Abstract
microRNAs (miRNAs) are small endogenous non-coding RNAs that function as the universal specificity factors in post-transcriptional gene silencing. Discovering miRNAs, identifying their targets and further inferring miRNA functions have been a critical strategy for understanding normal biological processes of miRNAs and their roles in the development of disease. In this review, we focus on computational methods of inferring miRNA functions, including miRNA functional annotation and inferring miRNA regulatory modules, by integrating heterogeneous data sources. We also briefly introduce the research in miRNA discovery and miRNA-target identification with an emphasis on the challenges to computational biology.
Keywords: miRNA, functional annotation, functional miRNA–mRNA regulatory modules
INTRODUCTION
The genetic material of an organism, or genome [1], plays a central role in encoding both the cellular fabric and the regulatory machinery that controls cell homeostasis and internal functions, such as DNA replication and response to environmental signals. While the genome is encoded by DNA, the complex biological processes derived from genome involve a myriad of interacting and co-functioning RNA molecules and diverse protein structures. These co-functioning groups of molecules described as gene regulatory modules are essential components in biological systems. In order to understand the composition of these modules and their roles in an organism, detailed investigation of gene structures, functions and activities must be determined within individual cells and in various tissues throughout development. However, since gene structure and function are relatively constant from one cell to another or from one species to another, it is the patterns of gene expression and its regulation or dysregulation that have the greatest consequence in normal biology and diseases.
While gene expressions can be influenced by many factors, post-transcriptional gene regulation involving microRNAs (miRNAs) is particularly fascinating because of the breadth of their interactions facilitated by their synergistic/combinatorial relationships with target genes. miRNAs are characterized by a growing class of ∼22 nt long non-protein-coding RNAs [2, 3]. They are expressed from longer transcripts encoded in animals, plants, viruses and single-celled eukaryotes. miRNAs are also an attractive topic for system modelling and computer science because of their roles as guide strands for mRNA degradation and translational inhibition to a large extent through the logic of complementary base pairing [4].
Increasing evidence suggests that miRNAs are pivotal regulators of development and cellular homeostasis through their control of diverse biological processes. miRNAs regulate target mRNAs and make fine-scale adjustments to protein outputs. Consequently, dysregulation of miRNA functions can lead to human diseases. Recent studies have reported differentially regulated miRNAs in diverse cancer types, such as breast cancer [5], lung cancer [6], prostate cancer [7], colon cancer [8], ovarian cancer [9] and head and neck cancer [10]. miRNAs are also implicated in a number of neurological disorders including Alzheimer’s disease [11], multiple sclerosis [12] and schizophrenia [13]. Thus, identifying miRNAs, targets and their functional regulatory networks are critical in understanding normal biological processes of miRNAs and their roles in the development of disease [14].
Great efforts have been made to discover miRNAs, identify miRNA targets and infer miRNA functions with both biological methods and computational approaches in recent years. These endeavours have drastically increased the amount of miRNA and mRNA data at both expression and sequence levels. However, it is unfeasible to explore all the complexity and diversity of miRNAs and their targets empirically with biological methods in a combinatorial matrix due to the laborious tasks involved. Fortunately, computational methods shed lights on biological research [15] as they facilitate experimental validation by producing statistically significant hypotheses from the large amount of biological measurements.
In this review, we briefly address bioinformatics approaches to miRNA discovery and target identification with an emphasis on the challenges to computational biology as these methodologies have been extensively reviewed elsewhere [16–20]. More attention will be devoted to computational methods of miRNA functional annotation and inferring miRNA regulatory modules (MRMs). This exciting and challenging new development in integrated genomics has been the potential to provide more robust and tangible functional annotation of miRNA and miRNA-associated gene networks.
miRNA DISCOVERY
miRNAs were first identified through genetic approach in the Caenorhabditis elegans through research investigating heterochronic mutants that affect developmental timing. One of these genes, lin-4, did not encode a protein but contained a small segment of homology to multiple motifs in the 3′-untranslated region (3′-UTR) of another heterochronic gene lin-14 which does encode protein [21]. The lin-4 sequence was poorly conserved and for some years this appeared to be an isolated case until the discovery of another miRNA gene, again in C. elegans, known as let-7. The broad conservation among metazoans created significant excitement about let-7 and the prospect of miRNA generally that led to a rapid discovery process both through molecular cloning and bioinformatic approaches [22]. Both of these discoveries were also enhanced by developments in our understanding of the biochemistry of RNA interference and miRNA biogenesis.
Briefly, we now know that miRNAs are initially produced in the nucleus as long primary transcripts (pri-miRNAs) by RNA polymerase II, typically from their own non-coding gene or from the introns of protein-coding genes. The pri-miRNAs fold into hairpins, which bind to two members of the RNase III families of enzymes, Drosha and Dicer. Drosha forms the microprocessor complex with DGCR8 in the nucleus and cleaves the primary transcript to liberate the ∼70 nt miRNA precursor (pre-miRNA) hairpin. After being exported to the cytoplasm by exportin-5, dicer further processes the transcript to produce the mature ∼20 bp miRNA/miRNA* duplex. miRNA discovery approaches, both biological and bioinformatics, have now yielded many thousands of miRNAs. This process continues with new miRNA appearing daily in various databases and compiled officially as the miRBase (http://www.mirbase.org/) [23], which is the primary online repository for published miRNA sequence and annotation (stored in miRBase database) as well as for novel miRNA genes prior to publication (stored in miRBase registry). Each entry in the database represents a predicted hairpin portion of a miRNA transcript with information on the location and sequence of the mature miRNA sequence.
With bioinformatic methods, putative miRNAs are first predicted in genome sequences based on the structural features of miRNA. These algorithms essentially identify hairpin structures in non-coding and non-repetitive regions of the genome that are characteristic of miRNA precursor sequences. The candidate miRNAs are then filtered by their evolutionary conservation in different species. Known miRNA precursors play important roles in searching algorithms because structures of known miRNA are used to train the learning processes to discriminate between true predictions and false positives [16, 24]. Many algorithms, for example, miRScan [25], miRSeeker [26], miRank [27], miRDeep [28], miRDeep2 [29] and miRanalyzer [30], have been proposed. Once predicted, experimental techniques such molecular cloning, sequencing or hybridization are typically used to validate the predictions.
These approaches have also led the discovery process, with experimental methods particularly high-throughput sequencing producing a small RNAs’ expression profile, which can be followed up by bioinformatics to identify RNAs whose structures meet the miRNA criteria. This approach is significantly faster than the classic forward genetics used to identify novel miRNAs. Forward genetics was used to discover the first known miRNA, lin-4, in C. elegans in 1993 [21]. The advantage of directional cloning is that it can be applied to any organism even when little or no genomic information is available. With the advance of next-generation sequencing (NGS), deep sequencing has been also used to discover miRNAs systematically at a phenomenal rate [28, 31], and predicted miRNAs from deep sequencing have been incorporated into miRNA databases [23].
These biological approaches to miRNA discovery have complemented discoveries made through computational approaches, which predict miRNA from genomic DNA sequence. Collectively very large number of miRNAs have been identified and predicted in a very short time frame [24, 32]. The latest miRBase, release 19, contains 21 264 hairpin precursor miRNAs, expressing 25 141 mature miRNA products, in 193 species [23]. Each upgrade refines the prediction continually. Compared with release 18, miRBase were added 3171 more new hairpin sequences and 3625 novel mature products, while over 130 misannotated and duplicate sequences have been deleted. This success in miRNA discovery has rapidly led to an even more daunting challenge in functional annotation, or in other words, what are these molecules doing in cells and what are the functional implications for their dysregulation in pathophysiology of diseases? While these questions have also been addressed both biologically and computationally, the sheer magnitude of this task particularly from an empirical perspective has driven significant development in the bioinformatics of miRNA-target prediction and systems-based analysis of miRNA function.
miRNA-TARGET PREDICTION
In the absence of high-throughput biological approaches to identify miRNA targets, many computational methods, such as miRanda [33], mirSVR [34], PicTar [35], TargetScan [36], TargetScanS [37], RNA22 [38], PITA [39], RNAhybird [40] and DIANA-microT [41], were developed relatively quickly to identify putative miRNA targets. In most cases, these algorithms were developed in conjunction with a limited amount of empirical evidence from a few experimentally validated target sites for a small selection of miRNAs [42].
miRNAs target mRNAs through complementary base pairing, in either complete or incomplete fashion. It has been generally believed that miRNAs bind to the 3′-UTRs of the target transcripts in at least one of two classes of binding patterns [17]. One class of target sites has perfect Watson–Crick complementarity to the 5′-end of the miRNAs, referred as ‘seed region’ which positions at 2–7 of miRNAs. The seed region has been shown that it is sufficient for miRNAs to suppress their targets without requiring significant further base pairings at the 3′-end of the miRNAs. On the contrary, the second class of target sites has imperfect complementary base pairing at the 5′-end of the miRNAs, but it is compensated via additional base pairings in the 3′-end of the miRNAs. However, the 3′-UTR boundaries are not clearly defined in many species and it is still an ongoing project to characterize the location, extent or splice variation of 3′-UTRs in a variety of species [18]. In addition, it has been demonstrated that a transcript can contain multiple target sites for a single miRNA and a transcript can have target sites for several miRNAs. The multiple-to-multiple relations between miRNAs and mRNAs lead to the even more complex miRNA regulatory mechanisms. Regardless of the binding sites, the short length of miRNAs lacks the power to be detected significantly by most statistical techniques in standard sequence analysis, such as Karlin–Altschul statistics [43]. Therefore, most algorithms apply the cross-species conservation requirement to reduce the number of false positives, despite some risk of increasing false negatives as some miRNAs, such as miR-430 [44], lack conserved targets. Overall, the complex features of miRNA pose great challenges on the computational approaches for miRNA-target prediction.
