Abstract
Oleuropein, the major phenolic compound found in olive leaves and oil, exerts antioxidant, anti-inflammatory and anti-atherogenic effects and suppresses the adipocyte differentiation in vitro. Herein, we characterized molecular mechanisms underlying the anti-adipogenic effects of oleuropein on 3T3-L1 cells and adipocytes derived from stromal-vascular fraction of dorsolumbar and gonadal fat dissected from mice. We found that oleuropein (>100 μM) decreased viability of preadipocytes proliferating in vitro and did not exerted any cytotoxic effects in post-confluent cells after induction of differentiation. Oleuropein (>100 μM) inhibited adipocyte differentiation, suppressed gene expression of peroxisome proliferator-activated receptor γ (PPARγ), CCAAT-/enhancer-binding protein α, sterol regulatory element-binding transcription factor 1c and fatty acid synthase. Furthermore, we tested ability of oleuropein to regulate of PPARγ-, PPARα- or PPARβ-/PPARδ-mediated β-lactamase expression in appropriate reporter gene assays. Oleuropein between 10 and 400 μM concentrations did not affect activity of PPARα or PPARβ/δ. Contrary, PPARγ activity, either basal or rosiglitazone activated, was inhibited by oleuropein. Our data suggest that oleuropein exerts anti-adipogenic effect through direct inhibition of PPARγ transcriptional activity.
Electronic supplementary material
The online version of this article (doi:10.1007/s12263-013-0376-0) contains supplementary material, which is available to authorized users.
Keywords: Oleuropein, Adipogenesis, PPARγ, Olive oil and leaf extracts, Catalase
Introduction
Plant-derived polyphenols, secondary plant products, are abundant micronutrients present in the human diet. These natural bioactive compounds have been discovered for based on their beneficial effects in the prevention of chronic disorders such a cancer, cardiovascular diseases, inflammatory and metabolic diseases including obesity. Many experiments with the polyphenols demonstrated substantial modulation of physiological and molecular pathway involved in energy metabolism and adiposity (reviewed in (Meydani and Hasan 2010)). Catechins from green tea (Lin et al. 2005), resveratrol from red grapes (Floyd et al. 2008) and curcumin from curcuma (Ejaz et al. 2009) suppressed adipogenesis on 3T3-L1 cell line through several mechanisms including attenuation of peroxisome proliferator-activated receptor γ (PPARγ) regulatory pathway (Manach et al. 2004). Also, the olive tree, botanically known as Olea europaea, and its products are a source of at least 30 phenolic compounds, and particularly, extra-virgin olive oil contains considerable amounts of phenolic compounds, e.g., oleuropein and its hydrolysis product hydroxytyrosol, which are responsible for its peculiar taste and high stability. Oleuropein concentration varies with cultivar and climate and is several times higher in the olive leaf than the oil (Omar 2010). Beneficial effects of olive leaf extract have been described in vivo, including a reduction in adiposity and hyperlipidemia in dietary obese rat (Oi-Kano et al. 2008), improvement of alloxan-induced diabetes in rabbits (Al-Azzawie and Alhamdani 2006) and improvement of insulin sensitivity in overweight humans (de Bock et al. 2013). Purified oleuropein considerably reduced the infarct size in both normal and hypercholesterolemic rabbits that were subjected to ischemia/reperfusion, protected the reperfused myocardium from the oxidative damage and decreased total cholesterol and triglyceride levels (Andreadou et al. 2006). In vitro, oleuropein reduced the expression of PPARγ, inhibited adipogenesis and enhanced osteoblastogenesis in stem cells derived from human bone marrow (Santiago-Mora et al. 2011). Oleuropein (in concentration higher than 100 μM) acted on 3T3-L1 cells to reduce preadipocyte differentiation and lipid accumulation and thus regulated the size of fat cells. The expression of key transcriptional modulator of adipocyte differentiation genes PPARγ, CCAAT-/enhancer-binding protein α (C/EBP1α) and sterol regulatory element-binding transcription factor 1c (SREBP-1c) and their downstream target genes were suppressed by oleuropein in dose depended manner during the differentiation process (Drira et al. 2011). The aim of current study is to further characterize molecular mechanisms underlying anti-adipogenic effects of oleuropein on 3T3-L1 cells and differentiated adipocytes derived from stromal-vascular fraction (SVF) of murine dorsolumbar and gonadal fat.
