Abstract
Although cell culture studies have provided landmark discoveries in the basic and applied life sciences, it is often under-appreciated that cells grown in culture are prone to generating artifacts. Here, we introduce the genotype status (exemplified by apolipoprotein E) of human-derived cells as a further important parameter that requires attention in cell culture experiments. Epidemiological and clinical studies indicate that variations from the main apolipoprotein E3/E3 genotype might alter the risk of developing chronic diseases, especially neurodegeneration, cardiovascular disease, and cancer. Whereas the apolipoprotein E allele distribution in human populations is well characterized, the apolipoprotein E genotype of human-derived cell lines is only rarely considered in interpreting cell culture data. However, we find that primary and immortalized human cell lines show substantial variation in their apolipoprotein E genotype status. We argue that the apolipoprotein E genotype status and corresponding gene expression level of human-derived cell lines should be considered to better avoid (or at least account for) inconsistencies in cell culture studies when different cell lines of the same tissue or organ are used and before extrapolating cell culture data to human physiology in health and disease.
Keywords: APOE, Cell culture artifacts, Human-derived cell lines, Genotyping, Cardiovascular disease, Neurodegeneration
Cell lines derived from humans or other animals are commonly used tools which dominate many research fields in the basic and applied life sciences. Some of the main reasons for conducting cell culture experiments, rather than in vivo studies, are their comparatively low input requirement in terms of time, labor, and financial resources (Freshney 2005). Indeed, cell culture studies have provided landmark discoveries, such as the identification of the tumor suppressor protein p53 and the description of telomeres as important regulators of the cell cycle and cell senescence machinery (Olovnikov 1973; Lane and Crawford 1979; Shampay et al. 1984). At the same time, however, it is important to remember that cells grown in culture have not only been removed from their natural environment but are often prone to generating cell culture artifacts. We know, for example, that the oxygen concentrations in most mammalian organs and tissues range from 1 to 6 % (with up to 14 % in arterial blood) to ensure physiological tissue function but at the same time to minimize the production of potentially detrimental reactive oxygen and nitrogen species (Roy et al. 2003; Sullivan et al. 2006; Shay and Wright 2007; Halliwell and Gutteridge 2007; Oze et al. 2012). Yet, most cell culture experiments are conducted in an atmosphere unnaturally rich in oxygen due to the cells’ exposure to normal air which contains 21 % oxygen (Halliwell 2003). There is good experimental evidence that such a high oxygen concentration often exerts detrimental effects on cell metabolism and subsequently cell growth and survival (Alaluf et al. 2000; Busuttil et al. 2003; Estrada et al. 2012; Long and Halliwell 2012). Other important contributors to artifactual data are the use of misidentified cell lines (Nardone 2008; American Type Culture Collection Standards Development Organization Workgroup ASN-0002 2010) and the instability of test compounds added into cell culture media. Whereas the reaction of autoxidizable test compounds (e.g., ascorbate, epigallocatechin-3-gallate, quercetin) with components of cell culture media can produce hydrogen peroxide (Long et al. 2000; Halliwell 2003; Long and Halliwell 2012), some test compounds (e.g., curcumin and resveratrol) rapidly decompose into agents often with unknown biological activities (Long et al. 2010). Genomic instability of cells in long-term culture is another area of concern (Gazdar et al. 2010; Skrobot et al. 2007). Depending on the experimental conditions, an approximately 30-fold variation in the spontaneous mutation rate has been determined for the mouse lymphoma cell line, GRSL13 (Boesen et al. 1994), for example. Such cell culture-induced genetic alterations might not only affect immortalized cell lines, but also, for instance, embryonic stem cells (Skrobot et al. 2007; Wu et al. 2011; Amps et al. 2011).
Here, we would like to introduce the genotype status of human-derived cells as another important parameter that requires attention in cell culture experiments. The genetic profile of cells is mechanistically important as it largely determines their phenotype and thus their response pattern to environmental changes (Desiere 2004; Ferguson 2008; Sharp et al. 1997) both in vivo and, as we will argue, also in vitro. In this regard, the apolipoprotein E (APOE) genotype is probably one of the best-known genetic factors with respect to onset and progression of several common chronic diseases (Davignon et al. 1988; Mrkonjic et al. 2009; Arold et al. 2012). From two SNPs, three alleles arise (APOE2, APOE3 and APOE4) which encode the main protein isoforms apoE2, apoE3, and apoE4 (Weisgraber et al. 1981). Various tissues express APOE, most notably liver (about 2/3 of total apoE synthesis), immune cells (macrophages, neutrophils), brain (especially astrocytes), spleen and kidney (Zhang et al. 2011). In addition, APOE expression has been detected in heart, testis, prostate, pancreas, and several other organs (Zannis et al. 1985; Law et al. 1997). Functionally, the apoE protein acts as a key regulator of cholesterol and lipid metabolism (Fazio et al. 2000).