Different miRNA-target prediction algorithms predict targets with different techniques and criteria including base pairing, target accessibility and evolutionary conservation of target site. Table 1 gives some basic features of the selected miRNA-target prediction methods, while the comprehensive review for these methods can be found in [33, 45–47].
Table 1:
miRNA-target prediction algorithm
| Algorithm | Regions scanned | Species conservation | Species | Brief description of the prediction method | Implementation | Download/web server | Reference |
|---|---|---|---|---|---|---|---|
| miRanda | 3′-UTR | Yes | Human, mouse, rat, fly and worm | Predict targets based on rules: (i) sequence complementarity, (ii) binding energy and (iii) evolutionary conservation. | C/open source | http://www.microrna.org | [33] |
| mirSVR | No restriction | Yes | Human, mouse, rat, fly and worm | To score and rank miRanda-predicted miRNA-target sites with a supervised vector regression (SVR) model for features including secondary structure accessibility of the site and conservation. | C/open source | http://www.microrna.org | [34] |
| PicTar | 3′-UTR | Yes | Vertebrates, fly and worm | Filter alignments according to the thermodynamic stability, then score and rank the predicted target by hidden Markov model maximum-likelihood fit approach. | Web-driven application | http://pictar.mdc-berlin.de/ | [35] |
| TargetScan | 8mer and 7mer sites, and open reading frames | Yes | Human, mouse, rate, dog and chicken | Predict targets by searching for the presence of conserved 8mer and 7mer sites that match the seed region. Predictions are ranked by a combinatorial score based on site number, site type and site context. | PerlScript/open source | http://www.targetscan.org/ | [36] |
| TargetScanS | 3′-UTR | Yes | Human, mouse, rate, dog and chicken | Predict targets that have a conserved 6 nt seed match flanked by either a m8 match or a t1A anchor. | Web-driven application | http://genes.mit.edu/tscan/targetscanS2005.html | [37] |
| RNA22 | No restriction | No restriction | Any | Use the patterns discovered from the known mature miRNAs for predicting candidate miRNA-target sites in a sequence. | Web-driven application | http://cm.jefferson.edu/rna22v1.0/ | [38] |
| PITA | 3′-UTR | Yes | Human, mouse, worm and fly | Predict miRNA targets using a non-parameter model that computes the difference between the free energy gained from the formation of the miRNA-target duplex and the energetic cost of unpairing the target to make it accessible to the miRNA. | PerlScript/open source | http://genie.weizmann.ac.il/pubs/mir07/ | [39] |
| RNAhybird | 3′-UTR and coding sequence | No restriction | Any | A tool to identify mRNA secondary structure and energetically favourable hybridization between miRNA and target mRNA. | Web-driven application | http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html | [40] |
| DIANA-microT | 3′-UTR and CDS | No restriction | Human and mouse | The fifth version of microT algorithm which is specifically trained on a positive and negative set of miRNA recognition elements located in both the 3′-UTR and CDS region. The conserved and non-conserved miRNA recognition elements are combined into a final prediction score. | Web-driven application | http://diana.cslab.ece.ntua.gr/microT/ | [41] |
Coding DNA sequence, CDC.
The binding patterns and criteria used by miRNA-target prediction algorithms generally have great influence on the outputs and performance of different algorithms. A small variation in the criteria for selection can lead to large discrepancy in the prediction [33]. However, each approach produce widely different lists of predictions with significant false-positive and false-negative rates [48]. In a study using mass spectrometry to measure the global impact of deletion of a single miRNA, hundreds of proteins were found to respond, but the best performing target prediction algorithms, TargetScan [36] and PicTar [49], which restrict their predictions primarily to conserved sites in 3′-UTRs, were reported to nevertheless have false-positive rates of ∼68% [50]. Although the false-positive rate might not be accurate because this work did not take into account the indirect influence on proteins by knocking down a miRNA, it demonstrated biologically that all target prediction algorithms suffer high false-positive errors.
Another assessment was conducted by Alexiou et al. [51] who compared the performance for eight widely used target prediction programs, including EIMMo [52], miRanda [33], miRBase [53], PicTar [49], PITA [39], RNA22 [54] and TargetScan 5.0 [36], for the human and mouse genome, using experimentally validated targets in Selbach et al. [55]. They found these algorithms have a precision of ∼50% with a sensitivity that ranges from 6 to 12%. These assessments arrived at similar while different conclusions. One possible explanation could be that the selected benchmark sets favour certain algorithms while underestimating others, particularly when the number of validated miRNA targets is still relatively small. However, it highlights the problem of validating miRNA-target interactions at a scale that is insufficient to provide an un-biased assessment of the performance of miRNA-target prediction algorithms.
Despite their limitations, these programmes have been broadly adopted and their prediction of miRNA targets in a broad spectrum of species has been prepared for genome data and stored in databases for download or query by biologists or medical scientists, in some cases with very limited understanding of how they were derived. Some of these databases provide additional features that provide some further insight into the strength and conservation of the putative interaction. For example, TargetScan searches for the presence of conserved 8mer and 7mer sites that match the seed region of each miRNA as well as predicts non-conserved sites. Since version 6.0, TargetScan has extended context score contributions to include seed-pairing stability and target-site abundance and all 3′-UTRs from RefSeq rather than just the longest UTR from each gene.
Recently, some novel biochemical approaches have been developed for miRNA-target identification, providing an extensive insight into the miRNA-binding sites. For example, Chi et al. [56] developed a technique, known as high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP), to identify direct miRNA targets. This technique was applied to mouse brain [56] and C. elegans [57]. Consequently, compelling data have been generated on the location of miRNA-binding sites within both the 3′-UTR and coding region, allowing the genome-wide interaction maps for specific miRNA to be depicted with a high specificity and low false discovery rate compared with previous computational methods [56]. A modified HITS-CLIP, termed photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitaion (PAR-CLIP) [58], is able to deliver more efficient ultraviolent crosslinking which in turn improves RNA recovery up to 1000-fold compared to its predecessor. It can also achieve more precise localization of binding sites between the RNA and protein [58].
By analysing HITS-CLIP/PAR-CLIP data, alternative modes of miRNA-target recognition have been identified. Ellwanger et al. [59] demonstrated that most conserved miRNAs interact with target sites endowed with short seed matches (6mer seeds) and a substantial fraction (40%) of all functional target sites are not conserved. In contrast, common miRNA-target prediction algorithms focus mainly on conserved seed of length seven or eight. Furthermore, Chi et al. [60] demonstrated that over 15% of Ago–miRNA interactions with G-bulge sites in mouse brain cannot be explained by canonical seed match, suggesting a novel mode of miRNA-target recognition. These analyses provide a qualitative change in our understanding and assessment of miRNA–mRNA regulation, which in turn may transform the miRNA prediction algorithms in the near future.
