Abstract
Given that the genetic risk for schizophrenia is highly polygenic and the effect sizes, even for rare or de novo events, are modest at best, it has been suggested that multiple biological pathways are likely to be involved in the etiopathogenesis of the disease. Most efforts in understanding the cellular basis of schizophrenia have followed a “neuron-centric” approach, focusing on alterations in neurotransmitter systems and synapse cytoarchitecture. However, multiple lines of evidence coming from genetics and systems biology approaches suggest that apart from neurons, oligodendrocytes and potentially other glia are affected from schizophrenia risk loci. Neurobiological abnormalities linked with genetic association signal could identify abnormalities that are more likely to be primary, versus environmentally induced changes or downstream events. Here, we summarize genetic data that support the involvement of oligodendrocytes in schizophrenia, providing additional evidence for a causal role with the disease. Given the undeniable evidence of both neuronal and glial abnormalities in schizophrenia, we propose a neuro-glial model that invokes abnormalities at the node of Ranvier as a functional unit in the etiopathogenesis of the disease.
Keywords: systems biology, polygenic, node of Ranvier, disconnectivity, GWAS
INTRODUCTION
The core symptoms of schizophrenia include the presence of delusions and hallucinations (positive symptoms), apathy, and social withdrawal (negative symptoms), as well as, stable impairments in specific domains of cognitive function. Despite our best effort, we still lack a basic understanding of the etiopathogenesis of schizophrenia and therefore tools for curative treatment or prevention do not exist (Insel, 2010). Given the high prevalence of schizophrenia (1% or more) the need for greater understanding is urgent.
Most efforts in understanding the cellular basis of schizophrenia have followed a “neuron-centric” approach, focusing exclusively on the role(s) of neurons. This has led to findings of alterations in neuronal function, such as neurotransmission and synapse cytoarchitecture, often overlooking the fact that neuronal function and neurotransmission is inextricably dependent on interactions between neurons and glia. Increasing recognition of the importance of neuro-glial elements as basic functional units (Allen and Barres, 2009) that participate in CNS function and dysfunction combined with multiple lines of evidence showing abnormalities in oligodendrocytes and myelination have implicated oligodendrocytes as highly relevant to schizophrenia. While the approach of grouping neurons and glia into neuro-glial functional units introduces significant complexity into the analysis of the neurobiology of schizophrenia, it encourages an integrative approach that favors the examination of the interplay between neurons and the more abundant glial cell types of the CNS.
Here, we first outline genetic evidence derived from candidate gene association studies or genome-wide approaches that support oligodendroglial and myelin related (OMR) abnormalities in schizophrenia as a primary contributor. In an attempt to provide a mechanistic and integrative interpretation of the neuronal and OMR changes in schizophrenia, we introduce a neuro-oligodendroglial model that involves abnormalities at the nodes of Ranvier (NOR) as a functional unit implicated in the etiopathogenesis of the disease.
MYELIN ASSOCIATED ABNORMALITIES IN SCHIZOPHRENIA
Myelination can increase action potential transmission speed and decrease refractory time, which increases the number of action potentials that can be transmitted per unit time. There is accumulating evidence supporting the notion of schizophrenia as a disorder of disconnectivity (Konrad and Winterer, 2008). While disconnectivity can be caused by different mechanisms, such as synaptic malfunction, at its core, it implicates inadequate or failed information transfer among neurons. OMR abnormalities in schizophrenia impair the saltatory conduction and information conduction from one neuron to others.
Growing evidence, accumulated over the last 15 years, provide strong support for OMR abnormalities in schizophrenia. The majority of those data is derived from neuroimaging and human postmortem studies (for review, see Sequeira et al., 2012; Fitzsimmons et al., 2013). Irrespective of the level of care and control, different confounding factors, such as antipsychotic medication, could contribute to the detected OMR neuroimaging, gene expression, and proteome abnormalities in schizophrenia. Therefore, incorporating genetic data, as discussed below, can help distinguish whether OMR changes are a primary contributor to disease pathophysiology vs. secondary changes associated with other primary lesions existing in schizophrenia or mere epiphenomena.
GENETIC ASSOCIATION OF OMR CANDIDATE GENES IN SCHIZOPHRENIA
Multiple candidate studies have provided evidence that OMR genes are genetically associated with schizophrenia (Table 1). It is interesting to note, that the products of these same genes have been implicated in numerous direct studies of OMR gene and protein expression in the brains of persons with schizophrenia, as well as, neuroimaging studies, providing a stronger support for causality with the disease (Bartzokis, 2011; Takahashi et al., 2011). The OMR genes with the strongest support for genetic association with schizophrenia are described below:
Table 1.
OMR genes implicated in schizophrenia.
The largest PGC-SWE (Ripke et al., 2013) in the form of summary statistic p-values was obtained from public access website (https://pgc.unc.edu/). For each gene (±100 kb from 5′ and 3′ ends of gene), the strongest P value association is provided in the “PGC-SWE P” column. The “Rank” column indicates the ranking of the illustrated genes based on the strength of association with the schizophrenia GWAS data set.
