Abstract
Orthodox seeds are living organisms that survive anhydrobiosis and may display dormancy, an inability to germinate at harvest. Seed germination potential can be acquired during a prolonged period of dry storage called after-ripening. The aim of this work was to determine if gene transcription is an underlying regulatory mechanism for dormancy alleviation during after-ripening. To identify changes in gene transcription strictly associated with the acquisition of germination potential but not with storage, we used seed storage at low relative humidity that maintains dormancy as control. Transcriptome profiling was performed using DNA microarray to compare change in gene transcript abundance between dormant (D), after-ripened non-dormant (ND) and after-ripened dormant seeds (control, C). Quantitative real-time polymerase chain reaction (qPCR) was used to confirm gene expression. Comparison between D and ND showed the differential expression of 115 probesets at cut-off values of two-fold change (p<0.05). Comparisons between both D and C with ND in transcript abundance showed that only 13 transcripts, among 115, could be specific to dormancy alleviation. qPCR confirms the expression pattern of these transcripts but without significant variation between conditions. Here we show that sunflower seed dormancy alleviation in the dry state is not related to regulated changes in gene expression.
Introduction
Seeds contribute to the survival and persistence of domesticated and non-domesticated plants in managed and natural ecosystems. Therefore, the completion of germination represents a key process that permits the maintenance of plant species. Seed germination can be modulated by dormancy, which provides a mechanism for delaying this process to survive unfavorable conditions for seedling establishment. Seed dormancy is defined as the failure of viable mature seeds to germinate under apparently favorable conditions [1]. Dormancy can be caused by the embryo itself and/or tissues surrounding the embryo, specified as embryo dormancy and seed coat imposed dormancy, respectively. Dormancy is released during after-ripening (dry storage), which results in changes in physiological status that induce seed germination [1], [2]. After-ripening is particularly an intriguing phenomenon since the physiological changes that allow the shift from non-permissive to permissive status for germination occur at low moisture content (MC), generally <0.10 g H2O/g dry weight (DW) [1], [3]. Under these conditions, water is presumed to be not available for biochemical reactions especially transcription and translation [4]. However, it has been shown that after-ripening trigger gene abundance changes in seeds of Nicotiana tabacum [5], Nicotiana plumbaginifolia [6], barley [7] and wheat [8]. Leubner-Metzger [5] hypothesized that the presence of hydrated pockets within cells of mature dry seeds would allow an active transcription. However, the possibility of hydrated areas within desiccated tissues is subjected to caution and is not supported by major thermodynamic laws of water equilibrium and binding properties in living tissues [9]. Moreover, the molecular mobility that is required for biochemical reactions is dramatically restricted in dry seeds whose cytoplasm displays a visco-elastic solid state, i.e. a glass [10]. Until now, there has been no experimental evidence supporting or rejecting the occurrence of regulated gene expression during alleviation of seed dormancy in anhydrobiosis, mainly because chemical biology of dry living organs is hardly investigable. In order to bring the first lines of evidence to this unsolved biological question, we have investigated the transcriptomic changes that are likely to occur in dormant dry sunflower seeds (MC close to 0.040 g H2O g DW−1) and after their storage under 2 relative humidity (RH) regimes, one allowing dormancy alleviation and the other not (60 and 5 % RH, respectively). Such experimental design allows differentiating between change in gene expression associated with the achievement of germination capacity and those associated with storage only.
Materials and Methods
Plant Material and After-ripening Treatments
Sunflower (Helianthus annuus cv LG5665) seeds produced in the year 2010 in “Drôme” (region of France) were purchased from Limagrain. After harvest, seeds are dormant (D), and they were stored immediately at −23°C until further use in order to maintain their dormancy. Dormant seeds were also stored at 20°C and 60 % RH for two months for after-ripening treatment (ND). Seeds used for storage control (C) were stored for the same duration at 20°C in tightly closed jars over saturated solution of ZnCl2 that gave RH of 5 % [3], [11].
Germination Tests
Germination assays were performed with embryos (i.e. seeds without pericarp) at 10°C in darkness on a layer of cotton wool moistened with deionized water in 9 cm Petri dishes (25 seeds per dish, four replicates). An embryo was considered as germinated when the radicle was elongated by 1 mm. Germination counts were done over a period of 10 days after imbibition.
