucleotide binding protein (G protein), alpha 15 (Gq class) | 0.037599143 | 11 | 8 | 3 | Up |
| MAPK13 | Mitogen-activated protein kinase 13 | 0.033950795 | 9 | 6 | 3 | Up |
| ARRB2 | Arrestin, beta 2 | 0.033861892 | 8 | 7 | 8 | Up |
| F2R | Coagulation factor II (thrombin) receptor | 0.032585932 | 3 | 2 | 2 | Up |
| VCAM1 | Vascular cell adhesion molecule 1 | 0.031990127 | 7 | 2 | 5 | Up |
| F11R | F11 receptor | 0.03029902 | 6 | 6 | 3 | Up |
| MLLT4 | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 | 0.028330801 | 4 | 4 | 4 | Down |
| STAT1 | Signal transducer and activator of transcription 1, 91 kDa | 0.027727017 | 7 | 2 | 5 | Up |
| ACTN2 | Actinin, alpha 2 | 0.027721674 | 5 | 5 | 5 | Down |
| SSX2IP | Synovial sarcoma, X breakpoint 2 interacting protein | 0.026250152 | 2 | 2 | 2 | Down |
| PRKCB | Protein kinase C, beta | 0.02479466 | 12 | 5 | 8 | Down |
| PRKCA | Protein kinase C, alpha | 0.024281711 | 11 | 5 | 10 | Up |
| PRKX | Protein kinase, X-linked | 0.018822441 | 12 | 4 | 9 | Up |
| ITGA4 | Integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) | 0.016703185 | 15 | 14 | 3 | Up |
| SOCS3 | Suppressor of cytokine signaling 3 | 0.01636563 | 21 | 2 | 19 | Up |
| ITGA9 | Integrin, alpha 9 | 0.016034992 | 14 | 13 | 3 | Down |
| CXCL1 | Chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | 0.015128782 | 7 | 1 | 6 | Up |
| IL8 | Interleukin 8 | 0.015128782 | 7 | 1 | 6 | Up |
| NCF4 | Neutrophil cytosolic factor 4, 40 kDa | 0.014984515 | 2 | 1 | 1 | Up |
| CYBB | Cytochrome b-245, beta polypeptide | 0.014984515 | 2 | 1 | 1 | Up |
| CXCL10 | Chemokine (C-X-C motif) ligand 10 | 0.014593392 | 7 | 1 | 6 | Up |
| PLCB4 | Phospholipase C, beta 4 | 0.01422471 | 7 | 6 | 2 | Down |
| PLCB1 | Phospholipase C, beta 1 (phosphoinositide-specific) | 0.01422471 | 7 | 6 | 2 | Down |
| PLCB2 | Phospholipase C, beta 2 | 0.01422471 | 7 | 6 | 2 | Up |
| LYN | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | 0.011954016 | 20 | 3 | 17 | Up |
| GRIA2 | Glutamate receptor, ionotropic, AMPA 2 | 0.010419088 | 3 | 1 | 2 | Down |
| STAT4 | Signal transducer and activator of transcription 4 | 0.008717991 | 4 | 1 | 3 | Up |
| CCR7 | Chemokine (C-C motif) receptor 7 | 0.008388993 | 12 | 11 | 2 | Up |
| CCR4 | Chemokine (C-C motif) receptor 4 | 0.008388993 | 12 | 11 | 2 | Up |
| CCR2 | Chemokine (C-C motif) receptor 2 | 0.008388993 | 12 | 11 | 2 | Up |
| CX3CR1 | Chemokine (C-X3-C motif) receptor 1 | 0.008388993 | 12 | 11 | 2 | Up |
| CCR1 | Chemokine (C-C motif) receptor 1 | 0.008388993 | 12 | 11 | 2 | Up |
| CXCR4 | Chemokine (C-X-C motif) receptor 4 | 0.008388993 | 12 | 11 | 2 | Up |
| CSF2RA | Colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) | 0.005161547 | 4 | 3 | 2 | Up |
| ITGB2 | Integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) | 0.004818256 | 7 | 4 | 5 | Up |
| SPI1 | Spleen focus forming virus (SFFV) proviral integration oncogene spi1 | 0.003699643 | 3 | 3 | 2 | Up |
| PGF | Placental growth factor | 0.003543193 | 5 | 2 | 3 | Up |
| GRIN2A | Glutamate receptor, ionotropic, N-methyl D-aspartate 2A | 0.003531713 | 5 | 5 | 2 | Down |
| VAV1 | Vav 1 guanine nucleotide exchange factor | 0.002788411 | 11 | 11 | 5 | Up |
| RYR2 | Ryanodine receptor 2 (cardiac) | 0.002768397 | 7 | 7 | 6 | Down |
| NFATC2 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | 0.002250563 | 3 | 1 | 2 | Up |
| SYK | Spleen tyrosine kinase | 0.002031231 | 17 | 8 | 12 | Up |
| PIK3CG | Phosphoinositide-3-kinase, catalytic, gamma polypeptide | 0.001985436 | 23 | 21 | 5 | Up |
| PIK3R5 | Phosphoinositide-3-kinase, regulatory subunit 5 | 0.001985436 | 23 | 21 | 5 | Up |
| PIK3R1 | Phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | 0.001985436 | 23 | 21 | 5 | Down |
| ITGAL | Integrin, alpha L [antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide] | 0.001811094 | 6 | 3 | 4 | Up |
| FZD3 | Frizzled homolog 3 (Drosophila) | 0.001625406 | 8 | 4 | 5 | Down |
| FZD4 | Frizzled homolog 4 (Drosophila) | 0.001625406 | 8 | 4 | 5 | Down |
| PTPN6 | Protein tyrosine phosphatase, non-receptor type 6 | 0.001615789 | 26 | 2 | 26 | Up |
| PDE3B | Phosphodiesterase 3B, cGMP-inhibited | 0.001519611 | 2 | 2 | 2 | Down |
