Abstract
In Atlantic salmon aquaculture, early sexual maturation represents a major problem for producers. This is especially true for grilse, which mature after one sea winter before reaching a desirable harvest weight, rather than after two sea winters. Salmon maturing as grilse have a much lower market value than later maturing individuals. For this reason, most companies desire fish that grow fast and mature late. Marker-assisted selection has the potential to improve the efficiency of selection against early maturation and for late sexual maturation; however, studies identifying age of sexual maturation-related genetic markers are lacking for Atlantic salmon. Therefore, we used a 6.5K single-nucleotide polymorphism (SNP) array to genotype five families from the Mainstream Canada broodstock program and search for SNPs associated with early (grilsing) or late sexual maturation. There were 529 SNP loci that were variable across all five families, and this was the set that was used for quantitative trait loci (QTL) analysis. GridQTL identified two chromosomes, Ssa10 and Ssa21, containing QTL related to grilsing. In contrast, only one QTL, on Ssa18, was found linked to late maturation in Atlantic salmon. Our previous work on these five families did not identify genome-wide significant growth-related QTL on Ssa10, Ssa21, or Ssa18. Therefore, taken together, these results suggest that both grilsing and late sexual maturation are controlled independently of one another and also from growth-related traits. The identification of genomic regions associated with grilsing or late sexual maturation provide an opportunity to incorporate this information into selective breeding programs that will enhance Atlantic salmon farming.
Keywords: Atlantic salmon, Sexual maturation, Grilsing, QTL, SNP array
Introduction
The Atlantic salmon (Salmo salar) exhibits a great deal of variability in age and size at sexual maturation. This variation is observed both between and within strains and year classes (Garcia de Leaniz et al. 2007; Taranger et al. 2010). The timing of sexual maturation is controlled by a complex process that involves genetic and environmental components (Thorpe and Metcalfe 1998). Internal factors, such as age and lipid reserves, together with external factors, like light abundance, temperature, and food intake, seem to have an impact on the initiation of sexual maturation (Gardner 1976; Herbinger and Friars 1992; Simpson 1992). The interactions of these factors result in the enormous variability in the age of sexual maturation, especially in male Atlantic salmon, which are able to reach maturity at 1 to 7 years of age (Simpson 1992). There is also a genetic component to age at sexual maturation in Atlantic salmon, and estimates of heritabilities (h 2) for this trait in Atlantic salmon range widely from 0.09–0.17 (Gjerde et al. 1994; Wild et al. 1994; Gjedrem 2000) to 0.39 (Gjerde and Gjedrem 1984) to 0.48 (Gjerde 1984). These observations strongly indicate that age at sexual maturity in Atlantic salmon is a heritable trait. Thus, it should be possible to select for Atlantic salmon that do not become sexually mature as grilse and also for late sexually maturing fish.
In salmon aquaculture, early sexual maturation can be a major economic problem. This is particularly costly in terms of feed and cage space if fish mature as grilse (i.e., after 1 year in a sea cage; Gjedrem 2000). Salmon maturing as grilse have a much lower market value than later maturing individuals (Saunders et al. 1983). The maturation process is energetically expensive, and this is reflected in a decrease in growth rate, lower meat quality, and increased mortality through susceptibility to pathogens (Gjerde 1984; Thorpe 1994). Late-maturing salmon are desired in commercial operations, and to reduce the frequency of grilsing, early maturing Atlantic salmon are discarded as potential breeding candidates (Gjedrem 2012). Selection for the economically important production traits of fast growth and late sexual maturation has been considered problematic as it has been suggested that there is a correlation between the phenotypes of fast growth and early sexual maturation (Thorpe et al. 1983).
Marker-assisted selection has the potential to improve the efficiency of selection for traits such as age of sexual maturation and growth in Atlantic salmon breeding. Genomic regions linked to sexual maturation have been identified in other salmonid species such as rainbow trout and Arctic charr (Easton et al. 2011; Haidle et al. 2008; Küttner et al. 2011; Martyniuk et al. 2003; Moghadam et al. 2007). However, such studies in Atlantic salmon are lacking. The identification of genetic markers independently related to either early or late sexual maturation, especially in male Atlantic salmon, would enable the implementation of selective breeding based on improved genetic selection practices by identifying animals with favorable genotypes. We previously identified QTL for growth at different stages of the production cycle in five families from the Mainstream Canada broodstock (Gutierrez et al. 2012). Here we report QTL for grilsing and late sexual maturation in the same families and show that the genomic locations of these QTL are independent of one another as well as the QTL associated with growth.