INFERRING miRNA FUNCTIONS
As many miRNAs have been identified, and a large number of miRNA targets have been predicted, research has quickly shifted to inferring miRNA functions, which generally include functional annotation and inferring miRNA regulatory mechanisms in specific biological conditions. We will review the methods of inferring miRNA functions in this section.
miRNA functional annotation
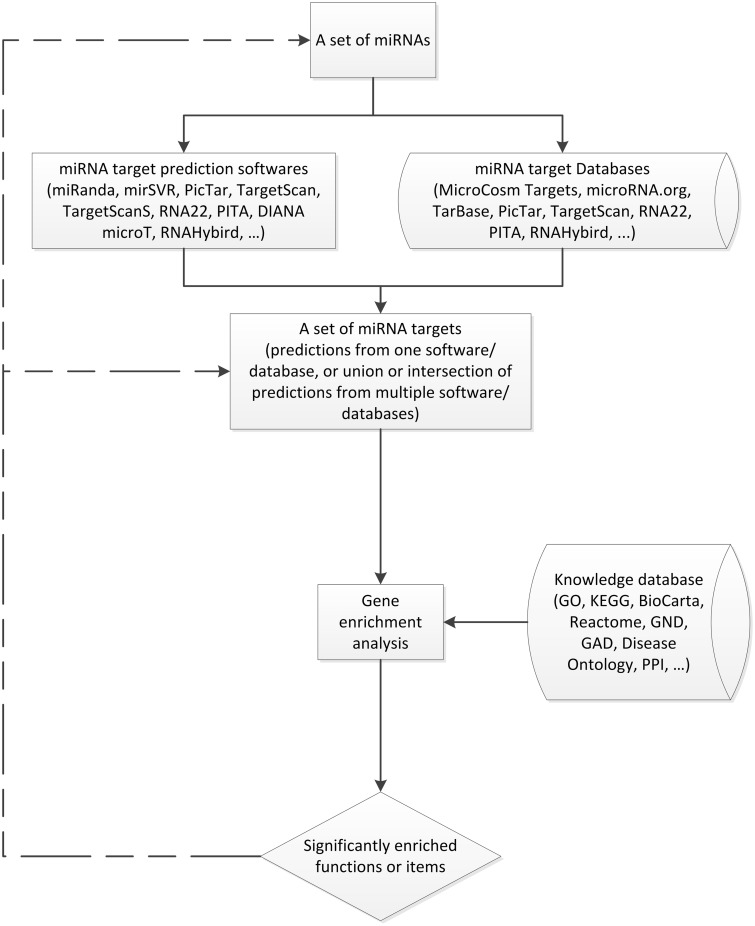
The most straight-forward approach of miRNA functional annotation is through functional enrichment analysis using the miRNA-target genes (Figure 1). This approach assumes that miRNAs have similar functions to their target genes given a large amount of knowledge of genes have been accumulated in the last few decades. Therefore, it is practical to assign the functions, which are significantly enriched with the targeted genes, to miRNAs. When a list of miRNA targets is available well-developed gene functional annotation resources such as DAVID [61] and WebGestalt [62] can be easily used to assign the functions of the target mRNAs to the group of miRNAs. Functional annotation for miRNAs gives great insights into the general functions of miRNAs. For the first time, enabled the mysteries of miRNA functions to be revealed in large scale.
Figure 1:
A framework of miRNA functional annotation.
Similarly, miRGator [63], miRDB [64], miRò [65], MAGIA [66] and FAME [67] have been developed with target prediction an built-in functional annotation. These are freely accessible databases with user-friendly web interfaces providing miRNA functional annotation with the similar strategy (Table 2). miRGator [63] infers miRNA functions from a list of target genes predicted by miRanda, PicTar and TargetScanS. As an option, the list of target genes can be the union or intersection of prediction from these three programmes. Statistical enrichment test of target genes in each term is carried out for Gene Ontology (GO), pathway and disease annotations. miRDB [64] uses MirTarget2 [68] for miRNA-target prediction using a machine learning method (support vector machines), with public microarray data sets. This method also adopts the Wiki model for functional annotation, which is an open environment allowing anyone with internet access to make contributions. miRò [65] associates miRNAs with phenotypes by integrating miRNA annotation and target databases, such as miRBase, miRNA Atlas, TargetScan, PicTar and miRecords, with multiple online biological knowledge databases including Gene and Nucleotide Database (GND, http://www.ncbi.nlm.nih.gov), GO and Genetic Association Database (GAD, http://geneticassociationdb.nih.gov, a database of human genetic association studies of complex diseases and disorders). Validated data are highlighted in the databases indicating the most significant associations. MAGIA [66] supports multiple algorithms of miRNA-target prediction, such as miRanda, PITA and TargetScan, and multiple statistical methods to infer the relationship between miRNA–mRNA pairs and biological processes or diseases. Different from other methods, FAME [67] annotates miRNA functions by incorporating the expression profiles of miRNAs/mRNA with the miRNA-target prediction. It uses a co-expressed subset of miRNA-target genes, which were considered to be the designated target set based on the parameters extracted from TargetScan, such as context score, for functional enrichments. Few other tools, such as miR2Disease [69] and miReg [70], manually curate annotation of miRNA functions and focus on the association with human diseases based on literature. They have a very limited scale due to the current knowledge about miRNAs. All these tools are similar in the strategy for functional annotation while they differ in the databases involved and enrichment methods used.
Table 2:
Tools for miRNA functional annotation
| Tool | Target databases | Use expression | Knowledge databases | Link | Reference |
|---|---|---|---|---|---|
| DAVID | N/A | No | GO, KEGG, BioCarta, GAD, OMIM Disease, PPI, etc. | http://david.abcc.ncifcrf.gov/ | [61] |
| WebGestalt | N/A | No | GO, KEGG, IPI, Pathway Commons, Wikipathways, MGI, SGD, MSigDB, NCBI dbSNP | http://bioinfo.vanderbilt.edu/webgestalt/ | [62] |
| miRGator | miRanda, PicTar and TargetScanS | No | GO, KEGG/GenMapp/BioCarta, Disease Ontology | http://genome.ewha.ac.kr/miRGator/ | [63] |
| miRDB | MirTarget2 | Yes | Wiki model | http://mirdb.org/miRDB/ | [64] |
| miRò | miRanda, PITA and TargetScan | Yes | NCBI Gene Database, NCBI Nucleotide Database, GO, GAD | http://ferrolab.dmi.unict.it/miro/ | [65] |
| MAGA | miRanda, PITA and TargetScan | Yes | Through DAVID APIs | http://gencomp.bio.unipd.it/magia/start/ | [66] |
| FAME | TargetScan | Yes | Experimentally verified miRNA-pathway and miRNA-process associations | http://acgt.cs.tau.ac.il/fame/ | [67] |
| miR2Disease | N/A | No | Manually curated database containing 1939 relationship between 299 human miRNAs and 94 human diseases | http://www.mir2disease.org/ | [69] |
| miReg | N/A | No | Manually curated database containing 47 human miRNAs, 85 proteins, 115 upstream regulators, 165 targets, 38 diseases, 295 reactions and 70 biological processes | http://www.iioab-mireg.webs.com/ | [70] |
Not applicable, N/A.
miRNA functional annotation heavily relies on the miRNA-target prediction as most of the approaches are based on the predicted targets. As discussed above, the target prediction varies greatly among different algorithms and with even a small change to parameters used by the algorithms. Furthermore, some evidence had shown that miRNAs may target mRNA outside of the 3′-UTR. Mature miRNAs can alter the expression of genes by binding to the 5′-UTR [71, 72]. Other regions, known as extended seed and delta seed regions, also contribute to the target selection [73]. Obviously, most prediction algorithms will miss those targets because they focus on the 3′-UTR [17]. Moreover, it is possible that the target sites for different miRNAs in the same 3′-UTR indicate that the mRNAs are regulated by tissue-specific or development-specific miRNAs [17]. It is not reasonable to group them together for the functional annotation. Therefore, more sophisticated methods are expected to infer miRNA functions.
Inferring miRNA regulatory mechanism
In order to gain global and yet specific insights into the functions of miRNAs in a broad layer of post-transcriptional control, methods beyond searching for the base pairing between miRNAs and mRNAs have been proposed. In the last few years, many studies have been conducted to infer the miRNA regulatory mechanisms by incorporating target prediction with other genomics data, such as the expression profiles of miRNAs and mRNAs.
Largely, the inference of miRNA regulatory mechanism can be regarded as a question of data integration in the functional analysis of miRNAs. Few data sets are available for inferring miRNA regulatory mechanism, such as (i) miRNA-target information from target prediction algorithms or miRNA-target databases, (ii) sample-matched expression profiles of miRNAs and mRNAs from microarray experiments or NGS techniques and (iii) biological conditions or diseases related with different samples.
miRNA-target information from target prediction algorithms or miRNA-target database provides a way to build the potential relationships between miRNAs and mRNAs. Several miRNA-target databases, such as TargetScan [36], PicTar [35], TarBase [74], miRecords [75] and miRWalk [76], store computationally predicted miRNA targets as well as few biologically validated ones. The miRNA-target information is usually presented as a table where each row indicates a target pair of miRNA and mRNA. Besides miRNA and its target mRNA, other information, such as the sequence of miRNA/mRNA, the binding score between miRNA and mRNA and number of conserved and non-conserved sites, may be also presented in each row depending on the miRNA-target prediction algorithms used.
For the expression profiles of miRNAs/mRNAs, they are usually organized as 2D tables where the columns are samples from different biological conditions and the rows are miRNAs/mRNAs. Each cell of the table is an expression value of certain miRNA/mRNA in a sample either from microarrays or estimated from NGS techniques. Microarray technology is a powerful method for routine studies of selected target sequences, while NGS data enable a more detailed inspection on gene diversity because it allows wider applications as well as provides better sensitivity, accuracy and dynamic range than microarrays. It is worth noting that in general there is good concordance between the platforms [77, 78] particularly in terms of the biological interpretation [79]. In general, the expression profiles are subjected to a series of pre-processing, such as background subtraction and normalization, before they can be further used for a variety of downstream analysis.
