Abstract
Ubiquitination has emerged as an essential signaling mechanism in eukaryotes. Deubiquitinases (DUBs) counteract the activities of the ubiquitination machinery and provide another level of control in cellular ubiquitination. Not surprisingly, DUBs are subjected to stringent regulations. Besides regulation by the noncatalytic domains present in the DUB sequences, DUB-interacting proteins are increasingly realized as essential regulators for DUB activity and function. This review focuses on DUBs that are associated with WD40-repeat proteins. Many human ubiquitin-specific proteases (USPs) were found to interact with WD40-repeat proteins, but little is known as to how this interaction regulates the activity and function of USPs. In recent years, significant progress has been made in understanding a prototypical WD40-repeat protein-containing DUB complex that comprises USP1 and USP1-associated factor 1 (UAF1). It has been shown that UAF1 activates USP1 through a potential active-site modulation, and the complex formation between USP1 and UAF1 is regulated by serine phosphorylation. Recently, human USPs have been recognized as a promising target class for inhibitor discovery. Small molecule inhibitors targeting several human USPs have been reported. USP1 is involved in two major DNA damage response pathways, DNA translesion synthesis and the Fanconi anemia pathway. Inhibiting the USP1/UAF1 deubiquitinase complex represents a new strategy to potentiate cancer cells to DNA- crosslinking agents and to overcome resistance that has plagued clinical cancer chemotherapy. The progress in inhibitor discovery against USPs and the WD40-repeat protein-containing USP complex will be discussed.
The Ubiquitin System
Ubiquitin, a 76 amino acid protein, has emerged as a critical signaling protein in many eukaryotic cellular processes. Arguably the best known function of ubiquitin is the proteasome-mediated protein degradation signaled by K48-linked polyubiquitin chain. However, in recent years the non-degradative roles of ubiquitin have emerged as essential components for cell signaling in a diverse array of cellular processes including DNA damage response, chromatin remodeling, cell cycle regulation, and kinase signaling(1).
Ubiquitination is mediated by an enzyme cascade consisting of E1, E2, and E3. E1 is the ubiquitin-activating enzyme that activates ubiquitin by forming a thioester bond between the C terminus of ubiquitin and the active site cysteine of E1. E2 is the ubiquitin-conjugating enzyme that receives the activated ubiquitin from E1. E3, the ubiquitin ligase, mediates the ubiquitination of a target protein at a lysine residue (Figure 1) (2-4). Two E1s, close to 40 E2s and more than 600 E3s have been identified in humans(1, 5). Ubiquitin contains seven lysine residues, i.e. K6, K11, K27, K29, K33, K48, and K63 (Figure 2A). These lysine residues together with the ubiquitin N-terminal methionine can be further ubiquitinated to form polyubiquitin chains (Figure 2B) (6-8), and recent studies also uncovered branched ubiquitin chains(7, 9, 10). These different chain linkages are believed to elicit specific cellular responses. Among them, the role of K48-linked polyubiquitin chain is by far the best studied. Our knowledge on other ubiquitin chain linkages has expanded quickly in recent years. The K63-linked ubiquitin chain is known to signal protein trafficking, endocytosis, inflammation response, and DNA repair. Chain linkages that involve K48 and K11 usually signal for protein degradation(11). Linkages through K6, K27, K29, and K33 are still poorly defined. Investigations on the non-K48-linked polyubiquitin chain and the branched ubiquitin chain are needed for a full understanding of ubiquitin's function in cell biology.
Figure 1.

The ubiquitin cycle. Ubiquitin (Ub) is activated by the ubiquitin-activating enzyme (E1) and forms a thioester linkage with the E1 active site cysteine residue in an ATP-dependent manner. The ubiquitin is then transferred to the ubiquitin-conjugating enzyme (E2). E2 with the ubiquitin ligase (E3) targets the substrate and attaches Ub to the substrate's Lys residue forming an isopeptide bond. Additional Ub can be attached to the same ubiquitin on the ubiquitin by the E1-E2-E3 cascade. Deubiquitinating enzymes (DUBs) are involved in the reverse process in editing or removing Ub from target protein.
Figure 2.

The structures of ubiquitin and polyubiquitin. A. The ubiquitin structure displays the seven lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) involved in forming polyubiquitin chains. B. Structures (depicted as surface) of monoubiquitin (PDB ID: 1UBQ), diubiquitin (PDB IDs: Lys6-linked diubiquitin, 2XK5; Lys11-linked diubiquitin, 2XEW; Lys48-linked diubiquitin, 3M3J; Lys63-linked diubiquitin, 2JF5), and tetraubiquitin (PDB IDs: Lys48-linked tetraubiquitin, 2O6V; Lys63-linked tetraubiquitin, 3HM3).
Classification of deubiquitinating enzymes
Ubiquitin modification is a reversible process that is mediated by a set of proteins collectively known as deubiquitinating enzymes or deubiquitinases (DUBs)(12, 13). DUBs play important roles in the ubiquitin system. They function to remove the ubiquitin moiety from mono- and polyubiquitinated proteins. They can edit the ubiquitin chain length and structure in conjunction with ubiquitin ligase(14). DUBs also rescue ubiquitin from proteins that are targeted for degradation by the proteasome. Thus, deubiquitination of proteins by DUBs allows for proper regulation of biological and cellular functions in the cell. Genetic deficiencies in DUBs have been associated with human diseases including cancer and neurodegeneration(12, 15, 16).
The human genome encodes close to 100 DUBs. DUBs are grouped into six subclasses according to their domain structure and sequence similarity (Figure 3). The six subclasses are: ubiquitin-specific proteases (USP), the ubiquitin C-terminal hydrolases (UCH), Otubain proteases (OTU), Machado-Joseph disease proteases (MJD), JAB1/MPN/Mov34 metalloenzymes (JAMM), and monocyte chemotactic protein-induced protein (MCPIP)(13, 15). Among them USPs, UCHs, OTUs, MJDs, and MCPIPs are cysteine proteases that contain an active site cysteine in the catalytic core. JAMMs are zinc-dependent metalloproteases(13, 17, 18). Of the six families of DUBs, the USP family is the largest with more than 50 members. UCH and MJD are smaller families with four and five members respectively(13).
Figure 3.
Classification of DUBs and the representative DUB structures. DUBs are grouped into six subclasses: Ubiquitin-Specific Proteases (USP), Machado-Joseph Domain (MJD), JAB1/MPN/Mov34 Metalloenzyme (JAMM), Ovarian tumor protease (OTU), Ubiquitin C-Terminal Hydrolase (UCH), and Monocyte Chemotactic Protein-Induced Protein (MCPIP). USPs encompass the majority of DUBs with over 50 members. USP, MJD, OTU, UCH, and MCPIP are cysteine proteases, JAMM is zinc-dependent metalloenzyme. Structures of the representative DUBs from each of the six families are shown. The catalytic triad residues are shown as spheres, and the active site Zn2+ in the JAMM structure is shown as a grey sphere. (PDB IDs: USP7, 1NBF; Ataxin-3, 1YZB; OTU2, 1TFF; UCH-L3, 1XD3; MCPIP1, 3V33; JAMM, 1R5X)
Regulation of DUBs by post-translational modification
Given their critical function, DUBs are heavily regulated in cells. In recent years it was found that many of the DUBs undergo post-translational modifications including phosphorylation, ubiquitination, and SUMOylation(19). An example is CYLD which suppress the ubiquitination of TNF receptor–associated factor 2 (TRAF2). IκB kinase (IKK), with regulatory subunit IKKγ, phosphorylates CYLD and results in the loss of its deubiquitinating function towards TRAF2(20). Phosphorylation of CYLD serves as a mechanism that inactivates its DUB activity. Besides phosphorylation, ubiquitination and SUMOylation have also been found in DUBs. Monoubiquitination of UCH-L1, a highly expressed DUB that is linked to Parkinson's disease, at a lysine residue near the active site prevents ubiquitin binding thus halting its DUB activity(21). USP25 was found to be SUMOylated within its ubiquitin interaction motif. This SUMOylation on USP25 impairs the binding and hydrolysis of target ubiquitin chains(22). Conversely post-translational modifications of DUBs have also been shown to activate DUB activity. For Ataxin-3 and DUBA, ubiquitination and phosphorylation were found to enhance the catalytic activity of both DUBs and allowed for the removal of the ubiquitin from their respective substrates(23, 24). Phosphorylation of USP1 has been shown as a key regulator in its interaction with the protein partner USP1-associated factor 1 (UAF1)(25). UAF1 is required for the activation of USP1. Three phosphoserine residues, i.e. Ser42, Ser67 and Ser313, have been reported in USP1(26-29). Phosphorylation of Ser313 in USP1 has been shown to be required for its proper interaction with UAF1(25). Mutation of Ser313 to Ala on USP1 abolished the complex formation between USP1 and UAF1, thus prevented the activation of USP1 by UAF1(25).