Neuregulin 1 (NRG1) – ERBB4 signaling
NRG1 risk genotypes or haplotypes have been associated with schizophrenia (Stefansson et al., 2002). Genetic evidence also supports ERBB4 – the NRG1 receptor – as a candidate susceptibility gene and suggests positive epistatic interactions between NRG1 and ERBB4 in schizophrenia (Norton et al., 2006). The potential pathophysiologic role of NRG1 is further supported by its diverse neurobiological functions, including neuro-glial trophic effects and myelination (Harrison and Law, 2006). Nevertheless, given the fact that the NRG1 – ERBB4 signaling has multiple effects in nervous system development (including neuronal migration and modulation of neurotransmission), this pathway may play a role in schizophrenia pathogenesis through other mechanisms aside from OMR effects. A direct way to assess the pathophysiologic effects of genetic variants would be to examine human subjects with risk variants for symptoms of schizophrenia. Along these lines, many groups have reported association of the NRG1 – ERBB4 risk variants with neurocognitive (Stefanis et al., 2007), electrophysiological (Roussos et al., 2011) and neuroimaging schizophrenia-related outcome variables, including altered fronto-temporal brain function (Hall et al., 2006) and white matter density and integrity (McIntosh et al., 2008; Konrad et al., 2009). Finally, several groups have assessed gene expression and proteins of NRG1 and ERBB4 in postmortem brains and have reported differences between schizophrenia and healthy control groups (Hashimoto et al., 2004; Law et al., 2006).
Disrupted-in-schizophrenia 1
Disrupted-in-schizophrenia 1 (DISC1) is a strong candidate gene for schizophrenia and was initially identified in a large Scottish pedigree (Millar et al., 2000) followed by additional genetic evidence for association with sporadic cases of schizophrenia (Chubb et al., 2008). Recent studies in zebrafish (Drerup et al., 2009) and transgenic mice (Katsel et al., 2011) with forebrain restricted expression of mutant human DISC1 have suggested a critical role for DISC1 in oligodendroglial differentiation during neurodevelopment. In human postmortem analyses, DISC1 transcripts that are more abundant during fetal development are upregulated in the hippocampus of patients with schizophrenia and their expression levels are associated with schizophrenia risk DISC1 polymorphisms (Nakata et al., 2009). One of those risk DISC1 SNPs is also associated with white matter integrity as measured by DTI (Sprooten et al., 2011).
Reticulon 4 receptor
Reticulon 4 receptor (RTN4R) is a myelin-associated protein that inhibits the outgrowth of neurites and nerve terminals (Chen et al., 2000). RTN4R is upregulated in the brains of patients with schizophrenia (Novak et al., 2002). Genetic evidence supports a role of RTN4R in the etiopathogenesis of schizophrenia (Novak et al., 2002; Budel et al., 2008). Interestingly, in vitro experiments demonstrated that cultured neurons expressing the schizophrenia-associated RTN4R variants failed to respond to the growth inhibiting activity of myelin, and functioned as a dominant negative to disrupt endogenous RTN4R (Budel et al., 2008).
Other genes
Oligodendrocyte lineage transcription factor 2 (OLIG2) encodes a transcription factor central to oligodendrocyte development (Ross et al., 2003). Genetic association analysis showed that OLIG2 is associated with schizophrenia and demonstrated an epistatic effect with 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNP) and ERBB4 (Georgieva et al., 2006). Furthermore, OLIG2 expression significantly correlated in cerebral cortex with CNP and ERBB4, suggesting that variation in OLIG2 confers susceptibility to schizophrenia as part of a network of genes implicated in oligodendrocyte function. Similarly, a genetic association of the CNP gene was found for schizophrenia (Peirce et al., 2006). The CNP risk polymorphism was associated with lower gene expression which is consistent with CNP gene expression downregulation in schizophrenia.
GENETIC ASSOCIATION OF OMR GENES IN SCHIZOPHRENIA DERIVED FROM GENOME-WIDE APPROACHES
Significant progress has been made over the last decade through large genome-wide association studies (GWAS). The GWAS design has provided evidence in an agnostic manner that specific common DNA genetic variants among people influence their genetic susceptibility to multiple different complex disorders, including schizophrenia (Ripke et al., 2013). Furthermore, we have learned that the genetic risk for schizophrenia is highly polygenic (Ripke et al., 2013), where the phenotype is influenced by multiple genetic variants, each one with small effect sizes. Here, we present results for association of each OMR gene with schizophrenia according to the largest published schizophrenia GWAS data set (PGC-SWE;https://pgc.unc.edu/; Ripke et al., 2013). No OMR genetic association reached genome-wide significance (Table 1); however, a handful of OMR genes (ANK3, ERBB4, and NRG1) show suggestive association (P < 5 × 10-5). Future GWAS with increased sample sizes will determine whether OMR genetic variants are true associations at reduced thresholds, but without the needed power to reach genome-wide significance.