Several studies describe an increased risk for cardiovascular disease (CVD) in APOE4 carriers (Davignon et al. 1988), probably originating from elevated levels of LDL cholesterol, although the exact mechanisms underlying the APOE4-CVD-risk associations are likely to be more complex (Minihane et al. 2007). Similarly, the APOE genotype exerts strong effects on the pathological processes leading to neurodegenerative diseases (Maezawa et al. 2006; Kornecook et al. 2010; Arold et al. 2012). Indeed, the prevalence of Alzheimer’s disease dramatically increases with the number of APOE4 alleles (Corder et al. 1993), while lifespan is shortened in APOE4 carriers (Smith 2002; Christensen et al. 2006). Furthermore, recent evidence indicates that possession of the APOE4 genotype may not only significantly affect the levels of biomarkers of oxidative stress and inflammation in animal models and humans (Dietrich et al. 2005; Vitek et al. 2009; Graeser et al. 2012), but may also regulate their vitamin D levels as well as vitamin E uptake (Huebbe et al. 2009, 2011). Consequently, variations in the APOE genotype are frequently considered in the analysis of clinical data (Corder et al. 1993; Minihane et al. 2007). In contrast, the APOE genotype status has rarely been taken into account in the plethora of published studies conducted in human-derived cell lines (Dupont-Wallois et al. 1997; Riddell et al. 2008; Jeannesson et al. 2009). We suggest that it is time to rethink.
First, the impact that different APOE genotypes can exert on the metabolism of cells in culture and their phenotypic behavior is well established. In agreement with in vivo data, cells of animal origin transfected with human APOE and those obtained from human APOE-targeted replacement mice show distinct APOE genotype-dependent differences in their cellular responses. Several studies suggest significantly increased levels of oxidative stress and inflammatory biomarkers in apoE4-synthesizing cells (Colton et al. 2002; Jofre-Monseny et al. 2007). In addition, considerable evidence demonstrates that apoE4 can, at least under certain circumstances, induce mitochondrial dysfunction in cells (Chen et al. 2011). Possible explanations for these findings are apoE isoform-dependent differences in apoE degradation and in the expression level of disease-modifying proteins, such as NFκB, Nrf2, and metallothionein (Ophir et al. 2005; Elliott et al. 2011; Graeser et al. 2011, 2012). Furthermore, apoE isoform- and dose-dependent effects, for example, on hydrogen peroxide scavenging and metal chelation, have been reported (Miyata and Smith 1996). The exact pathological mechanisms of apoE4 at the cellular level, however, remain to be elucidated.
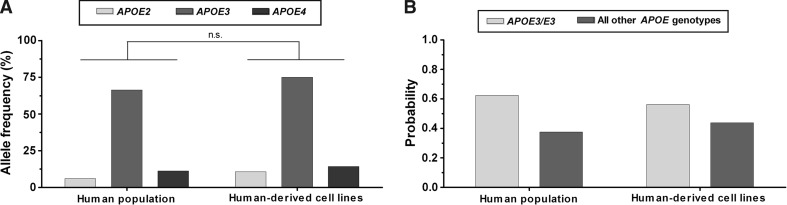
Second, a screen conducted in our two laboratories of all locally available human-derived cell lines reveals substantial variations in their APOE genotype status (Table 1). Among those cell lines which were hetero- or homozygous for the APOE2 or APOE4 allele (and thus diverge from the most common APOE3/E3 genotype) are popular cancer and immortalized cell lines, such as HaCat (APOE2/E4), HeLa (APOE3/E4), PC-3 (APOE2/E2), and U937 (APOE4/E4) as well as several primary cell lines obtained from human donors (Table 1). Despite our comparatively small sample size, examples of cell lines diverging from the main APOE3/E3 genotype were found for all human organs and tissues of origin (e.g., blood, bone, brain, cervix, colon, heart, kidney, liver, prostate, and skin) included here, with the exception of human lung and breast tissue-derived cells (as well as the fusion cell line EA.hy926; kindly provided by Prof. C. J. Edgell, University of North Carolina at Chapel Hill). Of note, from the APOE allele distributions depicted in Fig. 1a, which was not significantly different between human-derived cell lines and published population data, it follows that the probability of selecting a human-derived cell line carrying the most commonly found APOE3/E3 genotype is about 50–60 %. Conversely, scientists who randomly select a cell line originating from a human donor have an approximately 40–50 % chance of conducting experiments with cells whose genotype deviates from the main APOE3/E3 genotype (Fig. 1b).