Materials and Methods
Mapping Families and Phenotype Data
Families were part of a commercial broodstock program developed by Mainstream Canada and based on the Mowi strain of Atlantic salmon, which was derived from a breeding program established using Norwegian populations (Gjedrem et al. 1991). In November/December 2005, 130 single-pair mating families were established. At the fry stage (February 2006), 120 offspring from each family were pooled (15,600 fish in total) and grown communally at the Oceans Farms Hatchery, Vancouver Island. In September/October 2006, 5,000 of the fish were passive integrated transponder (PIT)-tagged, and phenotypic measurements were taken until early 2009. Maturation times in Atlantic salmon were classified as: precocious (≤12 months of age), grilse (36 months of age, at first sea winter (SW)), normally maturing (48 to 60 months of age at second SW or third SW), and late-maturing fish (>60 months) (Gjedrem 2000; Taranger et al. 2010). Sex was recorded during the confirmation of maturation status. The sex of an individual in the late-maturing group could not be ascertained phenotypically; therefore, we predicted the sex of the late-maturing Atlantic salmon using a polymerase chain reaction test, developed by Eisbrenner et al. (2013) that amplifies two exons of the sdY gene, which has been shown to be the sex-determining gene in rainbow trout and probably other members of the Salmoninae (Yano et al. 2012, 2013). It should be noted that the precocious Atlantic salmon that matured before 1 year of age were discarded during a standard grading procedure, and so no samples were available from this group.
DNA Extraction and Parental Assignments
These procedures were as described by Gutierrez et al. (2012). Briefly, DNA was extracted from the adipose fins of the PIT-tagged progeny and the parents used to produce the 130 families (Withler et al. 2005). The DNA was then genotyped using eight microsatellite markers, and parental assignment was carried out as described by Withler et al. (2007).
SNP Array and Linkage Mapping
The single-nucleotide polymorphism (SNP) data used for this analysis have been described previously (Gutierrez et al. 2012). Five families were selected for SNP genotyping at CIGENE, Norwegian University of Life Sciences, Ås, Norway, using an Atlantic salmon 6.5K Illumina iSelect SNP-array (Kent et al. 2009). Analyses were based on an Atlantic salmon linkage map, which contains ∼5,650 SNP markers and was constructed using genotyping data from 143 families comprising 3,297 fish (Lien et al. 2011). This map contains 29 linkage groups, which were assigned to their specific chromosome number according to the nomenclature established by Phillips et al. (2009). All non-informative markers were removed from the datasets, and subsequently, independent male and female linkage maps were manually created for each family and adjusted to the distances on the SNP map of Lien et al. (2011). Given the differences between the sets of informative markers present in each family, for this study, we restricted the analysis to the 529 markers which are informative in all of the five families. These markers are distributed across all 29 Atlantic salmon chromosomes. We have previously commented on some of the possible reasons why only ∼10% of the markers were variable in all of the five families (Gutierrez et al. 2012). It may reflect the starting population for the Mainstream Canada broodstock or past selection (e.g., primarily for growth in terms of days in salt water to reach 5 kg), or it may reflect the nature of biallelic SNPs. We note that many QTL studies in salmonids have traditionally used microsatellite markers, and these studies tend to have employed fewer variable markers and also fewer families. Even when SNPs were used as the genetic marker, the overall number tested for QTL analysis has been less than 350, and not all of these were variable in each family. Therefore, we consider this study to be robust in terms of what is currently available in the salmonid aquaculture literature.
QTL Analyses
QTL analyses were performed using a regression-interval mapping method available through the GridQTL portlet (Seaton et al. 2006). GridQTL is a portlet environment (available at http://www.gridqtl.org.uk/) that allows the analysis of extensive datasets. The datasets were analyzed as sib-pairs using the combined genotypic data from the 529 informative markers shared by all families. For the grilsing analysis, given the absence of female grilse, only the genotypic information from males was used. The phenotypic data were scored as a binary trait, i.e., male fish recorded as grilse were scored 0, and male fish recorded as maturing at second or third SW were scored as 1. The same procedure was used for the analysis of late sexual maturation, but both male and female fish were used in this case. Atlantic salmon that did not show evidence of gonadal development at 60 months were considered “late-maturing” and scored as 0. The results were illustrated using the female map as it provides a greater resolution than the male genetic map.
The percentage of phenotypic variance (PEV) explained by the QTL in the sib-pair analysis was calculated as a proportion of the phenotypic variance obtained from the observations. F-statistic values were calculated at 1 cM intervals on each chromosome for each analytical approach to yield the most likely position of the QTL. Empirical chromosome-wide significance thresholds were determined by permutation tests (Churchill and Doerge 1994). The 10,000 permutations were performed at the chromosome-wide level in order to establish the F-value thresholds for a p < 0.05 and p < 0.01. F-critical values corresponding to 0.05 > p > 0.01 were considered as “suggestive” QTL, whereas those corresponding to p < 0.01 were considered “significant” QTL. Those chromosomes which contained QTL that were found to be “significant” were tested for their significance at a genome-wide level of p < 0.05 by performing a 1,000 permutation test.