WD40 domain in the deubiquitinase complex
DUBs have been found to interact with a large group of cellular proteins. A recent proteomic analysis has identified over 770 proteins associated with 75 DUBs analyzed(30). Remarkably, a large number of USPs were found to be associated with WD40-repeat proteins (Figure 4). Some USPs interact with more than one WD40-repeat protein (e.g. USP7, USP12 and USP46) and some WD40-repeat proteins (such as WDR48 or UAF1) are able to bind multiple USPs (Figure 4). The WD40 domain is one of the most abundant domain structures in eukaryotes (31). The first WD40 domain was identified in bovine β-transducin, a subunit of the trimeric G protein transducin complex. Crystal structure of the β-transducin WD40 domain revealed a seven-bladed β-propeller fold with a four-stranded anti-parallel β-sheet in each blade(32, 33). The name WD40 was derived from the conserved tryptophan-aspartate (WD) dipeptide repeat at the end of strand C (Figure 5A) (33, 34). These antiparallel sheets form a strong hydrogen bond network that stabilizes the β-propeller structure. In many WD40-repeat proteins, the last blade is formed by the three β-strands in the C-terminus and the outer most β-strand is donated by the N-terminus of the WD40 domain (Figure 5B). This Velcro-like closure contributes to the stability of the fold. Not all WD40-repeat proteins contain such a Velcro closure. In DDB1 each individual β-propeller is stabilized by the intersheet hydrophobic interactions. Most WD40-repeat proteins contain 7 to 8 β-propeller blades while a range from 4 to 8 blades has been observed(35). It was predicted that the most ideal geometry for the WD40 domain is a 7-bladed β-propeller(36).
Figure 4.

Known interactions between USPs and WD40-repeat proteins in humans. Among the USPs, several are able to interact with multiple WD40-repeats. Several WD40-repeats can interact with more than one USPs.
Figure 5.
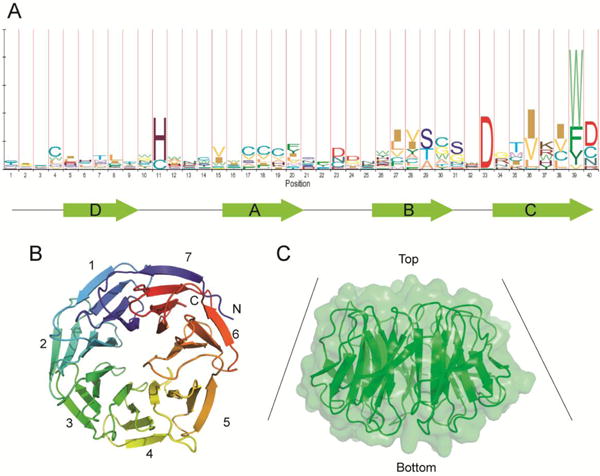
Structure and sequence of WD40-repeat domain. A. Sequence logo of WD40-repeats created using HMMlogo (144). WD40-repeat domains of know structures are used to create a sequence alignment. The letter plot represents conserved amino acid residues at each position, with larger letters representing more conservation. The β-strands within a repeat are depicted below the sequence. B. Structure of a representative WD40-repeat protein (WDR5, PDB ID: 2H14). Each repeat contains four antiparallel β-strands. The seventh blade is constructed by the first β-strand in the N-terminal end and three β-strands at the C-terminal end. C. Surface view of a WD40-repeat domain. WD40 repeats typically have a funnel-like shape with a narrow end defined as the top, and a wider part defined as the bottom.
The structure of WD40 domain is unique in that it can interact with many proteins using several surfaces of the β-propeller structure. The surfaces include the “top” surface conventionally defined as the narrow end of the funnel-shaped structure (Figure 5C). This end primarily comprises loop regions that connect strand D and A in each blade. WD40 domain also contains a wider “bottom” surface, and “side” or “circumference” in the propeller structure. The loop regions in the top and bottom surfaces allow for a rich functional diversity in protein-protein interactions. The side of WD40 domain provides more interaction sites with proteins and peptides(31, 37).
WD40 domains in the ubiquitin pathway
WD40 domains are involved in many cellular functions that include signal transduction, vesicular trafficking, cell cycle control, chromatin dynamics, and DNA damage response(31, 38, 39). The WD40 domains are found in many proteins and protein complexes that are involved in protein-protein interactions, especially those involved in scaffolding, assembly, and regulation of protein complexes.
The WD40 domain also play an important role in the ubiquitin-proteasome system. Many ubiquitin ligases are known to contain a WD40 domain. In the SCF (Skp, Cullin, and F-box protein) ubiquitin-ligase complex, the WD40 domain is mainly found in the protein binding domain, the F-box protein(40). The SCF ubiquitin ligase β-TrCP1 contains a WD40 domain that forms a seven-bladed β-propeller and binds to the protein substrate β-catenin, a part of the Wnt signaling pathway(41-43). Structural determination showed that β-catenin binds to the top face of the β-propeller. The interaction is held together by extensive hydrogen binding and electrostatic interactions between β-catenin and the WD40 domain (44). The CUL4-DDB1-ROC1 ubiquitin ligase, which is involved in cell-cycle regulation, DNA replication, and damage response, interacts with multiple WD40-repeat proteins, including WDR5, L2DTL, and ESC(45). The WDR5 and ESC are known to be involved in histone methylation, and L2DTL helps to regulate CDT1 proteolysis after DNA damage through CUL4-DDB1 interaction(46-48).
Interestingly some WD40-repeats also bind to ubiquitin. The yeast protein Doa1 contains a seven-bladed WD40 domain encompassing 300 amino acids in the N-terminal region. The top surface of this WD40 domain interacts with the β sheets of ubiquitin. A patch on the top surface of Doa1's WD40 domain containing the residues D15, F222, W265, and D281 was identified to interact with ubiquitin. Mutation in these residues abolished Doa1's function in vivo. A large patch on ubiquitin containing the residues L8, R42, I44, V70, and R72 were observed to form interactions with the WD40 domain. Mutational studies on the surface patch of ubiquitin on residues L8, R42 and I44 reduced the binding to Doa1 dramatically(49).
Cellular functions of WD40-associated USPs
USPs regulate a broad rang of cellular processes including protein quality control, trafficking, transcription, RNA processing, DNA damage response, and protein degradation (30, 50-52). Many of the functions have been identified through siRNA screening, topology classification, subcellular localization, and enzymatic activity studies. Notably, the global proteomic analysis has identified DUB-associated proteins and elucidated their potential biological functions(30). Over twenty WD40-associated USPs exist in humans and their cellular functions have been linked to a wide range of cellular processes (Table 1)(53-91).
Table 1. Known functions of the WD40-repeat protein associated USPs.