Materials and methods
Oleuropein
The oleuropein used in this study was isolated from Olea europaea leaves as described before (Andreadou et al.). Briefly, air-dried and pulverized leaves were extracted with mechanical stirring for 12 h with acetone. The extract was evaporated to dryness and washed with a mixture of dichloromethane/methanol (98:2). The insoluble material was separated, dried and submitted to medium pressure liquid chromatography with Si gel 60 Merck, using dichloromethane/methanol gradient as the eluent to provide pure oleuropein. The purified oleuropein (95 %) was entirely in the glucoside form and was free of any aglycone forms. The structure elucidation of oleuropein was carried out using spectroscopic and spectrometric methods, as well as comparison with literature data (Zoidou et al. 2010). Specifically, 1D and 2D NMR spectra (COSY, COSYLR, HSQC-DEPT, HMBC) were recorded in deuterated methanol (CD3OD-Merck), on a Bruker Advance III spectrometer (Bruker Biospin GmbH, Reinsteten, Germany) operating at 600.11 MHz for 1H and at 150.11 MHz for 13C, with a 5-mm inverse detection probe. The residual 1H (3.33 ppm) and 13C (49.50 ppm) signals of CD3OD were used as internal standard. One- and two-dimensional NMR experiments were performed with standard pulse programs, at room temperature. HRMS & HRMS/MS data were obtained by direct infusion method using a hybrid LTQ-Orbitrap Discovery Mass Spectrometer (Thermo Scientific, Bremen, Germany) equipped with an ESI probe, in positive mode. For each experiment, oleuropein was freshly dissolved in sterile water and directly diluted in appropriate supplemented cell culture medium (Dulbecco’s modified Eagle’s medium with 4.5 g glucose; DMEM).
Murine 3T3-L1 preadipocytes
Cells were plated and grown until 2 days post confluence in DMEM supplemented with 10 % calf serum and penicillin/streptomycin in a humidified atmosphere of 10 % CO2 at 37 °C. Cells differentiation was induced using a medium containing 10 % fetal bovine serum, 2 μM dexamethasone, 0.1 μM rosiglitazone (BRL 49653, in dimethyl sulfoxide) and 5 μg/ml insulin. This medium was containing or not catalase (100 units/ml, catalase from bovine liver, Sigma-Aldrich, Co., USA), and it was present for 2 days. Afterward the cells were maintained in DMEM containing 10 % fetal bovine serum and 5 μg/ml insulin in the presence or absence of catalase (100 units/ml).
Isolation, plating and culture condition of adipose precursor cells/primary SVF cultures
Male C57BL/6J mice were used for the preparation of primary SVF cultures. Adipose depots (subcutaneous dorsolumbar and abdominal gonadal) were dissect under sterile conditions, minced with scissors and digested using collagenase (3 mg/ml, type II collagenase; Sigma-Aldrich, Co., USA; Cat. # C-6885) in modified Krebs–Ringer bicarbonate buffer (118.5 mM NaCl, 4.8 mM KCl, 2.7 mM CaCl2, 1.2 mM KH2PO4, 1.1 mM MgSO4·7H2O, 25 mM NaHCO2, 5 mM glucose and 4 % (w/v) bovine serum albumin fraction V; pH 7.4). The cell suspension was filtered through 250-μm nylon mesh, floating mature adipocytes were discarded, and the infranatant (SVF) was filtered again through 25-μm nylon mesh. The filtered infranatant was centrifuged at 700×g for 10 min. The pellet was resuspended in D-MEM F12 (containing 5 μg/ml insulin and gentamycin) and centrifuged again. The cells were resuspended in medium with 10 % fetal bovine serum, counted in Bürker chamber and plated at a density of 2,000 cells/cm2. Cells were grown at 37 °C in atmosphere of 5 % CO2. The medium (D-MEM F12 containing 10 % fetal bovine serum, 5 μg/ml insulin and gentamycin) was changed two times during a week. After 5–7 days, rosiglitazone was added to the cultivation medium at final concentration of 100 nM. The cells were kept for 7 days until full adipocyte differentiation with or without catalase (100 units/ml) added to the medium.
Cell viability assays
The viability of 3T3-L1 cells was tested with or without catalase (100 units/ml); see above.
Trypan blue assay
Cells were detached using trypsin–EDTA solution in phosphate-buffered saline (pH 7.4) for 10 min at room temperature and immediately stained with 0.5 % trypan blue dye (Sigma-Aldrich, Co., USA), and the cell number was evaluated using a Vi-CELL XR analyzer (Beckman Coulter, USA). Three samples were used for each experimental group, and 50 measurements were performed on each sample (i.e., 150 measurements in total).
WST-1 assay
Cells were cultivated in microplates (96 wells) in a final volume of 100 μl culture medium/well. Cell Proliferation Reagent WST-1 (10 μl/well; Roche Diagnostics GmbH, Germany) was added, and the cells were incubated for 4 h. Then, the formazan product was quantified at 420–480 nm using a microplate absorbance reader (Wallac Victor 1420, Perkin-Elmer, USA).
Oil Red O staining
After differentiation, 3T3-L1 cells or primary SVF cultures were washed twice with phosphate-buffered saline (pH 7.4), fixed with 4 % paraformaldehyde at room temperature for 12 h and then washed twice with sterile water and once with 60 % isopropanol. Subsequently, cell were stained with 5 g/L Oil Red O (in 60 % isopropanol) at room temperature for 2 h and washed with 60 % isopropanol and sterile water. Pictures were taken using an inverted microscope (Nicon TE 300, Nicon, Japan) equipped with digital camera (Penguin 600 CL, Pixera, USA). The plates with stained cells were measured using a multiwell plate reader (Wallac Victor 1420, Perkin-Elmer, USA) at the absorbance 490 nm. Alternatively, captured pictures were analyzed using AIDA 3.28 software (Raytest, Germany).