Table 1.
APOE genotype status of human-derived cell lines
| No. | Tissue of origin | Cell line | Cell line description | Primary cell line (yes/no) | Donor detailsa | APOE genotype | ||
|---|---|---|---|---|---|---|---|---|
| Age (years) | Gender | Ethnicity | ||||||
| 1 | Aorta | VSMC | Vascular smooth muscle | Yes | 11 Months | F | Caucasian | E3/E4 |
| 2 | Blood | HL 60 | Human acute promyelocytic leukemia | No | 36 | F | Caucasian | E3/E3 |
| 3 | Jurkat Clone E6-1 | Human acute T lymphocytes leukemia | No | 14 | M | – | E3/E3 | |
| 4 | THP-1 | Human acute monocytic leukemia | No | 1 | M | – | E3/E3 | |
| 5 | U937 | Human histiocytic lymphoma | No | 37 | M | Caucasian | E4/E4 | |
| 6 | Bone | HTB 85/Saos-2 | Human bone osteosarcoma | No | 11 | F | Caucasian | E2/E3 |
| 7 | HTB 94/SW1353 | Human bone chondrosarcoma | No | 72 | F | Caucasian | E2/E3 | |
| 8 | HAC | Human articular chondrocytes | Yes | – | – | – | E3/E3 | |
| 9 | Brain | CCF-STTG1 | Human astrocytoma | No | 68 | F | Caucasian | E3/E4 |
| 10 | Kelly cells | Human neuroblastoma | No | – | – | – | E3/E4 | |
| 11 | SH-SY5Y | Human neuroblastoma | No | 4 | F | – | E3/E3 | |
| 12 | U87 | Human glioblastoma/astrocytoma | No | 44 | – | Caucasian | E3/E3 | |
| 13 | U118 | Human glioblastoma/astrocytoma | No | 50 | M | Caucasian | E2/E4 | |
| 14 | Breast | MCF-7 | Human breast adenocarcinoma | No | 69 | F | Caucasian | E3/E3 |
| 15 | Cervix | HeLa | Human cervix adenocarcinoma | No | 31 | F | African | E3/E4 |
| 16 | Colon | CaCo-2 | Human colorectal adenocarcinoma | No | 72 | M | Caucasian | E3/E3 |
| 17 | HCT 116 | Human colorectal carcinoma | No | Adult | M | – | E3/E3 | |
| 18 | HT 29 | Human colorectal adenocarcinoma | No | 44 | F | Caucasian | E3/E3 | |
| 19 | RKO | Human colon carcinoma | No | – | – | – | E3/E4 | |
| 20 | CCD33Co | Human normal colon fibroblasts | Yes | 7 | M | Caucasian | E2/E4 | |
| 21 | Fusion cell line | EA.hy926 | Somatic cell hybrid of primary HUVEC and thio-guanine resistant clone A549 | No | – | – | – | E3/E3 |
| 22 | Kidney | HEK 293 | Human embryonic kidney | No | Fetus | – | – | E3/E3 |
| 23 | HK-2 | Immortalized human normal kidney proximal tubule epithelial cells | No | Adult | M | – | E3/E3 | |
| 24 | RPTEC | Human normal renal proximal tubular epithelial cells | Yes | – | – | – | E2/E3 | |
| 25 | Liver | HepG2 | Human hepatocellular carcinoma | No | 15 | M | Caucasian | E3/E3 |
| 26 | HuH-7 | Human hepatocellular carcinoma | No | 57 | M | Japanese | E3/E3 | |
| 27 | Kyn 2 | Human pleomorphic hepatocellular carcinoma | No | 52 | M | Japanese | E3/E3 | |
| 28 | HFH | Human fetal hepatocytes | Yes | – | – | – | E3/E4 | |
| 29 | Lung | A549 | Human alveolar basal carcinoma | No | 58 | M | Caucasian | E3/E3 |
| 30 | 16HBE140 | Human bronchial epithelial cells | Yes | – | – | – | E3/E3 | |
| 31 | IMR90 | Human normal lung fibroblasts | Yes | 16 Weeks | F | Caucasian | E3/E3 | |
| 32 | Prostate | PC-3 | Human prostate carcinoma | No | 62 | M | Caucasian | E2/E2 |
| 33 | DU145 | Human prostate carcinoma (from brain) | No | 69 | M | Caucasian | E3/E3 | |
| 34 | LNCaP | Human prostate carcinoma (from lymph node) | No | 50 | M | Caucasian | E3/E3 | |
| 35 | Skin | HaCat | Immortalized human normal keratinocytes | No | 62 | M | Caucasian | E2/E4 |