Results
QTL Analysis for Male Grilsing
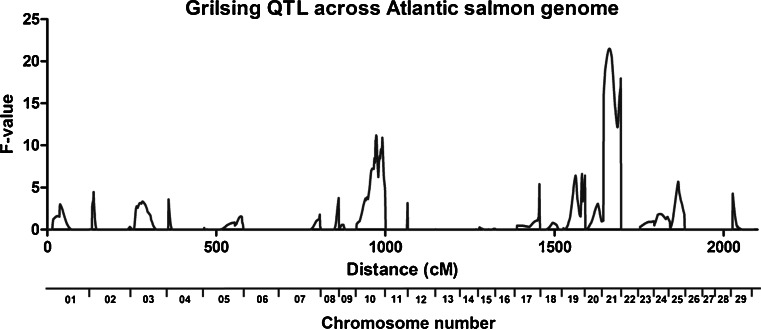
As described previously (Gutierrez et al. 2012), five families were chosen from the 2005 year class of the Mainstream Canada selective breeding program for analysis. Each family contained from five to eight male grilse. No female grilse were observed in any of these families (Table 1). As shown in Fig. 1 and Table 2, QTL analyses restricted to male grilsing revealed the presence of two statistically meaningful QTL, one on chromosome 10 (Ssa10) and the other on chromosome 21 (Ssa21). The QTL on Ssa21 was considered significant (p < 0.01), whereas the other on Ssa10 only reached the suggestive level of significance (0.05 > p > 0.01). Neither of these QTL reached genome-wide significance, which is in keeping with the relatively small sample size as well as the rather low h 2 estimates in Atlantic salmon for age at sexual maturation (Gjerde 1984; Gjerde et al. 1994; Wild et al. 1994). The PEV explained by these QTL was 16.7% and 25% in Ssa10 and Ssa21, respectively.
Table 1.
Number of grilse, normally maturing and late-maturing individuals per family used in this study
| Family | # Grilse individuals | # Normally maturing indiv. | # Late maturing indiv. | Total | ||||
|---|---|---|---|---|---|---|---|---|
| Male | Female | Male | Female | Male | Female | Grilsing analysis | Sexual mat. analysis | |
| F7 | 5 | 0 | 8 | 25 | 2 | 5 | 13 | 40 |
| F23 | 5 | 0 | 16 | 24 | 3 | 5 | 21 | 48 |
| F76 | 8 | 0 | 9 | 17 | 3 | 3 | 17 | 32 |
| F88 | 5 | 0 | 8 | 20 | 3 | 2 | 13 | 33 |
| F107 | 8 | 0 | 9 | 19 | 6 | 8 | 17 | 42 |
| Total | 31 | 0 | 50 | 105 | 17 | 23 | 81 | 195 |
Fig. 1.
QTL detection for male grilsing across the 29 Atlantic salmon chromosomes
Table 2.
Chromosomes containing male grilsing and late sexual maturation QTL in Atlantic salmon
| Trait | Chromosome | Position | F-value | LOD | %PEV | RT | AC |
|---|---|---|---|---|---|---|---|
| Grilsing | Ssa10a | 60 cM | 11.18 | 2.43 | 16.7 | 27 MT | 4 MT |
| Grilsing | Ssa21b | 17 cM | 21.49 | 4.67 | 25.0 | 5 MT | 36 BW, K |
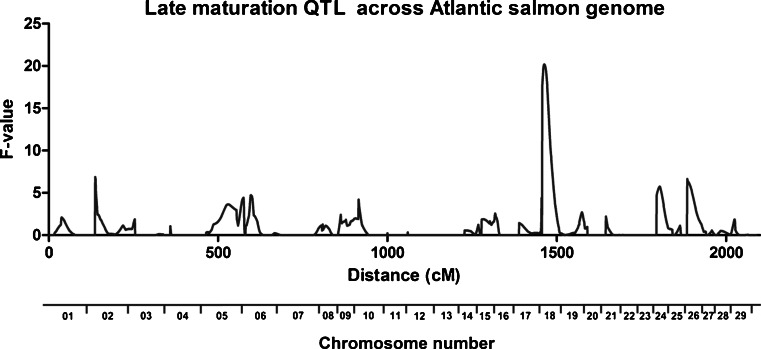
| Late maturation | Ssa18 b | 6 cM | 20.17 | 4.38 | 14.7 | – | – |
RT indicates homologous linkage group in rainbow trout, AC indicates homologous linkage group in Arctic charr, MT indicates a QTL associated with maturation, BW indicates a QTL associated with body weight, K indicates a QTL associated with condition factor
aIndicates suggestive QTL
bIndicates significant QTL
QTL Analysis of Late Sexual Maturation
From the 195 individuals used in the analysis of late sexual maturation, 40 were fish that did not show gonadal development after their third SW. The sex of the individuals in this latter group was determined based on the presence of two of the four exons of the sdY gene (Eisbrenner et al. 2013). Seventeen of the late-maturing 40 fish were male and 23 were female (Table 1). The GridQTL analysis revealed the presence of only one statistically significant (p < 0.01) QTL on chromosome 18 (Ssa18), which did not reach genome-wide significance, even though it was the only QTL found for this particular trait (Fig. 2). This QTL explained 14.7 % of the PEV associated with late sexual maturation in these Atlantic salmon families.
Fig. 2.