| THY1 | Thy-1 cell surface antigen | 0.001513199 | 2 | 2 | 2 | Up |
| PIP5K1B | Phosphatidylinositol-4-phosphate 5-kinase, type I, beta | 0.001487551 | 3 | 1 | 3 | Down |
| LCK | Lymphocyte-specific protein tyrosine kinase | 0.001429845 | 4 | 3 | 3 | Up |
| CSF1R | Colony stimulating factor 1 receptor | 0.001429845 | 3 | 3 | 2 | Up |
| RYR3 | Ryanodine receptor 3 | 0.001408472 | 6 | 6 | 6 | Down |
| TYROBP | TYRO protein tyrosine kinase binding protein | 0.001384962 | 2 | 1 | 1 | Up |
| IFNAR2 | Interferon (alpha, beta and omega) receptor 2 | 0.001350765 | 4 | 2 | 2 | Up |
| LCP2 | Lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76 kDa) | 0.001191975 | 6 | 5 | 4 | Up |
| CD4 | CD4 molecule | 0.001077192 | 9 | 9 | 1 | Up |
| PVRL2 | Poliovirus receptor-related 2 (herpesvirus entry mediator B) | 0.000865601 | 3 | 1 | 3 | Up |
| FCGR3A | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | 0.00085349 | 5 | 5 | 3 | Up |
| CCL5 | Chemokine (C-C motif) ligand 5 | 0.000801482 | 7 | 1 | 6 | Up |
| SORBS1 | Sorbin and SH3 domain containing 1 | 0.000795071 | 2 | 2 | 1 | Down |
| TNF | Tumor necrosis factor | 0.000756599 | 2 | 1 | 1 | Up |
| EGFR | Epidermal growth factor receptor | 0.000745058 | 15 | 10 | 5 | Down |
| ITK | IL2-inducible T-cell kinase | 0.00071535 | 8 | 7 | 2 | Up |
| ITGA7 | Integrin, alpha 7 | 0.000649735 | 13 | 13 | 2 | Down |
| PDGFRB | Platelet-derived growth factor receptor, beta polypeptide | 0.00059812 | 12 | 7 | 5 | Up |
| KDR | Kinase insert domain receptor (a type III receptor tyrosine kinase) | 0.000530261 | 10 | 5 | 5 | Down |
| BTK | Bruton agammaglobulinemia tyrosine kinase | 0.000391704 | 6 | 6 | 4 | Up |
| CACNB2 | Calcium channel, voltage-dependent, beta 2 subunit | 0.000371888 | 2 | 2 | 1 | Down |
| CACNA2D3 | Calcium channel, voltage-dependent, alpha 2/delta subunit 3 | 0.000371888 | 2 | 2 | 1 | down |
| IFNGR2 | Interferon gamma receptor 2 (interferon gamma transducer 1) | 0.000368682 | 4 | 2 | 2 | Up |
| IFNGR1 | Interferon gamma receptor 1 | 0.000368682 | 4 | 2 | 2 | Up |
| FLNC | Filamin C, gamma | 0.00036227 | 3 | 3 | 3 | Down |
| CACNA1H | Calcium channel, voltage-dependent, T type, alpha 1H subunit | 0.000281511 | 3 | 3 | 3 | Up |
| PARD3 | Par-3 partitioning defective 3 homolog (C. elegans) | 0.000278053 | 3 | 2 | 1 | Down |
| RASGRP1 | RAS guanyl releasing protein 1 (calcium and DAG-regulated) | 0.000218003 | 3 | 3 | 3 | Up |
| CD8A | CD8a molecule | 0.000134649 | 2 | 2 | 1 | Up |
| IGF1 | Insulin-like growth factor 1 (somatomedin C) | 0.000121825 | 10 | 1 | 9 | Down |
| HIF1A | Hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | 0.000115413 | 3 | 1 | 3 | Up |
| FGFR2 | Fibroblast growth factor receptor 2 | 0.000105796 | 9 | 6 | 3 | Down |
| CSF2RB | Colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | 0.000100025 | 6 | 2 | 4 | Up |
| GNAO1 | Guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O | 7.69423E-05 | 7 | 2 | 5 | Down |
| CD28 | CD28 molecule | 7.69423E-05 | 5 | 1 | 4 | Up |
| ERBB4 | v-erb - a erythroblastic leukemia viral oncogene homolog 4 (avian) | 6.35843E-05 | 7 | 2 | 5 | Down |
| BLNK | B-cell linker | 5.79918E-05 | 3 | 3 | 2 | Up |
| HCK | Hemopoietic cell kinase | 4.51375E-05 | 10 | 1 | 9 | Up |
| PLCE1 | Phospholipase C, epsilon 1 | 2.88534E-05 | 2 | 2 | 2 | Down |
| HLA-DMB | Major histocompatibility complex, class II, DM beta | 1.92356E-05 | 9 | 7 | 9 | Up |
| HLA-DMA | Major histocompatibility complex, class II, DM alpha | 1.92356E-05 | 9 | 7 | 9 | Up |
| PIK3AP1 | Phosphoinositide-3-kinase adaptor protein 1 | 1.28237E-05 | 4 | 4 | 3 | Up |
| LRP6 | Low density lipoprotein receptor-related protein 6 | 1.28237E-05 | 2 | 2 | 2 | Down |
| CD247 | CD247 molecule | 9.61779E-06 | 3 | 2 | 2 | Up |
| ITGAM | Integrin, alpha M (complement component 3 receptor 3 subunit) | 8.54915E-06 | 3 | 2 | 2 | Up |
| INHBA | Inhibin, beta A | 6.41186E-06 | 2 | 1 | 1 | Up |
| TRPC1 | Transient receptor potential cation channel, subfamily C, member 1 | 5.80121E-06 | 2 | 2 | 2 | Down |
| BST1 | Bone marrow stromal cell antigen 1 | 5.80121E-06 | 2 | 2 | 2 | Up |