| USP | WD40 | Funtions | Reference |
|---|---|---|---|
| USP1 | WDR48 (UAF1) | DNA damage response through TLS, FA pathway, and homologous recombination. Preserves stem cell state. | 53-56, 92, 102, 104, 116-118 |
| USP3 | WDTC1 | Regulates the H2A and H2B ubiquitination and involved in the response to DNA double-strand break. | 57 |
| USP4 | PRPF4 | mRNA splicing and Wnt signalling pathway. | 58, 59 |
| USP7 | BUB3, WDR21A, RAE1 | Regulates polycomb complex and factors associated with transcription, including p53 and Mdm2. Scilences the homeotic genes in Drosophila. | 60-67 |
| USP11 | BUB3, RAE1 | Participates in DNA damage repair by regulating DNA-associated proteins ncluding BRCA2, p53 and IkK. Regulates Polycomb complex. | 67, 68 |
| USP12 | WDR20, WDR48 (UAF1). DMWD | Deubiquitinates both H2A and H2B and involved in xenopus embryonic development through regulation of histone ubiquitination. Negatively regulates Notch signaling pathway. | 69, 70 |
| USP15 | PRPF4, CSTF1 | Participates in the regulation of Wnt and TGF-β signalling pathways. | 72, 91 |
| USP19 | DMWD | A membrane-anchored DUB and involved in the turnover of ERAD substrate. | 73 |
| USP22 | TAF5L | A component of the TFTC/STAGA. Deubiquitinates both H2A and H2B and participates in the activation of androgen receptor gene. | 74-76 |
| USP36 | WDR3, WDR36 | Controls p62-dependent selective autophagy activation. Might control the stability of RNA Pol I and regulate the production of rRNA. | 77, 78 |
| USP37 | BTRC, FBXW11 | Regulates oncogenic fusion protein PLZF/RARA stability and cell transformation. | 79 |
| USP39 | WDR57, PRPF4 | Functions in mRNA processing and essential for mototic spindle chekpoint integrity. | 80 |
| USP42 | WDR18 | Regulates p53-dependent transcription and p53-dependent cell-cycle arrest. USP42, fused to RUNX1, is involved in pathogenesis of acute leukemia | 81-83 |
| USP43 | WDR3 | - | |
| USP44 | TBL2 | Regulates stem cell differentiation through deubiquitinating H2Bub1. A key regulator of mitotic checkpoint. | 84, 85 |
| USP45 | CORO1C | - | |
| USP46 | WDR20, WDR48 (UAF1), DMWD | Deubiquitinates both H2A and H2B. Plays a role in GABAergic signaling and nervous system development. | 70, 71, 86, 87 |
| USP47 | BTRC, FBXW11 | Regulates base excision repair by deubiquitinating Pol ß. As a ß-TRCP interactor, regulates cell growth and survival. | 88, 89 |
| USP49 | COPA | - | |
| USP50 | PRPF4 | Be involved in G2/M checkpoint and regulates Wee1 stability through HSP9-dependent mechanism. | 90 |
As a prototypical USP/WD40 complex, USP1/UAF1 has been subjected to extensive studies. Herein, we review the regulation, catalysis, and inhibition of the USP1/UAF1 complex. It should be noted that besides USP1, UAF1 also interacts with two other human USPs, i.e. USP12 and USP46, although less is known about these two USP/UAF1 complexes(92, 93).
The biological functions of USP1/UAF1 complex
USP1/UAF1 plays important roles in human DNA damage response. By screening a library of 220 shRNAs targeting 55 human DUBs, USP1 was identified as a regulatory DUB in the Fanconi anemia (FA) pathway by deubiquitinating Fanconi anemia complementation group D2 (FANCD2) (53). Later studies showed that USP1 also deubiquitinates proliferating cell nuclear antigen (PCNA) (54) and Fanconi anemia complementation group I (FANCI) (94, 95). Normal DNA damage response requires both monoubiquitination and deubiquitination of PCNA and FANCD2/FANCI, which are implicated in DNA translesion synthesis and the FA pathway, respectively.
FA is a genetic disorder that prevents the repair of DNA interstrand crosslink (ICL) and predisposes the patients to cancer(96). 15 FA proteins have been identified in the regulation of the FA pathway. Eight of the FA proteins (FANCA/B/C/E/F/G/L/M) form a FA core complex that monoubiquitinates FANCD2 and FANCI(94, 95, 97). As an essential step in the DNA damage response, FANCD2 and FANCI are monoubiquitinated at Lys561 and Lys523, respectively, and then directed to the nuclear DNA damage foci, where the ubiquitinated FANCD2/FANCI complex binds to BRCA1 and RAD51 recombinase and interacts with the downstream FA proteins (FANCD1, FANCN, FANCJ) (98-100). The monoubiquitinated FANCD2/FANCI complex likely serves as a landing pad on DNA for multiple DNA nucleases such as FA- associated nuclease 1 (FAN1) and FA complementation group P/SLX4 protein that function in ICL repair(101, 102). To complete the DNA damage repair, timely deubiquitination of the modified FANCD2/FANCI is required(101). USP1 was identified as the DUB responsible for deubiquitinating FANCD2 from its monoubiquitinated form(53). It was found that knockdown of USP1 resulted in the increased level of monoubiquitinated FANCD2. USP1 is also predominantly localized to the chromatin in both mitomycin C (MMC)-treated and untreated HEK293T cells, suggesting a functional role of USP1 in ICL repair(53, 103). Together, these evidences support the notion that deubiquitination of FANCD2 and FANCI is important for the normal response to ICL through the FA pathway.
Translesion synthesis (TLS) is a DNA damage tolerance process, which allows DNA replication past a variety of DNA lesions caused by both endogenous and exogenous factors. Due to the error-prone nature of TLS, it is tightly regulated to avoid unwanted mutation. Ubiquitination of PCNA plays an important role in regulating TLS(104-107). When the progression of the replication fork is blocked by a lesion, PCNA undergoes monoubiquitination by RAD6 and RAD18, which allows for the recruitment of the specialized DNA polymerases to carry out lesion-bypass DNA synthesis(107-109). Equally important to the recruitment of TLS polymerases to the DNA damage site is the timely removal of TLS polymerases following the lesion-bypass synthesis. This control ensures that normal DNA replication is restored following the usually error-prone TLS to avoid excessive mutations caused by the extensive DNA synthesis carried out by the TLS polymerases. This notion is supported by an in vitro experiment showing in a reconstituted yeast TLS system that removal of ubiquitin from PCNA is required for the reverse polymerase switch between Polη and Polδ(107). Furthermore, studies in cells also pointed out that deubiquitination of PCNA following replication past the lesion can initiate the displacement of TLS polymerases by normal DNA polymerases(54, 110-114). Deubiquitination of PCNA by USP1 can also limit the access of TLS Polκ to the replication fork and ensure the low mutation frequency of DNA replication(115).
USP1 has been found to be essential for the human DNA damage response. Initially by analyzing the chromosomal breakage in the metaphase of cells treated with siRNA-targeting USP1 in combination with MMC, it was found that the number of chromosomal aberrations per cell decreased by 50 % compared to the cells treated with MMC alone. It was thus suggested that inhibition of USP1 could protect cells from DNA damage reagent, such as MMC(53). However, later works demonstrated an opposite effect of USP1 deficiency. Both disruption of USP1 in chicken DT40 cells(103) and knockout of the murine USP1 gene(116) resulted in increased sensitivity to DNA cross-linkers and chromosome instability that resembled a FA phenotype. Inhibition of USP1/UAF1 by small molecule inhibitors also sensitized non-small cell lung cancer (NSCLC) cells to cisplatin(117), providing further support of the indispensible role of USP1/UAF1 in proper DNA damage response. Moreover, evidence showed that loss of USP1 leads to aberrant recruitment of Polκ to replication folk, resulting in slower speed of replication fork progression and more micronuclei formation, a marker for genomic instability(115). However, to date the exact role of USP1 in the FA pathway and TLS remain to be fully elucidated. Further studies will be needed to uncover the molecular basis of the sensitivity to DNA damage in the absence of USP1 in human cells.
The role of UAF1 in DNA damage response
UAF1 is a WD40 domain-containing protein, which is required for the proper function of USP1. Several studies showed that UAF1 forms a complex with USP1 and stimulates the deubiquitination activity of USP1 both in vitro and in vivo(55, 117-119). Knockout of USP1 alone, UAF1 alone, or USP1 and UAF1, resulted in comparable level of increased monoubiquitinated FANCD2 and PCNA, as well as similar sensitivity to DNA cross-linking agent MMC, suggesting an epistatic relationship between USP1 and UAF1(55). The association of USP1 and UAF1 in cells is likely regulated in response to DNA damage. UV-irradiation induces autocleavage of USP1, and the resulting fragments can be bridged by UAF1 to form an enzyme complex(118). Recent work showed that phosphorylation of USP1 by kinase CDK1 promotes the complex formation between USP1 and UAF1(119). USP1 is likely activated by the interaction with UAF1 under genotoxic stress through USP1 Ser313 phosphorylation-mediated complex formation(29, 119).
The role of USP1 in the preservation of stem cell state
Recently, inhibitor of DNA-binding (ID) proteins were identified as substrates of USP1(56). IDs inhibit differentiation of stem cells by antagonizing basic-helix-loop-helix (bHLH) transcription factors. It has been shown that IDs are short lived in most tissues because of proteasome-mediated degradation(120). Deubiquitination of IDs by USP1 promotes ID protein stability and prevents stem cell differentiation (56). In a subset of primary osteosarcoma tumors, USP1 and ID2 were found coordinately overexpressed. USP1 was shown to promote oncogenic transformation in NIH 3T3 cells and in a xenograft mouse model. Thus, USP1 was proposed to belong to a family of ‘caulo-oncogenes’ that promote tumorigenesis by subverting normal stem cell biology(56).
