Abstract
Self-interaction of an antibody may lead to aggregation, low solubility or high viscosity. Rapid identification of highly developable leads remains challenging, even though progress has been made with the introduction of techniques such as self-interaction chromatography (SIC) and cross-interaction chromatography (CIC). Here, we report a high throughput method to detect antibody clone self-interaction (CSI) using bio-layer interferometry (BLI) technology. Antibodies with strong self-interaction responses in the CSI-BLI assay also show delayed retention times in SIC and CIC. This method allows hundreds of candidates to be screened in a matter of hours with minimal material consumption.
Keywords: self-interaction, bio-layer interferometry, developability, solubility, monoclonal antibody, aggregation
Self-interaction of a therapeutic monoclonal antibody may cause aggregation, high viscosity, or low solubility, which limits its likelihood of being successfully developed as a drug (i.e., the developability of the drug).1-5 Ideally, antibodies with self-interaction tendancies could be rapidly eliminated early in the discovery stage, with minimum material consumption, to minimize downstream risks. This screening process is usually performed by low to medium throughput assays such as self-interaction chromatography (SIC)6-13 or cross-interaction chromatography (CIC).14-16 SIC measures the retention time of an antibody as it flows across a column conjugated with the same antibody of interest (or “self”). A longer retention time results from stronger self-interaction of the molecule of interest, and this in turn is usually correlated with lower solubility. Material consumption and throughput, however, greatly limit the general applicability of this assay to a range of tens to hundreds of candidate molecules. Similar to SIC, CIC measures the retention time of an antibody as it flows across a column conjugated with polyclonal human serum antibodies. Later elution of an antibody in CIC indicates exposure of surfaces prone to formation of non-specific interactions, and this usually serves as an indicator of undesirable solution properties, including lower solubility.14 CIC is a more attractive option in that a single column can be used to screen multiple antibodies, which minimizes sample consumption and individual column preparation and improves throughput. Some antibodies do show strong binding to the unconjugated, quenched SIC or CIC columns, which limits analyses of them by either assay. Recently, non-chromatographic methods such as self-interaction nanoparticle spectroscopy (SINS)17 and affinity capture (AC)-SINS18 using gold nanoparticles, and surface plasmon resonance (SPR) based assays,19 have been reported to predict or confirm antibody self-interaction. Concurrently, bio-layer interferometry has emerged as a technology for the detection of biomolecular interactions using label-free biosensors.20-22 Here, we describe a high throughput method to detect antibody clone self-interaction by bio-layer interferometry (CSI-BLI) with low material consumption.
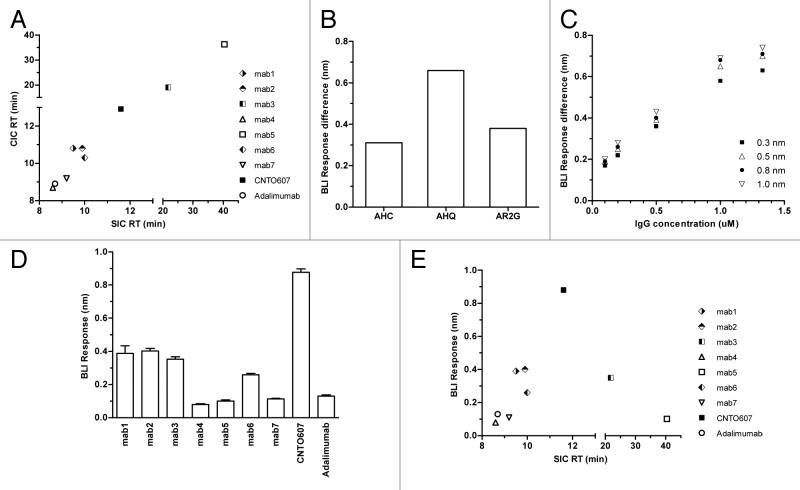
Nine antibodies, including seven human IgG1 antibodies against multiple targets that were discovered in-house and two control antibodies with known solubility in phosphate buffered saline (PBS) were used in this study. The control antibodies, CNTO607 (negative solubility control, ~13 mg/ml as reported by Wu et al.23,24) and adalimumab (positive solubility control), were made recombinantly as human IgG1 from published V region sequences and expressed in HEK293 cells. The in-house discovered antibodies were pre-selected based on SIC and CIC assays to represent different levels of self-reactivity. The SIC column was prepared by coupling > 1 mg of each antibody to a 1 ml HiTrap column (GE Healthcare # 17-0716-01), followed by ethanolamine quenching. Approximately 5 ug of that antibody was tested at a flow