RNA Extraction
Embryonic axes were isolated from dry seeds (D, ND and C) and were frozen in liquid nitrogen and stored at −80°C until subsequent use. For each sample, 25 axes were ground to a fine powder in liquid nitrogen, and total RNA was extracted by a hot phenol procedure as previously described by Oracz et al. [12], according to Verwoerd et al. [13]. RNA concentration was determined at 260 nm using Nanovue spectrophotometer (GE healthcare).
Affymetrix Array Hybridization
RNA hybridization (two biological and two technical repetitions) was performed using Affymetrix Sunflower Gene WT Chip [14], [15] at the affymetrix platform at INRA-URGV, Evry, France. RNA samples were checked for their integrity on a bioanalyzer (Agilent Technologies, Waldbroon, Germany). One hundred ng of total RNA were used for biotin-labelled cRNA synthesis using Genechip® WT cDNA synthesis and amplification kit (Affymetrix, Santa Clara, CA) following the manufacturer’s instructions. The raw and normalized microarray data are available in the database (AFFY_Dormance_totalRNA_Sunflower, Gagnot et al. [16]) and the Gene Expression Omnibus (GEO) repository of the National Center for Biotechnology Information (NCBI), accession number GSE23046 [17].
Data Analysis
The data were normalized with the Robust Multi-array Average algorithm [18]. Two group t-test was performed to determine differential expression between genes. The raw P values were adjusted by the Bonferroni method, which controls the Family Wise Error Rate [19]. A Bonferroni P-Value less than 0.05 was required for significant variation in gene expression.
Real-time Quantitative RT–PCR
Total RNA (4 µg) was treated with DNase I (Sigma), reverse transcribed with Revertaid Reverse Transcriptase (Fermentas) in a 25 µl reaction volume and amplified with Mastercycler ep Realplex (Eppendorf) using 5 µl of 30-fold diluted cDNA solution. Primers were designed with primer3 software and their respective sequences are shown in Table 1. Real-time PCRs were performed with the MaximaTM SYBR Green qPCR Master Mix (Fermentas) and 0.23 mM of each primer (Fermentas) in a 15 µl reaction. Cycle thresholds (Cts) were calculated using the Realplex 2.0 software (Eppendorf). For each plate and each gene, a standard curve made with dilutions of cDNA pools was used to calculate the reaction efficiencies, and the relative expression was calculated according to Hellemans et al. [20] with HaEF1, Haβ-tubulin [12] and HaS19 as reference genes (Table 1). An arbitrary value of 1 was assigned to dormant seed samples, which were used as control samples for normalization [21]. Results presented are the means ± SD of three biological replicates.
Table 1. Primer sequences of genes used in the present work.
| sunflower chips’s genereference | Heliagene accession number(www.heliagene.org) | Primer sequences |