| P2RX4 | Purinergic receptor P2X, ligand-gated ion channel, 4 | 5.80121E-06 | 2 | 2 | 2 | Up |
| P2RX1 | Purinergic receptor P2X, ligand-gated ion channel, 1 | 5.80121E-06 | 2 | 2 | 2 | Up |
| FCGR2A | Fc fragment of IgG, low affinity IIa, receptor (CD32) | 3.91836E-06 | 3 | 3 | 1 | Up |
| PRKAR2B | Protein kinase, cAMP-dependent, regulatory, type II, beta | 3.20593E-06 | 2 | 1 | 2 | Down |
| SHC3 | SHC (Src homology 2 domain containing) transforming protein 3 | 0 | 9 | 9 | 0 | Down |
| SHC2 | SHC (Src homology 2 domain containing) transforming protein 2 | 0 | 9 | 9 | 0 | Down |
| HLA-DQA2 | Major histocompatibility complex, class II, DQ alpha 2 | 0 | 8 | 7 | 8 | Up |
| HLA-DPA1 | Major histocompatibility complex, class II, DP alpha 1 | 0 | 8 | 7 | 8 | Up |
| HLA-DRA | Major histocompatibility complex, class II, DR alpha | 0 | 8 | 7 | 8 | Up |
| HLA-DPB1 | Major histocompatibility complex, class II, DP beta 1 | 0 | 8 | 7 | 8 | Up |
| HLA-DOA | Major histocompatibility complex, class II, DO alpha | 0 | 8 | 7 | 8 | Up |
| HLA-DOB | Major histocompatibility complex, class II, DO beta | 0 | 8 | 7 | 8 | Up |
| RXRG | Retinoid X receptor, gamma | 0 | 7 | 0 | 7 | Down |
| FGR | Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog | 0 | 7 | 0 | 7 | Up |
| GNG7 | Guanine nucleotide binding protein (G protein), gamma 7 | 0 | 6 | 0 | 6 | Down |
| CXCL2 | Chemokine (C-X-C motif) ligand 2 | 0 | 6 | 0 | 6 | Up |
| CCL4L1 | Chemokine (C-C motif) ligand 4-like 1 | 0 | 6 | 0 | 6 | Up |
| CCL3 | Chemokine (C-C motif) ligand 3 | 0 | 6 | 0 | 6 | Up |
| CCL2 | Chemokine (C-C motif) ligand 2 | 0 | 6 | 0 | 6 | Up |
| CXCL14 | Chemokine (C-X-C motif) ligand 14 | 0 | 6 | 0 | 6 | Down |
| CXCL16 | Chemokine (C-X-C motif) ligand 16 | 0 | 6 | 0 | 6 | Up |
| WWP1 | WW domain containing E3 ubiquitin protein ligase 1 | 0 | 5 | 0 | 5 | Down |
| CD74 | CD74 molecule, major histocompatibility complex, class II invariant chain | 0 | 5 | 5 | 0 | Up |
| PGR | Progesterone receptor | 0 | 4 | 0 | 4 | Down |
| PLA2G2A | Phospholipase A2, group IIA (platelets, synovial fluid) | 0 | 4 | 4 | 0 | Down |
| FCER1G | Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide | 0 | 4 | 3 | 3 | Up |
| CCND2 | Cyclin D2 | 0 | 4 | 4 | 0 | Down |
| PLA2G5 | Phospholipase A2, group V | 0 | 4 | 4 | 0 | Up |
| RASSF5 | Ras association (RalGDS/AF-6) domain family member 5 | 0 | 4 | 0 | 4 | Up |
| PDGFA | Platelet-derived growth factor alpha polypeptide | 0 | 4 | 0 | 4 | Up |
| WNT5A | Wingless-type MMTV integration site family, member 5A | 0 | 4 | 2 | 2 | Down |
| LIFR | Leukemia inhibitory factor receptor alpha | 0 | 3 | 2 | 1 | Down |
| CD226 | CD226 molecule | 0 | 3 | 3 | 0 | Up |
| LAMB3 | Laminin, beta 3 | 0 | 3 | 0 | 3 | Up |
| SPP1 | Secreted phosphoprotein 1 | 0 | 3 | 0 | 3 | Up |
| HGF | Hepatocyte growth factor (hepapoietin A; scatter factor) | 0 | 3 | 0 | 3 | Up |
| LAMA2 | Laminin, alpha 2 | 0 | 3 | 0 | 3 | Down |
| SFRP1 | Secreted frizzled-related protein 1 | 0 | 3 | 0 | 3 | Down |
| SFRP2 | Secreted frizzled-related protein 2 | 0 | 3 | 0 | 3 | Down |
| FGF9 | Fibroblast growth factor 9 (glia-activating factor) | 0 | 3 | 0 | 3 | Down |
| IL13RA1 | Interleukin 13 receptor, alpha 1 | 0 | 3 | 2 | 1 | Up |
| GHR | Growth hormone receptor | 0 | 3 | 2 | 1 | Down |
| ICAM1 | Intercellular adhesion molecule 1 | 0 | 3 | 3 | 0 | Up |
| IL6R | Interleukin 6 receptor | 0 | 3 | 2 | 1 | Up |
| IL7R | Interleukin 7 receptor | 0 | 3 | 2 | 1 | Up |
| AMPH | Amphiphysin | 0 | 3 | 3 | 0 | Down |
| COL6A6 | Collagen, type VI, alpha 6 | 0 | 3 | 0 | 3 | Down |
| IL10RB | Interleukin 10 receptor, beta | 0 | 3 | 2 | 1 | Up |
| THBS1 | Thrombospondin 1 | 0 | 3 | 0 | 3 | Up |
| THBS2 | Thrombospondin 2 | 0 | 3 | 0 | 3 | Up |
| THBS4 | Thrombospondin 4 | 0 | 3 | 0 | 3 | Down |
| FGF10 | Fibroblast growth factor 10 | 0 | 3 | 0 | 3 | Down |
| TGFB1 | Transforming growth factor, beta 1 | 0 | 3 | 3 | 0 | Up |
| FGF20 | Fibroblast growth factor 20 | 0 | 3 | 0 | 3 | Up |
| IL11RA | Interleukin 11 receptor, alpha | 0 | 3 | 2 | 1 | Down |
| HCST | Hematopoietic cell signal transducer | 0 | 3 | 0 | 3 | Up |
| RELN | Reelin | 0 | 3 | 0 | 3 | Down |
| COL1A1 | Collagen, type I, alpha 1 | 0 | 3 | 0 | 3 | Up |