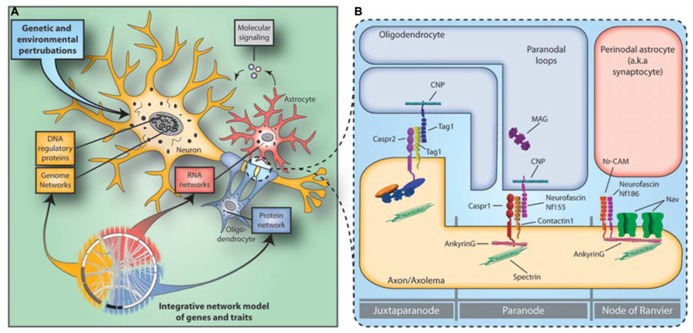
Systems biology approaches allow the integration of multi-scale datasets gained from genetic imaging and gene and protein expression studies. The risk SNPs are unlikely to be randomly distributed, but cluster in molecular subnetworks affecting specific cellular populations and molecular processes (Roussos, 2012; Figure 1A). Recent findings support the notion that polygenic risk for schizophrenia affects OMR gene networks and molecular pathways (Roussos et al., 2012b; Goudriaan et al., 2013). Perturbations in specific subnetworks related to neuronal function, myelination, immune response, and energy production were identified in a multi-regional system-level analysis of the schizophrenia brain transcriptome (Roussos et al., 2012b). The subnetworks related to neuronal function and myelination were enriched for genetically associated variants in schizophrenia, providing independent support for the neuro-oligodendroglial interactions as a potential causal functional unit in schizophrenia. No enrichment for genetic variants and immune response and energy production related subnetworks was found, suggesting that these changes may represent secondary or bystander phenomena or be attributable to environmental influences. Similar results were reported in an independent study, in which the genetic evidence for primary roles of specific glial cell type functions and pathways was examined based on gene set analysis (Goudriaan et al., 2013). In conclusion, recent systems biology approaches support the notion that schizophrenia involves hundreds of genetic loci that in combination with environmental factors converge to perturb a more selective/restrictive set of molecular/cellular networks and integrative functions.
FIGURE 1.
Neuro-glial functional unit and the disconnectivity syndrome in schizophrenia. (A) Constellations of multiple genetic and environmental perturbations affect molecular states of gene coexpression and protein–protein interaction networks in the neuro-glial functional unit that in turn increase the risk for schizophrenia. (B) The NOR is the site of intimate neuroglial cell–cell interactions in the nervous system. Abnormalities in the expression of NOR genes and proteins in schizophrenia hamper saltatory conduction by affecting NOR formation, maintenance and integrity and lead to the disconnectivity syndrome.
FUNCTIONAL NEURO-GLIAL UNITS AND THE NODES OF RANVIER
The above observations suggest that genetic alterations underlying OMR cell type functions increase susceptibility to schizophrenia and provide evidence that the “neuron-centric” hypothesis of schizophrenia should be extended to include a role for glia in the etiopathogenesis of the disease. The NOR is one of the best example of the neuroglial cell–cell interactions in the central nervous system. The NORs are myelin-sheath-free segments along myelinated axons, where sodium ion exchange takes place propagating action potentials from the axon initial segment to the axon terminal. NOR and OMR gene and protein expression abnormalities can hamper saltatory conduction by affecting NOR formation, maintenance, and integrity, which in turn leads to failures of saltatory conduction and disconnection of higher-order association areas (Nave, 2010).
Different and specific neuronal and oligodendroglial reciprocal interactions (for review, see Poliak and Peles, 2003) ensure the high concentration and anchoring of voltage gated Na+ channels (NaV; NaV1.6 in adults – SCN8A) to the NOR and the maintenance of adherens junctions between the axolemma and paranodal myelin loops (Figure 1B). The voltage gated Na+ channel cytoplasmic loops are linked to the underlying spectrin-actin cytoskeleton through the anchoring protein Ankyrin G (ANK3). ANK3 plays an important role in the NOR complex by anchoring not only the NaV to the node but also because several cis acting nodal and paranodal adherens junction proteins, such as neurofacin (NFASC), neuronal cell adhesion molecule (NRCAM), contactin 1 and caspr, are also associated with it. Finally, contactin 2 (CNTN2), along with Caspr interact in trans with cell adhesion molecules of oligodendroglial paranodal loops that include an isoform of NFASC (Nf155), and CNTN2 to ensure the integrity of the myelin-axolemma tight junctions, which form a barrier that prevents the diffusion of Nav from the nodal region.
