Abstract
The NimC1 molecule has been described as a phagocytosis receptor, and is being used as a marker for professional phagocytes, the plasmatocytes, in Drosophila melanogaster. In studies including tumor-biology, developmental biology, and cell mediated immunity, monoclonal antibodies (P1a and P1b) to the NimC1 antigen are used. As we observed that these antibodies did not react with plasmatocytes of several strains and genetic combinations, a molecular analysis was performed on the structure of the nimC1 gene. In these strains we found 2 deletions and an insertion within the nimC1 gene, which may result in the production of a truncated NimC1 protein. The NimC1 positivity was regained by recombining the mutation with a wild-type allele or by using nimC1 mutant lines under heterozygous conditions. By means of these procedures or using the recombined stock, NimC1 can be used as a marker for phagocytic cells in the majority of the possible genetic backgrounds.
Keywords: innate immunity, Drosophila, plasmatocyte, phagocytosis receptor, marker
Introduction
The classification of Drosophila hemocytes initially relied on morphological distinction between hemocyte types. To facilitate the identification of the different effector cell classes, an array of molecular markers was created. One of these markers is NimrodC1 (NimC1), which was identified on the surface of the Drosophila phagocytes (plasmatocytes) by the mixture of two monoclonal antibodies (P1a and P1b).1 Functional assessment of NimC1 in plasmatocytes and S2 cells proved that it plays a role in the phagocytosis of bacteria.1 These results were further underlined by in vitro analysis, which showed the direct binding of NimC1 to fluorescently labeled bacteria.2 Structural investigation of the protein revealed that it contains a set of characteristic EGF-like domains (NIM domains), which can also be found in a set of proteins potentially related to phagocytosis; these proteins constitute the Nimrod protein-family.3
In the past years, NimC1 has become the standard marker for the identification of Drosophila plasmatocytes.4-9 However, it was recently observed that there is a variation in the expression of the NimC1 receptor among Drosophila lines. As NimC1 is broadly used as a marker for plasmatocytes, we performed an analysis involving the nimC1 gene to find an explanation for this phenomenon.
Results and Discussion
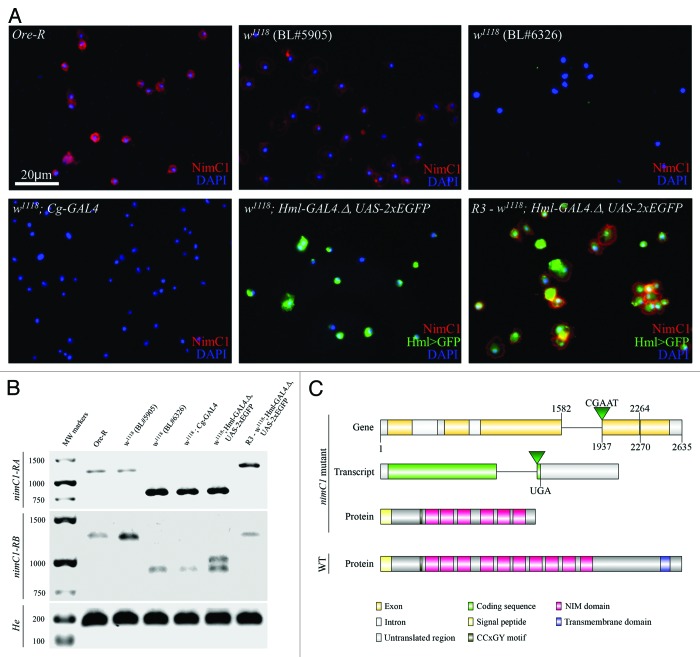
To find out the reason for the lack of the NimC1 antigen in some stocks and genetic combinations (Fig. 1A), we analyzed the expression of the nimC1 gene in NimC1 negative and NimC1 positive strains using RT-PCR. According to the data available on FlyBase (http://flybase.org/), 2 transcripts, nimC1-RA and nimC1-RB are annotated; therefore, we used isoform specific primer pair sets to identify the predicted transcripts. We detected both transcripts in all of the investigated lines, however, in P1 negative lines (w1118 [BL#6326], w1118; Cg-Gal4 [BL#7011] and w1118; Hml-Gal4.Δ, UAS-2XEGFP [BL#30140]) the transcripts were 350 bp shorter than in the P1 positive ones (Oregon-R) and w1118 (BL#5905) (Fig. 1B). Hemese10 specific primers gave the appropriate sized products in each sample.