rate of 0.1 ml/min using PBS as a mobile phase on an Agilent 1100 series HPLC system. During the SIC assay, Mab4 and 7 showed similar retention times to adalimumab while Mab1, 2, 6 and CNTO607 showed delayed retention time (Fig. 1A). Mab3 and 5 showed significant column binding, whereas retention time on the blank column was greater than that on the SIC column. The same set of antibodies was also tested by CIC. The CIC column was prepared by coupling ~30 mg of human serum polyclonal antibodies (Sigma #I4506) to a 1 ml HiTrap column, followed by quenching with ethanolamine. Other experimental conditions were identical to those of SIC. Good correlation was observed between these two assays (Fig. 1A). Presumably the polyclonal human serum antibodies conjugated to the CIC column provide a sufficiently diverse sampling population to include the type(s) of interaction responsible for the original antibody self-interaction. These antibodies were later characterized by DLS at ~50 mg/mL in PBS (except CNTO607). Detectable large particles were observed for Mab1, 2, 3, and 6, but not for Mab4, 5, 7 and adalimumab (data not shown).
Figure 1. (A) Comparison of retention time for 9 antibodies on CIC vs. SIC. (B) Self-binding response difference between adalimumab and CNTO607 on AHC, AHQ and AR2G sensors. (C) Self-binding response difference between CNTO607 and adalimumab under varied AHQ sensor loading densities (0.3−1.0 nm) and solution antibody concentrations (0.1−1.3 μM). (D) Self- binding response for 9 antibodies under optimized conditions. Measurements were made in quadruplicate. (E) Comparison of BLI self-binding response vs. SIC retention time for 9 antibodies.
To further improve throughput and reduce material consumption, CSI-BLI using the Octet® RED384 was developed to assess antibody self-interaction. In this assay, the antibody of interest was loaded to a BLI biosensor to measure any significant self-binding response. High solubility antibodies such as adalimumab generally have minimal self-interaction and generate a low self-binding response in this assay. On the other hand, antibodies with poor solubility, such as CNTO607, have stronger self-interaction, and we postulate that avidity effects further increase the binding response because this weak self-interaction is usually not observable when monovalent Fab is used in solution. To test which biosensor type generates the greatest response difference between adalimumab and CNTO607, anti-human Fc capture (AHC), anti-human IgG quantitation (AHQ), and amine reactive second-generation (AR2G) biosensors were evaluated. Control antibodies were loaded onto AHC or AHQ sensors, followed by blocking with an in-house produced human IgG1 Fc. The antibodies were also covalently coupled to AR2G biosensors followed by quenching. The self-binding test was performed with 1 μM antibody in 100 μl PBSF (0.1% BSA in PBS) solution on the Octet® RED384. The AHQ biosensor generated the greatest response difference (Fig. 1B), and thus was chosen for further signal optimization in biosensor loading density and solution antibody concentration. This observation aligned with our previous experience during kinetic screening of antigen binding to antibody-loaded sensors. Even though AHC sensors allowed higher IgG loading capacity than AHQ sensors, the antigen binding responses were usually lower. AHC and AHQ sensors are constructed in a very similar fashion, both using goat anti-human Fc polyclonal antibodies for human IgG capturing, but with different linker types and lengths. A shorter linker is used and there is no cross-linking of polyclonal goat anti-human Fc antibodies to AHQ sensors. In the case of AHC sensors, a longer linker and cross-linking treatment provide higher loading capacity and a more stable baseline for kinetic screening. Crowding effects, however, may lower the active surface binding concentration of IgGs on the sensor and decrease antigen binding response.