| CNTFR | Ciliary neurotrophic factor receptor | 0 | 3 | 2 | 1 | Down |
| DOCK2 | Dedicator of cytokinesis 2 | 0 | 3 | 3 | 0 | Up |
| IL12RB1 | Interleukin 12 receptor, beta 1 | 0 | 3 | 2 | 1 | Up |
| ICOS | Inducible T-cell co-stimulator | 0 | 3 | 0 | 3 | Up |
| MYLK | Myosin light chain kinase | 0 | 3 | 3 | 0 | Down |
| IL21R | Interleukin 21 receptor | 0 | 3 | 2 | 1 | Up |
| PAK7 | p21 protein (Cdc42/Rac)-activated kinase 7 | 0 | 3 | 0 | 3 | Down |
| PAK3 | p21 protein (Cdc42/Rac)-activated kinase 3 | 0 | 3 | 0 | 3 | Down |
| IL2RG | Interleukin 2 receptor, gamma | 0 | 3 | 2 | 1 | Up |
| PTPRC | Protein tyrosine phosphatase, receptor type, C | 0 | 3 | 0 | 3 | Up |
| IL2RB | Interleukin 2 receptor, beta | 0 | 3 | 2 | 1 | Up |
| TNXB | Tenascin XB | 0 | 3 | 0 | 3 | Down |
| IL2RA | Interleukin 2 receptor, alpha | 0 | 3 | 2 | 1 | Up |
| COL4A6 | Collagen, type IV, alpha 6 | 0 | 3 | 0 | 3 | Down |
| BTC | Betacellulin | 0 | 2 | 0 | 2 | Down |
| FAS | Fas (TNF receptor superfamily, member 6) | 0 | 2 | 2 | 0 | Up |
| PTPRF | Protein tyrosine phosphatase, receptor type, F | 0 | 2 | 0 | 2 | Down |
| PIM1 | Pim-1 oncogene | 0 | 2 | 2 | 0 | Up |
| GNAZ | Guanine nucleotide binding protein (G protein), alpha z polypeptide | 0 | 2 | 0 | 2 | Down |
| KCNMA1 | Potassium large conductance calcium-activated channel, subfamily M, alpha member 1 | 0 | 2 | 2 | 0 | Down |
| GZMA | Granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) | 0 | 2 | 0 | 2 | Up |
| ARHGEF6 | Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 | 0 | 2 | 2 | 0 | Up |
| ICAM3 | Intercellular adhesion molecule 3 | 0 | 2 | 0 | 2 | Up |
| MMP14 | Matrix metallopeptidase 14 (membrane-inserted) | 0 | 2 | 2 | 0 | Up |
| RUNX1 | Runt-related transcription factor 1 | 0 | 2 | 1 | 2 | Up |
| LIMK1 | LIM domain kinase 1 | 0 | 2 | 2 | 0 | Up |
| LIPE | Lipase, hormone-sensitive | 0 | 2 | 2 | 0 | Down |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | 0 | 1 | 1 | 0 | Up |
| ADCY5 | Adenylate cyclase 5 | 0 | 1 | 0 | 1 | Down |
| FST | Follistatin | 0 | 1 | 0 | 1 | Up |
| LHCGR | Luteinizing hormone/choriogonadotropin receptor | 0 | 1 | 0 | 1 | Down |
| PRKG1 | Protein kinase, cGMP-dependent, type I | 0 | 1 | 0 | 1 | Down |
| AGTR1 | Angiotensin II receptor, type 1 | 0 | 1 | 0 | 1 | Down |
| PPP1R1A | Protein phosphatase 1, regulatory (inhibitor) subunit 1A | 0 | 1 | 1 | 0 | Down |
| MAP2K6 | Mitogen-activated protein kinase kinase 6 | 0 | 1 | 0 | 1 | Down |
| LILRB3 | Leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 | 0 | 1 | 1 | 1 | Up |
| GRAP2 | GRB2-related adaptor protein 2 | 0 | 1 | 1 | 1 | Up |
| DUSP10 | Dual specificity phosphatase 10 | 0 | 1 | 0 | 1 | Up |
| CD72 | CD72 molecule | 0 | 1 | 1 | 1 | Up |
| CTSL1 | Cathepsin L1 | 0 | 1 | 0 | 1 | Up |
| AVPR1A | Arginine vasopressin receptor 1A | 0 | 1 | 0 | 1 | Down |
| AREG | Amphiregulin | 0 | 1 | 0 | 1 | Up |
| WAS | Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | 0 | 1 | 1 | 0 | Up |
| CARD11 | Caspase recruitment domain family, member 11 | 0 | 1 | 1 | 0 | Up |
| DAPP1 | Dual adaptor of phosphotyrosine and 3-phosphoinositides | 0 | 1 | 1 | 0 | Up |
| PLIN1 | Perilipin 1 | 0 | 1 | 1 | 0 | Down |
| GRIA3 | Glutamate receptor, ionotrophic, AMPA 3 | 0 | 1 | 0 | 1 | Down |
| RPS6KA6 | Ribosomal protein S6 kinase, 90kDa, polypeptide 6 | 0 | 1 | 1 | 0 | Down |
| DUSP2 | Dual specificity phosphatase 2 | 0 | 1 | 0 | 1 | Up |
| RPS6KA1 | Ribosomal protein S6 kinase, 90kDa, polypeptide 1 | 0 | 1 | 1 | 0 | Up |
| ARNT2 | Aryl-hydrocarbon receptor nuclear translocator 2 | 0 | 1 | 1 | 1 | Down |
| CDKN2B | Cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 0 | 1 | 0 | 1 | Up |
| CDKN2C | Cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | 0 | 1 | 0 | 1 | Up |
| MS4A2 | Membrane-spanning 4-domains, subfamily A, member 2 (Fc fragment of IgE, high affinity I, receptor for; beta polypeptide) | 0 | 1 | 1 | 0 | Up |
| HLA-E | Major histocompatibility complex, class I, E | 0 | 1 | 0 | 1 | Up |
| CYSLTR2 | Cysteinyl leukotriene receptor 2 | 0 | 1 | 0 | 1 | Up |
| ANGPTL4 | Angiopoietin-like 4 | 0 | 1 | 1 | 0 | Up |