Figure 1. Expression and sequence analysis of NimC1 (A) NimC1 (P1a/P1b) staining of larval hemocytes (red) in different Drosophila lines. In the case of transgenic lines, GRP expression was amplified with anti-GFP (green) (B) Agarose gel electrophoresis of RT-PCR products gained with primer pairs specific for nimC1-RA (1138 bp or 788 bp), nimC1-RB (1268 bp or 918 bp), and Hemese (175 bp) transcripts. In case of w1118; Hml-Gal4.Δ, UAS-2XEGFP strain, genomic DNA contamination was observed; we detected a 1016 bp band besides the cDNA specific 910 bp fragment. (C) Map of the nimC1 open reading frame, the transcripts and the predicted truncated NimC1 proteins of the P1 negative strains, compared with the wild-type NimC1. Since the size of nimC1-RA and nimC1-RB transcripts differ in only 6 bp, the difference between the isoforms is not indicated. The deleted segments are indicated by their respective start and end positions, connected by a horizontal line.
To reveal the grounds for the difference in size of nimC1 transcripts, we performed a genome-sequence analysis. We found 2 independent deletions in the nimC1 gene of the P1 negative lines. One of them is a 6 bp microdeletion from nucleotides 2264 to 2270 (2L:13974483-2L:13974489), while the other one covers 355 bps from nucleotides 1582 to 1937 (2L:13974817-2L:13975172), accompanied by a 5 bp long insertion (at position 2L:13974817) (Fig. 1C). The latter deletion and the insertion are presumable results of the excision of a mobile element, which are known to leave footprints in the genome after remobilization.
The 355 bp deletion and the 5 bp insertion generate a frameshift mutation in both nimC1 transcripts. As a result of the frameshift, the insertion is followed by a newly arisen sequence of 12 codons and a UGA stop codon (Fig. 1C). These alterations suggest that the mutant NimC1 proteins may be truncated. Instead of the normal 620 and 622 amino acid residues, the mutant NimC1 proteins are composed of only 307 and 309 amino acids in the case of the NimC1-PA and the NimC1-PB, respectively. The truncated NimC1 proteins, therefore, lack their C-terminal regions, including their intracellular and transmembrane domains, as well as four NIM repeats from the extracellular region (Fig. 1C). As the deletions do not affect the N-terminal signal peptide, the truncated proteins of the mutant lines may be secreted to the hemolymph, and in this particular case, resemble the NimB proteins, the secreted members of the Nimrod protein family.1,3 As the NimC1 antigen was not detected in the hemolymph in the tested mutant lines (data not shown), the specific epitopes for the P1a and P1b monoclonal antibodies may be located on the missing protein regions.
In order to assess whether P1 positivity can be regained by recombination, we replaced the mutant nimC1 gene with the wild-type allele of a P1 positive line. We successfully restored the P1 positivity of the w1118; Hml-GAL4.Δ, UAS-2xEGFP12 line by exchanging the chromosomal region in which NimC1 is located (34E5) using black (34D1) as a marker gene for recombination (the resulting R3-w1118; Hml-Gal4.Δ, nimC1+, UAS-2XEGFP stock is available for the scientific community upon request, from VH).
Our results imply that some of the P1 negative lines used in the laboratories are nimC1 mutants, while the majority of the laboratory stocks express NimC1. It should be noted, therefore, that multistep genetic crosses, such as lineage tracing combinations, may results in the introduction of nimC1 mutant chromosomes. For this reason, a rigorous test for NimC1 expression is necessary.
Investigation of hemocyte differentiation requires the detection of NimC1 expression. Basic genetic analysis demonstrated that the occurrence of the P1-negative phenotype is consistent with the presence of a recessive mutation on the second chromosome. Thus, when it is suspected that P1 negativity is the result of the genetic background (as in the case of progeny emerging from GAL4/UAS crosses), it is advisable to check the parental chromosomes for P1 staining. This can be done by test crossing the parental lines to known P1-negative and P1-positive lines, and checking for P1 expression in the trans-heterozygous F1 progeny. Therefore, we recommend using the nimC1 mutants in heterozygous conditions, and if unavoidable, the nimC1 mutant locus can be recombined with the black marker gene to gain a P1 positive line. For a list of confirmed P1 negative stocks, please refer to Table S2.