| 36 | NEB-1 | Immortalized human normal Epidemolysis-Bullosa cells | No | – | – | – | E3/E3 | |
| 37 | CRL 2115 | Human normal skin fibroblasts | Yes | 27 | M | Caucasian | E3/E3 | |
| 38 | GM16678 | Human RCP-3 skin fibroblasts | Yes | 4 Months | F | Caucasian/Lebanese | E3/E3 | |
| 39 | HDFn | Human dermal fibroblasts | Yes | Neonatal | – | – | E3/E4 | |
| 40 | KF116 | Human keloid fibroblasts | Yes | – | – | – | E3/E3 | |
| 41 | KF112 | Human dermal fibroblasts (from breast) | Yes | 35 | F | Chinese | E2/E3 | |
| 42 | PHK | Human keratinocytes | Yes | – | – | – | E3/E3 | |
The APOE genotype was determined either by RFLP analysis (Singapore) or by TaqMan® method (Germany). A subset of data was taken from (Dupont-Wallois et al. 1997; Riddell et al. 2008; Jeannesson et al. 2009)
a Donor details were obtained from various cell line repositories [Japanese Collection of Research Bioresources (http://www.cellbank.nibio.go.jp), ATCC (http://www.atcc.org), NIGMS Human Genetics Cell Repository, Coriell Institute (http://ccr.coriell.org/Sections/Collections/NIGMS/?SsId=8), Cell Lines Service (http://www.cell-lines-service.de/content/index_eng.html); all accessed electronically on 28/11/2011]; details for cell lines KF116 and KF112 were obtained from the Wound Healing and Stem Cell Research Group (NUS)
Fig. 1.
APOE comparison between human world population (Singh et al. 2006) and human-derived cell lines. a Allele frequency for APOE2, APOE3, and APOE4. Selected data sets were analyzed by χ 2 test (n.s., non-significant). b Probability to select an APOE3/E3 compared to any of the other APOE genotypes (i.e., APOE2/E2, APOE2/E3, APOE3/E4, APOE4/E4, APOE2/E4)
Third, the actual impact that a particular polymorphism exerts on cell physiology and disease susceptibility depends on the level of gene expression (Jeannesson et al. 2009). As mentioned before, various tissues express APOE, although at different levels (Zannis et al. 1985; Law et al. 1997; Zhang et al. 2011). However, one must be careful of automatically assuming that cells obtained from human APOE-expressing tissue show similar levels of APOE gene expression in vitro. As an immune cell, the human macrophage-like U937 cell line (APOE4/E4) is expected to express APOE. Previous studies, however, failed to detect APOE mRNA in this cell line cultured both under standard conditions or the presence of APOE expression inducers such as dexamethasone, thus limiting their suitability for studying CVD and other disease mechanisms (Zannis et al. 1985). Similarly, no endogenous APOE gene expression has been found in HeLa cells (APOE3/E4) (Smith et al. 1988). In a direct comparison of three human prostate carcinoma cell lines, APOE was highly expressed in PC-3 cells (APOE2/E2), detectable in DU145 cells (APOE3/E3) but absent in LNCaP cells (APOE3/E3) (Venanzoni et al. 2003), thus again highlighting the importance of determining both APOE genotype and expression level for minimizing biased data interpretation. Interestingly, PC-3 cells were highly tumorigenic, DU-145 cells were moderately tumorigenic, and LNCaP cells were only weakly tumorigenic (Venanzoni et al. 2003). It is tempting to speculate that the tumorigenic potential of the three human prostate carcinoma cell lines was influenced not only by the actual apoE expression level but also by the differences in the APOE genotype status.