| Heli060118_st | HuCL00003C004 | Left:AAAAGCCAATGCTTACATAACAA |
| Right: GAACTCGTGATTCAGGGCTAG | ||
| Heli052463_st | HuCL00117C002 | Left:TATCGGGAAAATCGGTGA |
| Right: GAGGGGAAGTGGTGTGTGTT | ||
| Heli058062_st | HuCL21481C001 | Left: CCATAGGAGCCAAAAAGCAC |
| Right: GCATAGCAGTCGGGAAGAAA | ||
| Heli093833_st | HuCX944051 | Left: GTCCCCTTGGGCTATCAACT |
| Right: TAGAGGCGGGAAAAGTAGCA | ||
| Heli022775_st | HuCL12355C001 | Left:TCTCTCGTAAGCAGTCATTTTG |
| Right: CAGGGGAAGTCATAGCCAAT | ||
| Heli059723_st | HuCL11935C002 | Left:TGATGAGCAAATACAGCAATCT |
| Right: GGGTGCGAAGAAGAAACCAT | ||
| Name of H. annnusL. constitutive gene | CGPDB EST accession(http://cgpdb.ucdavis.edu/) | Primer sequences |
| HaEF1 | QH_CA_Contig2764 | Left: TCTCCACTCCTCCAACAC |
| Right: CTCAATCACTCGCTACACC | ||
| Haβ-tubulin | QH_CA_Contig4019 | Left: GGCGTCTACCTTCATTGGT |
| Right: TCCATCTCATCCATTCCTTC | ||
| HaS19 | QHG12B09.yg.ab1 | Left:ACACACTCACCCCCACCAC |
| Right:GGAAAGCACCAACACCAAG |
Results
After-ripening Induced Dormancy Breaking is Dependent on Relative Humidity
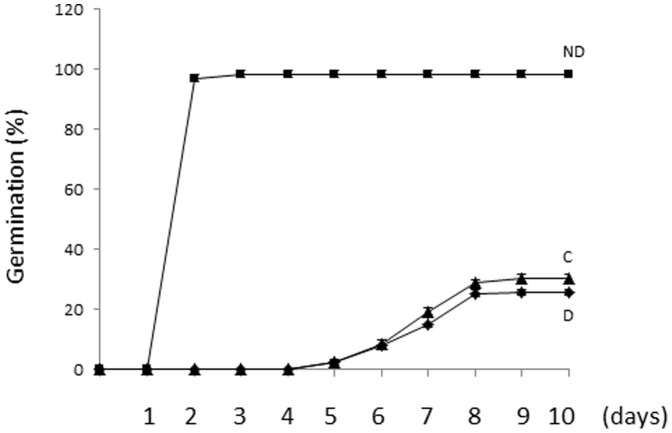
Figure 1 shows that only 25 % of dormant (D) embryos (after harvest) germinated after 10 days (d) of imbibition at 10°C while 96.6 % of embryos after-ripened at 20°C and 60% RH for 2 months germinated within 2 d at the same temperature (non-dormant, ND, Fig. 1). After 2 months of storage at 5 % RH, only 30.3 % of embryos germinated, demonstrating that low MC prevented dormancy release (storage control, C, Fig. 1).
Figure 1. Germination percentage of dormant (D), after-ripened control (C) and non-dormant (ND) seeds.
Small Change in Expression Profile at Dry State
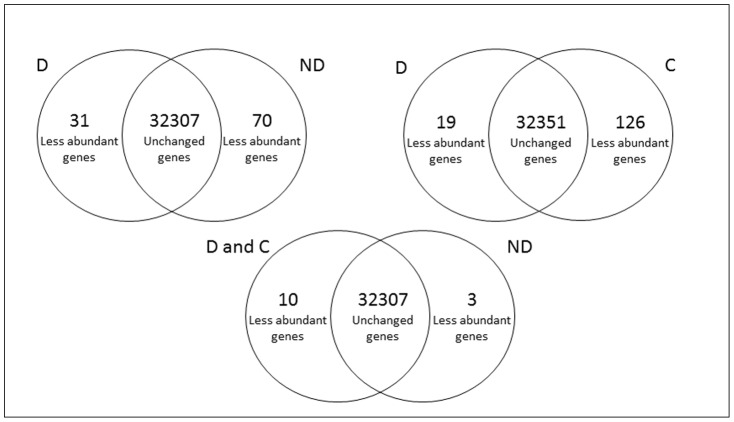
We compared mRNA species in dry dormant with after-ripened non-dormant and control embryos using microarray analysis in order to identify dry storage induced transcriptomic changes related to dormancy alleviation. Almost all transcripts represented on the sunflower Affymetrix GenechipR arrays were detected in the embryos from the different conditions. Comparison between D and ND embryos (see Fig. 2) showed that after-ripening triggered differential abundance of 101 probesets at cut-off values of two-fold change (70 less abundant in ND and 31 less abundant in D, Table S1), which represent a small percentage (0.3 %) of total probeset on the GenechipsR array. Comparison between D and C samples showed differential abundance of 145 genes (126 less abundant in C and 19 less abundant in D, Table S1). These results show that seed storage was associated with variation in gene expression corresponding mainly to a decrease in transcript abundance.
Figure 2. Number of genes with altered expression during after-ripening (≥ 2-fold; p<0.05).
Change in gene transcript abundance between dormant (D), after-ripened control (C) and non-dormant (ND) seeds.