A recently study demonstrated that several NOR genes and proteins, including ANK3, NFASC, NRCAM, CNTN2, and SCN8A are affected in schizophrenia (Roussos et al., 2012a). The importance of ANK3 to serious mental illness is further supported by recent GWAS implicating ANK3 in schizophrenia (Athanasiu et al., 2010) and bipolar disorder (Sklar et al., 2011). Furthermore, an ANK3 risk polymorphism is associated with decreased ANK3 gene expression, which is consistent with ANK3 gene expression downregulation in schizophrenia and neurocognitive and neuroimaging abnormalities (Roussos et al., 2012a). Overall, these data highlight that in addition to OMR changes in schizophrenia, abnormalities in how myelin sheaths interact with the axon to form the NOR are also present and identify the NOR as a functional neuro-glial unit where neuronal and OMR genetic perturbations could potentially converge.
Conclusion
There are multiple lines of evidence derived from neuroimaging, molecular and genetic data that support a role of OMR genes and molecular pathways in schizophrenia. Recent systems biology and pathway analyses that integrate gene expression and genetic data support the notion that oligodendroglial abnormalities are among the primary deficits in the disorder. Future studies need to integrate the larger GWAS and determine the polygenic effect of OMR genes in schizophrenia. While most efforts in understanding the cellular basis of schizophrenia have followed a “neuron-centric” approach, there is an emerging need to conduct more complex neurobiological studies and examine the neuro-glial unit as a whole. The data to date provide evidence for abnormalities in the NOR as a plausible biological substrate for the disconnectivity syndrome in schizophrenia. Recent evidence suggesting that antipsychotics may influence OMR function beneficially provides clues for improved treatment strategies (Ren et al., 2013). Therefore, a more thorough understanding of the role of OMR and NOR pathophysiology is necessary since it holds the potential for providing new insights into the treatment of this disease.
Author Contributions
Panos Roussos and Vahram Haroutunian drafted and revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Georgios Voloudakis for figures. We apologize for not citing some of papers due to space limitation. Work in the authors’ laboratory has been supported by NIH grants MH066392 and MH064673 and Veterans Administration MIRECC.
REFERENCES
- Aberg K., Saetre P., Jareborg N., Jazin E. (2006a). Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 103 7482–7487 10.1073/pnas.0601213103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberg K., Saetre P., Lindholm E., Ekholm B., Pettersson U., Adolfsson R., et al. (2006b). Human QKI, a new candidate gene for schizophrenia involved in myelination. Am. J. Med. Genet. B Neuropsychiatr. Genet. 141B 84–90 10.1002/ajmg.b.30243 [DOI] [PubMed] [Google Scholar]
- Agim Z. S., Esendal M., Briollais L., Uyan O., Meschian M., Martinez L. A., et al. (2013). Discovery, validation and characterization of Erbb4 and Nrg1 haplotypes using data from three genome-wide association studies of schizophrenia. PLoS ONE 8:e53042 10.1371/journal.pone.0053042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen N. J., Barres B. A. (2009). Neuroscience: glia – more than just brain glue. Nature 457 675–677 10.1038/457675a [DOI] [PubMed] [Google Scholar]
- Athanasiu L., Mattingsdal M., Kahler A. K., Brown A., Gustafsson O., Agartz I., et al. (2010). Gene variants associated with schizophrenia in a Norwegian genome-wide study are replicated in a large European cohort. J. Psychiatr. Res. 44 748–753 10.1016/j.jpsychires.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalew M., Le-Niculescu H., Levey D. F., Jain N., Changala B., Patel S. D., et al. (2012). Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol. Psychiatry 17 887–905 10.1038/mp.2012.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barley K., Dracheva S., Byne W. (2009). Subcortical oligodendrocyte- and astrocyte-associated gene expression in subjects with schizophrenia, major depression and bipolar disorder. Schizophr. Res. 112 54–64 10.1016/j.schres.2009.04.019 [DOI] [PubMed] [Google Scholar]
- Bartzokis G. (2011). Neuroglialpharmacology: white matter pathophysiologies and psychiatric treatments. Front. Biosci. (Landmark Ed) 16:2695–2733 10.2741/3881 [DOI] [PubMed] [Google Scholar]