Each sample is usually related with a biological condition, such as cancer or normal, which provides the class information when inferring the function of miRNAs. Microarray experiments and NGS are commonly designed in a comparative fashion in which the samples are extracted from different biological conditions in order to find biological differences among conditions. It is critical information for guiding inference of miRNA functions. This information is usually a class label tagged to each sample.
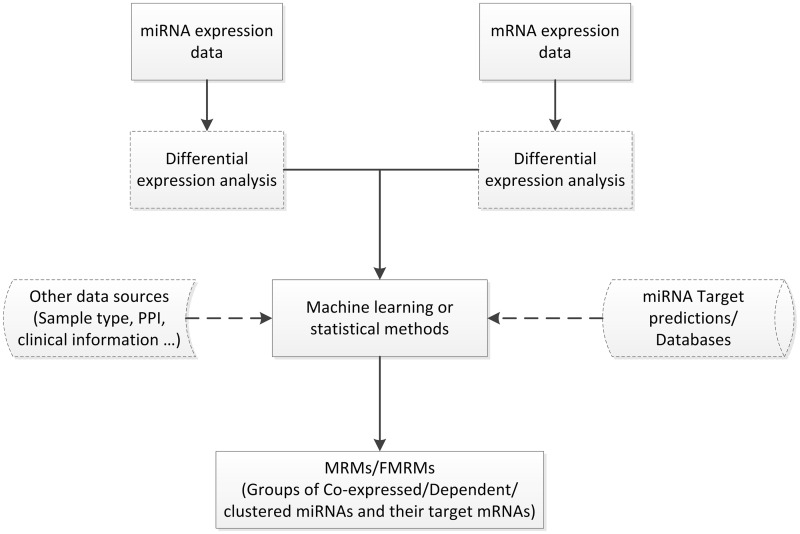
Depending on what information is involved, we classify the computational methods of inferring miRNA regulatory mechanisms into two categories: (i) predicting MRMs, that is, to identify a group of co-expressed miRNAs and mRNAs, either at the sequence level [80] or by integrating sequence and expression profiles of miRNAs and mRNAs [81–84] and (ii) inferring functional miRNA–mRNA regulatory modules (FMRMs), which are regulatory networks of miRNAs and their target mRNAs in specific biological processes [85–88]. MRMs suggest a broader control of miRNAs to mRNAs in terms of general functions, while FMRMs focus on more detailed miRNA regulatory mechanism in specific biological conditions. Figure 2 illustrates the general framework of inferring MRMs/FMRMs.
Figure 2:
A framework of inferring MRMs/FMRMs.
Predicting MRMs
The first attempt of predicting MRMs was conducted by Yoon and De Micheli [80] who modelled the miRNA co-target relationship with the graph theory. In this approach, a MRM is defined as a special bipartite graph, named biclique, where two sets of nodes are connected by edges. Every node of the first set representing miRNA is connected to every node of the second set representing mRNAs by edges with similar weights. The weights of edges correspond to the miRNA–mRNA binding strength inferred from target prediction algorithms, such as the methods described in Lewis et al. [36] and John et al. [33] where the strength of miRNA-target binding can be quantified. The biological observation, in which the strength of each binding is not too strong or weak but modest and similar when multiple binding sites exist on a target from Lai [89], is formulated in the method. Potential miRNA-target relationships are first constructed as weighted bipartite graphs based on the sequence binding between miRNAs and mRNAs. Then, a graph-mining method is proposed to discover bicliques in which all the edges have similar weights in the given bipartite graphs. Statistically significant MRMs are selected by calculating the probability of finding a module by chance. This is the first method that explicitly searches for the multiple-to-multiple relationships among miRNAs and their target genes. The limitation is that it models the miRNA co-target relationship at the sequence level only, thus the miRNA regulatory patterns at the expression level are not characterized.
Recent methods have integrated the analysis of expression profiles of miRNAs and mRNAs in conjunction with the predicted miRNA targets. Most of the integrative methods of MRM discovery are based on the assumption that miRNA negatively regulate their target mRNAs to the effect that an inverse relationship should exist between the expression of a specific miRNA and its targets.
Huang et al. [81, 90] applied the Bayesian network (BN) parameter learning to infer miRNA–mRNA interactions using both the miRNA–mRNA sequence binding information and the sample-matched expression profiles of miRNAs and mRNAs. An initial network representing the putative target relationships between miRNAs and mRNAs is constructed according to the target information predicted for sequence binding. Then the observation, miRNAs down-regulate their target mRNAs, is encoded in the network at the expression level. It models the expression of a mRNA, which is assumed to follow a Gaussian distribution, as the negative of a sum of weighted expression values of their regulator miRNAs. The Gaussian BN parameter learning is used to infer the likelihood of miRNAs regulating target mRNAs at the expression level. This method explicitly encodes the inverse expression patterns between miRNAs and their target mRNAs in the interaction network. Furthermore, this model searches for co-expressed miRNAs and mRNAs which are presumed to function together. Thus, this model can potentially detect co-functional miRNAs and mRNAs besides refine miRNA-target predictions at the expression level.
Joung et al. [82] proposed a probabilistic method to integrate the miRNA-target binding information and expression profiles of miRNAs and mRNAs for MRMs. In this method, a MRM is defined as a group of miRNAs and mRNAs with coherent expression patterns in terms of 3D, miRNA–mRNA, miRNA–miRNA and mRNA–mRNA, across all biological conditions. Two heterogeneous data sources, miRNA-target prediction scores based on target binding and the sample-matched expression profiles of miRNA and mRNA, are integrated to extract the coherent miRNA–mRNA modules. To describe the coherence of miRNA–mRNA modules in the above 3D, the means of Pearson’s correlation coefficients between all miRNA or mRNA pairs are aggregated with the mean binding scores of all miRNA–mRNA pairs. It is characterized by a fitness function in which a co-evolutionary learning and estimation-of-distribution algorithms are used to mine the optimal groups of miRNAs and mRNAs, which give the best fitness scores in an iterative fashion. Thus, this method allows detection of correlated miRNA–mRNA modules from multiple data sources by using a balanced fitness function. It was demonstrated with a human cancer data set and two miRNA–mRNA modules found, which are highly correlated with respect to their expression and biological functions.
Tran et al. [83] proposed a rule-based learning method to predict MRMs. It is based on an assumption that genes regulated by the same miRNAs show similar expression profiles. This method first utilizes the miRNAs and their targets, which were predicated by PicTar, to construct miRNA–mRNA relationships at the sequence level. This target relationship is denoted as a target binary matrix where rows are mRNAs and columns are miRNAs, and the element of the matrix is 1 if the miRNA in the column targets the mRNA in the row, otherwise 0. For each mRNA expression value in the given data set, this method calculates the Pearson’s correlation coefficients between it and every other mRNA. The level of correlation is then denoted as ‘Similarity’ or ‘Dissimilarity’ using a pre-set arbitrary threshold. This similarity information is added to the target binary matrix as an extra column to construct a regulatory decision table, which is then fed into a CN2-SD [91], a rule induction method. The CN2-SD searches the regulatory decision table for a set of 1 in the columns with ‘Similarity’ denoted at the rows. That is, a group of correlated mRNAs co-targeted by a group of miRNAs. The Pearson’s correlation coefficients are further calculated for miRNAs output from CN2-SD, and only highly correlated miRNAs are maintained for final MRMs. This procedure is repeated for every mRNA to find all MRMs in the given data sets. This method was demonstrated with a public data set. Several MRMs with high correlation in expression patterns of miRNAs and mRNAs were found. They also showed that the mRNAs included in the same modules share similar biological functions. However, it is not completely true that genes regulated by the same miRNAs show similar expression profiles, which was used as the basic assumption for this method. Thus, it can be misleading and many potential MRMs may be missed.
Peng et al. [84] developed an approach from both Yoon’s [80] and Tran’s [83] methods. In this work, MRMs are bicliques where miRNAs co-target mRNAs predicted at the sequence level while the miRNAs and mRNAs are negatively correlated at the expression level. At the expression level, pair-wise correlations between the differentially expressed miRNAs and mRNAs are calculated with Pearson’s correlation across all matched samples. A correlation threshold is determined by a desired false detection rate, which is the percentage of miRNA–mRNA pairs out of the total number of selected pairs that would have the same or better correlation just by chance. By applying this threshold, a 2D correlation matrix is constructed where elements stand for the miRNAs in columns negatively correlated with the mRNAs in rows. In parallel, for the same set of miRNAs and mRNA, a 3D miRNA-target matrix is built by examining if miRNA–mRNA pairs match in the seed region. Then, a miRNA–mRNA regulatory matrix is constructed by multiplying the binary correlation matrix with the miRNA-target matrix in the dot product fashion. The miRNA–mRNA regulatory matrix is further represented as bipartite graphs in which a fast searching algorithm is used to enumerate all the bicliques. The statistically significant bicliques are the final MRMs through a permutation test. This method was applied to a data set of human liver biopsy samples for hepatitis C virus study and identified 38 MRMs that were associated with the hepatitis C virus infection.