The biological function of other USP/UAF1 complexes
Two other human USPs, i.e. USP12 and USP46, also interact with UAF1. A strong homology has been identified between USP12 and USP46 in humans. Recent studies suggest that USP12 and USP46 regulate deubiquitination of histone proteins H2A and H2B(70). Histone deubiquitination in the mesoderm by USP12, but not USP46, regulates Xenopus development during the gastrula stage(70). In addition, USP12 was identified as the negative regulator of Notch signaling by regulating the quantity of full-length Notch at the cell surface(69). USP46 has also been implicated in the degradation of glutamate receptor GLR-1 in C. elegans (71), and pre- and post-synaptic GABAergic signaling in mice(86, 87, 121).
USPs are subjected to regulation through active site modulation
USPs usually contain multiple domains including a conserved catalytic core domain. Sequence analysis in conjunction with structure determination has revealed a diverse array of domains either flanking the catalytic core domain or being inserted within the catalytic core(13, 122, 123). USPs contain the conserved finger, thumb, and palm subdomains as revealed by the crystal structures of several USP catalytic core(122, 124-128) (Figure 6A). The extended finger domain together with palm and thumb domains form a binding pocket for ubiquitin. The C-terminal tail of ubiquitin interacts with the catalytic cleft formed between palm and thumb domains, while the N-terminal end of ubiquitin contacts the USP finger domain. The catalytic core domain of USPs contains a conserved cysteine catalytic triad. Common to the USP sequences are the Cys and His boxes that contain the catalytic cysteine and histidine respectively, and a third box containing either Asp or Asn. Notably, a small group of USPs (USP16, USP30 and USP45) appears to utilize serine as the third catalytic residue(129) although biochemical characterization is required to confirm the catalytic role of the serine residue.
Figure 6.

Plasticity of the USP active sites. A. Overall structure of the catalytic core of USP using USP7 (PBD ID 1NB8) as an example. The catalytic core structure comprises three subdomains, i.e. finger (blue), palm (green), and thumb (pink) domains. The active site cysteine catalytic triad is located between the palm and the thumb domains. B. Comparison of the apo USP7 (left, PBD ID 1NB8) and the USP7 bound with ubiquitin-aldehyde (right, PDB ID: INBF). The catalytic residues are shown as stick. The binding of ubiquitin-aldehyde to USP7 brings the distance between the catalytic cysteine and histidine from 9.7 Å to 3.6 Å. C. Comparison of the apo USP14 (left, PDB ID: 2AYN) and the USP14 bound with ubiquitin-aldehyde (right, PDB ID: 2AYO). In USP14, the ubiquitin-binding cleft leading to the active site is blocked by two surface loops BL1 (yellow) and BL2 (purple) in the apo structure. These two loops undergo conformational changes to allow the C terminus of ubiquitin to bind to the cleft.
An intriguing observation regarding the USP active site is that a conformation change is often observed in the USP catalytic site upon the binding of ubiquitin, suggesting a plasticity of the USP active site (Figure 6B). In USP7, the binding of ubiquitin aldehyde leads to a productive active site conformation by bringing the distance between the catalytic cysteine and histidine from 9.7 Å (as in the apo USP7 structure) to 3.6 Å that is close to a hydrogen bond distance(125). In USP14, the ubiquitin binding cleft leading to the active site is blocked by two surface loops (BL1 and BL2) in the absence of ubiquitin (Figure 6C). Upon ubiquitin binding these two loops undergo conformational changes to allow the C terminus of ubiquitin to bind to the active site cleft(127). In the catalytic domain of apo USP4, both BL1 and BL2 as well as a third blocking loop (BL3) have been observed to prevent the ubiquitin binding(122). A major conformation change in the active site loops in USP4 is likely required for the binding of ubiquitin.
Regulation of the DUB activity of USP1 by UAF1
USP1 alone has a low catalytic activity. The formation of USP1/UAF1 complex greatly stimulates the DUB activity of USP1(118, 119). UAF1, like may other WD40-repeat proteins, does not harbor enzymatic activity. The stimulation of USP1's catalytic activity by UAF1 has been subjected to detailed biochemical studies. We demonstrated that the binding of UAF1 to USP1 does not affect the binding of ubiquitin to USP1. By probing the active site residues we revealed that the USP1 active site can be modulated by the binding of UAF1(119). The exact mode of interaction between UAF1 and USP1 remains to be determined. We envision that UAF1 can interact with the active site directly or at a region distant from the USP1 active site. The latter evokes an allosteric mechanism for activation. Indeed, it has been observed in other DUBs that the binding of a protein partner can effect a rearrangement of the DUB active site into an “on state”. For example, Ubp8 in the SAGA deubiquitination module is shown to be inactive in the absence of its protein partners Sgf11, Sus1, and Sgf73(130-132). Upon the formation of a stable DUB complex, the zinc finger of Sgf11 interacts with the Ubp8 active site cysteine to attain a catalytically competent form of Ubp8(130). Another example of a DUB that undergoes active site modulation when bound to a protein partner is USP7. USP7 contains a C-terminal sequence that has five ubiquitin-like domains (ULDs) with the last two domains being involved in activating USP7. The ULDs in USP7 were found to interact with the active site of USP7. The interaction helps to switch the DUB into an active state by rearranging the active site into a catalytically competent state(133). Besides the ULDs, a protein partner of USP7, GMP synthetase (GMPS), also helps to further increase the activity of USP7 by binding to the ULDs on USP7, thus promoting the active state of USP7 catalytic site(133).
As stated earlier, UAF1 interacts with two other USPs, USP12 and USP46. These two proteins are about half the size of USP1. The mode of interaction of USP12 and USP46 with UAF1 is still unclear. Whether the interaction site for UAF1 is the same for USP1, USP12, and USP46 still needs to be determined. A comparison of the three USP/WD40 repeat complexes will provide rich information on the regulation of USP by WD40-repeat proteins.
Remarkably, for USP12 and USP46, UAF1 is not the only interacting WD40-repeat protein. USP12 and USP46 have been shown to bind WDR20(30, 92, 93). Interestingly, the addition of WDR20 to the USP12/UAF1 complex further increased USP12's DUB activity. However, the addition of USP12 alone did not increase its activity (93). Moreover, WDR20 is specific for USP12 and it did not stimulate the DUB activity of the USP1/UAF1 complex (93). These observations raised an interesting possibility that a USP can interact with more than one WD40-repeat protein and the interaction may contribute to the regulation of specific USPs.
Targeting USPs for inhibitor discovery
Because of ubiquitin's important roles in proteasomal protein degradation and many other cellular pathways, human USPs have been increasingly recognized as a promising target class for inhibitor discovery(52, 134, 135). In fact, several small molecule inhibitors have been identified targeting USP1, USP7, USP8, USP10, USP13, and USP14 (117, 135-143) (Table 2). In 2009, HBX 41108, was first identified as a uncompetitive reversible small-molecule inhibitor against USP7 with submicromolar IC50 in vitro and an effect on p53 ubiquitination in cells(137). Reverdy et al. later reported another USP7 inhibitor HBX 19818, which is selective and irreversible(138). Specific and potent autophagy inhibitor-1 (Spautin-1) was identified to inhibit USP10 and USP13, which deubiquitinate the Beclin 1 subunit of Vsp34 complex, and thus promoted the degradation of Vsp34 PI3 kinase complex(140). IU1 inhibits USP14 in the proteasome specifically and reversibly with a low-micromolar IC50 in vitro and accelerated substrate degradation by proteasome in cells(141). More recently, we reported the first small molecule inhibitors targeting the USP/WD40-repeat protein complex. Through a high-throughput screening campaign, several known compounds were identified and validated as selective USP1/UAF1 inhibitors(117). The most potent inhibitors, pimozide and GW7647, inhibited USP1/UAF1 reversibly with a micromolar IC50. Both compounds inhibited USP1/UAF1 through a noncompetitive mechanism. Importantly, the inhibitors are selective against different classes of DUBs and unrelated cysteine proteases. Notably pimozide demonstrated no inhibition against USP46/UAF1. An on-target effect of the USP1/UAF1 inhibitors was also demonstrated by detecting the cellular level of monoubiquitinated PCNA and FANCD2, two essential proteins in DNA damage response.