P1-negative chromosomes harboring FRT elements for mitotic clone generation represent a special case. Mitotic clones are generated to examine the phenotypic effects of homozygous mutation of a given gene. If the P1-negative locus resides on the same arm, then the resulting FRT clones in blood cells (in the lymph gland or circulating hemocytes) will also be P1-negative, while surrounding heterozygous cells, often used as an internal experimental control, will be P1-positive. Without knowing the P1 status of the chromosome beforehand, the P1-negative phenotype may be erroneously attributed to the mutant locus under study. The test crossing mentioned above helps to avoid such interpretational errors.
Functional tests, such as phagocytosis assay, can also be used to monitor plasmatocyte differentiation in P1 negative genetic lines and crosses11; however, NimC1 is still the simplest and one of the most appropriate tools to detect the phagocytic cells in the majority of the possible genetic backgrounds.
Materials and Methods
Drosophila stocks and genetic crosses
Wild-type Oregon-R, 2 w1118strains (NimC1 positive BL#5905 and NimC1 negative BL#6326), w1118; Cg-Gal4 (BL#7011), w1118; Hml-Gal4.Δ, UAS-2XEGFP (BL#30140) and a meiotic mapping line containing the b1 mutation, (a gift from Dr László Sipos, BRC, Szeged, Hungary) were used. The P1 positive R3-w1118; Hml-Gal4.Δ, UAS-2XEGFP line was generated in 2 recombination steps. First, the region containing the b1 marker (located in the 34D1 region) was recombined into the Hml-Gal4.Δ, UAS-2XEGFP chromosome, then it was exchanged with the respective genomic region of a wild-type (b+) chromosome. The flies were kept on a standard cornmeal-yeast diet at 25°C.
Isolation of RNA and cDNA synthesis
The RNA was isolated from third instar larvae using TRIzol Reagent (Invitrogen). The contaminating DNA was eliminated by DNase I treatment (Fermentas): 1 µg RNA was treated with 1 u DNase I and then reverse transcribed using RevertAid First Strand cDNA Synthesis Kit (Fermentas).
Analysis of nimC1 expression
The primers used for the analysis are described in Table S1. To detect the transcription of nimC1, we performed reverse transcription polymerase chain reactions (RT-PCR) with isoform-specific primers using 1 µl undiluted cDNA as template. The P1A2 forward and the P1del.utanA,B reverse primers were used to amplify a 1138 bp fragment of the nimC1-RA transcript. Samples eventually contaminated with genomic DNA would result in the synthesis of a 1735 bp PCR fragment. As the nimC1 specific forward primer is inappropriate to amplify the nimC1-RB transcript the P1isoBfw forward primer was used in combination with the P1del.utanA,B reverse primer. This primer pair amplifies a 1268 bp fragment in case if cDNA and a 1365 bp fragment if genomic DNA is used as template. As a positive control, the Hemese10 specific Hemesefw forward and Hemeserev reverse primers were used, which generated a 175 bp fragment. In case of genomic DNA contamination, a 354 bp PCR fragment was expected.
Immunostaining and hemocyte imaging
Larvae were dissected in 20 µl of Shields and Sang’s medium (Sigma) on a multispot microscope slide (SM-011, Hendley); the released hemocytes were allowed to settle and adhere at room temperature for 45 min. The samples were fixed with paraformaldehyde (2% paraformaldehyde in PBS, 12 min), washed and blocked for 15 min with PBS containing 0.1% BSA (Sigma), incubated with the respective monoclonal antibodies for 1 h at room temperature, washed three times with PBS and reacted with anti-mouse Alexa Fluor 568 (Sigma) immunoglobulin at 1:1000 dilution for 1 h. The GFP signal was enhanced with rabbit anti-GFP antibody (Sigma, 1:2000 dilution) in combination with Alexa Fluor 488-conjugated anti-rabbit immunoglobulin (Sigma, 1:1000 dilution). Nuclei were stained with DAPI (Sigma). The microscopic analysis was performed with a Zeiss Axioskope 2MOT epifluorescent microscope.
Genome sequence analysis
Genomic DNA was isolated from female adult flies using the GenEluteTM Mammalian Genomic DNA Miniprep Kit (Sigma). For each reaction, 100 ng DNA was used as template. To amplify a 752 bp long fragment including the 5′ end of the gene and a short intergenic region, we used the P11exonF and P11exonR primers. Proceeding toward the 3′ end of the nimC1 gene the following primer sets were used: P1isoBfw-P1B600rev (which amplify a 579 bp fragment), P11189fw-P11189rev (which amplify a 1254 bp fragment), and P1utan600fw-P1utan300rev to amplify the 3′ end of the gene and a short intergenic region (yielding a 593 bp fragment). The sequences of the primers are indicated in Table S1. Sequence analysis of the obtained PCR fragments was performed using BigDye Terminator v3.1 Cycle Sequencing Kit (Invitrogen) and a 3500-Genetic Analyzer (Applied Biosystems).