Control antibodies were loaded on an AHQ biosensor to 0.3, 0.5, 0.8, or 1.0 nm (by controlling loading time), then tested for self-binding in a 0.1, 0.2, 0.5, 1.0, or 1.3 μM antibody solution after blocking the biosensor with human IgG1 Fc. As shown in Figure 1C, a greater response difference was obtained at higher biosensor loading densities. More importantly, the antibody concentration in solution had a larger effect on the CSI response difference between the positive and negative control antibodies. A loading density of ~0.8 nm and 1 μM antibody solution concentration were chosen as the final screening conditions.
Under these optimized conditions, CSI-BLI was used to test self-interaction in quadruplicate for the set of nine human monoclonal antibodies discussed above and the results were compared with the SIC and CIC data. As shown in Figure 1D, Mab4 and 7 have responses similar to that of adalimumab, while Mab1, 2, 6 and CNTO607 have responses significantly greater than that of adalimumab. These four antibodies also show delayed retention times on SIC and CIC (Fig. 1A), indicating that CSI-BLI has a positive correlation with these column-based chromatographic assays (Fig. 1E). When performing DLS for Mab1, 2, and 6 at 50 mg/mL in PBS, large particles were also observed, confirming their aggregation propensity (data not shown). Moreover, CSI-BLI also works well with antibodies that are incompatible with SIC or CIC, such as Mab3 and 5, where severe blank column binding is observed. In CSI-BLI, Mab5 showed a low self-binding response and Mab3 showed a higher self-binding response (Fig. 1D), which correlates well with the DLS data (data not shown).
In this study, all antibodies were formulated in PBS buffer at pH 7.4 prior to SIC, CIC or CSI-BLI assays. Buffer composition is a critical factor that influences antibody self-interaction, and consequently developability attributes, including solubility. If an antibody shows minimal self-interaction in a relatively harsh formulation buffer such as PBS, it usually suggests good developability properties. Adalimumab was chosen as a positive control for developability, and antibodies with significantly higher self-binding responses (greater than 0.1 nm) in CSI-BLI than adalimumab are considered potentially problematic downstream. The choice of response cutoff value is dependent upon selection output. A less stringent response cutoff may be used when fewer biologically relevant clones are available.
In CSI-BLI, the Fc region of the antibody is directionally captured on the biosensor so that the Fab region is accessible for binding by the Fc or Fab portion of an antibody in solution19,25-28 via hydrogen bond pairing, ionic interaction, or hydrophobic interaction.29 Rather than relying on cumulative effects on retention time in the chromatographic methods to detect these weak interactions, sensitive BLI technology22 allows for direct monitoring of antibody self-binding. CSI-BLI has unparalleled throughput compared with CIC or SIC in that a plate of 96 antibodies can be tested for self-binding within 2 h, rather than a few days. This high throughput is contributed partly by the design of the Octet® RED384, where 16 channels are available for data collection simultaneously. Furthermore, only 15 ug of each antibody is necessary for this assay, making CSI-BLI even more attractive for early-stage discovery screening, where a large number of candidates are produced at low quantity.
Acknowledgments
All of the in-house antibodies used in this study were discovered by Adimab’s Antibody Discovery group, sequence confirmed by the Core and produced by the High Throughput Expression department. We appreciate the manuscript editing and critical review by Kristin Rookey, Michael Ruse, Piotr Bobrowicz, Max Vasquez, Errik Anderson, Dane Wittrup, and Tillman Gerngross.