- Budel S., Padukkavidana T., Liu B. P., Feng Z., Hu F., Johnson S., et al. (2008). Genetic variants of Nogo-66 receptor with possible association to schizophrenia block myelin inhibition of axon growth. J. Neurosci. 28 13161–13172 10.1523/JNEUROSCI.3828-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum J. D., Georgieva L., Young J. J., Plescia C., Kajiwara Y., Jiang Y., et al. (2008). Molecular dissection of NRG1-ERBB4 signaling implicates PTPRZ1 as a potential schizophrenia susceptibility gene. Mol. Psychiatry 13 162–172 10.1038/sj.mp.4001991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. S., Huber A. B., Van Der Haar M. E., Frank M., Schnell L., Spillmann A. A., et al. (2000). Nogo-A is a myelin-associated neurite outgrowth inhibitor and an antigen for monoclonal antibody IN-1. Nature 403 434–439 10.1038/35000219 [DOI] [PubMed] [Google Scholar]
- Chong V. Z., Thompson M., Beltaifa S., Webster M. J., Law A. J., Weickert C. S. (2008). Elevated neuregulin-1 and ErbB4 protein in the prefrontal cortex of schizophrenic patients. Schizophr. Res. 100 270–280 10.1016/j.schres.2007.12.474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb J. E., Bradshaw N. J., Soares D. C., Porteous D. J., Millar J. K. (2008). The DISC locus in psychiatric illness. Mol. Psychiatry 13 36–64 10.1038/sj.mp.4002106 [DOI] [PubMed] [Google Scholar]
- Cruz D. A., Weaver C. L., Lovallo E. M., Melchitzky D. S., Lewis D. A. (2009). Selective alterations in postsynaptic markers of chandelier cell inputs to cortical pyramidal neurons in subjects with schizophrenia. Neuropsychopharmacology 34 2112–2124 10.1038/npp.2009.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debono R., Topless R., Markie D., Black M. A., Merriman T. R. (2012). Analysis of the DISC1 translocation partner (11q14.3) in genetic risk of schizophrenia. Genes Brain Behav. 11 859–863 10.1111/j.1601-183X.2012.00832.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dracheva S., Davis K. L., Chin B., Woo D. A., Schmeidler J., Haroutunian V. (2006). Myelin-associated mRNA and protein expression deficits in the anterior cingulate cortex and hippocampus in elderly schizophrenia patients. Neurobiol. Dis. 21 531–540 10.1016/j.nbd.2005.08.012 [DOI] [PubMed] [Google Scholar]
- Drerup C. M., Wiora H. M., Topczewski J., Morris J. A. (2009). Disc1 regulates foxd3 and sox10 expression, affecting neural crest migration and differentiation. Development 136 2623–2632 10.1242/dev.030577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimmons J., Kubicki M., Shenton M. E. (2013). Review of functional and anatomical brain connectivity findings in schizophrenia. Curr. Opin. Psychiatry 26 172–187 10.1097/YCO.0b013e32835d9e6a [DOI] [PubMed] [Google Scholar]
- Friedman J. I., Vrijenhoek T., Markx S., Janssen I. M., Van Der Vliet W. A., Faas B. H., et al. (2008). CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol. Psychiatry 13 261–266 10.1038/sj.mp.4002049 [DOI] [PubMed] [Google Scholar]
- Georgieva L., Moskvina V., Peirce T., Norton N., Bray N. J., Jones L., et al. (2006). Convergent evidence that oligodendrocyte lineage transcription factor 2 (OLIG2) and interacting genes influence susceptibility to schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 103 12469–12474 10.1073/pnas.0603029103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudriaan A., De Leeuw C., Ripke S., Hultman C. M., Sklar P., Sullivan P. F., et al. (2013). Specific glial functions contribute to schizophrenia susceptibility. Schizophr. Bull. 10.1093/schbul/sbt109 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakak Y., Walker J. R., Li C., Wong W. H., Davis K. L., Buxbaum J. D., et al. (2001). Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 98 4746–4751 10.1073/pnas.081071198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Whalley H. C., Job D. E., Baig B. J., McIntosh A. M., Evans K. L., et al. (2006). A neuregulin 1 variant associated with abnormal cortical function and psychotic symptoms. Nat. Neurosci. 9 1477–1478 10.1038/nn1795 [DOI] [PubMed] [Google Scholar]
- Harrison P. J., Law A. J. (2006). Neuregulin 1 and schizophrenia: genetics, gene expression, and neurobiology. Biol. Psychiatry 60 132–140 10.1016/j.biopsych.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Hashimoto R., Straub R. E., Weickert C. S., Hyde T. M., Kleinman J. E., Weinberger D. R. (2004). Expression analysis of neuregulin-1 in the dorsolateral prefrontal cortex in schizophrenia. Mol. Psychiatry 9 299–307 10.1038/sj.mp.4001434 [DOI] [PubMed] [Google Scholar]
- Insel T. R. (2010). Rethinking schizophrenia. Nature 468 187–193 10.1038/nature09552 [DOI] [PubMed] [Google Scholar]