Reporter gene assay
Cellular activation of PPAR nuclear receptors was assessed in reporter gene assays according to the manufacturer’s protocol (GeneBLAzer® PPARα UAS-bla HEK293T, PPAR β/δ HEK 293T DA and PPARγ 293H DA, Life Technologies Corporation, USA). In brief, cells were stably expressing GAL4-specific PPAR ligand-binding domain fusion protein and UAS-β-lactamase reporter gene. Cells were incubated with tested compound or antagonist for 30 min before adding agonist and then incubated in a humid 37 °C/5 % CO2 incubator for 16 h. Fluorescence intensity at 460 and 530 nm emission following excitation at 406 nm was measured using a TECAN Safire2 (Tecan Group Ltd., Switzerland) with optimal gain settings determined by the instrument. After subtraction of fluorescence background from cell-free wells, the ratio of fluorescence intensity at 460 versus 530 nm (designated as 460:530 nm) was calculated. T0070907 (Cayman CHEMICAL, Cayman Europe, Estonia) was used as specific PPARγ inhibitor (Lee et al. 2002).
Quantitative real-time PCR
Total RNA isolated using TRI reagent (Sigma-Aldrich, Co., USA). Levels of various transcripts were evaluated using LightCycler 480 II instrument (Roche Diagnostic Ltd., Switzerland) and LightCycler 480 SYBR Green I Master kit (Roche Diagnostic Ltd., Germany). PCR condition were 95 °C for 5 min and 45 cycles of 95 °C for 10 s, 55–60 °C for 10 s and 72 °C for 20 s. Specificity of the amplified PCR product was assessed by performing a melting curve analysis. Lasergene 9 software (DNASTAR, Inc., USA) was used to design primers. To correct for inter-sample variation, levels of the transcript were normalized using geometrical mean of two reference genes—CYPHβ (Gene ID: 19035) and EEF2 (Gene ID: 13628). For primer sequences, see Hensler et al. (2011). Primers for PPARγ (Gene ID: 19016)—GCCTTGCTGTGGGGATGTCTC/CCTCGCCTTGGCTTTGGTCAG, fatty acid synthase (FAS, Gene ID: 14104)—GGCTGCCTCCGTGGACCTTATC/GTCTAGCCCTCCCGTACACTCACTCGT, C/EBPα (Gene ID: 12606)—CGCTGGCCGGCCTCTTCCCCTACC/GCCCCGCAGCGTGTCCAGTTCA, SREBP-1c (Gene ID: 20787)—GCTTCCGGCCTGCTATGA/CTCGTGTGGCTCCGGGAC.
Statistical analysis