Taken together, we think that the data and arguments summarized above warrant a more widespread consideration of the APOE genotype status by research groups working with human-derived cell lines. In addition to immortalized cell lines, this approach is particularly important when working with primary cells as their APOE genotype might vary with each new donor. Of course, as the cause of most diseases is not monogenetic, a cell’s response to experimental stimuli depends not only on the presence of a particular APOE genotype, but on the presence and interplay of various risk genes in a given environment (Jeannesson et al. 2009). Considering that many chronic disease processes are driven, at least partly, by inflammation and oxidative stress, we suggest that SNPs of genes associated with both phenomena are probably also worth monitoring. Screening human-derived cell lines for the presence of polymorphisms in the tumor necrosis factor alpha (Elahi et al. 2009; Qidwai and Khan 2011) and paraoxonase-1/-2 genes (Shih and Lusis 2009; Schrader and Rimbach 2011) might be particularly interesting in this context. Ultimately, it might be necessary to genotype a particular human-derived cell line for several, and sometimes even all, gene variants that are known to be important in cell metabolism. As discussed for U937 and the set of human prostate carcinoma cells, the obtained genotype data should be ideally supplemented with information regarding the actual gene expression (i.e., effective protein level). Obviously, the final course of action should always be context-driven.
From a practical point of view, genotyping can be accomplished comparatively easily by RFLP or TaqMan® probe analysis (Table 1) or by one of the other methods published in the recent literature (Hixson and Vernier 1990; Calero et al. 2009; Rihn et al. 2009). Similarly to cell line authentication data, information regarding the genotype status of human-derived cell lines should be made easily available, perhaps in the form of an online database. We believe that the genotype status and the corresponding gene expression level of human-derived cell lines could help the scientific community to better avoid (or at least account for) inconsistencies in cell culture studies when different cell lines of the same tissue or organ are used and before extrapolating cell culture data to human physiology in health and disease.
Acknowledgments
BH, SS, and VL are most grateful for financial support provided by the Biomedical Research Council of Singapore (BMRC Grant 09/1/21/19/598).
Conflict of interest
None.
References
- Alaluf S, Muir-Howie H, Hu HL, et al. Atmospheric oxygen accelerates the induction of a post-mitotic phenotype in human dermal fibroblasts: the key protective role of glutathione. Differentiation. 2000;66:147–155. doi: 10.1046/j.1432-0436.2000.660209.x. [DOI] [PubMed] [Google Scholar]
- American Type Culture Collection Standards Development Organization Workgroup ASN-0002 Cell line misidentification: the beginning of the end. Nat Rev Cancer. 2010;10:441–448. doi: 10.1038/nrc2852. [DOI] [PubMed] [Google Scholar]
- Amps K, Andrews PW, Anyfantis G, et al. Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat Biotechnol. 2011;29:1132–1144. doi: 10.1038/nbt.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arold S, Sullivan P, Bilousova T, et al. Apolipoprotein E level and cholesterol are associated with reduced synaptic amyloid beta in Alzheimer’s disease and apoE TR mouse cortex. Acta Neuropathol. 2012;123:39–52. doi: 10.1007/s00401-011-0892-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesen JJ, Niericker MJ, Dieteren N, et al. How variable is a spontaneous mutation rate in cultured mammalian cells? Mutat Res. 1994;307:121–129. doi: 10.1016/0027-5107(94)90284-4. [DOI] [PubMed] [Google Scholar]
- Busuttil RA, Rubio M, Dolle ME, et al. Oxygen accelerates the accumulation of mutations during the senescence and immortalization of murine cells in culture. Aging Cell. 2003;2:287–294. doi: 10.1046/j.1474-9728.2003.00066.x. [DOI] [PubMed] [Google Scholar]