Recently, Zhang et al. [92] proposed a method to infer MRMs by integrating miRNA-target predictions, expression profiles of miRNA and mRNA and the topological structures of protein–protein interactions (PPIs). This method uses the miRNA-target predictions as the basic structures, while the expression profiles of miRNA and mRNA are applied on the structures to find the co-expressed miRNAs and mRNAs, then PPIs are further used to refine the structures. A novel machine learning method sparse network regularized multiple non-negative matrix factorization (SNMNMF) was developed in this work to integrate three heterogeneous data sources. They tested this method on a data set of ovarian cancer samples, and 49 significant MRMs were identified, where the miRNA modules are enriched with miRNAs clusters in their chromosomal locations and the gene modules are enriched with known function gene sets.
The above methods (Table 3) aim at exploring general miRNA–mRNA regulatory modules by integrating miRNA-target prediction on sequence with expression profiles of miRNA and mRNA or other data sources. They have archived variant successes on different trial data sets. However, they identify groups of co-expressed miRNAs and mRNAs without considering the biological conditions of the samples. Therefore, no implications regarding the functions of MRMs in specific biological conditions can be identified. The functions of MRMs in terms of biological processes usually are unclear until a functional enrichment analysis is conducted by querying the identified target genes against the GO or other similar annotation databases [80, 83]. Those biological conditions are very important in biological experimental design, and hence, some conditionally related MRMs may be omitted if we do not take into account the conditions. This question, however, is of great interest in understanding the biological pathways of MRMs in more detail.
Table 3:
Summary of methods for inferring MRMs
| Method | Data sources | miRNA- target database used | Differential gene analysis | Key features | Availability of software | References |
|---|---|---|---|---|---|---|
| Yoon and De Micheli | miRNA-target binding information | TargetScan | N/A | Sequence level method; searching for bicliques with modest or similar binding strength of miRNAs and mRNAs. | Upon request | [80] |
| Huang et al. | Sample-matched expression profiles of miRNA and mRNA, and miRNA-target prediction | Any | No | BN parameter learning-based method; the inverse patterns of expression between miRNAs and mRNAs are encoded in the network. | GenMiR++, http://www.psi.toronto.edu/genmir/ | [81, 90] |
| Joung et al. | miRNA-target prediction scores and the sample-matched expression profiles of miRNA and mRNA | miRBase | No | A machine learning method to capture co-expressed miRNAs and mRNAs. | Upon request | [82] |
| Tran et al. | miRNA-target prediction and the sample-matched expression profiles of miRNA and mRNA | PicTar | No | A rule-based method to capture groups of mRNA with similar expression patterns targeted by groups of miRNA with similar expression patterns. | Upon request | [83] |
| Peng et al. | miRNA-target prediction and the sample-matched expression profiles of miRNA and mRNA | Any | Yes | Combination of Yoon’s and Tran’s methods. | Upon request | [84] |
| Zhang et al. | miRNA-target prediction, expression profiles of miRNA and mRNA, and topological structures of PPI | MicroCosm | Yes | A computational framework integrating expression profiles of miRNA/mRNA, PPI, DNA–protein interaction and miRNA–mRNA targeting. | http://zhoulab.usc. edu/SNMNMF/ | [92] |
Not applicable, N/A.
Inferring FMRMs
In order to resolve the limitation of MRMs and gain understanding of miRNA functions in specific biological processes, the concept of FMRMs [85] was proposed. FMRMs explicitly characterize how groups of miRNAs regulate their target mRNAs and how they co-act together to form pathways in complex regulatory networks for specific conditions. Many methods have been proposed to infer FMRMs (Table 4) since then.
Table 4:
Summary of methods for inferring FMRMs
| Method | Data sources | miRNA-target database use | Differential gene analysis | Key features | Availability of software | Reference |
|---|---|---|---|---|---|---|
| Liu et al. | miRNA-target predictions, expression profiles of miRNA and mRNA, and sample information | Any | Yes | A rule-based method; searching for bicliques with inversed miRNA–mRNA pairs associating with biological conditions. | Upon request | [85] |
| Joung et al. | miRNA-target prediction and expression profiles of mRNA | Any | No | A hierarchical clustering method to identify the co-expressed miRNAs/mRNAs. | Upon request | [86] |
| Liu et al. | miRNA-target predictions, expression profiles of miRNA and mRNA, and sample information | Any | Yes | A BN structure learning-based method; sample information is incorporated into the network. | BNSA, upon request | [87] |
| Nunez-Iglesias et al. | miRNA-target predictions, expression profiles of miRNA and mRNA, and sample information | Any | Yes | The miRNA–mRNA pairs are identified by searching for the highest difference in the standardized correlation of miRNAs and mRNAs between case and control sample. | Upon request | [96] |
| Bonnet et al. | Expression profiles of miRNA and mRNA, and sample information | N/A | No | A two-step method: (i) two-way clustering identify the tight clustered genes in all conditions and (ii) a fuzzy decision tree model is applied to each cluster to identify the regulation programs. | LeMoNe, http://bioinformatics.psb.ugent.be/software/details/LeMoNe | [97] |
| Liu et al. | Expression profiles of miRNA and mRNA with or without miRNA-target predictions | Any | No | A probabilistic graphical model based on Corr-LDA. | Corr-LDA, upon request | [88] |
Not applicable, N/A.
Liu et al. [85] proposed an approach to infer FMRMs by combining graph theory and association rule mining. This method consists of two steps at both sequence and expression levels. At the sequence level, putative miRNA regulatory networks are constructed as bipartite graphs where a connection is made between a specific miRNA and its predicated target mRNAs based on the miRNA-target predictions algorithms or target databases. A fast biclique searching algorithm, modular input consensus algorithm (MICA) [93], is then applied to enumerate all bicliques given the bipartite graphs. At the expression level, association rule mining is used to discover the significant associations between specific biological conditions and the inverse expression patterns of miRNAs and mRNAs on all enumerated bicliques. Finally, the association relationships among miRNAs, mRNAs and conditions are merged to be the final FMRMs. This method was demonstrated on a publicly available prostate cancer data set where two modules are identified. Those modules are associated with cancer and normal conditions, respectively. This study was the first published work to explicitly discover FMRMs.
Joung et al. [86] used a generative model to predict FMRMs adopted from the Author-Topic model [94] which was proposed initially in information retrieval. This method models the miRNA–mRNA regulatory mechanism as hierarchical steps in which the FMRMs are defined as functional clusters of miRNAs and target mRNAs involved in the same biological processes. In this generative model, the expression value of an mRNA is regarded as the number of times an event of the mRNA expressed in a sample. Each mRNA has events of its expression in a specific condition that is likely to be associated with its regulator miRNAs given by miRNA-target predictions. A hierarchical generative process hypothesizes that a miRNA is sampled from a multinomial distribution over FMRMs, and then the sampled miRNA is used to sample mRNAs which have a multinomial distribution over conditions. An approximate method, Gibbs sampling, is used to infer the parameters of the generative model, which can characterize FMRMs. This method integrates data sets, including miRNA-target information and expression profiles of mRNAs. It predicted several biological processes related to miRNA–mRNA modules on an Arabidopsis data set. The drawback of this method is that it does not use the expression profiles of miRNAs. Thus, the regulatory relationships of miRNAs and mRNAs largely rely on the miRNA-target information predicated at the sequence level.