Table 2. Reported inhibitors against DUBs in the ubiquitin-specific protease (USP) family.
| Name | Structure | Target USPS | IC50(μM) | Reported off-target activity IC50 (μM) | Reversibility | Inhibition mechanism | Reference |
|---|---|---|---|---|---|---|---|
| Pimozide |

|
USP1/UAF1 | 2 | NA | reversible | noncompetitive | 117 |
| GW7647 |
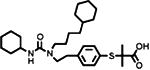
|
USP1/UAF1 | 5 | USP7 (44) USP46/UAF1 (12) |
reversible | noncompetitive | 117 |
| HBX 41,108 |

|
USP7 | 0.4 | Cathepsin B (>1) Cathepsin L (>1) Cathepsin S (>1) UCH-L1 (>1) |
reversible | uncompetitive | 137 |
| HBX 19,818 |
|
USP7 | 28.1 | NA | irreversible | - | 138 |
| P022077 |

|
USP7 | 8 | NA | - | - | 139 |
| P005091 |

|
USP7 USP47 |
4.2 4.3 |
NA | - | - |
142 143 |
| compound 14 |

|
USP7 USP47 |
0.4 1.0 |
NA | - | - | 142 |
| HBX 90,397 |

|
USP8 | 0.5 | UCH-L3 (>10) | - | - | 135 |
| Spautin-1 |

|
USP10 USP13 |
0.6 0.7 |
- | - | - | 140 |
| IU1 |

|
USP14 | 4.7 | NA | reversible | - | 141 |
| b-AP15 |

|
USP14 | - | UCHL5 | - | - | 136 |
NA, either no inhibition was observed for other USPs or cysteine proteases tested or the IC50 against the other USPs or cysteine protease was at least 20 folds higher than that against the target USP.
The role of USP1 in the two major DNA damage response pathways suggest USP1/UAF1 as a promising therapeutic target for cancer. It has been shown that both disruption of USP1 or UAF1 in chicken DT40 cells and knockout of the murine USP1 gene resulted in hypersensitivity to DNA cross-linker(103, 116, 118). With the identified USP1/UAF1 inhibitors, we demonstrated that inhibition of USP1/UAF1 activity by small molecules sensitized non-small cell lung cancer cells to the DNA damaging agent cisplatin(117). Moreover, USP1 has been more recently shown to stabilize ID proteins and promote tumorigenesis. Differentiation treatments for cancer have been proven to be successful for lethal cancers, such as acute promyelocytic leukemia(56). Thus targeting USP1/UAF1 may represent an effect treatment for osteogenic sarcoma (56).
Other UAF1-interacting USPs, i.e. USP12 and USP46, are related to protein trafficking and protein quality control(69, 71). In addition, USP12 and USP46 display deubiquitination activity toward H2A and H2B. It is demonstrated that USP12 regulates Xenopus development during gastrula stages by histone deubiquitination(70). USP46 functions in nervous system and is involved in several behavioral processes, including basal immobility, the anti-immobility effects of imipramine and nest building(86). The understanding of how USP46 functions in the nervous system underlying the mental disorder may contribute to the discovery of a potential therapeutic route for some mental illnesses.
Another strategy to inhibit the USP/WD40-repeat protein complex is to disrupt the formation of the USP complex. Such a protein-protein interaction disruptor has not been reported to date. The success in inhibiting USP/WD40-repeat protein complexes relies on an in-depth understanding of how WD40-repeat proteins interact with USPs and how USP's activity is regulated by WD40-repeat proteins. In-depth mechanistic and structural investigations of the USP/WD40-repeat protein complex will fuel the discovery of more potent and selective small molecule USP inhibitors that can eventually lead to new therapeutics.
Future Directions
Despite recent progress in understanding the catalysis and regulation of USPs, more studies are needed to uncover the molecular details of the regulation of USPs by a variety of binding partners. USP/WD40-repeat protein complexes represent a valuable system to understand the multi-level regulation of USP's function and activity. Particularly obtaining a better understanding of how USPs and WD40-repeat proteins interact is highly desirable. A major task is to map out the interaction site(s) between USPs and WD40 domains through both low- and high-resolution techniques. Also important is to understand how the USP active site is influenced by USP's association with WD40-repeat proteins. To that end, sophisticated enzymological and structural approaches will be needed. The mechanistic details uncovered will add to our understanding of the USP catalysis and regulation in general. The knowledge obtained in the above-mentioned studies will eventually aid the effort in targeting USPs for inhibitor discovery. One unique aspect of inhibiting USP/WD40-repeat protein complex is the potential allosteric site that may be targeted to achieve better selectivity than targeting the USP active site. Moreover, finding small molecules or peptide mimetics that can disrupt the interaction between USPs and WD40-repeat proteins also represents a future direction in DUB inhibitor discovery given the wide-spread interactions between DUBs and a large number of WD40-repeat proteins. What is learned from targeting USP/WD40-repeat protein complex will also help targeting other USP complexes that have been implicated in various human diseases.
Acknowledgments
This work was supported by a grant from the US National Institutes of Health to Z. Zhuang (R01GM097468).
References
- 1.Komander D. The emerging complexity of protein ubiquitination. Biochemical Society transactions. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 4.Fang S, Weissman AM. A field guide to ubiquitylation. Cellular and molecular life sciences : CMLS. 2004;61:1546–1561. doi: 10.1007/s00018-004-4129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Veen AG, Ploegh HL. Ubiquitin-like proteins. Annu Rev Biochem. 2012;81:323–357. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- 6.Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. A ubiquitin ligase complex assembles linear polyubiquitin chains. Embo J. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 8.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iphofer A, Kummer A, Nimtz M, Ritter A, Arnold T, Frank R, van den Heuvel J, Kessler BM, Jansch L, Franke R. Profiling ubiquitin linkage specificities of deubiquitinating enzymes with branched ubiquitin isopeptide probes. Chembiochem : a European journal of chemical biology. 2012;13:1416–1420. doi: 10.1002/cbic.201200261. [DOI] [PubMed] [Google Scholar]
- 10.Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- 11.Chen PC, Na CH, Peng J. Quantitative proteomics to decipher ubiquitin signaling. Amino acids. 2012;43:1049–1060. doi: 10.1007/s00726-012-1286-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochimica et biophysica acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Komander D, Rape M. The ubiquitin code. Annual review of biochemistry. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 15.Fraile JM, Quesada V, Rodriguez D, Freije JM, Lopez-Otin C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31:2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 16.Hussain S, Zhang Y, Galardy PJ. DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell cycle. 2009;8:1688–1697. doi: 10.4161/cc.8.11.8739. [DOI] [PubMed] [Google Scholar]
- 17.Ambroggio XI, Rees DC, Deshaies RJ. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS biology. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang J, Saad Y, Lei T, Wang J, Qi D, Yang Q, Kolattukudy PE, Fu M. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J Exp Med. 2010;207:2959–2973. doi: 10.1084/jem.20092641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kessler BM, Edelmann MJ. PTMs in conversation: activity and function of deubiquitinating enzymes regulated via post-translational modifications. Cell Biochem Biophys. 2011;60:21–38. doi: 10.1007/s12013-011-9176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reiley W, Zhang M, Wu X, Granger E, Sun SC. Regulation of the deubiquitinating enzyme CYLD by IkappaB kinase gamma-dependent phosphorylation. Mol Cell Biol. 2005;25:3886–3895. doi: 10.1128/MCB.25.10.3886-3895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meray RK, Lansbury PT., Jr Reversible monoubiquitination regulates the Parkinson disease-associated ubiquitin hydrolase UCH-L1. J Biol Chem. 2007;282:10567–10575. doi: 10.1074/jbc.M611153200. [DOI] [PubMed] [Google Scholar]
- 22.Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol Cell. 2008;30:610–619. doi: 10.1016/j.molcel.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 23.Todi SV, Winborn BJ, Scaglione KM, Blount JR, Travis SM, Paulson HL. Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3. Embo J. 2009;28:372–382. doi: 10.1038/emboj.2008.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang OW, Ma X, Yin J, Flinders J, Maurer T, Kayagaki N, Phung Q, Bosanac I, Arnott D, Dixit VM, Hymowitz SG, Starovasnik MA, Cochran AG. Phosphorylation-dependent activity of the deubiquitinase DUBA. Nat Struct Mol Biol. 2012;19:171–175. doi: 10.1038/nsmb.2206. [DOI] [PubMed] [Google Scholar]