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Olga Kovalcsik, Szilvia Tápai, and Anita Balázs for technical help. We acknowledge the constructive criticism of the two unnamed referees. This research was supported by grants from the Hungarian Science Foundation (OTKA grant NK 101730), TÁMOP 4.2.2.A-11/1KONV-2012-0035 (IA), a National Institutes of Health grant R01HL67395 (UB), and GCs had a Junior Scientist Fellowship from the Hungarian Academy of Sciences.
Supplemental Materials
Supplemental materials may be found here: http://www.landesbioscience.com/journals/fly/article/25654
Footnotes
Previously published online: www.landesbioscience.com/journals/fly/article/25654
References
- 1.Kurucz E, Márkus R, Zsámboki J, Folkl-Medzihradszky K, Darula Z, Vilmos P, et al. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol. 2007;17:649–54. doi: 10.1016/j.cub.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 2.Zsámboki J, Csordás G, Honti V, Pintér L, Bajusz I, Galgóczy L, et al. Drosophila Nimrod proteins bind bacteria. Cent. Eur. J. Biol. 2013;7:633–45. doi: 10.2478/s11535-013-0183-4. [DOI] [Google Scholar]
- 3.Somogyi K, Sipos B, Pénzes Z, Kurucz E, Zsámboki J, Hultmark D, et al. Evolution of genes and repeats in the Nimrod superfamily. Mol Biol Evol. 2008;25:2337–47. doi: 10.1093/molbev/msn180. [DOI] [PubMed] [Google Scholar]
- 4.Mandal L, Martinez-Agosto JA, Evans CJ, Hartenstein V, Banerjee UA. A Hedgehog- and Antennapedia-dependent niche maintains Drosophila haematopoietic precursors. Nature. 2007;446:320–4. doi: 10.1038/nature05585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinenko SA, Hung T, Moroz T, Tran QM, Sidhu S, Cheney MD, et al. Genetic manipulation of AML1-ETO-induced expansion of hematopoietic precursors in a Drosophila model. Blood. 2010;116:4612–20. doi: 10.1182/blood-2010-03-276998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makki R, Meister M, Pennetier D, Ubeda JM, Braun A, Daburon V, et al. A short receptor downregulates JAK/STAT signalling to control the Drosophila cellular immune response. PLoS Biol. 2010;8:e1000441. doi: 10.1371/journal.pbio.1000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cordero JB, Macagno JP, Stefanatos RK, Strathdee KE, Cagan RL, Vidal M. Oncogenic Ras diverts a host TNF tumor suppressor activity into tumor promoter. Dev Cell. 2010;18:999–1011. doi: 10.1016/j.devcel.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tokusumi T, Tokusumi Y, Hopkins DW, Shoue DA, Corona L, Schulz RA. Germ line differentiation factor Bag of Marbles is a regulator of hematopoietic progenitor maintenance during Drosophila hematopoiesis. Development. 2011;138:3879–84. doi: 10.1242/dev.069336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Márkus R, Laurinyecz B, Kurucz É, Honti V, Bajusz I, Sipos B, et al. Sessile hemocytes as a hematopoietic compartment in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2009;106:4805–9. doi: 10.1073/pnas.0801766106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurucz E, Zettervall CJ, Sinka R, Vilmos P, Pivarcsi A, Ekengren S, et al. Hemese, a hemocyte-specific transmembrane protein, affects the cellular immune response in Drosophila. Proc Natl Acad Sci U S A. 2003;100:2622–7. doi: 10.1073/pnas.0436940100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honti V, Csordás G, Márkus R, Kurucz E, Jankovics F, Andó I. Cell lineage tracing reveals the plasticity of the hemocyte lineages and of the hematopoietic compartments in Drosophila melanogaster. Mol Immunol. 2010;47:1997–2004. doi: 10.1016/j.molimm.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 12.Sinenko SA, Mathey-Prevot B. Increased expression of Drosophila tetraspanin, Tsp68C, suppresses the abnormal proliferation of ytr-deficient and Ras/Raf-activated hemocytes. Oncogene. 2004;23:9120–8. doi: 10.1038/sj.onc.1208156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.