QTL detection for late sexual maturation across the 29 Atlantic salmon chromosomes
Discussion
Previous studies performed in rainbow trout and Arctic charr have shown an apparent link between QTL for sexual maturation and growth (Haidle et al. 2008; Martyniuk et al. 2003; Moghadam et al. 2007); however, our results indicate a different scenario for Atlantic salmon in the Mainstream Canada breeding program. Here we only detected two QTL associated with male grilsing in Atlantic salmon, on Ssa10 and Ssa21. Previously using the same families, Gutierrez et al. (2012) found genome-wide significant growth-related QTL on chromosomes Ssa02, Ssa07, Ssa13, Ssa09, Ssa17, and Ssa26. Although Gutierrez et al. (2012) found some suggestive evidence for growth-related QTL on Ssa10 and Ssa21, neither reached genome-wide significance, and they mapped to different regions of the chromosomes compared with the grilsing QTL. Moreover, the results of QTL analyses for growth and sexual maturation in the Mainstream Canada broodstock families indicate that sexual maturation is controlled by fewer genomic regions than growth in Atlantic salmon. Therefore, it should be possible to tease apart these traits genetically and so select for each independently using specific genetic markers. A different situation was observed for the QTL for late sexual maturation detected on Ssa18. To date, no growth-related QTL have been reported on this chromosome in Atlantic salmon or on homologous chromosomes in other salmonids (Gutierrez et al. 2012).
Correlations between the phenotypes of growth (as estimated by body weight) and early sexual maturation have been reported in Atlantic salmon (Gjerde 1984; Gjerde et al. 1994; Wild et al. 1994). Moreover, considerable genetic variation in age at sexual maturity has been described in farmed fish species. For example, in Atlantic salmon h 2 estimates range from 0.09 to 0.17 (Gjerde 1984; Gjerde et al. 1994; Wild et al. 1994) to 0.39 (Gjerde and Gjedrem 1984) to 0.48 (Gjerde 1984) whereas in rainbow trout h 2 estimates range from 0.12 to 0.35 (Gjerde and Schaeffer 1989; Kause et al. 2003). Nevertheless, selective breeding programs have been effective in increasing body size while also controlling undesired early sexual maturity in farmed fish (Gjedrem 2000), giving clues that these traits are influenced by different genes. Our previous results showed that growth-related QTL were spread across the Atlantic salmon genome (Gutierrez et al. 2012); however, none of the QTL associated with grilsing or late sexual maturation were found in the same chromosomal positions and with the same statistical significance indicating that different genes are associated with these traits.
Comparative genetic mapping in salmonid species has revealed homologous regions of the genomes of Atlantic salmon, rainbow trout, and Arctic charr (Danzmann et al. 2005, 2008; Phillips et al. 2009). Ssa21 corresponds to rainbow trout linkage group 5 (RT-5) (equivalent to chromosome 22) (Phillips et al. 2009) and Arctic charr linkage group 18 (AC-18) (Danzmann et al. 2005), whereas Ssa10 corresponds to RT-8q and RT-27q (equivalent to chromosomes 5q and 2q, respectively) (Phillips et al. 2009) and Arctic charr linkage groups 4, 16, and 40 (AC-4, AC-16, and AC-40) (Danzmann et al. 2005). Genome-wide significant QTL for early male sexual maturation were mapped to RT-8, RT-17, and RT-24, whereas chromosome-wide QTL for this trait was found on RT-3 and RT-19 (Haidle et al. 2008). Martyniuk et al. (2003) did report an association (p = 0.043) between microsatellite locus OmyRGT1TUF on RT-5 and precocious maturation and noted that one allele was associated with both a higher body mass and precociousness. However, this association was not considered significant after a sequential Bonferroni correction. Similarly, no p values less than 0.01 were identified for microsatellite marker alleles segregating with male sexual maturation in Arctic charr, although SSOSL32 on AC-4 gave a suggestive value of 0.014 (Moghadam et al. 2007).
It is believed that sexual maturation in salmonids depends on the ability of the fish to reach certain developmental thresholds during a “critical period” over the winter/spring (Adams and Thorpe 1989; Thorpe 1994), in a direct relationship with somatic growth and/or energy storage during this period (Taranger et al. 1999). Further investigation of genotype–environment interactions are required to improve Atlantic salmon breeding programs. Photoperiod manipulation using artificial light exposure has proven effective as a means of preventing Atlantic salmon from reaching an anticipated sexual maturation (Endal et al. 2000; Oppedal et al. 2006), and this procedure is used with many other farmed fish (reviewed by Taranger et al. 2010). A clear link between feeding and age of sexual maturation has been described in salmonids (Shearer et al. 2006; Silverstein et al. 1998; Taranger et al. 2010; Thorpe et al. 1990), suggesting that by restricting rations, energy storage and, in particular, lipid stores decrease, thereby delaying sexual maturation (Duston and Saunders 1999; Rowe et al. 1991; Shearer et al. 2006; Shearer and Swanson 2000). This also explains the increasing use of tetradecylthioacetic acid to reduce early sexual maturation in Atlantic salmon (Arge et al. 2012) as it affects the β-oxidation of fatty acids, reducing levels of plasma lipids and adipose lipid stores, and enhancing transport of fatty acids to the liver.
Candidate Genes
To identify candidate genes associated with the traits of early or late sexual maturation, we made use of the currently available information from the Atlantic salmon genome sequencing project (Davidson et al. 2010), which is publicly available at ASalBase (www.asalbase.org). Most of the SNPs in the 6.5K array correspond to an expressed sequence tag (EST) or are linked to a specific genomic sequence, and these sequences were assigned to a specific whole genome shotgun (WGS) contig by sequence similarity searches. WGS contigs were then annotated using an in-house annotation pipeline (trutta.mbb.sfu.ca) (see Table 3).
Table 3.