- 25.Villamil MA, Liang Q, Chen J, Choi YS, Hou S, Lee KH, Zhuang Z. Serine Phosphorylation Is Critical for the Activation of Ubiquitin-Specific Protease 1 and Its Interaction with WD40-Repeat Protein UAF1. Biochemistry. 2012 doi: 10.1021/bi300845s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gauci S, Helbig AO, Slijper M, Krijgsveld J, Heck AJ, Mohammed S. Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal Chem. 2009;81:4493–4501. doi: 10.1021/ac9004309. [DOI] [PubMed] [Google Scholar]
- 28.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 29.Cotto-Rios XM, Jones MJ, Huang TT. Insights into phosphorylation-dependent mechanisms regulating USP1 protein stability during the cell cycle. Cell Cycle. 2011;10:4009–4016. doi: 10.4161/cc.10.23.18501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stirnimann CU, Petsalaki E, Russell RB, Muller CW. WD40 proteins propel cellular networks. Trends in biochemical sciences. 2010;35:565–574. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Wall MA, Coleman DE, Lee E, Iniguez-Lluhi JA, Posner BA, Gilman AG, Sprang SR. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 33.Fong HK, Hurley JB, Hopkins RS, Miake-Lye R, Johnson MS, Doolittle RF, Simon MI. Repetitive segmental structure of the transducin beta subunit: homology with the CDC4 gene and identification of related mRNAs. Proc Natl Acad Sci U S A. 1986;83:2162–2166. doi: 10.1073/pnas.83.7.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 35.Paoli M. Protein folds propelled by diversity. Progress in biophysics and molecular biology. 2001;76:103–130. doi: 10.1016/s0079-6107(01)00007-4. [DOI] [PubMed] [Google Scholar]
- 36.Murzin AG. Structural principles for the propeller assembly of beta-sheets: the preference for seven-fold symmetry. Proteins. 1992;14:191–201. doi: 10.1002/prot.340140206. [DOI] [PubMed] [Google Scholar]
- 37.Chen CK, Chan NL, Wang AH. The many blades of the beta-propeller proteins: conserved but versatile. Trends in biochemical sciences. 2011;36:553–561. doi: 10.1016/j.tibs.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Xu C, Min J. Structure and function of WD40 domain proteins. Protein & cell. 2011;2:202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith TF. Diversity of WD-repeat proteins. Sub-cellular biochemistry. 2008;48:20–30. doi: 10.1007/978-0-387-09595-0_3. [DOI] [PubMed] [Google Scholar]
- 40.Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-5-reviews3002. REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hart M, Concordet JP, Lassot I, Albert I, del los Santos R, Durand H, Perret C, Rubinfeld B, Margottin F, Benarous R, Polakis P. The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr Biol. 1999;9:207–210. doi: 10.1016/s0960-9822(99)80091-8. [DOI] [PubMed] [Google Scholar]
- 42.Liu C, Kato Y, Zhang Z, Do VM, Yankner BA, He X. beta-Trcp couples beta-catenin phosphorylation-degradation and regulates Xenopus axis formation. Proc Natl Acad Sci U S A. 1999;96:6273–6278. doi: 10.1073/pnas.96.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K, Nakamichi I, Kikuchi A, Nakayama K, Nakayama K. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin. Embo J. 1999;18:2401–2410. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 45.Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol. 2006;8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- 46.Sims RJ, 3rd, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends in genetics : TIG. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 47.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 48.Higa LA, Banks D, Wu M, Kobayashi R, Sun H, Zhang H. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle. 2006;5:1675–1680. doi: 10.4161/cc.5.15.3149. [DOI] [PubMed] [Google Scholar]
- 49.Pashkova N, Gakhar L, Winistorfer SC, Yu L, Ramaswamy S, Piper RC. WD40 repeat propellers define a ubiquitin-binding domain that regulates turnover of F box proteins. Mol Cell. 2010;40:433–443. doi: 10.1016/j.molcel.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clague MJ, Liu H, Urbe S. Governance of endocytic trafficking and signaling by reversible ubiquitylation. Developmental cell. 2012;23:457–467. doi: 10.1016/j.devcel.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 51.Clague MJ, Coulson JM, Urbe S. Cellular functions of the DUBs. Journal of cell science. 2012;125:277–286. doi: 10.1242/jcs.090985. [DOI] [PubMed] [Google Scholar]
- 52.Todi SV, Paulson HL. Balancing act: deubiquitinating enzymes in the nervous system. Trends in neurosciences. 2011 doi: 10.1016/j.tins.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Molecular cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 54.Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D'Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nature cell biology. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 55.Murai J, Yang K, Dejsuphong D, Hirota K, Takeda S, D'Andrea AD. The USP1/UAF1 complex promotes double-strand break repair through homologous recombination. Molecular and cellular biology. 2011;31:2462–2469. doi: 10.1128/MCB.05058-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams SA, Maecker HL, French DM, Liu J, Gregg A, Silverstein LB, Cao TC, Carano RA, Dixit VM. USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell. 2011;146:918–930. doi: 10.1016/j.cell.2011.07.040. [DOI] [PubMed] [Google Scholar]
- 57.Nicassio F, Corrado N, Vissers JH, Areces LB, Bergink S, Marteijn JA, Geverts B, Houtsmuller AB, Vermeulen W, Di Fiore PP, Citterio E. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Current biology : CB. 2007;17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 58.Song EJ, Werner SL, Neubauer J, Stegmeier F, Aspden J, Rio D, Harper JW, Elledge SJ, Kirschner MW, Rape M. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes & development. 2010;24:1434–1447. doi: 10.1101/gad.1925010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao B, Schlesiger C, Masucci MG, Lindsten K. The ubiquitin specific protease 4 (USP4) is a new player in the Wnt signalling pathway. Journal of Cellular and Molecular Medicine. 2009;13:1886–1895. doi: 10.1111/j.1582-4934.2008.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 61.Felle M, Joppien S, Nemeth A, Diermeier S, Thalhammer V, Dobner T, Kremmer E, Kappler R, Langst G. The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic acids research. 2011;39:8355–8365. doi: 10.1093/nar/gkr528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saridakis V, Sheng Y, Sarkari F, Holowaty MN, Shire K, Nguyen T, Zhang RG, Liao J, Lee W, Edwards AM, Arrowsmith CH, Frappier L. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Molecular cell. 2005;18:25–36. doi: 10.1016/j.molcel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 63.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nature cell biology. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 64.Feroz Sarkari, Yi Sheng, Frappier L. USP7/HAUSP Promotes the Sequence-Specific DNA Binding Activity of p53. PloS one. 2010;5:e13040. doi: 10.1371/journal.pone.0013040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, Karch F, Verrijzer CP. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Molecular cell. 2005;17:695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 66.van der Knaap JA, Kozhevnikova E, Langenberg K, Moshkin YM, Verrijzer CP. Biosynthetic enzyme GMP synthetase cooperates with ubiquitin-specific protease 7 in transcriptional regulation of ecdysteroid target genes. Molecular and cellular biology. 2010;30:736–744. doi: 10.1128/MCB.01121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maertens GN, El Messaoudi-Aubert S, Elderkin S, Hiom K, Peters G. Ubiquitin-specific proteases 7 and 11 modulate Polycomb regulation of the INK4a tumour suppressor. The EMBO journal. 2010;29:2553–2565. doi: 10.1038/emboj.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schoenfeld AR, Apgar S, Dolios G, Wang R, Aaronson SA. BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Molecular and cellular biology. 2004;24:7444–7455. doi: 10.1128/MCB.24.17.7444-7455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moretti J, Chastagner P, Liang CC, Cohn MA, Israel A, Brou C. The Ubiquitin-specific Protease 12 (USP12) Is a Negative Regulator of Notch Signaling Acting on Notch Receptor Trafficking toward Degradation. The Journal of biological chemistry. 2012;287:29429–29441. doi: 10.1074/jbc.M112.366807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Joo HY, Jones A, Yang C, Zhai L, Smith ADt, Zhang Z, Chandrasekharan MB, Sun ZW, Renfrow MB, Wang Y, Chang C, Wang H. Regulation of histone H2A and H2B deubiquitination and Xenopus development by USP12 and USP46. The Journal of biological chemistry. 2011;286:7190–7201. doi: 10.1074/jbc.M110.158311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kowalski JR, Dahlberg CL, Juo P. The deubiquitinating enzyme USP-46 negatively regulates the degradation of glutamate receptors to control their abundance in the ventral nerve cord of Caenorhabditis elegans. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:1341–1354. doi: 10.1523/JNEUROSCI.4765-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang X, Langelotz C, Hetfeld-Pechoc BK, Schwenk W, Dubiel W. The COP9 signalosome mediates beta-catenin degradation by deneddylation and blocks adenomatous polyposis coli destruction via USP15. Journal of molecular biology. 2009;391:691–702. doi: 10.1016/j.jmb.2009.06.066. [DOI] [PubMed] [Google Scholar]