| FABP4 | Fatty acid binding protein 4, adipocyte | 0 | 1 | 1 | 0 | Down |
| CD79A | CD79a molecule, immunoglobulin-associated alpha | 0 | 1 | 1 | 0 | Up |
| ADRA1D | Adrenergic, alpha-1D-, receptor | 0 | 1 | 0 | 1 | Down |
| TACR1 | Tachykinin receptor 1 | 0 | 1 | 0 | 1 | Down |
| GSN | Gelsolin | 0 | 1 | 1 | 0 | Down |
| EGR1 | Early growth response 1 | 0 | 1 | 0 | 1 | Up |
| CTSS | Cathepsin S | 0 | 1 | 0 | 1 | Up |
| ACADL | Acyl-CoA dehydrogenase, long chain | 0 | 1 | 1 | 0 | Down |
| CD86 | CD86 molecule | 0 | 1 | 0 | 1 | Up |
| CD80 | CD80 molecule | 0 | 1 | 1 | 0 | Up |
| CTSB | Cathepsin B | 0 | 1 | 0 | 1 | Up |
| OXTR | Oxytocin receptor | 0 | 1 | 0 | 1 | Up |
| DCN | Decorin | 0 | 1 | 0 | 1 | Down |
| TNFRSF1B | Tumor necrosis factor receptor superfamily, member 1B | 0 | 1 | 1 | 0 | Up |
| OLR1 | Oxidized low density lipoprotein (lectin-like) receptor 1 | 0 | 1 | 1 | 0 | Up |
| OR51E2 | Olfactory receptor, family 51, subfamily E, member 2 | 0 | 1 | 1 | 0 | Up |
| ADIPOQ | Adiponectin, C1Q and collagen domain containing | 0 | 1 | 1 | 0 | Down |
| IGFBP3 | Insulin-like growth factor binding protein 3 | 0 | 1 | 0 | 1 | Up |
Discussion
Several clinical and therapeutic factors have been reported as significant to the occurrence of CTEPH. The pathophysiology of CTEPH remains incompletely understood. In most cases it is associated with a history of acute venous thromboembolism (29); however, in a small percentage of patients, thrombi do not resolve after an acute event and the reasons for this are unclear. The current knowledge is based on a triad of enhanced thrombosis (7,30), disturbed thrombolysis (31–33) and inflammation (34). A number of novel prognostic factors, such as cytological features, standard karyotyping, fluorescence in situ hybridization, centromeric probes, single nucleotide polymorphism and gene expression profiling have been investigated. Using the advanced and inexpensive technique of microarray, new genes which may affect the development of CTEPH can be identified.
In this study, to investigate variations in gene expression profiles and signaling pathways in CTEPH, 10 samples were divided into a normal (control) and CTEPH group to identify CTEPH-related differentially expressed genes. The expression of the upregulated genes was higher in the CTEPH group compared with the control group. Gene chips have become a useful tool for studying the development and progression of tumors owing to the high-throughout, but it remains difficult to predict patients with CTEPH, mainly due to the high number of variations in CTEPH and the great challenge in interpreting numerous complex data produced by the microarray (35) and determining the main responsible genes. The present study made use of bioinformatics the method to analyze functions and pathways of the differentially expressed genes, and further clarified their biological significance, and defined the key genes that affect the development of CTEPH.
More than 1,600 genes were differentially upregulated or downregulated in CTEPH in this study. The first upregulated differentially expressed gene in CTEPH, oxidized low density lipoprotein (lectin-like) receptor 1 (OLR1), has been studied in several cardiovascular diseases, such as atherosclerosis for its polymorphisms (36), and recently Wynants et al found that it is highly expressed CTEPH (5). The second most significantly upregulated gene, intereukin (IL)8, has been found to be associated with hemodynamic instability following PEA in patients with CTEPH (37). The role of the third most upregulated gene, secreted phosphoprotein 1 (SPP1), which encodes osteopontin, in CTEPH is poorly understood. Most of the genes that were strongly downregulated showed close associations with tumors, chemokines and lipids, including the gene for CXCL14, which is a type of chemokine ligand involved in the regulation of tumors (38,39). Heparanase 2 (HPSE2) encodes a specific enzyme and is associated with urofacial syndrome (40). None of these genes were found to be associated with CTEPH. As the sample number we used in this study was limited, we tried to investigate the microarray results using another method.