Genes Associated with Dormancy Alleviation
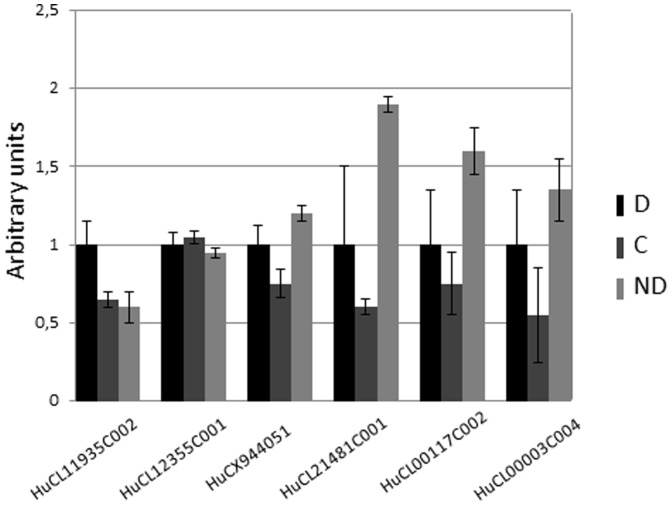
In order to compare changes in gene abundance between dormant (D and C that cannot germinate) and ND (that do germinate), we performed analysis consisting of selected genes that were expressed 2-fold or greater (p<0.05) in both D and C compared with ND. Interestingly, our analysis showed variation of only 13 transcripts, among 115 (Fig. 2). Among the 13 genes with altered transcription, seven show similarity with proteins from other species such as auxin-responsive protein, tify domain containing protein or transport protein particle component (Table 2). Genes whose abundance was altered during after-ripening were studied using real time quantitative PCR. As sequence length of some of the EST did not enable us to design primers for amplification, only 6 of the 13 transcripts were studied. As illustrated in Figure 3, the abundance pattern of these transcripts showed no significant variation between conditions although the pattern was generally confirmed. In fact, HuCL11935C002 was less abundant in ND comparing to D and C but its decrease did not exceed 0.5 fold and the increase in abundance of HuCX944051, HuCL21481C001, HuCL00117C002 and HuCL00003C004 transcripts in ND as compared to D was below 0,5 fold (Fig. 3). For HuCL12355C001, no variation in transcript abundance was observed (Fig. 3). It is important to note that differential abundance of these transcripts was also subtle in the microarray data (Table 2).
Table 2. Genes –more or less abundant (≥ 2-fold; p<0.05) in non-dormant (ND) as compared to dormant (D) or after-ripened control (C) sunflower seeds.
| Sunflower transcript | Arabidopsishomologue AGI | InterPro domain | Log2 (D/ND) | Log2 (ND/C) | |
| Affymetrix chip IDwww.heliagene.org | Reference transcriptome IDwww.heliagene.org/HaT13l | ||||
| HuCL12355C001 | HaT13l000709 | ALDH2B4AT3G48000.1 | Glutamate-5-semialdehyde dehydrogenase;Aldehyde/histidinoldehydrogenase | −1.74 | −1.28 |
| HuCL00003C004 | HaT13l002053 | ASN1AT3G47340.1 | Asparagine synthase, glutamine-hydrolyzingGlutamine amidotransferase,type II | 1.7 | 2.13 |
| HuCD855014 | HaT13l003958 | AT5G54750.1 | Transport protein particle (TRAPP)component; Heme-NO binding | 1.11 | 1.39 |
| HuCX943719 | HaT13l008585 | alpha/beta-Hydrolasessuperfamily proteinAT4G12230.1 | Alpha/beta hydrolase fold-1 | 1.07 | 1.06 |
| HuBU029639 | HaT13l010420 | nd | nd | −1.27 | −1.22 |
| HuCL06132C001 | HaT13l012873 | JAZ6AT1G72450.1 | Tify; CO/COL/TOC1, conserved site | 1.07 | 1.12 |
| HuCL00117C002 | HaT13l014495 | IAA14 (SOLITARY ROOT)AT4G14550.1 | AUX/IAA protein | 1.23 | 1.95 |
| HuCL11935C002 | HaT13l021935 | nd | nd | −1.23 | −1.07 |
| HuBU021987 | HaT13l031074 | TXP2AT5G37478.1 | Xklp2 targeting protein | 1.11 | 1.6 |
| HuCL21481C001 | HaT13l152236 | nd | nd | 1.21 | 1.44 |
| HuCX944051 | HaT13l152236 | nd | nd | 1.14 | 1.69 |
| HuDY906631 | not found | nd | nd | 1.12 | 0.96 |
| HuCX946398 | not found | nd | nd | 1.11 | 1.24 |
nd : not determined.
Figure 3. Real-time quantitative PCR analysis of transcripts altered during dry after-ripening.
Transcript abundance in after-ripened control (C) and non-dormant (ND) seeds relative to that found in dormant (D) embryos to which the value 1 is assigned. HaEF1, Haβ-tubulin [12] and HaS19 were used as reference genes. Bars represent means ± standard deviation of three biological replicates.