Disclosure of Potential Conflicts of Interest
No conflict of interest was disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/26186
References
- 1.Zhang J, Liu XY. Effect of protein–protein interactions on protein aggregation kinetics. J Chem Phys. 2003;119:10972. doi: 10.1063/1.1622380. [DOI] [Google Scholar]
- 2.Yadav S, Liu J, Shire SJ, Kalonia DS. Specific interactions in high concentration antibody solutions resulting in high viscosity. J Pharm Sci. 2010;99:1152–68. doi: 10.1002/jps.21898. [DOI] [PubMed] [Google Scholar]
- 3.Kanai S, Liu J, Patapoff TW, Shire SJ. Reversible self-association of a concentrated monoclonal antibody solution mediated by Fab-Fab interaction that impacts solution viscosity. J Pharm Sci. 2008;97:4219–27. doi: 10.1002/jps.21322. [DOI] [PubMed] [Google Scholar]
- 4.Saito S, Hasegawa J, Kobayashi N, Kishi N, Uchiyama S, Fukui K. Behavior of monoclonal antibodies: relation between the second virial coefficient (B (2)) at low concentrations and aggregation propensity and viscosity at high concentrations. Pharm Res. 2012;29:397–410. doi: 10.1007/s11095-011-0563-x. [DOI] [PubMed] [Google Scholar]
- 5.Connolly BD, Petry C, Yadav S, Demeule B, Ciaccio N, Moore JM, Shire SJ, Gokarn YR. Weak interactions govern the viscosity of concentrated antibody solutions: high-throughput analysis using the diffusion interaction parameter. Biophys J. 2012;103:69–78. doi: 10.1016/j.bpj.2012.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patro SY, Przybycien TM. Self-interaction chromatography: a tool for the study of protein-protein interactions in bioprocessing environments. Biotechnol Bioeng. 1996;52:193–203. doi: 10.1002/(SICI)1097-0290(19961020)52:2<193::AID-BIT2>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 7.Tessier PM, Vandrey SD, Berger BW, Pazhianur R, Sandler SI, Lenhoff AM. Self-interaction chromatography: a novel screening method for rational protein crystallization. Acta Crystallogr D Biol Crystallogr. 2002;58:1531–5. doi: 10.1107/S0907444902012775. [DOI] [PubMed] [Google Scholar]
- 8.Tessier PM, Lenhoff AM, Sandler SI. Rapid measurement of protein osmotic second virial coefficients by self-interaction chromatography. Biophys J. 2002;82:1620–31. doi: 10.1016/S0006-3495(02)75513-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.García CD, Hadley DJ, Wilson WW, Henry CS. Measuring protein interactions by microchip self-interaction chromatography. Biotechnol Prog. 2003;19:1006–10. doi: 10.1021/bp025788z. [DOI] [PubMed] [Google Scholar]
- 10.Le Brun V, Friess W, Bassarab S, Mühlau S, Garidel P. A critical evaluation of self-interaction chromatography as a predictive tool for the assessment of protein-protein interactions in protein formulation development: a case study of a therapeutic monoclonal antibody. Eur J Pharm Biopharm. 2010;75:16–25. doi: 10.1016/j.ejpb.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Ahamed T, Ottens M, van Dedem GWK, van der Wielen LAM. Design of self-interaction chromatography as an analytical tool for predicting protein phase behavior. J Chromatogr A. 2005;1089:111–24. doi: 10.1016/j.chroma.2005.06.065. [DOI] [PubMed] [Google Scholar]
- 12.Johnson DH, Parupudi A, Wilson WW, DeLucas LJ. High-throughput self-interaction chromatography: applications in protein formulation prediction. Pharm Res. 2009;26:296–305. doi: 10.1007/s11095-008-9737-6. [DOI] [PubMed] [Google Scholar]
- 13.Ahamed T, Esteban BNA, Ottens M, van Dedem GW, van der Wielen LA, Bisschops MA, Lee A, Pham C, Thömmes J. Phase behavior of an intact monoclonal antibody. Biophys J. 2007;93:610–9. doi: 10.1529/biophysj.106.098293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs SA, Wu S-J, Feng Y, Bethea D, O’Neil KT. Cross-interaction chromatography: a rapid method to identify highly soluble monoclonal antibody candidates. Pharm Res. 2010;27:65–71. doi: 10.1007/s11095-009-0007-z. [DOI] [PubMed] [Google Scholar]