- Calero O, Hortiguela R, Bullido MJ, et al. Apolipoprotein E genotyping method by real time PCR, a fast and cost-effective alternative to the TaqMan and FRET assays. J Neurosci Method. 2009;183:238–240. doi: 10.1016/j.jneumeth.2009.06.033. [DOI] [PubMed] [Google Scholar]
- Chen HK, Ji ZS, Dodson SE, et al. Apolipoprotein E4 domain interaction mediates detrimental effects on mitochondria and is a potential therapeutic target for Alzheimer disease. J Biol Chem. 2011;286:5215–5221. doi: 10.1074/jbc.M110.151084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K, Johnson TE, Vaupel JW. The quest for genetic determinants of human longevity: challenges and insights. Nat Rev Genet. 2006;7:436–448. doi: 10.1038/nrg1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colton CA, Brown CM, Cook D, et al. APOE and the regulation of microglial nitric oxide production: a link between genetic risk and oxidative stress. Neurobiol Aging. 2002;23:777–785. doi: 10.1016/S0197-4580(02)00016-7. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Davignon J, Gregg RE, Sing CF. Apolipoprotein E polymorphism and atherosclerosis. Arteriosclerosis. 1988;8:1–21. doi: 10.1161/01.ATV.8.1.1. [DOI] [PubMed] [Google Scholar]
- Desiere F. Towards a systems biology understanding of human health: interplay between genotype, environment and nutrition. Biotechnol Annu Rev. 2004;10:51–84. doi: 10.1016/S1387-2656(04)10003-3. [DOI] [PubMed] [Google Scholar]
- Dietrich M, Hu Y, Block G, et al. Associations between apolipoprotein E genotype and circulating F2-isoprostane levels in humans. Lipids. 2005;40:329–334. doi: 10.1007/s11745-006-1390-4. [DOI] [PubMed] [Google Scholar]
- Dupont-Wallois L, Soulie C, Sergeant N, et al. ApoE synthesis in human neuroblastoma cells. Neurobiol Dis. 1997;4:356–364. doi: 10.1006/nbdi.1997.0155. [DOI] [PubMed] [Google Scholar]
- Elahi MM, Asotra K, Matata BM, et al. Tumor necrosis factor alpha -308 gene locus promoter polymorphism: an analysis of association with health and disease. Biochim Biophys Acta. 2009;1792:163–172. doi: 10.1016/j.bbadis.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Elliott DA, Tsoi K, Holinkova S, et al. Isoform-specific proteolysis of apolipoprotein-E in the brain. Neurobiol Aging. 2011;32:257–271. doi: 10.1016/j.neurobiolaging.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Estrada JC, Albo C, Benguria A, et al. Culture of human mesenchymal stem cells at low oxygen tension improves growth and genetic stability by activating glycolysis. Cell Death Differ. 2012;19:743–755. doi: 10.1038/cdd.2011.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazio S, Linton MF, Swift LL. The cell biology and physiologic relevance of ApoE recycling. Trends Cardiovasc Med. 2000;10:23–30. doi: 10.1016/S1050-1738(00)00033-5. [DOI] [PubMed] [Google Scholar]
- Ferguson LR. Dissecting the nutrigenomics, diabetes, and gastrointestinal disease interface: from risk assessment to health intervention. OMICS. 2008;12:237–244. doi: 10.1089/omi.2008.0044. [DOI] [PubMed] [Google Scholar]
- Freshney RI. Culture of animal cells: a manual of basic technique. New York: Wiley; 2005. [Google Scholar]
- Gazdar AF, Gao B, Minna JD. Lung cancer cell lines: useless artifacts or invaluable tools for medical science? Lung Cancer. 2010;68:309–318. doi: 10.1016/j.lungcan.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeser AC, Boesch-Saadatmandi C, Lippmann J, et al. Nrf2-dependent gene expression is affected by the proatherogenic apoE4 genotype-studies in targeted gene replacement mice. J Mol Med. 2011;89:1027–1035. doi: 10.1007/s00109-011-0771-1. [DOI] [PubMed] [Google Scholar]
- Graeser AC, Huebbe P, Storm N, et al. Apolipoprotein E genotype affects tissue metallothionein levels: studies in targeted gene replacement mice. Genes Nutr. 2012;7:247–255. doi: 10.1007/s12263-012-0282-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B. Oxidative stress in cell culture: an under-appreciated problem? FEBS Lett. 2003;540:3–6. doi: 10.1016/S0014-5793(03)00235-7. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge J. Free radicals in biology and medicine. Oxford: Oxford University Press; 2007. [Google Scholar]
- Hixson JE, Vernier DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res. 1990;31:545–548. [PubMed] [Google Scholar]