All values are presented as mean ± SE. Logarithmic transformation was used to stabilize variance in cells when necessary. Data were evaluated by ANOVA (one-way or two-way) with Holm–Sidak post hoc test using Sigma Stat 3.5 statistical software. Comparisons were judged to be significant at p ≤ 0.05.
Results
Cytotoxic effect of oleuropein on proliferating cells is prevented by catalase
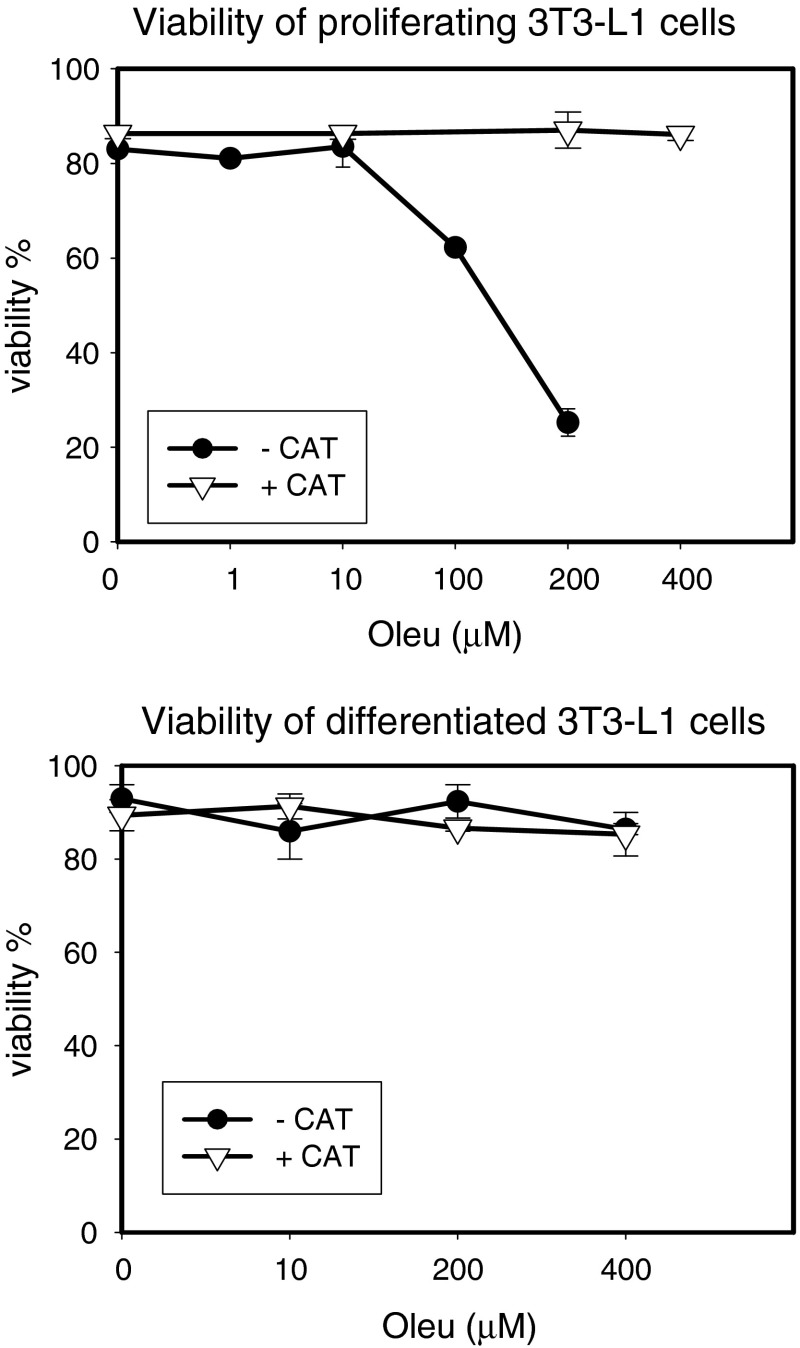
Exposure of proliferating 3T3-L1 cells to increasing concentrations of oleuropein (from 10 to 400 μM) for 24 h resulted in lower viability of the cells. When 100 units catalase/ml was added into medium for culturing for pre-confluent cells, the inhibitory effect was completely prevented (Fig. 1—upper panel). Interestingly, no cytotoxic effects were detected in post-confluent cells after induction of adipocyte differentiation (Fig. 1—lower panel). The results were confirmed by two independent approaches, trypan blue (Fig. 1) and WST-1 assays (data not shown).
Fig. 1.
Effect of oleuropein on viability of proliferating and differentiated 3T3-L1 cells. Cells were cultured in appropriate medium at 37 °C in a humidified 5 % CO2, in the absence or presence of oleuropein (Oleu—0, 1, 10, 100, 200 and 400 μM) and in the presence or absence of catalase (CAT; 100 units/ml)
Inhibition of differentiation of 3T3-L1 adipocytes and cells in primary SVF cultures
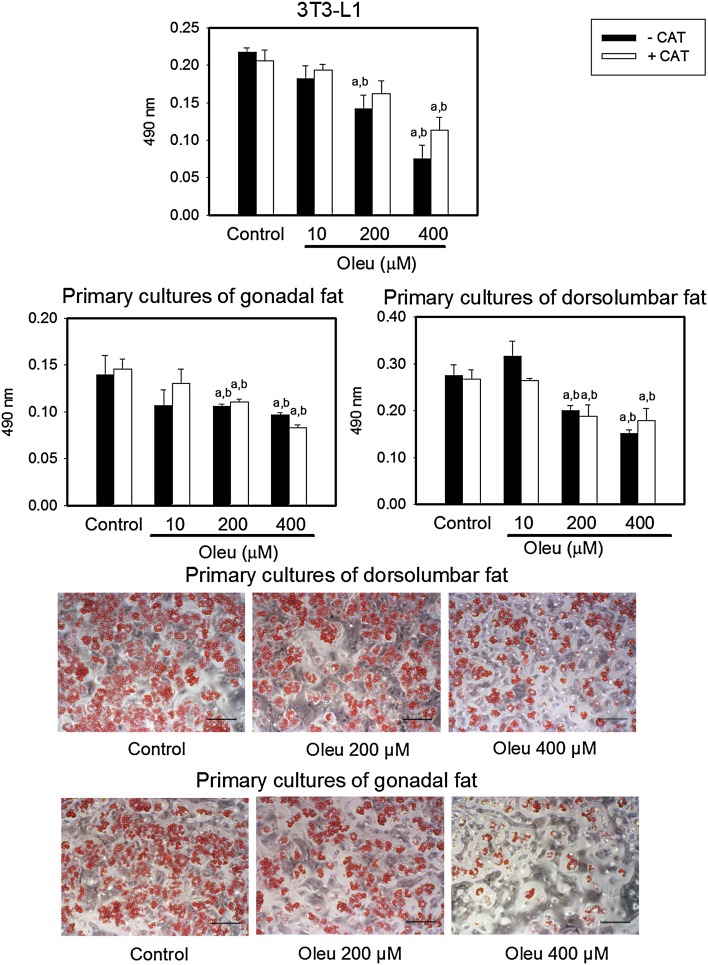
We verified that oleuropein, added to the medium simultaneously with induction of differentiation by dexamethasone and rosiglitazone, significantly inhibited 3T3-L1 adipocytes differentiation. Moreover, we next examined the effect of oleuropein in primary SVF cultures of dorsolumbar and gonadal fat, physiologically more relevant model of adipocytes in vitro. As shown in Fig. 2, oleuropein inhibited most adipocyte differentiation at 200 and 400 μM, respectively. To test whether the anti-adipogenic function of oleuropein was affected by compound oxidation, we incubated cells with catalase. Results from Oil Red O staining showed that oleuropein reduced total lipid content in a dose-dependent manner, in the same extent as in the absence of catalase (Fig. 2).