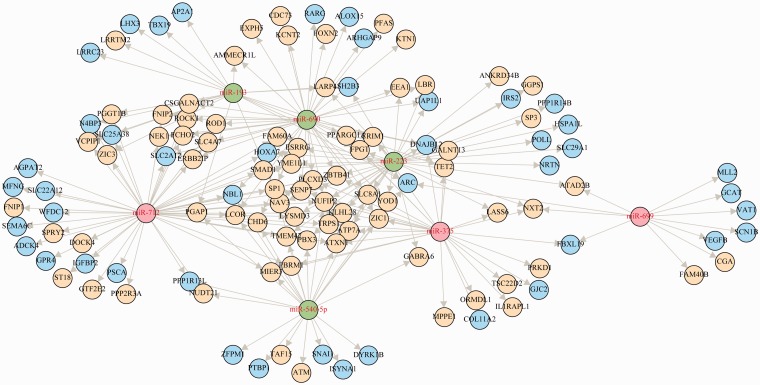
Liu et al. [87] proposed a BN-based method to identify complex miRNA–mRNA interactions for FMRMs, named Bayesian network with splitting-averaging (BNSA). They demonstrated that the conventional BNs are not able to identify all the interactions potentially existing in the data. Thus, BNSA was proposed to discover all possible miRNA–mRNA interactions, including the subtle ones undetectable for conventional BNs. This method integrates miRNA-target information, sample-matched expression profiles of miRNA and mRNA, and sample categories. In order to capture all possible interactions, this method groups expression profiles of miRNAs and mRNAs together according to their sample category and then learns BN structures on the expression profiles of miRNA and mRNA in each category, respectively. The miRNA-target information acts as a constraint to guide the structure learning, whereby the miRNAs represent the parent nodes while the mRNAs are the descendant nodes. The edges linking the parent nodes to descendant nodes can only be those defined in the miRNA-target predictions. Interaction networks learned on each category are then integrated by BN averaging procedure. To avoid statistically insignificant results due to the small size of data sets, it uses bootstrapping to achieve reliable inference and integration. This method was demonstrated on NCI-60 data sets [95] and used to characterize the FMRMs towards epithelial to mesenchymal transition (EMT). The results show that this method captured all possible types of miRNA–mRNA interactions, including both negatively correlated and positively correlated miRNA–mRNA pairs, from the data in terms of EMT. For the first time, it demonstrated that positively correlated expression patterns of miRNA–mRNA also widely exist in the data besides negatively correlated ones (Figure 3). Many interactions are of tremendous biological significance according to pathway analysis. Some discoveries have been validated by previous research, such as miR-200 family that negatively regulates ZEB1 and ZEB2 for EMT. Some are consistent with the literature, and many novel interactions are statistically significant and worthy of further investigation and validation.
Figure 3:
A FMRM identified by BNSA from analysis of schizophrenia subjects. It shows that miRNAs may up/down regulate their target mRNAs, either direct or indirect. Up-regulated miRNAs are coloured in red and down-regulated miRNAs are coloured in green. Up-regulated mRNAs are coloured in yellow, while down-regulated mRNAs are coloured in blue.
Nunez-Iglesias et al. [96] proposed a method to infer FMRMs by correlation tests with permutation. This method calculates the expression correlations between miRNAs and predicted target mRNAs with permutation tests, across all given samples (globally) and on case and control samples (locally). The correlation coefficients are then standardized, thus scores how well the miRNAs are concordant with mRNAs globally and locally. Then the miRNA–mRNA pairs are identified by searching for the highest difference in the scores between case and control sample. The identified miRNA–mRNA pairs merge to be the final FMRMs that have the differential power in different biological conditions. This method again identified that both negative and positive correlations widely exist in the expression of miRNAs and their target. Functional analysis further suggests that positively correlated miRNA–mRNA pairs have equally important functions as the negative ones.
In contrast to the above methods, Bonnet et al. [97] proposed to infer FMRMs using expression profiles of miRNA and mRNA only. Their method involves two steps. In the first step, it searches the mRNA expression profile for sets of tight clusters, which are groups of genes consistently clustered together under the different biological conditions. A Gibbs sampling approach is used to cluster the expression profiles in directions of both genes and conditions simultaneously. Multiple clustering solutions are generated by Gibbs samplers with a range of configurations. Then, a set of tight clusters is produced by averaging multiple clustering solutions. In the second step, this method assigns a set of regulators including miRNAs, transcription factors or signal transducers, to each tight cluster. This assignment is learned using a fuzzy decision tree model. In this model, the clustered conditions of each module output from the first step are first linked together with a hierarchical decision tree where each node of the tree is a split of two sets of conditions corresponding to the under- or over-expressed levels of mRNAs. Then, regulators are assigned to each node of the tree using a probabilistic score reflecting how well the expression levels of the regulator match the mRNA expression levels defined by the split value. In order to avoid over-fitting, multiple condition clusters are generated. Thus, there are multiple decision trees and multiple regulators assigned for each node of each hierarchical tree. Finally, FMRMs are extracted by an ensemble approach which is used to capture the regulators most frequently assigned to each condition, given a set of tight clusters of mRNAs. The algorithm was initially designed to infer the gene regulatory modules, and the open-source software package is named LeMoNe. Different from inferring the gene regulatory modules, miRNAs are assigned as candidate regulators in LeMoNe. This method demonstrated that FMRMs can be inferred from expression profiles of miRNA and mRNA only. Some researchers have suggested that algorithms that do not consider known targets, may avoid bias [36, 37, 98]. Thus, this method potentially avoids this problem incurred by miRNA-target predictions.
A method proposed by Liu et al. [88] allows more flexible choices where expression profiles of miRNA and mRNA are used to infer FMRMs with or without integrating the miRNA-target predictions. This approach is a probabilistic graphical model based on correspondence latent Dirichlet allocation (Corr-LDA) [99], in which FMRMs are defined as latent variables governing the expression values of miRNA and mRNA which in turn are associated with a variety of biological functions. Given k-latent FMRMs presented in the samples, this method models miRNAs and mRNAs as observations generated from a probabilistic process over the k-FMRMs. Therefore, each sample is a random mixture of miRNAs and mRNAs associated with k-modules. By inferring the probability distributions of the latent variables, this method captures the likelihood that samples, miRNAs and mRNAs, are associated with functional modules. A Gibbs sampling method was developed to infer the parameters of this model. Under this model, miRNAs can be associated with any functional modules, while mRNAs may only be associated with the modules that produce the miRNAs. In effect, it captures the hierarchical notion that miRNAs are generated under specific FMRMs, and mRNAs are regulated by the miRNAs in the given FMRMs. This model was applied to a mouse mammary data set. It effectively captured several biological process-specific modules involving miRNAs and their target mRNAs. Furthermore, without using prior target binding information, the identified miRNAs and mRNAs in each module show a large proportion of overlap with predicted miRNA-target relationships, suggesting that expression profiles of miRNA and mRNA are crucial for both target identification and discovery of FMRMs.
CONCLUSIONS AND OUTLOOKS
miRNAs have been recognized as pivotal factors in defining the specificity and sensitivity of post-transcriptional gene silencing. Identifying miRNA, their target genes from genome and further inferring their functions and regulatory mechanisms are critical in understanding biological processes of organisms and may shed light on deciphering their roles in the pathophysiology of disease.
While some validated miRNAs and their target genes have been collated in databases, such as TarBase [100] and miRecords [75], these in no way reflect the diversity and abundance of potential miRNA regulatory influences. It is unfeasible to explore empirically all the possibilities in this combinatorial matrix due to the laborious tasks involved. As such, a complete understanding of miRNA functions and their precise regulatory mechanisms remain elusive.
After many miRNAs have been identified and their targets have been predicted, research interests are moving to identify the functions of miRNAs and their regulatory mechanisms. However, characterizing these aspects represents a significant challenge because of complex and subtle features of miRNA and RNA-induced silencing complex which miRNAs might associate with. To gain global and yet specific insights into the functions and evolution of a broad layer of post-transcriptional control, it is particularly useful to integrate miRNA and miRNA sequence and expression profiles and compare these data with other comparative genomic information [17]. High-throughput technologies, such as microarray, mass spectrometry and especially the newly developed NGS, have provided tremendous potential for profiling variant molecules at several levels with unprecedented resolution, depth and speed. These features of technologies bring new bioinformatics opportunities as well as challenges.
In this review, we focused on the computational methods of inferring miRNA functions at miRNA–mRNA level and provide an introduction of miRNA discovery and miRNA-target prediction. The concepts applied by these methods can be largely regarded as integration of heterogeneous data sources in functional analysis of miRNAs. Depending on the data sources involved, we classify these methods into three categories: miRNA functional annotation, inferring MRMs and inferring FMRMs. Several methods have been proposed in the last few years. Some methods have been released and free for use, while others are available on request.
How effective these algorithms are at present is still difficult to determine with such a limited selection of data sets without extensive biological validation. Complex features of miRNAs make functional annotation and regulatory mechanisms even harder to evaluate, particularly different algorithms focus on slightly different aspects of miRNA–mRNA interactions. The selection of methodology will be dependent on the available information. If a list of miRNAs is the only available information, we are limited to miRNA functional annotation. If the miRNA-target prediction and the expression of mRNAs are available, then Jong et al.’s [86] method based on Author-Topic model is probably the most appropriate. If expression profiles of both miRNA and mRNA are available, it is possible to go further and infer the biological specific FMRMs through BNSA [87] or Corr-LDA [88]. While these methods are not directly comparable, they can complement each other.
