Abstract
l-Histidine biosynthesis is an ancient metabolic pathway present in bacteria, archaea, lower eukaryotes, and plants. For decades l-histidine biosynthesis has been studied mainly in Escherichia coli and Salmonella typhimurium, revealing fundamental regulatory processes in bacteria. Furthermore, in the last 15 years this pathway has been also investigated intensively in the industrial amino acid-producing bacterium Corynebacterium glutamicum, revealing similarities to E. coli and S. typhimurium, as well as differences. This review summarizes the current knowledge of l-histidine biosynthesis in C. glutamicum. The genes involved and corresponding enzymes are described, in particular focusing on the imidazoleglycerol-phosphate synthase (HisFH) and the histidinol-phosphate phosphatase (HisN). The transcriptional organization of his genes in C. glutamicum is also reported, including the four histidine operons and their promoters. Knowledge of transcriptional regulation during stringent response and by histidine itself is summarized and a translational regulation mechanism is discussed, as well as clues about a histidine transport system. Finally, we discuss the potential of using this knowledge to create or improve C. glutamicum strains for the industrial l-histidine production.
Introduction
Corynebacterium glutamicum is a well-established microorganism for biotechnological applications. Although it has been engineered for the production of various fine chemicals like succinate (Litsanov et al., 2012) or isobutanol (Blombach et al., 2011), it is still mainly employed for the production of l-amino acids (Becker and Wittmann, 2012). The most important amino acids are l-glutamate (flavour enhancer) and l-lysine (feed additive) based on production scales (Becker and Wittmann, 2011). Furthermore, there are also efforts to create efficient producers for other amino acids like l-leucine, l-serine, and l-methionine. These efforts are supported by a detailed insight into the corresponding amino acid biosynthetic pathways and their regulation in C. glutamicum and have been summarized in several reviews or book chapters (Eggeling and Bott, 2005; Wendisch, 2007; Blombach and Seibold, 2010; Brinkrolf et al., 2010). However, to date there is no review available about l-histidine biosynthesis and its regulation in this amino acid-producing microorganism. Here, we intend to summarize the current knowledge on histidine biosynthesis, its regulation and attempts for application in C. glutamicum. The published data are discussed critically and compared with the knowledge of histidine biosynthesis in Escherichia coli and Salmonella enterica serovar Typhimurium (S. typhimurium), the reference organisms regarding this particular pathway.
Properties of l-histidine
l-Histidine is one of the 20 standard proteinogenic amino acids present in proteins of all living organisms. In the following, we will use the term histidine instead, meaning its biologically active isomer l-histidine. Its side-chain is an imidazole ring and therefore has aromatic properties. Histidine is the only amino acid whose side-chain can switch from an unprotonated to a protonated state under neutral pH conditions due to the pKa value of 6.0 of its side-chain (Nelson and Cox, 2009). This characteristic enables histidine residues to act as both, a proton acceptor or a proton donor, in many cellular enzymatic reactions (Rebek, 1990; Polgár, 2005).
The histidine biosynthesis pathway
Since the late 1950s, the histidine biosynthesis pathway has been studied intensively in different organisms like yeasts, S. typhimurium, and E. coli. Initially, Ames and Martin elucidated the complete histidine pathway by identifying all metabolic intermediates and the enzymes catalysing the corresponding reactions in S. typhimurium (Brenner and Ames, 1971; Martin et al., 1971). At that time, last uncertainties remained regarding the reaction steps and intermediates at the interconnection to the pathway of de novo purine biosynthesis. These issues were finally elucidated by Klem and Davisson revealing the final number of catalytic reactions and intermediates (Klem and Davisson, 1993). Based on this knowledge, histidine biosynthesis is an unbranched pathway with ten enzymatic reactions, starting with phosphoribosyl pyrophosphate (PRPP) and leading to l-histidine (Fig. 1) (Alifano et al., 1996; Stepansky and Leustek, 2006). It turned out early that the histidine pathways of S. typhimurium and E. coli are identical. Moreover, histidine biosynthesis seems to be conserved in all organisms including archaea (Lee et al., 2008), Gram-positive bacteria (Chapman and Nester, 1969), lower eukaryotes (Fink, 1964), and plants (Stepansky and Leustek, 2006). The general histidine pathway and its regulation has already been reviewed in great detail, mainly focusing on E. coli, S. typhimurium, and plants (Brenner and Ames, 1971; Martin et al., 1971; Alifano et al., 1996; Winkler, 1996; Stepansky and Leustek, 2006). This work focuses on the histidine biosynthesis, the involved enzymes and its regulation in C. glutamicum, since there are some interesting differences in comparison to other organisms.