Candidate genes identified in the QTL regions associated with grilsing and late sexual maturation
| Trait | SNP_ID | Female map | Male map | WGS contig | BLASTn | Annotation |
|---|---|---|---|---|---|---|
| Grilsing | GCR_cBin22360_Ctg1_176 | 58.6 | 2.3 | AGKD01085592 | Salmo salar leukocyte elastase inhibitor (ileu), mRNA | Leukocyte elastase inhibitor (serpin-like) [Salmo salar] |
| Grilsing | GCR_cBin18557_Ctg1_186 | 58.6 | 2.3 | AGKD01261913 | Osmerus mordax clone omor-eva-507-083 GDP-L-fucose synthetase putative mRNA, complete cds | PREDICTED: GDP-L-fucose synthase-like [Oreochromis niloticus] |
| Grilsing | GCR_cBin51840_Ctg1_131 | 59.4 | 2.3 | AGKD01037205 | Salmo salar clone BHMS423 microsatellite sequence | only ESTs |
| Grilsing | GCR_cBin11397_Ctg1_74 | 61.7 | 2.3 | AGKD01171932 | No hits | Only ESTs |
| Late maturation | GCR_cBin5728_Ctg1_254 | 2.2 | 1 | AGKD01158387 | No hits | Only ESTs |
| Late maturation | GCR_cBin38765_Ctg1_214 | 8.4 | 1.3 | AGKD01032149 | Predicted: Oreochromis niloticus myopalladin-like (LOC100700984), mRNA | Predicted: myopalladin-like [Oreochromis niloticus] |
| Grilsing | GCR_cBin17226_Ctg1_170 | 7.5 | 1 | AGKD01023960 | No hits | Predicted: hypothetical protein LOC100002220 [Danio rerio] plus ESTs |
| Grilsing | ESTNV_25113_1234 | 7.8 | 1 | AGKD01008431 | Predicted: Oreochromis niloticus formimidoyltransferase-cyclodeaminase-like (LOC100693205), mRNA | Predicted: formimidoyltransferase-cyclodeaminase-like [Oreochromis niloticus] |
| Grilsing | GCR_cBin2393_Ctg1_480 | 26.2 | 1.1 | AGKD01000933 | Salmo salar clone ssal-rgf-503-163 E3 ubiquitin-protein ligase UBR3 putative mRNA, partial cds | E3 ubiquitin-protein ligase UBR3 [Salmo salar] |
| Grilsing | GCR_cBin2393_Ctg1_311 | 26.2 | 1.1 | AGKD01000933 | Salmo salar clone ssal-rgf-503-163 E3 ubiquitin-protein ligase UBR3 putative mRNA, partial cds | E3 ubiquitin-protein ligase UBR3 [Salmo salar] |
There are four SNPs linked to the QTL associated with grilsing on Ssa10, of which two could be linked to two genes by a BLASTn search (Altschul et al. 1990) and subsequent sequence annotation. These genes were a leukocyte elastase inhibitor (SERPIN-like) and a GDP-l-fucose synthase-like protein. We cannot speculate how these genes may be related to grilsing in Atlantic salmon. In the case of the SNPs linked to the grilsing QTL on Ssa21, one is associated with a gene encoding a formimidoyl transferase-cyclodeaminase, a protein related to metabolic processes. Two other SNPs (from the same EST) identified E3 ubiquitin-protein ligase (UBR3), a protein that has been shown to have a regulatory role in sensory pathways, including olfaction in mammals (Tasaki et al. 2007). The QTL detected for the late-maturation trait on Ssa18 was linked to two SNPs, but only one of them could be related to a known gene, namely, myopalladin-like gene. Studies in pig and cattle suggest that this gene is associated with meat quality and carcass traits including fat content (Chong et al. 2007; Jiao et al. 2010).
Conclusions
In this study, we were able to identify a limited number of QTL associated with age at sexual maturation in Atlantic salmon. Moreover, our results suggest that these traits are controlled by genes independently from growth-related traits. Age at sexual maturation is a trait that has been heavily selected and improved after several generations of selective breeding based on family selection and individual selection (Gjedrem 2000; Gjerde 1984; Gjøen and Bentsen 1997). However, these improvements have been based on phenotypic observations, and thus, the identification of genetic markers and genomic regions associated with grilsing and late sexual maturation provides new resources for Atlantic salmon selective breeding programs.
Acknowledgments
This study was funded through an NSERC strategic grant (STPGP/381479-2009 to WSD).