We are a long way from understand miRNA regulatory mechanisms on a large scale. The methodologies discussed in this review have the capacity to infer miRNA regulatory mechanism with miRNA-target predictions and expression profiles of miRNA and mRNA. Biological discovery has suggested that miRNA regulation can degrade mRNAs as well as inhibit protein translation. Although one-third of mRNAs repressed in the translation process display detectable destabilization, more are repressed without detectable changes in mRNA levels [50]. Thus, the global impact on protein outputs had not yet been determined in great detail. Furthermore, transcription factors also play important roles in translation. They may co-regulate genes with miRNAs. Exploration of the wiring of miRNA regulatory relationships together with known protein–protein interaction data, phenotypic data, transcriptional regulatory interactions and other functional genomics data may help to further elucidate the function of miRNAs at a system-wide level. Fortunately, some work falling in these categories is emerging [101–103]. In summary, it may be that by integrating genome-wide computational and experimental data we have the unprecedented opportunity to study functions and evolution of a broad layer of gene regulatory control mediated by miRNAs.
Key Points.
Discovering miRNAs, identifying their targets and further inferring miRNA functions have been a critical strategy of understanding normal biological processes of miRNAs and their roles in the development of disease.
The complexities of miRNAs pose great challenges on discovering their functions and regulatory mechanisms, while computational methods greatly advanced our understanding in miRNA functions by integrating heterogeneous data sources.
To reveal the functions of miRNAs, computational methods have been proposed to (i) annotate functions of miRNAs through their target mRNAs, (ii) identify the co-expressed miRNA/mRNAs groups (MRM) and (iii) infer the FMRMs.
By integrating genome-wide computational and experimental data, we have the unprecedented opportunity to study functions and evolution of a broad layer of gene regulatory control mediated by miRNAs.
FUNDING
M.C. is supported by the Schizophrenia Research Institute, the National Health and Medical Research Council Project Grant (631057). National Alliance for Research on Schizophrenia and Depression (NARSAD) Young Investigator Award, the Hunter Medical Research Institute, the Neurobehavioural Genetics Unit and New South Wales Department of Health project grant and an M.C. Ainsworth Research Fellowship in Epigenetics.
Biographies
Bing Liu is a bioinformatician at the Children’s Cancer Institute Australia (CCIA) for medical research. CCIA is the only independent medical research institute in Australia devoted to research into the causes, prevention, better treatment and ultimately a cure of childhood cancer. Dr Bing Liu received his BSc (1997) and MSc (2000) in electronics and computer engineering from the Yunnan University, China, and PhD (2010) in computer science from the University of South Australia. Before he joined CCIA, he has worked as a research fellow at the Centre for Cancer Biology, Adelaide and the University of Newcastle, Australia. His major research areas are in bioinformatics and data mining, particularly integrating heterogeneous data for biological and medical research.
Jiuyong Li is a professor at the School of Computer Science, University of South Australia. He received his BSc degree in physics and MPhil degree in information processing from the Yunnan University, China in 1987 and 1998, respectively, and received his PhD degree in computer science from the Griffith University, Australia (2002). His research interests are in the field of data mining, privacy preserving and data mining and their applications in medical and biological data. His research has been supported by three prestigious Australian Research Council Discovery grants since 2005.
Murray J. Cairns is a senior research fellow at the Schizophrenia Research Institute, and a conjoint is a senior lecturer at the School of Biomedical Sciences and Pharmacy, University of Newcastle, Australia. He received his PhD degree in biochemistry and molecular genetics from the University of New South Wales (1998). His major research area is in molecular neuroscience and post-transcriptional gene regulation. In recent years he has focused on the role of microRNA and the neuropsychiatric disorder schizophrenia.
References
- 1.Ridley M. Genome: The Autobiography of a Species in 23 Chapters. New York: Topeka Bindery; 2006. [Google Scholar]
- 2.Bartel D. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Zhao T, Li G, Mi S, et al. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 2007;21(10):1190–203. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 5.Iorio MV, Ferracin M, Liu C-G, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 6.Yanaihara N. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 7.Porkka KP, Pfeiffer MJ, Waltering KK, et al. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67(13):6130–5. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 8.Akao Y, Nakagawa Y, Naoe T. MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 2007;26(5):311–20. doi: 10.1089/dna.2006.0550. [DOI] [PubMed] [Google Scholar]
- 9.Yang H, Kong W, He L, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68(2):425–33. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Cairns M, Rose B, et al. Alterations in miRNA processing and expression in pleomorphic adenomas of the salivary gland. Int J Can. 2009;124(12):2855–63. doi: 10.1002/ijc.24298. [DOI] [PubMed] [Google Scholar]
- 11.Hébert SS, Horré K, Nicolaï L, et al. MicroRNA regulation of Alzheimer's amyloid precursor protein expression. Neurobiol Dis. 2009;33(3):422–8. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 12.Cox MB, Cairns MJ, Gandhi KS, et al. MicroRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PLoS ONE. 2010;5(8):e12132. doi: 10.1371/journal.pone.0012132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beveridge NJ, Gardiner E, Carroll AP, et al. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry. 2010;15(12):1176–89. doi: 10.1038/mp.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aluru S. Handbook of Computational Molecular Biology. Chapman & Hall; /CRC Computer and Information Science Series, 2006, Taylor & Francis Group, Boca Raton, FL USA. [Google Scholar]
- 16.Isaac B. Prediction and validation of microRNAs and their targets. FEBS Lett. 2005;579(26):5904–10. doi: 10.1016/j.febslet.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 17.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38:S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 18.Mazière P, Enright AJ. Prediction of microRNA targets. Drug Discov Today. 2007;12(11–12):452–8. doi: 10.1016/j.drudis.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Barbato C, Arisi I, Frizzo ME, et al. Computational challenges in miRNA target predictions: to be or not to be a true target? J Biomed Biotechnol. 2009 doi: 10.1155/2009/803069. doi:10.1155/2009/803069 (Advance Access publication 17 June 2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L, Xu J, Yang D, et al. Computational approaches for microRNA studies: a review. Mamm Genome. 2010;21(1):1–12. doi: 10.1007/s00335-009-9241-2. [DOI] [PubMed] [Google Scholar]
- 21.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 22.Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 23.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Suppl. 1):D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berezikov E, Cuppen E, Plasterk RHA. Approaches to microRNA discovery. Nat Genet. 2006;38:Suppl:S2–7. doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- 25.Lim LP, Glasner ME, Yekta S, et al. Vertebrate microRNA genes. Science. 2003;299(5612):1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 26.Lai E, Tomancak P, Williams R, et al. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Y, Zhou X, Zhang W. MicroRNA prediction with a novel ranking algorithm based on random walks. Bioinformatics. 2008;24(13):i50–8. doi: 10.1093/bioinformatics/btn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedlander MR, Chen W, Adamidi C, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotech. 2008;26(4):407–15. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 29.Friedländer MR, Mackowiak SD, Li N, et al. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2011;40(1):37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hackenberg M, Sturm M, Langenberger D, et al. miRanalyzer: a microRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 2009;37(Suppl. 2):W68–76. doi: 10.1093/nar/gkp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bar M, Wyman SK, Fritz BR, et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26(10):2496–505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Y, Zou Q, Wang S, et al. The discovery approaches and detection methods of microRNAs. Mol Biol Rep. 2011;38(6):4125–35. doi: 10.1007/s11033-010-0532-1. [DOI] [PubMed] [Google Scholar]
- 33.John B, Enright AJ, Aravin A, et al. Human microRNA targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Betel D, Koppal A, Agius P, et al. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11(8):R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krek A, Grun D, Poy M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 36.Lewis BP, Shih Ih, Jones-Rhoades MW, et al. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 37.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 38.Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 39.Kertesz M, Iovino N, Unnerstall U, et al. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–84. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 40.Rehmsmeier M, Steffen P, Höchsmann M, et al. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10(10):1507–17. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maragkakis M, Alexiou P, Papadopoulos G, et al. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics. 2009;10(1):295. doi: 10.1186/1471-2105-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Meth. 2006;3(11):881–6. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 43.Karlin S, Altschul SF. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc Natl Acad Sci USA. 1990;87(6):2264–8. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giraldez AJ, Mishima Y, Rihel J, et al. Zebrafish miR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312(5770):75–9. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 45.Chaudhuri K, Chatterjee R. MicroRNA detection and target prediction: integration of computational and experimental approaches. DNA Cell Biol. 2007;26(5):321–37. doi: 10.1089/dna.2006.0549. [DOI] [PubMed] [Google Scholar]