- 73.Hassink GC, Zhao B, Sompallae R, Altun M, Gastaldello S, Zinin NV, Masucci MG, Lindsten K. The ER-resident ubiquitin-specific protease 19 participates in the UPR and rescues ERAD substrates. EMBO reports. 2009;10:755–761. doi: 10.1038/embor.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Xy, Pfeiffer HK, Thorne AW, McMahon SB. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell cycle. 2008;7:1522–1524. doi: 10.4161/cc.7.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Molecular cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, Georgieva SG, Schule R, Takeyama K, Kato S, Tora L, Devys D. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Molecular cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 77.Taillebourg E, Gregoire I, Viargues P, Jacomin AC, Thevenon D, Faure M, Fauvarque MO. The deubiquitinating enzyme USP36 controls selective autophagy activation by ubiquitinated proteins. Autophagy. 2012;8:767–779. doi: 10.4161/auto.19381. [DOI] [PubMed] [Google Scholar]
- 78.Richardson LA, Reed BJ, Charette JM, Freed EF, Fredrickson EK, Locke MN, Baserga SJ, Gardner RG. A conserved deubiquitinating enzyme controls cell growth by regulating RNA polymerase I stability. Cell reports. 2012;2:372–385. doi: 10.1016/j.celrep.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang WC, Shih HM. The deubiquitinating enzyme USP37 regulates the oncogenic fusion protein PLZF/RARA stability. Oncogene. 2012 doi: 10.1038/onc.2012.537. [DOI] [PubMed] [Google Scholar]
- 80.van Leuken Renske J, Luna-Vargas Mark P, Sixma Titia K, Wolthuis RMF, Medema RH. Usp39 is essential for mitotic spindle checkpoint integrity and controls mRNA-levels of aurora B. Cell cycle. 2008;7:2710–2719. doi: 10.4161/cc.7.17.6553. [DOI] [PubMed] [Google Scholar]
- 81.Hock AK, Vigneron AM, Carter S, Ludwig RL, Vousden KH. Regulation of p53 stability and function by the deubiquitinating enzyme USP42. The EMBO journal. 2011;30:4921–4930. doi: 10.1038/emboj.2011.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jeandidier E, Gervais C, Radford-Weiss I, Zink E, Gangneux C, Eischen A, Galoisy AC, Helias C, Dano L, Cammarata O, Jung G, Harzallah I, Guerin E, Martzolff L, Drenou B, Lioure B, Tancredi C, Rimelen V, Mauvieux L. A cytogenetic study of 397 consecutive acute myeloid leukemia cases identified three with a t(7;21) associated with 5q abnormalities and exhibiting similar clinical and biological features, suggesting a new, rare acute myeloid leukemia entity. Cancer genetics. 2012;205:365–372. doi: 10.1016/j.cancergen.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 83.Giguere A, Hebert J. Microhomologies and topoisomerase II consensus sequences identified near the breakpoint junctions of the recurrent t(7;21)(p22;q22) translocation in acute myeloid leukemia. Genes, chromosomes & cancer. 2011;50:228–238. doi: 10.1002/gcc.20848. [DOI] [PubMed] [Google Scholar]
- 84.Fuchs G, Shema E, Vesterman R, Kotler E, Wolchinsky Z, Wilder S, Golomb L, Pribluda A, Zhang F, Haj-Yahya M, Feldmesser E, Brik A, Yu X, Hanna J, Aberdam D, Domany E, Oren M. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Molecular cell. 2012;46:662–673. doi: 10.1016/j.molcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Y, Foreman O, Wigle DA, Kosari F, Vasmatzis G, Salisbury JL, van Deursen J, Galardy PJ. USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. The Journal of clinical investigation. 2012;122:4362–4374. doi: 10.1172/JCI63084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tomida S, Mamiya T, Sakamaki H, Miura M, Aosaki T, Masuda M, Niwa M, Kameyama T, Kobayashi J, Iwaki Y, Imai S, Ishikawa A, Abe K, Yoshimura T, Nabeshima T, Ebihara S. Usp46 is a quantitative trait gene regulating mouse immobile behavior in the tail suspension and forced swimming tests. Nature genetics. 2009;41:688–695. doi: 10.1038/ng.344. [DOI] [PubMed] [Google Scholar]
- 87.Zhang W, Tian QB, Li QK, Wang JM, Wang CN, Liu T, Liu DW, Wang MW. Lysine 92 amino acid residue of USP46, a gene associated with ‘behavioral despair’ in mice, influences the deubiquitinating enzyme activity. PloS one. 2011;6:e26297. doi: 10.1371/journal.pone.0026297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Parsons JL, Dianova II, Khoronenkova SV, Edelmann MJ, Kessler BM, Dianov GL. USP47 is a deubiquitylating enzyme that regulates base excision repair by controlling steady-state levels of DNA polymerase beta. Molecular cell. 2011;41:609–615. doi: 10.1016/j.molcel.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 89.Peschiaroli A, Skaar JR, Pagano M, Melino G. The ubiquitin-specific protease USP47 is a novel beta-TRCP interactor regulating cell survival. Oncogene. 2010;29:1384–1393. doi: 10.1038/onc.2009.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aressy B, Jullien D, Cazales M, Marcellin M, Bugler B, Burlet-Schiltz O, Ducommun B. A screen for deubiquitinating enzymes involved in the G(2)/M checkpoint identifies USP50 as a regulator of HSP90-dependent Wee1 stability. Cell cycle. 2010;9:3815–3822. doi: 10.4161/cc.9.18.13133. [DOI] [PubMed] [Google Scholar]
- 91.Inui M, Manfrin A, Mamidi A, Martello G, Morsut L, Soligo S, Enzo E, Moro S, Polo S, Dupont S, Cordenonsi M, Piccolo S. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nature cell biology. 2011;13:1368–1375. doi: 10.1038/ncb2346. [DOI] [PubMed] [Google Scholar]
- 92.Cohn MA, Kee Y, Haas W, Gygi SP, D'Andrea AD. UAF1 Is a Subunit of Multiple Deubiquitinating Enzyme Complexes. J Biol Chem. 2009;284:5343–5351. doi: 10.1074/jbc.M808430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kee Y, Yang KL, Cohn MA, Haas W, Gygi SP, D'Andrea AD. WDR20 Regulates Activity of the USP12 center dot UAF1 Deubiquitinating Enzyme Complex. J Biol Chem. 2010;285:11252–11257. doi: 10.1074/jbc.M109.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sims AE, Spiteri E, Sims RJ, 3rd, Arita AG, Lach FP, Landers T, Wurm M, Freund M, Neveling K, Hanenberg H, Auerbach AD, Huang TT. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nature structural & molecular biology. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 95.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, Elledge SJ. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.D'Andrea AD. The Fanconi road to cancer. Genes & development. 2003;17:1933–1936. doi: 10.1101/gad.1128303. [DOI] [PubMed] [Google Scholar]
- 97.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nature reviews Genetics. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 98.Grompe M, van de Vrugt H. The Fanconi family adds a fraternal twin. Developmental cell. 2007;12:661–662. doi: 10.1016/j.devcel.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 99.Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D'Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 100.Wang X, Andreassen PR, D'Andrea AD. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol Cell Biol. 2004;24:5850–5862. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim H, D'Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes & development. 2012;26:1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Crossan GP, Patel KJ. The Fanconi anaemia pathway orchestrates incisions at sites of crosslinked DNA. The Journal of pathology. 2012;226:326–337. doi: 10.1002/path.3002. [DOI] [PubMed] [Google Scholar]
- 103.Oestergaard VH, Langevin F, Kuiken HJ, Pace P, Niedzwiedz W, Simpson LJ, Ohzeki M, Takata M, Sale JE, Patel KJ. Deubiquitination of FANCD2 is required for DNA crosslink repair. Molecular cell. 2007;28:798–809. doi: 10.1016/j.molcel.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen J, Bozza W, Zhuang Z. Ubiquitination of PCNA and its essential role in eukaryotic translesion synthesis. Cell biochemistry and biophysics. 2011;60:47–60. doi: 10.1007/s12013-011-9187-3. [DOI] [PubMed] [Google Scholar]