Fig. 2.
Effect of oleuropein on differentiation of 3T3-L1 adipocytes and SVF cells isolated from dorsolumbar and gonadal adipose tissue of C57BL/6J mice in primary cultures. Cultured cells were exposed for 11 days to differentiation medium containing or not oleuropein (Control or Oleu—0, 10, 200 and 400 μM) in the absence or presence of catalase (CAT; 100 units/ml) at 37 °C in a humidified 5 % CO2, respectively, 10 % CO2. Cells were stained with Oil Red O. a,bSignificantly different from Control withouta or withb catalase, *significant effect of catalase (two-way ANOVA)
Effect of oleuropein on gene expression during differentiation of 3T3-L1 adipocytes
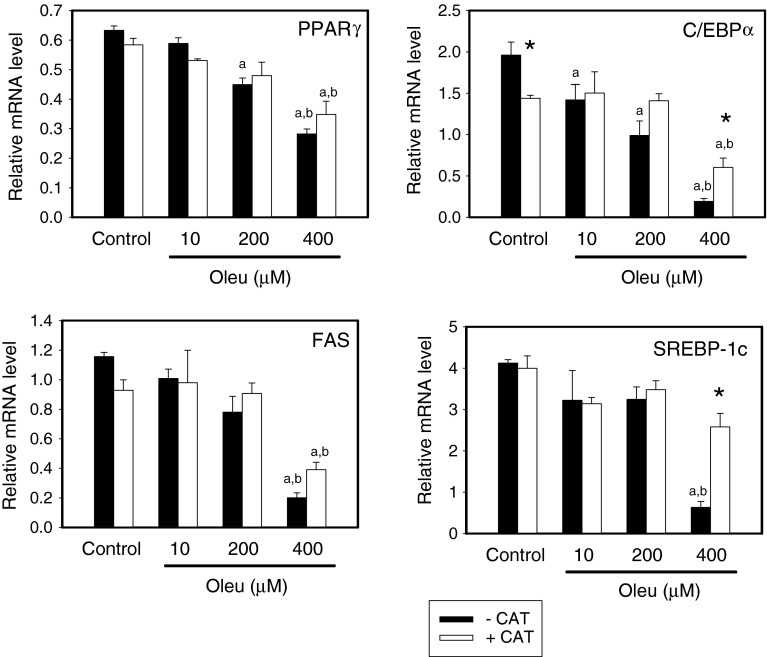
To confirm that oleuropein inhibits the adipocyte differentiation pathway, we analyzed the effect of oleuropein on the expression of the genes for PPARγ, FAS, C/EBPα and SREBP-1c using qRT-PCR. In accordance with the results of others (Drira et al. 2011), oleuropein in a dose-dependent manner (starting at a 10 μM concentration) reduced expression of the genes for the transcription factors PPARγ and C/EBPα, and pro-lipogenic SREBP-1c and its target gene FAS (Fig. 3). The presence of catalase (100 units/ml) in the medium during cell differentiation resulted in a decrease of C/EBPα transcript level in the control (non-treated by oleuropein) 3T3-L1 adipocytes, while it had no effect on the expression of the other genes studied. Catalase diminished the effect of 400 μM oleuropein (the highest concentration studied) on C/EBPα and SREBP-1c expression (Fig. 3).
Fig. 3.
Effect of oleuropein on expression of PPARγ, FAS, C/EBPα and SREBP-1c genes in 3T3-L1 adipocytes. Gene expression was evaluated in cells differentiated in vitro as described in legend to Fig. 2. The relative expression of the genes was quantified by qRT-PCR. a,bSignificantly different from Control withouta or withb catalase, *significant effect of catalase (two-way ANOVA)
Effect of oleuropein on activity of PPARα, β/δ and γ
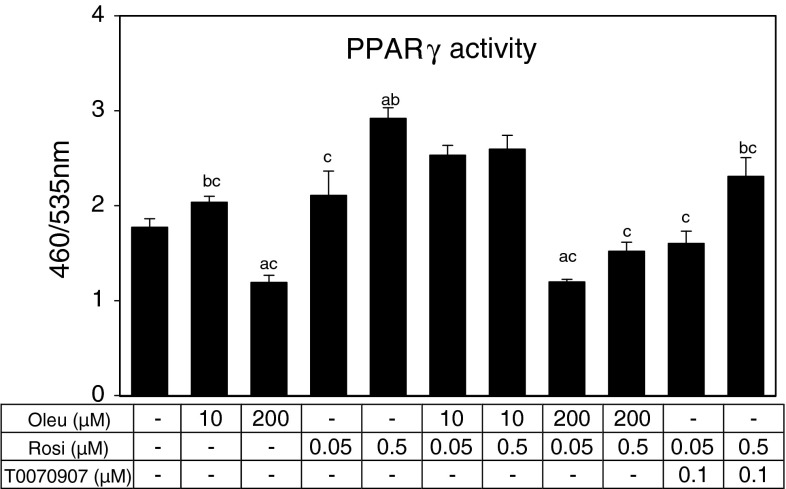
We used the cell-based reporter gene assay to test whether oleuropein directly affects the activity of nuclear receptors PPARα, PPARβ/δ and PPARγ. Oleuropein between 10 and 400 μM concentrations did not affect the activity PPARα or β/δ (see Supplemental Fig. 1). Contrary, PPARγ activity, either basal or rosiglitazone activated, was inhibited (by 30–50 %) by oleuropein (200 μM; Fig. 4). The inhibitory effect of oleuropein was stronger than that of the specific inhibitor of PPARγ—T0070907 (Lee et al. 2002) at concentration of 0.1 μM.