GO is widely recognized as the premier tool for the organization and functional annotation of molecular aspects (41). GO analysis was used to interpret each GO of the differentially expressed genes and analyze it statistically. By using the criteria of P<0.05, significant GO items and genes involved were identified. Guo et al used GO analysis to analyze miRNA microarrays and found that miR-15b and miR-16 may be indispensable for apoptosis by targeting Bcl-2 (42). GO terms regarding inflammatory response play an important role in CTEPH; a number of studies have reported CRP as a predictor of adverse outcome in pulmonary arterial hypertension (43), and IL-6-mediated systemic inflammatory cascades may also be involved in the regulation of peripheral vascular tone following pulmonary thromboendarterectomy (PTE) (44). Quarck et al reported that a proliferative phenotype of pulmonary arterial smooth muscle cells and endothelial cells contributed to proximal vascular remodeling in CTEPH (45). A number of studies have proven that patients with CTEPH can generate a pronounced inflammatory response with the release of pro-inflammatory and anti-inflammatory cytokines (44,46). Therefore, we hypothesized that the functions of other items listed may play a role in CTEPH, which has not been elucidated yet.
GO analysis is a classical method used to annotate gene function but is still inexact in some fields. Pathway analysis can reveal the distinct biological process and identify significant pathways that differentially expressed genes participate in; based on this, we can have a comprehensive understanding as to the interactions of genes, functions that they participate in and associations between up- and down-stream pathways, and identify genes involved in these significant pathways. The concordance of the MAPK signaling pathway, cytokine-cytokine receptor interaction and apoptosis with the GO terms confirmed their critical role in CTEPH. Wei et al demonstrated that JNK is a critical molecule in 5-HT-induced pulmonary artery smooth muscle cell (PASMC) proliferation and migration and may act at an important point for crosstalk of the MAPK and phosphoinositide 3-kinase (PI3K) pathways (47); however, to our knowledge, there is no study available on the role of the MAPK signaling pathway in CTEPH. Numerous studies have proven that focal adhesion and cytokine participate in the process of vascular remodeling, which is an important characteristic of CTEPH (48,49). A previous study on the role of CRP in proximal pulmonary endothelial cells and smooth muscle cells also demonstrated the effects of cell adhesion molecules on CTEPH (5). Since CTEPH is still a rare disease worldwide, information on the signaling pathways associated with its development is limited. We hypothesized that the other seemingly irrelevant pathways may play a role in CTEPH. However, this requires further investigation. Pathway analysis revealed equally important roles and functions as GO analysis.
In the investigation of genes involved in significant GO terms and pathways, 232 genes in common were found that may affect the development of CTEPH. JAK3 is an enzyme in the janus kinase family and has been implicated in cell signaling processes important in cancer and immune-inflammatory diseases (50). Recently, it was found to improve myocardial vascular permeability (51); however, its role in CTEPH remains unelucidated. The functions of GNA15 and ARRB2 have been less frequently reported. MAPK13 mainly participates in cholangiocarcinoma and increases cell migration; it may play a similar role in CTEPH (52). F2R, also known as PAR-1, affects vascular remodeling in the small intestine (53). Vascular cell adhesion molecule 1 (VCAM1) has been reported to modulate blood vessel endothelial cell-leukocyte interactions and increase the strength of cell adhesion (54). Although their functions have not been fully investigated, a number of genes may play a role in the development of CTEPH. Based on these data, further studies on the expression of these genes and protein functions are required using a greater sample number and using methods, such as reverse transcriptase-polymerase chain reaction and western blot analysis; moreover, the regulatory functions of the identified genes and proteins also requires investigation. Further studies may help to improve the clinical diagnosis and treatment of patients with CTEPH.
In conlcusion, the results presented in our study suggest that differences in gene expression exist between CTEPH and normal samples. These genes encode proteins involved in different GO items and signaling pathways, the disruption of which can affect the development of CTEPH. Several genes, such as JAK3, GNA15, MAPK13, F2R and VCAM1 provide potential candidates for distinguishing between CTEPH from healthy individuals in the future. This distinction may aid in the diagnosis and prevention of CTEPH, based on the different features.
Acknowledgements
This study was supported by the High-Level Technic Personnel Training Plan of Beijing Health System (no. 2011-3-017).