- Ji W., Li T., Pan Y., Tao H., Ju K., Wen Z., et al. (2013). CNTNAP2 is significantly associated with schizophrenia and major depression in the Han Chinese population. Psychiatry Res. 207 225–228 10.1016/j.psychres.2012.09.024 [DOI] [PubMed] [Google Scholar]
- Katsel P., Davis K. L., Haroutunian V. (2005). Variations in myelin and oligodendrocyte-related gene expression across multiple brain regions in schizophrenia: a gene ontology study. Schizophr. Res. 79 157–173 10.1016/j.schres.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Katsel P., Tan W., Abazyan B., Davis K. L., Ross C., Pletnikov M. V., et al. (2011). Expression of mutant human DISC1 in mice supports abnormalities in differentiation of oligodendrocytes. Schizophr. Res. 130 238–249 10.1016/j.schres.2011.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G., Grozeva D., Norton N., Ivanov D., Mantripragada K. K., Holmans P., et al. (2009). Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum. Mol. Genet. 18 1497–1503 10.1093/hmg/ddp043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad A., Vucurevic G., Musso F., Stoeter P., Dahmen N., Winterer G. (2009). ErbB4 genotype predicts left frontotemporal structural connectivity in human brain. Neuropsychopharmacology 34 641–650 10.1038/npp.2008.112 [DOI] [PubMed] [Google Scholar]
- Konrad A., Winterer G. (2008). Disturbed structural connectivity in schizophrenia primary factor in pathology or epiphenomenon? Schizophr. Bull. 34 72–92 10.1093/schbul/sbm034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law A. J., Kleinman J. E., Weinberger D. R., Weickert C. S. (2007). Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum. Mol. Genet. 16 129–141 10.1093/hmg/ddl449 [DOI] [PubMed] [Google Scholar]
- Law A. J., Lipska B. K., Weickert C. S., Hyde T. M., Straub R. E., Hashimoto R., et al. (2006). Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5′ SNPs associated with the disease. Proc. Natl. Acad. Sci. U.S.A. 103 6747–6752 10.1073/pnas.0602002103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Feng G., He L. (2009). Case-control study of association between the functional candidate gene ERBB3 and schizophrenia in Caucasian population. World J. Biol. Psychiatry 10 595–598 10.1080/15622970903304442 [DOI] [PubMed] [Google Scholar]
- Liu X., Qin W., He G., Yang Y., Chen Q., Zhou J., et al. (2005). A family-based association study of the MOG gene with schizophrenia in the Chinese population. Schizophr. Res. 73 275–280 10.1016/j.schres.2004.07.018 [DOI] [PubMed] [Google Scholar]
- Maeno N., Takahashi N., Saito S., Ji X., Ishihara R., Aoyama N., et al. (2007). Association of SOX10 with schizophrenia in the Japanese population. Psychiatr. Genet. 17 227–231 10.1097/YPG.0b013e3280ae6cd8 [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D., Gattaz W. F., Schmitt A., Maccarrone G., Hunyadi-Gulyas E., Eberlin M. N., et al. (2009a). Proteomic analysis of dorsolateral prefrontal cortex indicates the involvement of cytoskeleton, oligodendrocyte, energy metabolism and new potential markers in schizophrenia. J. Psychiatr. Res. 43 978–986 10.1016/j.jpsychires.2008.11.006 [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D., Gattaz W. F., Schmitt A., Rewerts C., Marangoni S., Novello J. C., et al. (2009b). Alterations in oligodendrocyte proteins, calcium homeostasis and new potential markers in schizophrenia anterior temporal lobe are revealed by shotgun proteome analysis. J. Neural. Transm. 116 275–289 10.1007/s00702-008-0156-y [DOI] [PubMed] [Google Scholar]
- Matthews P. R., Eastwood S. L., Harrison P. J. (2012). Reduced myelin basic protein and actin-related gene expression in visual cortex in schizophrenia. PLoS ONE 7:e38211 10.1371/journal.pone.0038211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullumsmith R. E., Gupta D., Beneyto M., Kreger E., Haroutunian V., Davis K. L., et al. (2007). Expression of transcripts for myelination-related genes in the anterior cingulate cortex in schizophrenia. Schizophr. Res. 90 15–27 10.1016/j.schres.2006.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh A. M., Moorhead T. W., Job D., Lymer G. K., Munoz Maniega S., Mckirdy J., et al. (2008). The effects of a neuregulin 1 variant on white matter density and integrity. Mol. Psychiatry 13 1054–1059 10.1038/sj.mp.4002103 [DOI] [PubMed] [Google Scholar]
- Millar J. K., Wilson-Annan J. C., Anderson S., Christie S., Taylor M. S., Semple C. A., et al. (2000). Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 9 1415–1423 10.1093/hmg/9.9.1415 [DOI] [PubMed] [Google Scholar]