Fig. 4.
Cell-based PPARγ reporter gene assay. Cells were incubated with oleuropein (Oleu) in concentration 10 and 200 μM. Rosiglitazone (Rosi) was used as an activator PPARγ in concentration 0.05 and 0.5 μM and T0070907 as specific inhibitor of PPARγ at concentration 0.1 μM. Cells were incubated with Oleu or T0070907 for 30 min before adding Rosi and then incubated at 37 °C in a humidified 5 % CO2 for 16 h. a,b,cSignificant difference (one-way ANOVA) compared with control, Oleu 200 μM and Rosi 0.5 μM, respectively
Discussion
Oleuropein and other polyphenols from olive were studied, among others, for their ability to protect against metabolic syndrome, type 2 diabetes, cardiac ischemia, and other disorders, both in humans (de Bock et al. 2013) and in experimental animal models (Andreadou et al. 2006); (Al-Azzawie and Alhamdani 2006); (Oi-Kano et al. 2008). As it has been already reported, oleuropein suppressed adipogenesis in a dose-dependent manner in murine 3T3-L1 cell culture model (Drira et al. 2011). Our current findings reveal that oleuropein suppressed not only the differentiation of 3T3-L1 cells, but also that of SVF cells derived from either dorsolumbar or gonadal adipose tissue. The SVF cultures with multiple cell types are more predictive of in vivo condition. These can be obtained from various depots and following various in vivo treatments.
The important feature of the adipocyte differentiation is the increases expression of the PPARγ gene. In general, bioactive plant polyphenols, such as green tea catechins, resveratrol, curcumin and also oleuropein, are known to decrease PPARγ expression, leading to the inhibition of adipocytes differentiation (see “Introduction”). Accordingly, in the current study increasing concentration of oleuropein in the cell culture medium resulted in a remarkable reduction in mRNA levels for the key transcription factors and markers of adipogenesis, including PPARγ, C/EBPα, SREB-1c and FAS. Moreover, we found, using the reporter gene assay, that oleuropein directly inhibits transcription activity of PPARγ. Contrary, we did not find any effects on transcriptional activity of other members of PPAR nuclear receptor family—PPARα and PPAR β/δ (see Supplemental Fig. 1). Oleuropein diminished PPARγ activity at a concentration (200 μM) shown to inhibit adipocyte differentiation in primary SVF cultures, and the inhibitory effect was even stronger when compared to that of specific inhibitor of PPARγ, T0070907 at 0.1 μM. It is important to note that the concentration of rosiglitazone used for induction of differentiation in 3T3-L1 or primary SVF cultures was 0.1 μM, i.e., within the concentrations of rosiglitazone used in the activity assay (0.05 and 0.5 μM).
PPARy is essential for the formation and function of adipocytes. Regulation of PPARγ activity in adipocytes provides a link between nutritional status, lipid metabolism and gene expression (Kliewer et al. 1994). PPARγ is also involved in regulating insulin sensitivity. Although activation of PPARγ by the thiazolidinediones class of insulin-sensitizing drugs (e.g., rosiglitazone) increases insulin sensitivity, heterozygous PPARγ-deficient mice are protected from the development of insulin resistance due to adipocyte hypertrophy (Kubota et al. 1999). Moreover, chronic activation of PPARγ by thiazolidinediones results in unwanted side effects of this therapy such as weight gain, possible risk of heart failure and bone loss (summarized in Kuda et al. (2009)).
It was shown previously that oleuropein exhibits anti-proliferative and anticancer effect in various cell lines (Acquaviva et al. 2012); (Odiatou et al. 2013). In a sharp contrast to the post-confluent cells after induction of adipocyte differentiation, viability of proliferating 3T3-L1 cells treated by oleuropein is also decreased, depending on the concentrations of this compound. The cytotoxic effects were totally prevented by addition of catalase, enzyme catalyzing the decomposition of hydrogen peroxide into water and oxygen. Oleuropein was identified recently (Odiatou et al. 2013) to produce hydrogen peroxide in culture media containing sodium bicarbonate (MEM, DMEM). In our current study, 3T3-L1 cells were cultured in serum-supplemented DMEM. It can be speculated that also in our conditions oleuropein generates sufficient amounts of hydrogen peroxide to diminish cell viability of proliferating 3T3-L1 cells. It should be noted that primary SVF cultures were grown in D-MEM F12 medium containing sodium pyruvate, which abrogates hydrogen peroxide produced by oleuropein (Odiatou et al. 2013). Results from Oil Red O staining showed that the anti-adipogenic effect of oleuropein was not affected by the presence of catalase, despite gene expression analysis on 3T3-L1 cells revealed that catalase interfered with the inhibitory effect of the highest concentration of oleuropein (400 μM) on C/EBPα and SREBP-1c genes transcription. Notable, C/EBPα transcript was also suppressed by catalase in the control (non-treated by oleuropein) 3T3-L1 adipocytes.