References
- 1.Hoeper MM, Mayer E, Simonneau G, Rubin LJ. Chronic thromboembolic pulmonary hypertension. Circulation. 2006;113:2011–2020. doi: 10.1161/CIRCULATIONAHA.105.602565. [DOI] [PubMed] [Google Scholar]
- 2.Mayer E, Klepetko W. Techniques and outcomes of pulmonary endarterectomy for chronic thromboembolic pulmonary hypertension. Proc Am Thorac Soc. 2006;3:589–593. doi: 10.1513/pats.200605-120LR. [DOI] [PubMed] [Google Scholar]
- 3.Freed DH, Thomson BM, Tsui SS, et al. Functional and haemodynamic outcome 1 year after pulmonary thromboendarterectomy. Eur J Cardiothorac Surg. 2008;34:525–530. doi: 10.1016/j.ejcts.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 4.Bonderman D, Jakowitsch J, Redwan B, et al. Role for staphylococci in misguided thrombus resolution of chronic thromboembolic pulmonary hypertension. Arterioscler Thromb Vasc Biol. 2008;28:678–684. doi: 10.1161/ATVBAHA.107.156000. [DOI] [PubMed] [Google Scholar]
- 5.Wynants M, Quarck R, Ronisz A, et al. Effects of C-reactive protein on human pulmonary vascular cells in chronic thromboembolic pulmonary hypertension. Eur Respir J. 2012;40:886–894. doi: 10.1183/09031936.00197511. [DOI] [PubMed] [Google Scholar]
- 6.Lang IM, Klepetko W. Chronic thromboembolic pulmonary hypertension: an updated review. Curr Opin Cardiol. 2008;23:555–559. doi: 10.1097/HCO.0b013e328311f254. [DOI] [PubMed] [Google Scholar]
- 7.Wolf M, Boyer-Neumann C, Parent F, et al. Thrombotic risk factors in pulmonary hypertension. Eur Respir J. 2000;15:395–399. doi: 10.1034/j.1399-3003.2000.15b28.x. [DOI] [PubMed] [Google Scholar]
- 8.Dartevelle P, Fadel E, Mussot S, et al. Chronic thromboembolic pulmonary hypertension. Eur Respir J. 2004;23:637–648. doi: 10.1183/09031936.04.00079704. [DOI] [PubMed] [Google Scholar]
- 9.Devaraj S, Singh U, Jialal I. The evolving role of C-reactive protein in atherothrombosis. Clin Chem. 2009;55:229–238. doi: 10.1373/clinchem.2008.108886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barton M, Yanagisawa M. Endothelin: 20 years from discovery to therapy. Can J Physiol Pharmacol. 2008;86:485–498. doi: 10.1139/Y08-059. [DOI] [PubMed] [Google Scholar]
- 11.Hoeper MM, Barbera JA, Channick RN, et al. Diagnosis, assessment, and treatment of non-pulmonary arterial hypertension pulmonary hypertension. J Am Coll Cardiol. 2009;54:S85–S96. doi: 10.1016/j.jacc.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Badesch DB, Champion HC, Sanchez MA, et al. Diagnosis and assessment of pulmonary arterial hypertension. J Am Coll Cardiol. 2009;54:S55–S66. doi: 10.1016/j.jacc.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 13.Auger WR, Fedullo PF, Moser KM, Buchbinder M, Peterson KL. Chronic major-vessel thromboembolic pulmonary artery obstruction: appearance at angiography. Radiology. 1992;182:393–398. doi: 10.1148/radiology.182.2.1732955. [DOI] [PubMed] [Google Scholar]
- 14.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 15.Yang H, Crawford N, Lukes L, Finney R, Lancaster M, Hunter KW. Metastasis predictive signature profiles pre-exist in normal tissues. Clin Exp Metastasis. 2005;22:593–603. doi: 10.1007/s10585-005-6244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke R, Ressom HW, Wang A, et al. The properties of high-dimensional data spaces: implications for exploring gene and protein expression data. Nat Rev Cancer. 2008;8:37–49. doi: 10.1038/nrc2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupuy D, Bertin N, Hidalgo CA, et al. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat Biotechnol. 2007;25:663–668. doi: 10.1038/nbt1305. [DOI] [PubMed] [Google Scholar]
- 20.Schlitt T, Palin K, Rung J, et al. From gene networks to gene function. Genome Res. 2003;13:2568–2576. doi: 10.1101/gr.1111403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi M, Horton JD, Cohen JC, Hobbs HH, Stephens RM. WholePathwayScope: a comprehensive pathway-based analysis tool for high-throughput data. BMC Bioinformatics. 2006;7:30. doi: 10.1186/1471-2105-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Draghici S, Khatri P, Tarca AL, et al. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jansen R, Greenbaum D, Gerstein M. Relating whole-genome expression data with protein-protein interactions. Genome Res. 2002;12:37–46. doi: 10.1101/gr.205602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C, Li H. Network-constrained regularization and variable selection for analysis of genomic data. Bioinformatics. 2008;24:1175–1182. doi: 10.1093/bioinformatics/btn081. [DOI] [PubMed] [Google Scholar]
- 26.Wei Z, Li H. A Markov random field model for network-based analysis of genomic data. Bioinformatics. 2007;23:1537–1544. doi: 10.1093/bioinformatics/btm129. [DOI] [PubMed] [Google Scholar]
- 27.Zhang JD, Wiemann S. KEGGgraph: a graph approach to KEGG PATHWAY in R and bioconductor. Bioinformatics. 2009;25:1470–1471. doi: 10.1093/bioinformatics/btp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci USA. 2003;100:12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pepke-Zaba J, Delcroix M, Lang I, et al. Chronic thromboembolic pulmonary hypertension (CTEPH): results from an international prospective registry. Circulation. 2011;124:1973–1981. doi: 10.1161/CIRCULATIONAHA.110.015008. [DOI] [PubMed] [Google Scholar]
- 30.Bonderman D, Turecek PL, Jakowitsch J, et al. High prevalence of elevated clotting factor VIII in chronic thromboembolic pulmonary hypertension. Thromb Haemost. 2003;90:372–376. doi: 10.1160/TH03-02-0067. [DOI] [PubMed] [Google Scholar]
- 31.Lang IM, Marsh JJ, Olman MA, Moser KM, Schleef RR. Parallel analysis of tissue-type plasminogen activator and type 1 plasminogen activator inhibitor in plasma and endothelial cells derived from patients with chronic pulmonary thromboemboli. Circulation. 1994;90:706–712. doi: 10.1161/01.cir.90.2.706. [DOI] [PubMed] [Google Scholar]