- 105.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 106.Stelter P, Ulrich HD. Control of spontaneous and damage- induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 107.Zhuang Z, Johnson RE, Haracska L, Prakash L, Prakash S, Benkovic SJ. Regulation of polymerase exchange between Poleta and Poldelta by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides polg to replication stalling sites through physical interaction and PCNA monoubiquitination. The EMBO journal. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kannouche P. Domain structure, localization, and function of DNA polymerase eta, defective in xeroderma pigmentosum variant cells. Genes & development. 2001;15:158–172. doi: 10.1101/gad.187501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Juhasz S, Balogh D, Hajdu I, Burkovics P, Villamil MA, Zhuang Z, Haracska L. Characterization of human Spartan/C1orf124, an ubiquitin-PCNA interacting regulator of DNA damage tolerance. Nucleic Acids Res. 2012;40:10795–10808. doi: 10.1093/nar/gks850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brown S, Niimi A, Lehmann AR. Ubiquitination and deubiquitination of PCNA in response to stalling of the replication fork. Cell Cycle. 2009;8:689–692. doi: 10.4161/cc.8.5.7707. [DOI] [PubMed] [Google Scholar]
- 113.Lee KY, Yang K, Cohn MA, Sikdar N, D'Andrea AD, Myung K. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through Its interactions with PCNA and USP1. J Biol Chem. 2010;285:10362–10369. doi: 10.1074/jbc.M109.092544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fox JT, Lee KY, Myung K. Dynamic regulation of PCNA ubiquitylation/deubiquitylation. FEBS Lett. 2011;585:2780–2785. doi: 10.1016/j.febslet.2011.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jones MJ, Colnaghi L, Huang TT. Dysregulation of DNA polymerase kappa recruitment to replication forks results in genomic instability. The EMBO journal. 2012;31:908–918. doi: 10.1038/emboj.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, Kutok JL, D'Andrea AD. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Developmental cell. 2009;16:314–320. doi: 10.1016/j.devcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen J, Dexheimer TS, Ai Y, Liang Q, Villamil MA, Inglese J, Maloney DJ, Jadhav A, Simeonov A, Zhuang Z. Selective and cell-active inhibitors of the USP1/UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chemistry & biology. 2011;18:1390–1400. doi: 10.1016/j.chembiol.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D'Andrea AD. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Molecular cell. 2007;28:786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 119.Villamil MA, Chen J, Liang Q, Zhuang Z. A noncanonical cysteine protease USP1 is activated through active site modulation by USP1-associated factor 1. Biochemistry. 2012;51:2829–2839. doi: 10.1021/bi3000512. [DOI] [PubMed] [Google Scholar]
- 120.Bounpheng MA, Dimas JJ, Dodds SG, Christy BA. Degradation of Id proteins by the ubiquitin-proteasome pathway. Faseb J. 1999;13:2257–2264. [PubMed] [Google Scholar]
- 121.Imai S, Mamiya T, Tsukada A, Sakai Y, Mouri A, Nabeshima T, Ebihara S. Ubiquitin-specific peptidase 46 (Usp46) regulates mouse immobile behavior in the tail suspension test through the GABAergic system. Plos One. 2012;7:e39084. doi: 10.1371/journal.pone.0039084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Luna-Vargas MP, Faesen AC, van Dijk WJ, Rape M, Fish A, Sixma TK. Ubiquitin-specific protease 4 is inhibited by its ubiquitin-like domain. EMBO Rep. 2011;12:365–372. doi: 10.1038/embor.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 123.Ye Y, Scheel H, Hofmann K, Komander D. Dissection of USP catalytic domains reveals five common insertion points. Molecular bioSystems. 2009;5:1797–1808. doi: 10.1039/b907669g. [DOI] [PubMed] [Google Scholar]
- 124.Renatus M, Parrado SG, D'Arcy A, Eidhoff U, Gerhartz B, Hassiepen U, Pierrat B, Riedl R, Vinzenz D, Worpenberg S, Kroemer M. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hu M, Li P, Li M, Li W, Yao T, Wu JW, Gu W, Cohen RE, Shi Y. Crystal Structure of a UBP-Family Deubiquitinating Enzyme in Isolation and in Complex with Ubiquitin Aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 126.Avvakumov GV, Walker JR, Xue S, Finerty PJ, Jr, Mackenzie F, Newman EM, Dhe-Paganon S. Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8) The Journal of biological chemistry. 2006;281:38061–38070. doi: 10.1074/jbc.M606704200. [DOI] [PubMed] [Google Scholar]
- 127.Hu M, Li P, Song L, Jeffrey PD, Chernova TA, Wilkinson KD, Cohen RE, Shi Y. Structure and mechanisms of the proteasome- associated deubiquitinating enzyme USP14. The EMBO journal. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ye Y, Akutsu M, Reyes-Turcu F, Enchev RI, Wilkinson KD, Komander D. Polyubiquitin binding and cross-reactivity in the USP domain deubiquitinase USP21. EMBO Rep. 2011;12:350–357. doi: 10.1038/embor.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang W, Sulea T, Tao L, Cui Q, Purisima EO, Vongsamphanh R, Lachance P, Lytvyn V, Qi H, Li Y, Menard R. Contribution of active site residues to substrate hydrolysis by USP2: insights into catalysis by ubiquitin specific proteases. Biochemistry. 2011;50:4775–4785. doi: 10.1021/bi101958h. [DOI] [PubMed] [Google Scholar]
- 130.Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science. 2010;328:1025–1029. doi: 10.1126/science.1190049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wilson MA, Koutelou E, Hirsch C, Akdemir K, Schibler A, Barton MC, Dent SY. Ubp8 and SAGA regulate Snf1 AMP kinase activity. Molecular and cellular biology. 2011;31:3126–3135. doi: 10.1128/MCB.01350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Molecular and cellular biology. 2005;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Faesen AC, Dirac AM, Shanmugham A, Ovaa H, Perrakis A, Sixma TK. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Molecular cell. 2011;44:147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]
- 134.Singhal S, Taylor MC, Baker RT. Deubiquitylating enzymes and disease. BMC biochemistry. 2008;9(Suppl 1):S3. doi: 10.1186/1471-2091-9-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Colland F. The therapeutic potential of deubiquitinating enzyme inhibitors. Biochemical Society transactions. 2010;38:137–143. doi: 10.1042/BST0380137. [DOI] [PubMed] [Google Scholar]
- 136.D'Arcy P, Brnjic S, Olofsson MH, Fryknas M, Lindsten K, De Cesare M, Perego P, Sadeghi B, Hassan M, Larsson R, Linder S. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nature medicine. 2011;17:1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 137.Colland F, Formstecher E, Jacq X, Reverdy C, Planquette C, Conrath S, Trouplin V, Bianchi J, Aushev VN, Camonis J, Calabrese A, Borg-Capra C, Sippl W, Collura V, Boissy G, Rain JC, Guedat P, Delansorne R, Daviet L. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Molecular Cancer Therapeutics. 2009;8:2286–2295. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- 138.Reverdy C, Conrath S, Lopez R, Planquette C, Atmanene C, Collura V, Harpon J, Battaglia V, Vivat V, Sippl W, Colland F. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chemistry & biology. 2012;19:467–477. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 139.Tian X, Isamiddinova NS, Peroutka RJ, Goldenberg SJ, Mattern MR, Nicholson B, Leach C. Characterization of selective ubiquitin and ubiquitin-like protease inhibitors using a fluorescence-based multiplex assay format. Assay and drug development technologies. 2011;9:165–173. doi: 10.1089/adt.2010.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, Jin M, Zhu Z, Wang H, Yu J, Hao Y, Choi A, Ke H, Ma D, Yuan J. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, Wilson SM, King RW, Finley D. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Weinstock J, Wu J, Cao P, Kingsbury WD, McDermott JL, Kodrasov MP, McKelvey DM, Suresh Kumar KG, Goldenberg SJ, Mattern MR, Nicholson B. Selective Dual Inhibitors of the Cancer-Related Deubiquitylating Proteases USP7 and USP47. ACS Medicinal Chemistry Letters. 2012;3:789–792. doi: 10.1021/ml200276j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R, McDermott JL, Leach CA, Fulcinniti M, Kodrasov MP, Weinstock J, Kingsbury WD, Hideshima T, Shah PK, Minvielle S, Altun M, Kessler BM, Orlowski R, Richardson P, Munshi N, Anderson KC. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer cell. 2012;22:345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schuster-Bockler B, Schultz J, Rahmann S. HMM Logos for visualization of protein families. BMC bioinformatics. 2004;5:7. doi: 10.1186/1471-2105-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]