- 46.Watanabe Y, Tomita M, Kanai A. Computational methods for microRNA target prediction. In: John JR, Gregory JH, editors. Methods in Enzymology. Vol. 427. New York: Academic Press; 2007. pp. 65–86. [DOI] [PubMed] [Google Scholar]
- 47.Min H, Yoon S. Got target?: computational methods for microRNA target prediction and their extension. Exp Mol Med. 2010;42(4):233–44. doi: 10.3858/emm.2010.42.4.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritchie W, Flamant S, Rasko JEJ. Predicting microRNA targets and functions: traps for the unwary. Nat Meth. 2009;6(6):397–8. doi: 10.1038/nmeth0609-397. [DOI] [PubMed] [Google Scholar]
- 49.Lall S, Grün D, Krek A, et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol. 2006;16(5):460–71. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 50.Baek D, Villen J, Shin C, et al. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alexiou P, Maragkakis M, Papadopoulos GL, et al. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics. 2009;25(23):3049–55. doi: 10.1093/bioinformatics/btp565. [DOI] [PubMed] [Google Scholar]
- 52.Gaidatzis D, van Nimwegen E, Hausser J, et al. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics. 2007;8(1):69. doi: 10.1186/1471-2105-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Griffiths-Jones S, Saini HK, van Dongen S, et al. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Suppl. 1):D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. 2006;126(6):1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 55.Selbach M, Schwanhausser B, Thierfelder N, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 56.Chi SW, Zang JB, Mele A, et al. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460(7254):479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zisoulis DG, Lovci MT, Wilbert ML, et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17(2):173–9. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ellwanger DC, Büttner FA, Mewes H-W, et al. The sufficient minimal set of miRNA seed types. Bioinformatics. 2011;27(10):1346–50. doi: 10.1093/bioinformatics/btr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chi SW, Hannon GJ, Darnell RB. An alternative mode of microRNA target recognition. Nat Struct Mol Biol. 2012;19(3):321–7. doi: 10.1038/nsmb.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols. 2008;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 62.Duncan D, Prodduturi N, Zhang B. WebGestalt2: an updated and expanded version of the Web-based Gene Set Analysis Toolkit. BMC Bioinformatics. 2010;11(Suppl. 4):P10. [Google Scholar]
- 63.Nam S, Kim B, Shin S, et al. miRGator: an integrated system for functional annotation of microRNAs. Nucleic Acids Res. 2008;36(Suppl. 1):D159–64. doi: 10.1093/nar/gkm829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14(6):1012–7. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laganà A, Forte S, Giudice A, et al. miRò: a miRNA knowledge base. Database. 2009 doi: 10.1093/database/bap008. 2009: article ID bap008, doi:10.1093/database/bap008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sales G, Coppe A, Bisognin A, et al. MAGIA, a web-based tool for miRNA and Genes Integrated Analysis. Nucleic Acids Res. 2010;38(Web Server issue):W352–9. doi: 10.1093/nar/gkq423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ulitsky I, Laurent LC, Shamir R. Towards computational prediction of microRNA function and activity. Nucleic Acids Res. 2010;38(15):e160. doi: 10.1093/nar/gkq570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24(3):325–32. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- 69.Jiang Q, Wang Y, Hao Y, et al. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37(Suppl. 1):D98–104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barh D, Bhat D, Viero C. miReg: a resource for microRNA regulation. J Integr Bioinform. 2010;7(1) doi: 10.2390/biecoll-jib-2010-144. doi: 10.2390/biecoll-jib-2010-144. [DOI] [PubMed] [Google Scholar]
- 71.Place RF, Li L-C, Pookot D, et al. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. 2008;105(5):1608–13. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tay Y, Zhang J, Thomson AM, et al. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 73.Grimson A, Farh KK-H, Johnston WK, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vergoulis T, Vlachos IS, Alexiou P, et al. TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 2012;40(D1):D222–9. doi: 10.1093/nar/gkr1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xiao F, Zuo Z, Cai G, et al. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37(Suppl. 1):D105–10. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dweep H, Sticht C, Pandey P, et al. miRWalk Database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44(5):839–47. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 77.Marioni JC, Mason CE, Mane SM, et al. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18(9):1509–17. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Git A, Dvinge H, Salmon-Divon M, et al. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA. 2010;16(5):991–1006. doi: 10.1261/rna.1947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Su Z, Li Z, Chen T, et al. Comparing next-generation sequencing and microarray technologies in a toxicological study of the effects of aristolochic acid on rat kidneys. Chem Res Toxicol. 2011;24(9):1486–93. doi: 10.1021/tx200103b. [DOI] [PubMed] [Google Scholar]
- 80.Yoon S, De Micheli G. Prediction of regulatory modules comprising microRNAs and target genes. Bioinformatics. 2005;21(Suppl. 2):ii93–100. doi: 10.1093/bioinformatics/bti1116. [DOI] [PubMed] [Google Scholar]
- 81.Huang JC, Babak T, Corson TW, et al. Using expression profiling data to identify human microRNA targets. Nat Meth. 2007;4(12):1045–9. doi: 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]
- 82.Joung J-G, Hwang K-B, Nam J-W, et al. Discovery of microRNA-mRNA modules via population-based probabilistic learning. Bioinformatics. 2007;23(9):1141–7. doi: 10.1093/bioinformatics/btm045. [DOI] [PubMed] [Google Scholar]
- 83.Tran D, Satou K, Ho T. Finding microRNA regulatory modules in human genome using rule induction. BMC Bioinformatics. 2008;9(Suppl. 12):S5. doi: 10.1186/1471-2105-9-S12-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peng X, Li Y, Walters K-A, et al. Computational identification of hepatitis C virus associated microRNA-mRNA regulatory modules in human livers. BMC Genomics. 2009;10(1):373. doi: 10.1186/1471-2164-10-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu B, Li J, Tsykin A. Discovery of functional miRNA-mRNA regulatory modules with computational methods. J Biomed Inform. 2009;42(4):685–91. doi: 10.1016/j.jbi.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 86.Joung J-G, Fei Z. Identification of microRNA regulatory modules in Arabidopsis via a probabilistic graphical model. Bioinformatics. 2009;25(3):387–93. doi: 10.1093/bioinformatics/btn626. [DOI] [PubMed] [Google Scholar]
- 87.Liu B, Li J, Tsykin A, et al. Exploring complex miRNA-mRNA interactions with Bayesian networks by splitting-averaging strategy. BMC Bioinformatics. 2009;10(1):408. doi: 10.1186/1471-2105-10-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu B, Liu L, Tsykin A, et al. Identifying functional miRNA–mRNA regulatory modules with correspondence latent Dirichlet allocation. Bioinformatics. 2010;26(24):3105–11. doi: 10.1093/bioinformatics/btq576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lai E. Predicting and validating microRNA targets. Genome Biol. 2004;5(9):115. doi: 10.1186/gb-2004-5-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang J, Morris Q, Frey B. Detecting microRNA targets by linking sequence, microRNA and gene expression data. Research in Computational Molecular Biology. 2006:114–29. [Google Scholar]
- 91.Lavrac N, Kavsek B, Flach P, et al. Subgroup discovery with CN2-SD. J Machine Learning Res. 2004;5:153–88. [Google Scholar]
- 92.Zhang S, Li Q, Liu J, et al. A novel computational framework for simultaneous integration of multiple types of genomic data to identify microRNA-gene regulatory modules. Bioinformatics. 2011;27(13):i401–9. doi: 10.1093/bioinformatics/btr206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Alexe G, Alexe S, Crama Y, et al. Consensus algorithms for the generation of all maximal bicliques. Discrete Appl Math. 2004;145(1):11–21. [Google Scholar]
- 94.Steyvers M, Smyth P, Rosen-Zvi M, et al. Proceedings of the Tenth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Seattle, WA, USA, 2004. Probabilistic author-topic models for information discovery. In: Seattle, WA, USA: ACM. [Google Scholar]
- 95.Gaur A, Jewell DA, Liang Y, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67(6):2456–68. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 96.Nunez-Iglesias J, Liu C-C, Morgan TE, et al. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer's disease cortex reveals altered miRNA regulation. PLoS ONE. 2010;5(2):e8898. doi: 10.1371/journal.pone.0008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bonnet E, Tatari M, Joshi A, et al. Module network inference from a cancer gene expression data set identifies microRNA regulated modules. PLoS ONE. 2010;5(4):e10162. doi: 10.1371/journal.pone.0010162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Blei DM, Jordan MI. Proceedings of the 26th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, Toronto, Canada, 2003. Toronto: Canada: ACM; Modeling annotated data. In: [Google Scholar]
- 100.Papadopoulos GL, Reczko M, Simossis VA, et al. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37(Suppl. 1):D155–8. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Su W-L, Kleinhanz RR, Schadt EE. Characterizing the role of miRNAs within gene regulatory networks using integrative genomics techniques. Mol Syst Biol. 2011;7:490. doi: 10.1038/msb.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Naeem H, Küffner R, Zimmer R. MIRTFnet: analysis of miRNA regulated transcription factors. PLoS ONE. 2011;6(8):e22519. doi: 10.1371/journal.pone.0022519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang J, Haubrock M, Cao K-M, et al. Regulatory coordination of clustered microRNAs based on microRNA-transcription factor regulatory network. BMC Syst Biol. 2011;5(1):199. doi: 10.1186/1752-0509-5-199. [DOI] [PMC free article] [PubMed] [Google Scholar]