- 32.Morris TA, Marsh JJ, Chiles PG, et al. High prevalence of dysfibrinogenemia among patients with chronic thromboembolic pulmonary hypertension. Blood. 2009;114:1929–1936. doi: 10.1182/blood-2009-03-208264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suntharalingam J, Goldsmith K, van Marion V, et al. Fibrinogen Aalpha Thr312Ala polymorphism is associated with chronic thromboembolic pulmonary hypertension. Eur Respir J. 2008;31:736–741. doi: 10.1183/09031936.00055107. [DOI] [PubMed] [Google Scholar]
- 34.Bonderman D, Wilkens H, Wakounig S, et al. Risk factors for chronic thromboembolic pulmonary hypertension. Eur Respir J. 2009;33:325–331. doi: 10.1183/09031936.00087608. [DOI] [PubMed] [Google Scholar]
- 35.Chen X, Wang L. Integrating biological knowledge with gene expression profiles for survival prediction of cancer. J Comput Biol. 2009;16:265–278. doi: 10.1089/cmb.2008.12TT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu Y, Liu Z, Li L, et al. OLR1, PON1 and MTHFR gene polymorphisms, conventional risk factors and the severity of coronary atherosclerosis in a Chinese Han population. Cell Physiol Biochem. 2013;31:143–152. doi: 10.1159/000343356. [DOI] [PubMed] [Google Scholar]
- 37.Lindner J, Maruna P, Kunstyr J, et al. Hemodynamic instability after pulmonary endarterectomy for chronic thromboembolic pulmonary hypertension correlates with cytokine network hyperstimulation. Eur Surg Res. 2009;43:39–46. doi: 10.1159/000218101. [DOI] [PubMed] [Google Scholar]
- 38.Zeng J, Yang X, Cheng L, et al. Chemokine CXCL14 is associated with prognosis in patients with colorectal carcinoma after curative resection. J Transl Med. 2013;11:6. doi: 10.1186/1479-5876-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu XL, Ou ZL, Lin FJ, et al. Expression of CXCL14 and its anticancer role in breast cancer. Breast Cancer Res Treat. 2012;135:725–735. doi: 10.1007/s10549-012-2206-2. [DOI] [PubMed] [Google Scholar]
- 40.Mahmood S, Beetz C, Tahir MM, et al. First HPSE2 missense mutation in urofacial syndrome. Clin Genet. 2012;81:88–92. doi: 10.1111/j.1399-0004.2011.01649.x. [DOI] [PubMed] [Google Scholar]
- 41.Lovering RC, Camon EB, Blake JA, Diehl AD. Access to immunology through the Gene Ontology. Immunology. 2008;125:154–160. doi: 10.1111/j.1365-2567.2008.02940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo CJ, Pan Q, Li DG, Sun H, Liu BW. miR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: an essential role for apoptosis. J Hepatol. 2009;50:766–778. doi: 10.1016/j.jhep.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 43.Quarck R, Nawrot T, Meyns B, Delcroix M. C-reactive protein: a new predictor of adverse outcome in pulmonary arterial hypertension. J Am Coll Cardiol. 2009;53:1211–1218. doi: 10.1016/j.jacc.2008.12.038. [DOI] [PubMed] [Google Scholar]
- 44.Langer F, Schramm R, Bauer M, Tscholl D, Kunihara T, Schafers HJ. Cytokine response to pulmonary thromboendarterectomy. Chest. 2004;126:135–141. doi: 10.1378/chest.126.1.135. [DOI] [PubMed] [Google Scholar]
- 45.Quarck R, Wynants M, Ronisz A, et al. Characterization of proximal pulmonary arterial cells from chronic thromboembolic pulmonary hypertension patients. Respir Res. 2012;13:27. doi: 10.1186/1465-9921-13-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Von Haehling S, von Bardeleben RS, Kramm T, et al. Inflammation in right ventricular dysfunction due to thromboembolic pulmonary hypertension. Int J Cardiol. 2010;144:206–211. doi: 10.1016/j.ijcard.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 47.Wei L, Liu Y, Kaneto H, Fanburg BL. JNK regulates serotonin-mediated proliferation and migration of pulmonary artery smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2010;298:L863–L869. doi: 10.1152/ajplung.00281.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stone JD, Narine A, Shaver PR, Fox JC, Vuncannon JR, Tulis DA. AMP-activated protein kinase inhibits vascular smooth muscle cell proliferation and migration and vascular remodeling following injury. Am J Physiol Heart Circ Physiol. 2013;304:H369–H381. doi: 10.1152/ajpheart.00446.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farkas L, Gauldie J, Voelkel NF, Kolb M. Pulmonary hypertension and idiopathic pulmonary fibrosis: a tale of angiogenesis, apoptosis, and growth factors. Am J Respir Cell Mol Biol. 2011;45:1–15. doi: 10.1165/rcmb.2010-0365TR. [DOI] [PubMed] [Google Scholar]
- 50.Sun Z, Luo Q, Ye D, Chen W, Chen F. Role of toll-like receptor 4 on the immune escape of human oral squamous cell carcinoma and resistance of cisplatin-induced apoptosis. Mol Cancer. 2012;11:33. doi: 10.1186/1476-4598-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JE, Lee AS, Kim DH, et al. Janex-1, a JAK3 inhibitor, ameliorates tumor necrosis factor-alpha-induced expression of cell adhesion molecules and improves myocardial vascular permeability in endotoxemic mice. Int J Mol Med. 2012;29:864–870. doi: 10.3892/ijmm.2012.920. [DOI] [PubMed] [Google Scholar]
- 52.Tan FL, Ooi A, Huang D, et al. p38delta/MAPK13 as a diagnostic marker for cholangiocarcinoma and its involvement in cell motility and invasion. Int J Cancer. 2010;126:2353–2361. doi: 10.1002/ijc.24944. [DOI] [PubMed] [Google Scholar]
- 53.Reinhardt C, Bergentall M, Greiner TU, et al. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature. 2012;483:627–631. doi: 10.1038/nature10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rafat M, Rotenstein LS, Hu JL, Auguste DT. Engineered endothelial cell adhesion via VCAM1 and E-selectin antibody-presenting alginate hydrogels. Acta Biomater. 2012;8:2697–2703. doi: 10.1016/j.actbio.2012.04.010. [DOI] [PubMed] [Google Scholar]