Although our data indicate that oleuropein, similarly to resveratrol and some other plant polyphenols, is able to decrease differentiation of adipocytes via PPARγ-mediated mechanism, additional data are needed to confirm that oleuropein can modulate PPARγ-mediated gene expression in adipose tissue in vivo. Recent studies based on metabolite-profiling approaches demonstrated that oleuropein is rapidly absorbed and distributed through the blood stream to various tissues in vivo (Serra et al. 2012) and entered to cell cytoplasm in vitro (Quirantes-Pine et al. 2013). In acute toxicity studies for oleuropein, no adverse effects were observed in mice at doses as high as 1 g/kg of body weight (Petkov and Manolov 1972). Taking into account concentration at which PPARγ inhibition was detected in vitro (200 μM) and reported level of oleuropein (Cmax 4 h: 24 μM) in rat plasma (Serra et al. 2012) after an acute intake of a phenolic extract from olive cake (3 g/kg body weight, oleuropein content ~0.1 mg/g of extract), oleuropein could potentially modulate PPARγ activity also in vivo, which needs to be demonstrated.
In conclusion, oleuropein in vitro exerts anti-adipogenic effect through inhibition of both expression and activity of PPARγ. As revealed in recent clinical trial (de Bock et al. 2013), dietary supplementation by polyphenols from olive leaf, and namely oleuropein, may be used as part of novel therapeutic strategies for prevention and treatment of obesity and insulin resistance.
Electronic supplementary material
Acknowledgments
This work was supported by the Czech Science Foundation (301/11/0226).
Abbreviations
- C/EBP1α
CCAAT-/enhancer-binding protein α
- DMEM
Dulbecco’s modified Eagle’s medium
- FAS
Fatty acid synthase
- FCS
Fetal calf serum
- PPARα/β/δ/γ
Peroxisome proliferator-activated receptor α/β/δ/γ
- SREBP-1
Sterol regulatory element-binding transcription factor 1c
- SVF
Stromal-vascular fraction
References
- Acquaviva R, Di GC, Sorrenti V, Galvano F, Santangelo R, Cardile V, Gangia S, D’Orazio N, Abraham NG, Vanella L. Antiproliferative effect of oleuropein in prostate cell lines. Int J Oncol. 2012;41(1):31–38. doi: 10.3892/ijo.2012.1428. [DOI] [PubMed] [Google Scholar]
- Al-Azzawie HF, Alhamdani MS. Hypoglycemic and antioxidant effect of oleuropein in alloxan-diabetic rabbits. Life Sci. 2006;78(12):1371–1377. doi: 10.1016/j.lfs.2005.07.029. [DOI] [PubMed] [Google Scholar]
- Andreadou I, Iliodromitis EK, Mikros E, Constantinou M, Agalias A, Magiatis P, Skaltsounis AL, Kamber E, Tsantili-Kakoulidou A, Kremastinos DT. The olive constituent oleuropein exhibits anti-ischemic, antioxidative, and hypolipidemic effects in anesthetized rabbits. J Nutr. 2006;Suppl 136(8):2213–2219. doi: 10.1093/jn/136.8.2213. [DOI] [PubMed] [Google Scholar]
- de Bock M, Derraik JG, Brennan CM, Biggs JB, Morgan PE, Hodgkinson SC, Hofman PL, Cutfield WS. Olive (Olea europaea L.) leaf polyphenols improve insulin sensitivity in middle-aged overweight men: a randomized, placebo-controlled, crossover trial. PLoS ONE. 2013;8(3):e57622. doi: 10.1371/journal.pone.0057622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drira R, Chen S, Sakamoto K. Oleuropein and hydroxytyrosol inhibit adipocyte differentiation in 3T3-L1 cells. Life Sci. 2011;89(19–20):708–716. doi: 10.1016/j.lfs.2011.08.012. [DOI] [PubMed] [Google Scholar]
- Ejaz A, Wu D, Kwan P, Meydani M. Curcumin inhibits adipogenesis in 3T3-L1 adipocytes and angiogenesis and obesity in C57/BL mice. J Nutr. 2009;139(5):919–925. doi: 10.3945/jn.108.100966. [DOI] [PubMed] [Google Scholar]
- Floyd ZE, Wang ZQ, Kilroy G, Cefalu WT. Modulation of peroxisome proliferator-activated receptor gamma stability and transcriptional activity in adipocytes by resveratrol. Metabolism. 2008;57(7 Suppl 1):S32–S38. doi: 10.1016/j.metabol.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensler M, Bardova K, Jilkova ZM, Wahli W, Meztger D, Chambon P, Kopecky J, Flachs P. The inhibition of fat cell proliferation by n-3 fatty acids in dietary obese mice. Lipids Health Dis. 2011;10:128. doi: 10.1186/1476-511X-10-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Forman BM, Blumberg B, Ong ES, Borgmeyer U, Mangelsdorf DJ, Umesono K, Evans RM. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc Natl Acad Sci USA. 1994;91(15):7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota N, Terauchi Y, Miki H, Tamemoto H, Yamauchi T, Komeda K, Satoh S, Nakano R, Ishii C, Sugiyama T, Eto K, Tsubamoto Y, Okuno A, Murakami K, Sekihara H, Hasegawa G, Naito M, Toyoshima Y, Tanaka S, Shiota K, Kitamura T, Fujita T, Ezaki O, Aizawa S, Kadowaki T. PPAR gamma mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol Cell. 1999;4(4):597–609. doi: 10.1016/S1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- Kuda O, Jelenik T, Jilkova Z, Flachs P, Rossmeisl M, Hensler M, Kazdova L, Ogston N, Baranowski M, Gorski J, Janovska P, Kus V, Polak J, Mohamed-Ali V, Burcelin R, Cinti S, Bryhn M, Kopecky J. n-3 Fatty acids and rosiglitazone improve insulin sensitivity through additive stimulatory effects on muscle glycogen synthesis in mice fed a high-fat diet. Diabetologia. 2009;52(5):941–951. doi: 10.1007/s00125-009-1305-z. [DOI] [PubMed] [Google Scholar]
- Lee G, Elwood F, McNally J, Weiszmann J, Lindstrom M, Amaral K, Nakamura M, Miao S, Cao P, Learned RM, Chen JL, Li Y. T0070907, a selective ligand for peroxisome proliferator-activated receptor gamma, functions as an antagonist of biochemical and cellular activities. J Biol Chem. 2002;277(22):19649–19657. doi: 10.1074/jbc.M200743200. [DOI] [PubMed] [Google Scholar]
- Lin J, Della-Fera MA, Baile CA. Green tea polyphenol epigallocatechin gallate inhibits adipogenesis and induces apoptosis in 3T3-L1 adipocytes. Obes Res. 2005;13(6):982–990. doi: 10.1038/oby.2005.115. [DOI] [PubMed] [Google Scholar]
- Manach C, Scalbert A, Morand C, Remesy C, Jimenez L. Polyphenols: food sources and bioavailability. Am J Clin Nutr. 2004;79(5):727–747. doi: 10.1093/ajcn/79.5.727. [DOI] [PubMed] [Google Scholar]
- Meydani M, Hasan ST. Dietary polyphenols and obesity. Nutrients. 2010;2(7):737–751. doi: 10.3390/nu2070737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odiatou EM, Skaltsounis AL, Constantinou AI. Identification of the factors responsible for the in vitro pro-oxidant and cytotoxic activities of the olive polyphenols oleuropein and hydroxytyrosol. Cancer Lett. 2013;330(1):113–121. doi: 10.1016/j.canlet.2012.11.035. [DOI] [PubMed] [Google Scholar]
- Oi-Kano Y, Kawada T, Watanabe T, Koyama F, Watanabe K, Senbongi R, Iwai K. Oleuropein, a phenolic compound in extra virgin olive oil, increases uncoupling protein 1 content in brown adipose tissue and enhances noradrenaline and adrenaline secretions in rats. J Nutr Sci Vitaminol. 2008;54(5):363–370. doi: 10.3177/jnsv.54.363. [DOI] [PubMed] [Google Scholar]
- Omar SH. Oleuropein in olive and its pharmacological effects. Sci Pharm. 2010;78(2):133–154. doi: 10.3797/scipharm.0912-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov V, Manolov P. Pharmacological analysis of the iridoid oleuropein. Arzneimittelforschung. 1972;22(9):1476–1486. [PubMed] [Google Scholar]
- Quirantes-Pine R, Zurek G, Barrajon-Catalan E, Bassmann C, Micol V, Segura-Carretero A, Fernandez-Gutierrez A. A metabolite-profiling approach to assess the uptake and metabolism of phenolic compounds from olive leaves in SKBR3 cells by HPLC-ESI-QTOF-MS. J Pharm Biomed Anal. 2013;72:121–126. doi: 10.1016/j.jpba.2012.09.029. [DOI] [PubMed] [Google Scholar]
- Santiago-Mora R, Casado-Diaz A, De Castro MD, Quesada-Gomez JM. Oleuropein enhances osteoblastogenesis and inhibits adipogenesis: the effect on differentiation in stem cells derived from bone marrow. Osteoporos Int. 2011;22(2):675–684. doi: 10.1007/s00198-010-1270-x. [DOI] [PubMed] [Google Scholar]
- Serra A, Rubio L, Borras X, Macia A, Romero MP, Motilva MJ. Distribution of olive oil phenolic compounds in rat tissues after administration of a phenolic extract from olive cake. Mol Nutr Food Res. 2012;56(3):486–496. doi: 10.1002/mnfr.201100436. [DOI] [PubMed] [Google Scholar]
- Zoidou E, Melliou E, Gikas E, Tsarbopoulos A, Magiatis P, Skaltsounis AL. Identification of Throuba Thassos, a traditional Greek table olive variety, as a nutritional rich source of oleuropein. J Agric Food Chem. 2010;58(1):46–50. doi: 10.1021/jf903405e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.