Discussion
Sunflower (Helianthus annuuus L.) seed displays both seed coat and embryo dormancy [22]. This study concerns embryo dormancy, and thus all experiments were performed with de-coated (naked) sunflower seeds. During after-ripening, sunflower embryo dormancy alleviates but can be modulated by the RH applied (Fig. 1). It is well established that both temperature and seed MC can alter the rate of dormancy release during after-ripening. Here we demonstrated that at a given temperature, MC determines dormancy release. Embryos stored at 5 or 60 % RH at 20 °C displayed a MC at equilibrium at 0.020 and 0.040 g H2O g DW−1, respectively. It is worth to underline that at such MC, water is considered as being structural and tightly bound on macromolecules [11]. It has been shown that the shift from dormant to non-dormant state occurs within the range of 0.04–0.2 g H2O g DW−1 [23], [24]. Based on sorption isotherm curve, such range of MC corresponds to weakly bound water [24]. These conditions do not allow biochemical reactions such as gene transcription and translation [4]. However, transcript variation has been reported during dry after-ripening [5], [6], [7], [8]. In this paper, we have used an after-ripening control to distinguish between storage associated- and dormancy alleviation specific- in gene transcription variation.
Transcriptomic analysis showed that almost all transcripts represented on the sunflower Affymetrix GenechipR arrays were detected in the embryos from the different conditions. It has been shown that mature seeds contain transcripts representing over 60 % of the genome of other species as Arabidopsis or rice [25], [26]. It is assumed that non dormant seeds contain stored transcripts ready for use upon imbibition to trigger germination [26].
Only very small percentage (0.3 %) of total probesets represented at a detectable level on the genechip array showed change in expression between dry dormant and after-ripened sunflower seeds. This result is in accordance with that of wheat seeds, as only 58 probesets representing 0.1 % of total probesets detected in wheat affymetrix array have been shown to be differentially present in dry D and after-ripened seeds [8]. These results show that dry after-ripening was associated with very small transcript variation, which rises the question of the occurrence of gene regulation. Finch-savage et al. [27] showed that the expression of all genes represented in Arabidopsis Affymetrix microarray of dry dormant and after-ripened non dormant seeds was similar. On the other hand, for different seed species, variation in gene expression between dormant and after-ripened seeds during imbibition represents a higher number, about 10 fold in wheat for example [8]. The latter proportion is in accordance with gene variation in other physiological processes such as during fruit development [28], response to ionizing radiation [29] or bud dormancy transitioning [30].
Interestingly, variation between D, ND and C transcriptome corresponds mainly to a decrease in transcript abundance (Fig. 2). This trend has been shown in all previous studies that reported changes in transcript abundance during dry after-ripening, using microarrays gene chips or cDNA AFLP [6], [7], [8], [27]. However, an after-ripening storage control has never been used to target genes strictly associated with the acquisition of germination potential. The use of a storage control in the array analysis is important in the field of seed dormancy since it allows ruling out the involvement of changes in gene expression during dry after-ripening. In particular, as it was shown here, seed storage per se induces change in transcripts abundance and this change is not related to dormancy alleviation. Consequently, our data suggest that the changes in transcript abundance cannot be related to a putative dormancy alleviation induced gene expression in the dry state. This is in accordance with an early study which reported that genes are not transcribed in vivo in dry seeds [31]. Indeed, when dormant seeds were stored at low RH (C), water is supposed to be tightly associated with macromolecules and forming a monolayer [32]. Such structural water is involved in the maintenance of structural integrity of macromolecules and absolutely not available for biochemical reactions, that excludes any transcriptional activity explaining the apparent change in gene expression. This is in accordance with previous indirect evidence, which showed that below an embryo MC of 0.1 g H2O g−1 DW, sunflower seed dormancy release was associated with negative activation energy corresponding to the occurrence of non-enzymatic reactions rather than metabolic ones [3]. Non-enzymatic reactions, such as lipid peroxidation or the Amadori and Maillard reactions associated with free radical production and oxidation processes are indeed likely to occur during dry storage of seeds. Interestingly, it has been demonstrated that sunflower seed dormancy alleviation is associated with ROS and oxidized products accumulation in embryos, suggesting that ROS might play a major role in dormancy alleviation [33], [34], [14]. Seed stored proteins and mRNA are targets of ROS, since a pool of them became specifically oxidized during after-ripening [34], [14]. It has been shown that mRNA oxidation is associated with dormancy release during after-ripening in sunflower and wheat [14], [35] suggesting cross-species evolutionary conservation of mRNA oxidation as a post-transcriptional seed dormancy-regulating mechanism [35]. Interestingly, mRNA oxidation results in artifacts in cDNA-amplified fragment length polymorphim analysis which may explain apparent expression change reported in dry seeds [14].