- 15.Tessier PM, Sandler SI, Lenhoff AM. Direct measurement of protein osmotic second virial cross coefficients by cross-interaction chromatography. Protein Sci. 2004;13:1379–90. doi: 10.1110/ps.03419204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spencer S, Bethea D, Raju TS, Giles-Komar J, Feng Y. Solubility evaluation of murine hybridoma antibodies. MAbs. 2012;4:319–25. doi: 10.4161/mabs.19869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sule SV, Sukumar M, Weiss WF, 4th, Marcelino-Cruz AM, Sample T, Tessier PM. High-throughput analysis of concentration-dependent antibody self-association. Biophys J. 2011;101:1749–57. doi: 10.1016/j.bpj.2011.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sule SV, Dickinson CD, Lu J, Chow C-K, Tessier PM. Rapid analysis of antibody self-association in complex mixtures using immunogold conjugates. Mol Pharm. 2013;10:1322–31. doi: 10.1021/mp300524x. [DOI] [PubMed] [Google Scholar]
- 19.Nishi H, Miyajima M, Wakiyama N, Kubota K, Hasegawa J, Uchiyama S, Fukui K. Fc domain mediated self-association of an IgG1 monoclonal antibody under a low ionic strength condition. J Biosci Bioeng. 2011;112:326–32. doi: 10.1016/j.jbiosc.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 20.Rich RL, Myszka DG. Higher-throughput, label-free, real-time molecular interaction analysis. Anal Biochem. 2007;361:1–6. doi: 10.1016/j.ab.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 21.Abdiche YN, Malashock DS, Pinkerton A, Pons J. Exploring blocking assays using Octet, ProteOn, and Biacore biosensors. Anal Biochem. 2009;386:172–80. doi: 10.1016/j.ab.2008.11.038. [DOI] [PubMed] [Google Scholar]
- 22.Concepcion J, Witte K, Wartchow C, Choo S, Yao D, Persson H, Wei J, Li P, Heidecker B, Ma W, et al. Label-free detection of biomolecular interactions using BioLayer interferometry for kinetic characterization. Comb Chem High Throughput Screen. 2009;12:791–800. doi: 10.2174/138620709789104915. [DOI] [PubMed] [Google Scholar]
- 23.Wu S-J, Luo J, O’Neil KT, Kang J, Lacy ER, Canziani G, Baker A, Huang M, Tang QM, Raju TS, et al. Structure-based engineering of a monoclonal antibody for improved solubility. Protein Eng Des Sel. 2010;23:643–51. doi: 10.1093/protein/gzq037. [DOI] [PubMed] [Google Scholar]
- 24.Bethea D, Wu S-J, Luo J, Hyun L, Lacy ER, Teplyakov A, Jacobs SA, O’Neil KT, Gilliland GL, Feng Y. Mechanisms of self-association of a human monoclonal antibody CNTO607. Protein Eng Des Sel. 2012;25:531–7. doi: 10.1093/protein/gzs047. [DOI] [PubMed] [Google Scholar]
- 25.Perchiacca JM, Tessier PM. Engineering aggregation-resistant antibodies. Annu Rev Chem Biomol Eng. 2012;3:263–86. doi: 10.1146/annurev-chembioeng-062011-081052. [DOI] [PubMed] [Google Scholar]
- 26.Dudgeon K, Rouet R, Kokmeijer I, Schofield P, Stolp J, Langley D, Stock D, Christ D. General strategy for the generation of human antibody variable domains with increased aggregation resistance. Proc Natl Acad Sci U S A. 2012;109:10879–84. doi: 10.1073/pnas.1202866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Singh SK, Kumar S. Potential aggregation-prone regions in complementarity-determining regions of antibodies and their contribution towards antigen recognition: a computational analysis. Pharm Res. 2010;27:1512–29. doi: 10.1007/s11095-010-0143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pepinsky RB, Silvian L, Berkowitz SA, Farrington G, Lugovskoy A, Walus L, Eldredge J, Capili A, Mi S, Graff C, et al. Improving the solubility of anti-LINGO-1 monoclonal antibody Li33 by isotype switching and targeted mutagenesis. Protein Sci. 2010;19:954–66. doi: 10.1002/pro.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saluja A, Kalonia DS. Nature and consequences of protein-protein interactions in high protein concentration solutions. Int J Pharm. 2008;358:1–15. doi: 10.1016/j.ijpharm.2008.03.041. [DOI] [PubMed] [Google Scholar]