References
- Adams CE, Thorpe JE. Photoperiod and temperature effects on early development and reproductive investment in Atlantic salmon (Salmo salar L.) Aquaculture. 1989;79:403–409. doi: 10.1016/0044-8486(89)90483-3. [DOI] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arge R, Thomassen MS, Berge RK, Zambonino-Infante JL, Terjesen BF, Oehme M, Rørvik K-A. Reduction of early sexual maturation in male S0 Atlantic salmon (Salmo salar L.) by dietary supplementation of tetradecylthioacetic acid (TTA) Aquac Res. 2012 [Google Scholar]
- Chong W, Zhi-hong H, Xi-quan Z, Zhi-peng Q, Jia-qi L, Wei F, Yao-sheng C. Analysis on associations of myopalladin gene polymorphisms with carcass traits in pigs. Acta Vet Zool Sin. 2007;38:760–764. [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danzmann RG, Cairney M, Davidson WS, Ferguson MM, Gharbi K, Guyomard R, Holm L-E, Leder E, Okamoto N, Ozaki A, Rexroad CE, III, Sakamoto T, Taggart JB, Woram RA, Traut W. A comparative analysis of the rainbow trout genome with 2 other species of fish (Arctic charr and Atlantic salmon) within the tetraploid derivative Salmonidae family (subfamily: Salmoninae) Genome. 2005;48:1037–1051. doi: 10.1139/g05-067. [DOI] [PubMed] [Google Scholar]
- Danzmann RG, Davidson EA, Ferguson MM, Gharbi K, Koop BF, Hoyheim B, Lien S, Lubieniecki KP, Moghadam HK, Park J, Phillips RB, Davidson WS. Distribution of ancestral proto-actinopterygian chromosome arms within the genomes of 4R-derivative salmonid fishes (rainbow trout and Atlantic salmon) BMC Genomics. 2008;9:557. doi: 10.1186/1471-2164-9-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson WS, Koop BF, Jones SJM, Iturra P, Vidal R, Maass A, Jonassen I, Lien S, Omholt SW. Sequencing the genome of the Atlantic salmon (Salmo salar) Genome Biol. 2010;11:403. doi: 10.1186/gb-2010-11-9-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duston J, Saunders RL. Effect of winter food deprivation on growth and sexual maturity of Atlantic salmon (Salmo salar) in seawater. Can J Fish Aquat Sci. 1999;56:201–207. doi: 10.1139/f98-165. [DOI] [Google Scholar]
- Easton AA, Moghadam HK, Danzmann RG, Ferguson MM. The genetic architecture of embryonic developmental rate and genetic covariation with age at maturation in rainbow trout Oncorhynchus mykiss. J Fish Biol. 2011;78:602–623. doi: 10.1111/j.1095-8649.2010.02881.x. [DOI] [PubMed] [Google Scholar]
- Eisbrenner WD, Botwright N, Cook M, Davidson EA, S Dominik, NG Elliott, J Henshall, SL Jones,PD Kube, KP Lubieniecki, S Peng and WS Davidson (2013) Evidence for multiple sex-determining loci in Tasmanian Atlantic salmon (Salmo salar). Heredity doi:10.1038/hdy.2013.55 [DOI] [PMC free article] [PubMed]
- Endal HP, Taranger GL, Stefansson SO, Hansen T. Effects of continuous additional light on growth and sexual maturity in Atlantic salmon, Salmo salar, reared in sea cages. Aquaculture. 2000;191:337–349. doi: 10.1016/S0044-8486(00)00444-0. [DOI] [Google Scholar]
- Garcia de Leaniz C, Fleming IA, Einum S, Verspoor E, Jordan WC, Consuegra S, Aubin-Horth N, Lajus D, Letcher BH, Youngson AF, Webb JH, Vøllestad LA, Villanueva B, Ferguson A, Quinn TP. A critical review of adaptive genetic variation in Atlantic salmon: implications for conservation. Biol Rev. 2007;82:173–211. doi: 10.1111/j.1469-185X.2006.00004.x. [DOI] [PubMed] [Google Scholar]
- Gardner MLG. A review of factors which may influence the sea-age and maturation of Atlantic salmon Salmo salar L. J Fish Biol. 1976;9:289–327. doi: 10.1111/j.1095-8649.1976.tb04680.x. [DOI] [Google Scholar]
- Gjedrem T. Genetic improvement of cold-water fish species. Aquac Res. 2000;31:25–33. doi: 10.1046/j.1365-2109.2000.00389.x. [DOI] [Google Scholar]
- Gjedrem T. Genetic improvement for the development of efficient global aquaculture: a personal opinion review. Aquaculture. 2012;344–349:12–22. doi: 10.1016/j.aquaculture.2012.03.003. [DOI] [Google Scholar]
- Gjedrem T, Gjøen HM, Gjerde B. Genetic origin of Norwegian farmed Atlantic salmon. Aquaculture. 1991;98:41–50. doi: 10.1016/0044-8486(91)90369-I. [DOI] [Google Scholar]
- Gjerde B. Response to individual selection for age at sexual maturity in Atlantic salmon. Aquaculture. 1984;38:229–240. doi: 10.1016/0044-8486(84)90147-9. [DOI] [Google Scholar]
- Gjerde B, Gjedrem T. Estimates of phenotypic and genetic parameters for carcass traits in Atlantic salmon and rainbow trout. Aquaculture. 1984;36:97–110. doi: 10.1016/0044-8486(84)90057-7. [DOI] [Google Scholar]
- Gjerde B, Schaeffer LR. Body traits in rainbow trout: II. Estimates of heritabilities and of phenotypic and genetic correlations. Aquaculture. 1989;80:25–44. doi: 10.1016/0044-8486(89)90271-8. [DOI] [Google Scholar]