Targeted protein and mRNA oxidation and degradation could play a role in the early steps of seed imbibition that govern the process toward germination. Breakdown of specific mRNAs has been implicated in seed dormancy alleviation, and might be a prerequisite to germination [26], [36]. It has been shown that de novo transcription is not essential for early stages of germination [37]. Consequently, the selective degradation of negative regulators of cell signaling might represent the major process involved in dormancy alleviation.
The present data strongly support the idea that regulated variation in gene expression does not occur during after-ripening, in agreement with the absence of metabolic activity in dry seeds, and that the apparent transcript abundance changes observed in these conditions are not related to dormancy alleviation. These data and the ones obtained previously [34], [3], [14], allow us to propose that non-enzymatic ROS production is the key event in loss of dormancy during after-ripening in sunflower seeds.
Supporting Information
List of probsets differentially abundant in dry seeds (≥ 2-fold; p<0.05) between dormant (D) and non-dormant (ND; D vs. ND), after-ripened control (C) and ND (C vs. ND) and D and C (D vs. C) samples.
(XLSX)
Funding Statement
This work has been supported by Ecoseed, KBBE project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bewley JD, Bradford KJ, Hilhorst HWM, Nonogaki H (2013) Seeds: Physiology of Development, Germination and Dormancy, 3rd Edition, Springer, 368–370.
- 2. Finch-Savage WE, Leubner-Metzger G (2006) Seed dormancy and the control of germination. New Phytol 171: 501–523. [DOI] [PubMed] [Google Scholar]
- 3. Bazin J, Batlla D, Dussert S, El-Maarouf-Bouteau H, Bailly C (2011) Role of relative humidity, temperature and water status in dormancy alleviation of sunflower seeds during dry after-ripening. J Exp Bot 62: 627–640.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vertucci CW, Farrant JM (1995) Seed development and germination. Kigel, J. and Galili, G. eds. New York, Marcel Dekker.
- 5. Leubner-Metzger G (2005) beta-1,3-glucanase gene expression in low-hydrated seeds as a mechanism for dormancy release during tobacco after-ripening. Plant J 41: 133–145. [DOI] [PubMed] [Google Scholar]
- 6. Bove J, Lucas P, Godin B, Ogé L, Jullien M, et al. (2005) Gene expression analysis by cDNA-AFLP highlights a set of new signaling networks and translational control during seed dormancy breaking in Nicotiana plumbaginifolia . Plant Mol Biol 57: 593–612. [DOI] [PubMed] [Google Scholar]
- 7. Leymarie J, Bruneaux E, Gibot-Leclerc S, Corbineau F (2007) Identification of transcripts potentially involved in barley seed germination and dormancy using cDNA-AFLP. J Exp Bot 58: 425–437. [DOI] [PubMed] [Google Scholar]
- 8. Gao F, Jordan MC, Ayele BT (2012) Transcriptional programs regulating seed dormancy and its release by after-ripening in common wheat (Triticum aestivum L.). Plant Biotech. J 10: 465–76. [DOI] [PubMed] [Google Scholar]
- 9. Crowe JH, Hoekstra FA, Crowe LM (1992) Anhydrobiosis. Ann Rev Physiol 54: 579–599. [DOI] [PubMed] [Google Scholar]
- 10. Buitink J, Leprince O (2004) Glass formation in plant anhydrobiotes: survival in the dry state. Cryobiol 48: 215–228. [DOI] [PubMed] [Google Scholar]
- 11. Vertucci CW, Roos EE (1993) Theoretical basis for seed storage II. The influence of temperature on optimal moisture level. Seed sci res 3: 201–213. [Google Scholar]
- 12. Oracz K, El-Maarouf-Bouteau H, Bogatek R, Corbineau F, Bailly C (2008) Release of sunflower seed dormancy by cyanide: cross-talk with ethylene signalling pathway. J Exp Bot 59: 2241–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Verwoerd TC, Dekker BMM, Hoekema A (1989) A small-scale procedure for the rapid isolation of plants RNAs. Nucleic Acids Res 17: 2362–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bazin J, Langlade N, Vincourt P, Arribat S, Balzergue S, et al. (2011) Targeted mRNA oxidation regulates sunflower seed dormancy alleviation during dry after-ripening. Plant Cell 23: 2196–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rengel D, Arribat S, Maury P, Martin-Magniette ML, Hourlier T, et al.. (2012) A gene-phenotype network based on genetic variability for drought responses reveals key physiological processes in controlled and natural environments. PLoS One 7, e45249. [DOI] [PMC free article] [PubMed]