- Huebbe P, Jofre-Monseny L, Rimbach G. Alpha-tocopherol transport in the lung is affected by the apoE genotype—studies in transgenic apoE3 and apoE4 mice. IUBMB Life. 2009;61:453–456. doi: 10.1002/iub.177. [DOI] [PubMed] [Google Scholar]
- Huebbe P, Nebel A, Siegert S, et al. APOE epsilon4 is associated with higher vitamin D levels in targeted replacement mice and humans. FASEB J. 2011;25:3262–3270. doi: 10.1096/fj.11-180935. [DOI] [PubMed] [Google Scholar]
- Jeannesson E, Siest G, Zaiou M, et al. Genetic profiling of human cell lines used as in vitro model to study cardiovascular pathophysiology and pharmacotoxicology. Cell Biol Toxicol. 2009;25:561–571. doi: 10.1007/s10565-008-9112-8. [DOI] [PubMed] [Google Scholar]
- Jofre-Monseny L, Pascual-Teresa S, Plonka E, et al. Differential effects of apolipoprotein E3 and E4 on markers of oxidative status in macrophages. Br J Nutr. 2007;97:864–871. doi: 10.1017/S0007114507669219. [DOI] [PubMed] [Google Scholar]
- Kornecook TJ, McKinney AP, Ferguson MT, et al. Isoform-specific effects of apolipoprotein E on cognitive performance in targeted-replacement mice overexpressing human APP. Genes Brain Behav. 2010;9:182–192. doi: 10.1111/j.1601-183X.2009.00545.x. [DOI] [PubMed] [Google Scholar]
- Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- Law GL, McGuinness MP, Linder CC, et al. Expression of apolipoprotein E mRNA in the epithelium and interstitium of the testis and the epididymis. J Androl. 1997;18:32–42. [PubMed] [Google Scholar]
- Long LH, Halliwell B. The effects of oxaloacetate on hydrogen peroxide generation from ascorbate and epigallocatechin gallate in cell culture media: potential for altering cell metabolism. Biochem Biophys Res Commun. 2012;417:446–450. doi: 10.1016/j.bbrc.2011.11.136. [DOI] [PubMed] [Google Scholar]
- Long LH, Clement MV, Halliwell B. Artifacts in cell culture: rapid generation of hydrogen peroxide on addition of (−)-epigallocatechin, (−)-epigallocatechin gallate, (+)-catechin, and quercetin to commonly used cell culture media. Biochem Biophys Res Commun. 2000;273:50–53. doi: 10.1006/bbrc.2000.2895. [DOI] [PubMed] [Google Scholar]
- Long LH, Hoi A, Halliwell B. Instability of and generation of hydrogen peroxide by, phenolic compounds in cell culture media. Arch Biochem Biophys. 2010;501:162–169. doi: 10.1016/j.abb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Maezawa I, Nivison M, Montine KS, et al. Neurotoxicity from innate immune response is greatest with targeted replacement of E4 allele of apolipoprotein E gene and is mediated by microglial p38MAPK. FASEB J. 2006;20:797–799. doi: 10.1096/fj.05-5423fje. [DOI] [PubMed] [Google Scholar]
- Minihane AM, Jofre-Monseny L, Olano-Martin E, et al. ApoE genotype, cardiovascular risk and responsiveness to dietary fat manipulation. Proc Nutr Soc. 2007;66:183–197. doi: 10.1017/S0029665107005435. [DOI] [PubMed] [Google Scholar]
- Miyata M, Smith JD. Apolipoprotein E allele-specific antioxidant activity and effects on cytotoxicity by oxidative insults and beta-amyloid peptides. Nat Genet. 1996;14:55–61. doi: 10.1038/ng0996-55. [DOI] [PubMed] [Google Scholar]
- Mrkonjic M, Chappell E, Pethe VV, et al. Association of apolipoprotein E polymorphisms and dietary factors in colorectal cancer. Br J Cancer. 2009;100:1966–1974. doi: 10.1038/sj.bjc.6605097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardone RM. Curbing rampant cross-contamination and misidentification of cell lines. Biotechniques. 2008;45:221–227. doi: 10.2144/000112925. [DOI] [PubMed] [Google Scholar]
- Olovnikov AM. A theory of marginotomy: the incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- Ophir G, Amariglio N, Jacob-Hirsch J, et al. Apolipoprotein E4 enhances brain inflammation by modulation of the NF-kappaB signaling cascade. Neurobiol Dis. 2005;20:709–718. doi: 10.1016/j.nbd.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Oze H, Hirao M, Ebina K, et al. Impact of medium volume and oxygen concentration in the incubator on pericellular oxygen concentration and differentiation of murine chondrogenic cell culture. In Vitro Cell Dev Biol Anim. 2012;48:123–130. doi: 10.1007/s11626-011-9479-3. [DOI] [PubMed] [Google Scholar]