- Gjerde B, Simianer H, Refstie T. Estimates of genetic and phenotypic parameters for body weight, growth rate and sexual maturity in Atlantic salmon. Livest Prod Sci. 1994;38:133–143. doi: 10.1016/0301-6226(94)90057-4. [DOI] [Google Scholar]
- Gjøen HM, Bentsen HB. Past, present, and future of genetic improvement in salmon aquaculture. ICES J Mar Sci. 1997;54:1009–1014. [Google Scholar]
- Gutierrez AP, Lubieniecki KP, Davidson EA, Lien S, Kent MP, Fukui S, Withler RE, Swift B, Davidson WS. Genetic mapping of quantitative trait loci (QTL) for body-weight in Atlantic salmon (Salmo salar) using a 6.5K SNP array. Aquaculture. 2012;358–359:61–70. doi: 10.1016/j.aquaculture.2012.06.017. [DOI] [Google Scholar]
- Haidle L, Janssen JE, Gharbi K, Moghadam HK, Ferguson MM, Danzmann RG. Determination of quantitative trait loci (QTL) for early maturation in rainbow trout (Oncorhynchus mykiss) Mar Biotechnol. 2008;10:579–592. doi: 10.1007/s10126-008-9098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbinger CM, Friars GW. Effects of winter temperature and feeding regime on the rate of early maturation in Atlantic salmon (Salmo salar) male parr. Aquaculture. 1992;101:147–162. doi: 10.1016/0044-8486(92)90239-H. [DOI] [Google Scholar]
- Jiao Y, Zan L, Liu Y, Wang H, Guo B. A novel polymorphism of the MYPN gene and its association with meat quality traits in Bos taurus. Genet Mol Res. 2010;9:1751–1758. doi: 10.4238/vol9-3gmr906. [DOI] [PubMed] [Google Scholar]
- Kause A, Ritola O, Paananen T, Mäntysaari E, Eskelinen U. Selection against early maturity in large rainbow trout Oncorhynchus mykiss: the quantitative genetics of sexual dimorphism and genotype-by-environment interactions. Aquaculture. 2003;228:53–68. doi: 10.1016/S0044-8486(03)00244-8. [DOI] [Google Scholar]
- Kent MP, Hayes B, Xiang Q, Berg PR, Gibbs RA, Lien S (2009) Development of a 6.5K SNP-Chip for Atlantic salmon. Paper presented at the Plant & Animal Genomes XVII Conference, San Diego, CA,
- Küttner E, Moghadam H, Skúlason S, Danzmann R, Ferguson M. Genetic architecture of body weight, condition factor and age of sexual maturation in Icelandic Arctic charr (Salvelinus alpinus) Mol Genet Genomics. 2011;286:67–79. doi: 10.1007/s00438-011-0628-x. [DOI] [PubMed] [Google Scholar]
- Lien S, Gidskehaug L, Moen T, Hayes B, Berg P, Davidson W, Omholt S, Kent M. A dense SNP-based linkage map for Atlantic salmon (Salmo salar) reveals extended chromosome homeologies and striking differences in sex-specific recombination patterns. BMC Genomics. 2011;12:615. doi: 10.1186/1471-2164-12-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyniuk CJ, Perry GML, Mogahadam HK, Ferguson MM, Danzmann RG. The genetic architecture of correlations among growth-related traits and male age at maturation in rainbow trout. J Fish Biol. 2003;63:746–764. doi: 10.1046/j.1095-8649.2003.00188.x. [DOI] [Google Scholar]
- Moghadam HK, Poissant J, Fotherby H, Haidle L, Ferguson MM, Danzmann RG. Quantitative trait loci for body weight, condition factor and age at sexual maturation in Arctic charr (Salvelinus alpinus): comparative analysis with rainbow trout (Oncorhynchus mykiss) and Atlantic salmon (Salmo salar) Mol Genet Genomics. 2007;277:647–661. doi: 10.1007/s00438-007-0215-3. [DOI] [PubMed] [Google Scholar]
- Oppedal F, Berg A, Olsen RE, Taranger GL, Hansen T. Photoperiod in seawater influence seasonal growth and chemical composition in autumn sea-transferred Atlantic salmon (Salmo salar L.) given two vaccines. Aquaculture. 2006;254:396–410. doi: 10.1016/j.aquaculture.2005.10.026. [DOI] [Google Scholar]
- Phillips RB, Keatley KA, Morasch MR, Ventura AB, Lubieniecki KP, Koop BF, Danzmann RG, Davidson WS. Assignment of Atlantic salmon (Salmo salar) linkage groups to specific chromosomes: Conservation of large syntenic blocks corresponding to whole chromosome arms in rainbow trout (Oncorhynchus mykiss) BMC Genet. 2009;10:46. doi: 10.1186/1471-2156-10-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe DK, Thorpe JE, Shanks AM. Role of fat stores in the maturation of male Atlantic salmon (Salmo salar) parr. Can J Fish Aquat Sci. 1991;48:405–413. doi: 10.1139/f91-052. [DOI] [Google Scholar]
- Saunders RL, Henderson EB, Glebe BD, Loudenslager EJ. Evidence of a major environmental component in determination of the grilse: larger salmon ratio in Atlantic salmon (Salmo salar) Aquaculture. 1983;33:107–118. doi: 10.1016/0044-8486(83)90391-5. [DOI] [Google Scholar]
- Seaton G, Hernandez J, Grunchec JA, White I, Allen J, De Koning D, Wei W, Berry D, Haley C, Knott S GridQTL: a grid portal for QTL mapping of compute intensive datasets. In: 8th World Congress on Genetics Applied to Livestock Production, Belo Horizonte, MG, Brasil, August 13-18 2006.