- Nakata K., Lipska B. K., Hyde T. M., Ye T., Newburn E. N., Morita Y., et al. (2009). DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc. Natl. Acad. Sci. U.S.A. 106 15873–15878 10.1073/pnas.0903413106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nave K. A. (2010). Myelination and support of axonal integrity by glia. Nature 468 244–252 10.1038/nature09614 [DOI] [PubMed] [Google Scholar]
- Norton N., Moskvina V., Morris D. W., Bray N. J., Zammit S., Williams N. M., et al. (2006). Evidence that interaction between neuregulin 1 and its receptor erbB4 increases susceptibility to schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 141B 96–101 10.1002/ajmg.b.30236 [DOI] [PubMed] [Google Scholar]
- Novak G., Kim D., Seeman P., Tallerico T. (2002). Schizophrenia and Nogo: elevated mRNA in cortex, and high prevalence of a homozygous CAA insert. Brain Res. Mol. Brain Res. 107 183–189 10.1016/S0169-328X(02)00492-8 [DOI] [PubMed] [Google Scholar]
- Peirce T. R., Bray N. J., Williams N. M., Norton N., Moskvina V., Preece A., et al. (2006). Convergent evidence for 2′,3′-cyclic nucleotide 3′-phosphodiesterase as a possible susceptibility gene for schizophrenia. Arch. Gen. Psychiatry 63 18–24 10.1001/archpsyc.63.1.18 [DOI] [PubMed] [Google Scholar]
- Poliak S., Peles E. (2003). The local differentiation of myelinated axons at nodes of Ranvier. Nat. Rev. Neurosci. 4 968–980 10.1038/nrn1253 [DOI] [PubMed] [Google Scholar]
- Prabakaran S., Swatton J. E., Ryan M. M., Huffaker S. J., Huang J. T., Griffin J. L., et al. (2004). Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol. Psychiatry 9 684-697 643 10.1038/sj.mp.4001511 [DOI] [PubMed] [Google Scholar]
- Qin W., Gao J., Xing Q., Yang J., Qian X., Li X., et al. (2005). A family-based association study of PLP1 and schizophrenia. Neurosci. Lett. 375 207–210 10.1016/j.neulet.2004.11.013 [DOI] [PubMed] [Google Scholar]
- Qu M., Yue W., Tang F., Wang L., Han Y., Zhang D. (2008). Polymorphisms of Transferrin gene are associated with schizophrenia in Chinese Han population. J. Psychiatr. Res. 42 877–883 10.1016/j.jpsychires.2007.10.005 [DOI] [PubMed] [Google Scholar]
- Rastogi A., Zai C., Likhodi O., Kennedy J. L., Wong A. H. (2009). Genetic association and post-mortem brain mRNA analysis of DISC1 and related genes in schizophrenia. Schizophr. Res. 114 39–49 10.1016/j.schres.2009.06.019 [DOI] [PubMed] [Google Scholar]
- Ratta-Apha W., Hishimoto A., Mouri K., Shiroiwa K., Sasada T., Yoshida M., et al. (2013). Association analysis of the DISC1 gene with schizophrenia in the Japanese population and DISC1 immunoreactivity in the postmortem brain. Neurosci. Res. 77 222–227 10.1016/j.neures.2013.08.010 [DOI] [PubMed] [Google Scholar]
- Ren Y., Wang H., Xiao L. (2013). Improving myelin/oligodendrocyte-related dysfunction: a new mechanism of antipsychotics in the treatment of schizophrenia? Int. J. Neuropsychopharmacol. 16 691–700 10.1017/S1461145712001095 [DOI] [PubMed] [Google Scholar]
- Ripke S., O’Dushlaine C., Chambert K., Moran J. L., Kahler A. K., Akterin S., et al. (2013). Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 45 1150–1159 10.1038/ng.2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S. E., Greenberg M. E., Stiles C. D. (2003). Basic helix-loop-helix factors in cortical development. Neuron 39 13–25 10.1016/S0896-6273(03)00365-9 [DOI] [PubMed] [Google Scholar]
- Roussos P. (2012). Transcription factor 4 as an important determinant of gating function in schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 109 5915–5916 10.1073/pnas.1203675109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P., Giakoumaki S. G., Adamaki E., Bitsios P. (2011). The influence of schizophrenia-related neuregulin-1 polymorphisms on sensorimotor gating in healthy males. Biol. Psychiatry 69 479–486 10.1016/j.biopsych.2010.09.009 [DOI] [PubMed] [Google Scholar]
- Roussos P., Katsel P., Davis K. L., Bitsios P., Giakoumaki S. G., Jogia J., et al. (2012a). Molecular and genetic evidence for abnormalities in the nodes of Ranvier in schizophrenia. Arch. Gen. Psychiatry 69 7–15 10.1001/archgenpsychiatry.2011.110 [DOI] [PubMed] [Google Scholar]
- Roussos P., Katsel P., Davis K. L., Siever L. J., Haroutunian V. (2012b). A system-level transcriptomic analysis of schizophrenia using postmortem brain tissue samples. Arch. Gen. Psychiatry 69 1205–1213 10.1001/archgenpsychiatry.2012.704 [DOI] [PubMed] [Google Scholar]
- Saetre P., Agartz I., De Franciscis A., Lundmark P., Djurovic S., Kahler A., et al. (2008). Association between a disrupted-in-schizophrenia 1 (DISC1) single nucleotide polymorphism and schizophrenia in a combined Scandinavian case-control sample. Schizophr. Res. 106 237–241 10.1016/j.schres.2008.08.024 [DOI] [PubMed] [Google Scholar]