- Qidwai T, Khan F. Tumour necrosis factor gene polymorphism and disease prevalence. Scand J Immunol. 2011;74:522–547. doi: 10.1111/j.1365-3083.2011.02602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddell DR, Zhou H, Atchison K, et al. Impact of apolipoprotein E (ApoE) polymorphism on brain ApoE levels. J Neurosci. 2008;28:11445–11453. doi: 10.1523/JNEUROSCI.1972-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihn BH, Berrahmoune S, Jouma M, et al. APOE genotyping: comparison of three methods. Clin Exp Med. 2009;9:61–65. doi: 10.1007/s10238-008-0012-2. [DOI] [PubMed] [Google Scholar]
- Roy S, Khanna S, Bickerstaff AA, et al. Oxygen sensing by primary cardiac fibroblasts: a key role of p21(Waf1/Cip1/Sdi1) Circ Res. 2003;92:264–271. doi: 10.1161/01.RES.0000056770.30922.E6. [DOI] [PubMed] [Google Scholar]
- Schrader C, Rimbach G. Determinants of paraoxonase 1 status: genes, drugs and nutrition. Curr Med Chem. 2011;18:5624–5643. doi: 10.2174/092986711798347216. [DOI] [PubMed] [Google Scholar]
- Shampay J, Szostak JW, Blackburn EH. DNA sequences of telomeres maintained in yeast. Nature. 1984;310:154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- Sharp MG, Kantachuvesiri S, Mullins JJ. Genotype and cardiovascular phenotype: lessons from genetically manipulated animals and diseased humans. Curr Opin Nephrol Hypertens. 1997;6:51–57. doi: 10.1097/00041552-199701000-00010. [DOI] [PubMed] [Google Scholar]
- Shay JW, Wright WE. Tissue culture as a hostile environment: identifying conditions for breast cancer progression studies. Cancer Cell. 2007;12:100–101. doi: 10.1016/j.ccr.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Shih DM, Lusis AJ. The roles of PON1 and PON2 in cardiovascular disease and innate immunity. Curr Opin Lipidol. 2009;20:288–292. doi: 10.1097/MOL.0b013e32832ca1ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PP, Singh M, Mastana SS. APOE distribution in world populations with new data from India and the UK. Ann Hum Biol. 2006;33:279–308. doi: 10.1080/03014460600594513. [DOI] [PubMed] [Google Scholar]
- Skrobot VN, Cukusic A, Ferenac KM, et al. Telomere dynamics and genome stability in the human pancreatic tumor cell line MIAPaCa-2. Cytogenet Genome Res. 2007;119:60–67. doi: 10.1159/000109620. [DOI] [PubMed] [Google Scholar]
- Smith JD. Apolipoproteins and aging: emerging mechanisms. Ageing Res Rev. 2002;1:345–365. doi: 10.1016/S1568-1637(02)00005-3. [DOI] [PubMed] [Google Scholar]
- Smith JD, Melian A, Leff T, et al. Expression of the human apolipoprotein E gene is regulated by multiple positive and negative elements. J Biol Chem. 1988;263:8300–8308. [PubMed] [Google Scholar]
- Sullivan M, Galea P, Latif S. What is the appropriate oxygen tension for in vitro culture? Mol Hum Reprod. 2006;12:653. doi: 10.1093/molehr/gal081. [DOI] [PubMed] [Google Scholar]
- Venanzoni MC, Giunta S, Muraro GB, et al. Apolipoprotein E expression in localized prostate cancers. Int J Oncol. 2003;22:779–786. [PubMed] [Google Scholar]
- Vitek MP, Brown CM, Colton CA. APOE genotype-specific differences in the innate immune response. Neurobiol Aging. 2009;30:1350–1360. doi: 10.1016/j.neurobiolaging.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisgraber KH, Rall SC, Jr, Mahley RW. Human E apoprotein heterogeneity. Cysteine-arginine interchanges in the amino acid sequence of the apo-E isoforms. J Biol Chem. 1981;256:9077–9083. [PubMed] [Google Scholar]
- Wu W, He Q, Li X, et al. Long-term cultured human neural stem cells undergo spontaneous transformation to tumor-initiating cells. Int J Biol Sci. 2011;7:892–901. doi: 10.7150/ijbs.7.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannis VI, Cole FS, Jackson CL, et al. Distribution of apolipoprotein A-I, C-II, C-III, and E mRNA in fetal human tissues. Time-dependent induction of apolipoprotein E mRNA by cultures of human monocyte-macrophages. Biochemistry. 1985;24:4450–4455. doi: 10.1021/bi00337a028. [DOI] [PubMed] [Google Scholar]
- Zhang H, Wu LM, Wu J. Cross-talk between apolipoprotein E and cytokines. Mediators Inflamm. 2011;2011:949072. doi: 10.1155/2011/949072. [DOI] [PMC free article] [PubMed] [Google Scholar]