- 16. Gagnot S, Tamby JP, Martin-Magniette ML, Bitton F, Taconnat L, et al. (2008) CATdb: a public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res 36: 986–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, et al. (2007) NCBI GEO: mining tens of millions of expression profiles database and tools update. Nucleic Acids Res 35: 760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- 19. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Corbineau F, Bagniol S, Côme D (1990) Sunflower (Helianthus-annuus) seed dormancy and its regulation by ethylene. Isr J Bot 39: 313–325. [Google Scholar]
- 23. Leopold AC, Glenister R, Cohn MA (1988) Relationship between water content and afterripening in red rice. Physiol Plant 74: 659–662. [Google Scholar]
- 24.Probert RJ (2000) The role of temperature in the regulation of seed dormancy and germination. In: Fenner M, ed. Seeds. The ecology of regeneration in plant communities. Oxon: CABI Publishing, 261–292.
- 25. Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709. [DOI] [PubMed] [Google Scholar]
- 26. Howell KA, Narsai R, Carroll A, Ivanova A, Lohse M, et al. (2009) Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol 149: 961–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Finch-Savage WE, Cadman CC, Toorop PE, Lynn JR, Hilhorst HWM (2007) Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J 51: 60–78. [DOI] [PubMed] [Google Scholar]
- 28. Deluc LG, Grimplet J, Wheatley MD, Tillett RL, Quilici DR, et al. (2007) Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics 8: 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim SH, Song M, Lee KJ, Hwang SG, Jang CS, et al. (2012) Genome-wide transcriptome profiling of ROS scavenging and signal transduction pathways in rice (Oryza sativa L.) in response to different types of ionizing radiation. Mol Biol Rep 39: 11231–48. [DOI] [PubMed] [Google Scholar]
- 30. Sreekantan L, Mathiason K, Grimplet J, Schlauch K, Dickerson JA, et al. (2010) Differential floral development and gene expression in grapevines during long and short photoperiods suggests a role for floral genes in dormancy transitioning. Plant Mol Biol 73: 191–205. [DOI] [PubMed] [Google Scholar]
- 31. Comai L, Harada JJ (1990) Transcriptional activities in dry seed nuclei indicate the timing of the transition from embryogeny to germination. Proc Natl Acad Sci USA 87: 2671–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. D’Arcy RL, Watt IC (1970) Analysis of sorption isotherms of non-homogeneous sorbents. Trans. Faraday Soc 66: 1236–1245. [Google Scholar]
- 33. El-Maarouf-Bouteau H, Job C, Job D, Corbineau F, Bailly C (2007) ROS signaling in seed dormancy alleviation. Plant Signal Behav 2: 362–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oracz K, El-Maarouf Bouteau H, Farrant JM, Cooper K, Belghazi M, et al. (2007) ROS production and protein oxidation as a novel mechanism for seed dormancy alleviation. Plant J 50: 452–465. [DOI] [PubMed] [Google Scholar]
- 35. Gao F, Rampitsch C, Chitnis VR, Humphreys GD, Jordan MC, et al. (2013) Integrated analysis of seed proteome and mRNA oxidation reveals distinct post-transcriptional features regulating dormancy in wheat (Triticum aestivum L.). Plant Biotech J 11: 921–32. [DOI] [PubMed] [Google Scholar]
- 36. Xu J, Chua NH (2009) Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell 21: 3270–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, et al. (2004) The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol 134: 1598–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of probsets differentially abundant in dry seeds (≥ 2-fold; p<0.05) between dormant (D) and non-dormant (ND; D vs. ND), after-ripened control (C) and ND (C vs. ND) and D and C (D vs. C) samples.
(XLSX)