- Sequeira P. A., Martin M. V., Vawter M. P. (2012). The first decade and beyond of transcriptional profiling in schizophrenia. Neurobiol. Dis. 45 23–36 10.1016/j.nbd.2011.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberberg G., Darvasi A., Pinkas-Kramarski R., Navon R. (2006). The involvement of ErbB4 with schizophrenia: association and expression studies. Am. J. Med. Genet. B Neuropsychiatr. Genet. 141B 142–148 10.1002/ajmg.b.30275 [DOI] [PubMed] [Google Scholar]
- Sklar P., Ripke S., Scott L. J., Andreassen O. A., Cichon S., Craddock N., et al. (2011). Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 43 977–983 10.1038/ng.943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Li W., Feng J., Heston L. L., Scaringe W. A., Sommer S. S. (2008). Identification of high risk DISC1 structural variants with a 2% attributable risk for schizophrenia. Biochem. Biophys. Res. Commun. 367 700–706 10.1016/j.bbrc.2007.12.117 [DOI] [PubMed] [Google Scholar]
- Sprooten E., Sussmann J. E., Moorhead T. W., Whalley H. C., Ffrench-Constant C., Blumberg H. P., et al. (2011). Association of white matter integrity with genetic variation in an exonic DISC1 SNP. Mol. Psychiatry 16685 688–689 10.1038/mp.2011.15 [DOI] [PubMed] [Google Scholar]
- Stefanis N. C., Trikalinos T. A., Avramopoulos D., Smyrnis N., Evdokimidis I., Ntzani E. E., et al. (2007). Impact of schizophrenia candidate genes on schizotypy and cognitive endophenotypes at the population level. Biol. Psychiatry 62 784–792 10.1016/j.biopsych.2006.11.015 [DOI] [PubMed] [Google Scholar]
- Stefansson H., Sigurdsson E., Steinthorsdottir V., Bjornsdottir S., Sigmundsson T., Ghosh S., et al. (2002). Neuregulin 1 and susceptibility to schizophrenia. Am. J. Hum. Genet. 71 877–892 10.1086/342734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Sakurai T., Davis K. L., Buxbaum J. D. (2011). Linking oligodendrocyte and myelin dysfunction to neurocircuitry abnormalities in schizophrenia. Prog. Neurobiol. 93 13–24 10.1016/j.pneurobio.2010.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W., Wang Y., Gold B., Chen J., Dean M., Harrison P. J., et al. (2007). Molecular cloning of a brain-specific, developmentally regulated neuregulin 1 (NRG1) isoform and identification of a functional promoter variant associated with schizophrenia. J. Biol. Chem. 282 24343–24351 10.1074/jbc.M702953200 [DOI] [PubMed] [Google Scholar]
- Tkachev D., Mimmack M. L., Ryan M. M., Wayland M., Freeman T., Jones P. B., et al. (2003). Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet 362 798–805 10.1016/S0140-6736(03)14289-4 [DOI] [PubMed] [Google Scholar]
- Voineskos A. N., De Luca V., Bulgin N. L., Van Adrichem Q., Shaikh S., Lang D. J., et al. (2008). A family-based association study of the myelin-associated glycoprotein and 2′,3′-cyclic nucleotide 3′-phosphodiesterase genes with schizophrenia. Psychiatr. Genet. 18 143–146 10.1097/YPG.0b013e3282fa1874 [DOI] [PubMed] [Google Scholar]
- Wan C., Yang Y., Feng G., Gu N., Liu H., Zhu S., et al. (2005). Polymorphisms of myelin-associated glycoprotein gene are associated with schizophrenia in the Chinese Han population. Neurosci. Lett. 388 126–131 10.1016/j.neulet.2005.06.051 [DOI] [PubMed] [Google Scholar]
- Wang K. S., Liu X. F., Aragam N. (2010). A genome-wide meta-analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophr. Res. 124 192–199 10.1016/j.schres.2010.09.002 [DOI] [PubMed] [Google Scholar]
- Weickert C. S., Tiwari Y., Schofield P. R., Mowry B. J., Fullerton J. M. (2012). Schizophrenia-associated HapICE haplotype is associated with increased NRG1 type III expression and high nucleotide diversity. Transl. Psychiatry 2 e104 10.1038/tp.2012.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. F., Qin W., Shugart Y. Y., He G., Liu X. M., Zhou J., et al. (2005). Possible association of the MAG locus with schizophrenia in a Chinese Han cohort of family trios. Schizophr. Res. 75 11–19 10.1016/j.schres.2004.11.013 [DOI] [PubMed] [Google Scholar]
- Yuan A., Yi Z., Sun J., Du Y., Yu T., Zhang C., et al. (2012a). Effect of SOX10 gene polymorphism on early onset schizophrenia in Chinese Han population. Neurosci. Lett. 521 93–97 10.1016/j.neulet.2012.05.040 [DOI] [PubMed] [Google Scholar]
- Yuan A., Yi Z., Wang Q., Sun J., Li Z., Du Y., et al. (2012b). ANK3 as a risk gene for schizophrenia: new data in Han Chinese and meta analysis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 159B 997–1005 10.1002/ajmg.b.32112 [DOI] [PubMed] [Google Scholar]