- Shearer KD, Swanson P. The effect of whole body lipid on early sexual maturation of 1+ age male Chinook salmon (Oncorhynchus tshawytscha) Aquaculture. 2000;190:343–367. doi: 10.1016/S0044-8486(00)00406-3. [DOI] [Google Scholar]
- Shearer K, Parkins P, Gadberry B, Beckman B, Swanson P. Effects of growth rate/body size and a low lipid diet on the incidence of early sexual maturation in juvenile male spring Chinook salmon (Oncorhynchus tshawytscha) Aquaculture. 2006;252:545–556. doi: 10.1016/j.aquaculture.2005.06.027. [DOI] [Google Scholar]
- Silverstein JT, Shearer KD, Dickhoff WW, Plisetskaya EM. Effects of growth and fatness on sexual development of Chinook salmon (Oncorhynchus tshawytscha) parr. Can J Fish Aquat Sci. 1998;55:2376–2382. doi: 10.1139/f98-111. [DOI] [Google Scholar]
- Simpson AL. Differences in body size and lipid reserves between maturing and nonmaturing Atlantic salmon parr, Salmo salar L. Can J Zool. 1992;70:1737–1742. doi: 10.1139/z92-241. [DOI] [Google Scholar]
- Taranger GL, Haux C, Hansen T, Stefansson SO, Björnsson BT, Walther BT, Kryvi H. Mechanisms underlying photoperiodic effects on age at sexual maturity in Atlantic salmon, Salmo salar. Aquaculture. 1999;177:47–60. doi: 10.1016/S0044-8486(99)00068-X. [DOI] [Google Scholar]
- Taranger GL, Carrillo M, Schulz RW, Fontaine P, Zanuy S, Felip A, Weltzien F-A, Dufour S, Karlsen Ø, Norberg B, Andersson E, Hansen T. Control of puberty in farmed fish. Gen Comp Endocrinol. 2010;165:483–515. doi: 10.1016/j.ygcen.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Tasaki T, Sohr R, Xia Z, Hellweg R, Hörtnagl H, Varshavsky A, Kwon YT. Biochemical and genetic studies of UBR3, a ubiquitin ligase with a function in olfactory and other sensory systems. J Biol Chem. 2007;282:18510–18520. doi: 10.1074/jbc.M701894200. [DOI] [PubMed] [Google Scholar]
- Thorpe JE. Reproductive strategies in Atlantic salmon, Salmo salar L. Aquac Res. 1994;25:77–87. doi: 10.1111/j.1365-2109.1994.tb00668.x. [DOI] [Google Scholar]
- Thorpe JE, Metcalfe NB. Is smolting a positive or a negative developmental decision? Aquaculture. 1998;168:95–103. doi: 10.1016/S0044-8486(98)00342-1. [DOI] [Google Scholar]
- Thorpe JE, Morgan RIG, Talbot C, Miles MS. Inheritance of developmental rates in Atlantic salmon, Salmo salar L. Aquaculture. 1983;33:119–128. doi: 10.1016/0044-8486(83)90392-7. [DOI] [Google Scholar]
- Thorpe JE, Talbot C, Miles MS, Keay DS. Control of maturation in cultured Atlantic salmon, Salmo salar, in pumped seawater tanks, by restricting food intake. Aquaculture. 1990;86:315–326. doi: 10.1016/0044-8486(90)90122-4. [DOI] [Google Scholar]
- Wild V, Simianer H, Gjøen HM, Gjerde B. Genetic parameters and genotype × environment interaction for early sexual maturity in Atlantic salmon (Salmo salar) Aquaculture. 1994;128:51–65. doi: 10.1016/0044-8486(94)90101-5. [DOI] [Google Scholar]
- Withler RE, Supernault KJ, Miller KM. Genetic variation within and among domesticated Atlantic salmon broodstocks in British Columbia, Canada. Anim Genet. 2005;36:43–50. doi: 10.1111/j.1365-2052.2004.01220.x. [DOI] [PubMed] [Google Scholar]
- Withler RE, Supernault J, Swift B, Peterson R, Fukui S. Microsatellite DNA assignment of progeny to parents enables communal freshwater rearing in an Atlantic salmon selective breeding program. Aquaculture. 2007;272:S318. doi: 10.1016/j.aquaculture.2007.07.201. [DOI] [Google Scholar]
- Yano A, Guyomard R, Nicol B, Jouanno E, Quillet E, Klopp C, Cabau C, Bouchez O, Fostier A, Guiguen Y. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr Biol. 2012;22:1423–1428. doi: 10.1016/j.cub.2012.05.045. [DOI] [PubMed] [Google Scholar]
- Yano A, Nicol B, Jouanno E, Quillet E, Fostier A, Guyomard R, Guiguen Y. The sexually dimorphic on the Y-chromosome gene (sdY) is a conserved male-specific Y-chromosome sequence in many salmonids. Evol Appl. 2013;6:486–496. doi: 10.1111/eva.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]