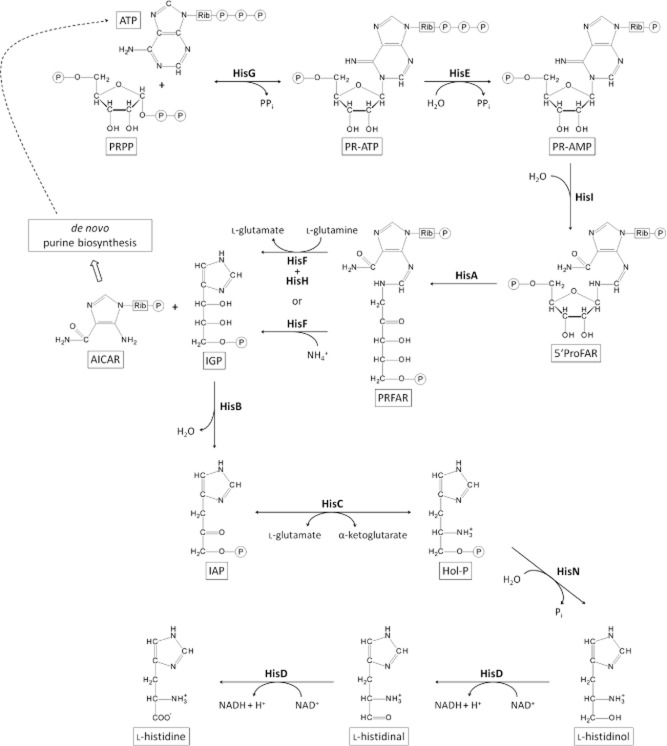
Fig. 1.
Histidine biosynthetic pathway in C. glutamicum. PRPP, phosphoribosyl pyrophosphate; ATP; adenosine triphosphate; PPi, pyrophosphate; PR-ATP, phosphoribosyl-ATP; PR-AMP, phosphoribosyl-AMP; 5′ProFAR, 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4 carboxamide; PRFAR, 5-[(5-phospho-1-deoxyribulos-1-ylamino)methylideneamino]-1-(5-phosphoribosyl)imidazole-4-carboxamide; IGP, imidazole-glycerol phosphate; AICAR, 1-(5′-phosphoribosyl)-5-amino-4-imidazolecarboxamide; IAP, imidazole-acetol phosphate; Hol-P, l-histidinol phosphate; Pi, phosphate; NAD+, oxidized nicotinamide adenine dinucleotide; NADH, reduced nicotinamide adenine dinucleotide; HisG, ATP phosphoribosyltransferase; HisE, phosphoribosyl-ATP pyrophosphatase; HisI, phosphoribosyl-AMP cyclohydrolase; HisA, 5′ProFAR isomerase; HisF, synthase subunit of IGP synthase; HisH, glutaminase subunit of IGP synthase; HisB, imidazoleglycerol-phosphate dehydratase; HisC, histidinol-phosphate aminotransferase; HisN, histidinol-phosphate phosphatase; HisD, histidinol dehydrogenase.
C. glutamicum as an amino acid producer
Corynebacterium glutamicum is a Gram-positive, aerobic, rod shaped, and non-sporulating soil bacterium. It is a member of the genus Corynebacterium, family Corynebacteriaceae, order Corynebacteriales (also containing Mycobacterium spp.), class Actinobacteria (also containing Streptomyces spp. and other filamentous bacteria) (Gao and Gupta, 2012; Goodfellow et al., 2012). It was originally isolated in Japan in the late 1950s during a screening for glutamic acid-secreting bacteria (Kinoshita et al., 1958). Already the unmodified type strain secretes up to 26 g l−1 l-glutamate in minimal medium under biotin-limited conditions and strains improved by classical strain development accumulate more than 100 g l−1 of this amino acid in the culture medium (Becker and Wittmann, 2012). Classical strain development played an important role in the beginnings of fermentative amino acid production. Since this technique has reached its limit to further increase productivity, nowadays metabolic engineering is used to further optimize l-glutamate production. At present these engineered strains do not reach the production titres of classical glutamate production strains (Sawada et al., 2010). However, there are promising results from metabolic engineering approaches with regard to the production of l-lysine. The implementation of 12 defined genome-based modifications enabled accumulation of 120 g l−1 l-lysine in the culture supernatant (Becker et al., 2011). These production titres are even higher than those reached with strains created by classical strain development with consecutive rounds of mutagenesis and selection (Becker and Wittmann, 2012). The intensive investigations on l-glutamate and l-lysine biosynthesis pathways and the understanding of their regulation and interconnection to the central metabolism of C. glutamicum helped to further improve production strains. Today, about 2.5 million tons of l-glutamate and 1.5 million tons of l-lysine are produced annually by Corynebacteria with estimated growth rates of 6–8% per year (Becker and Wittmann, 2011). There are also several strains available for the production of other amino acids which were created either by classical strain development, by metabolic engineering, or by a combination of both techniques. This includes strains for the production of l-isoleucine, l-tryptophan, l-phenylalanine, l-valine, l-alanine, and l-serine (Becker and Wittmann, 2012).
Corynebacterium glutamicum strains suitable for the industrial production of l-histidine have been established by means of combining classical strain development and metabolic engineering. Corynebacterium glutamicum mutants resistant to histidine analogues were reported to secrete 6–8 g l−1 l-histidine into the culture medium (Araki and Nakayama, 1971). The overexpression of a mutated ATP (adenosine triphosphate) phosphoribosyltransferase which is not inhibited by histidine analogues resulted in a C. glutamicum strain accumulating up to 23 g l−1 histidine (Mizukami et al., 1994). These or similar strains are still used for industrial l-histidine fermentation today (Ikeda, 2003; Becker and Wittmann, 2012).
Enzymes involved in histidine biosynthesis
Histidine biosynthesis genes in C. glutamicum
Corynebacterium glutamicum strain AS019, a derivative of C. glutamicum ATCC 13059, was used for the first genetic studies on histidine biosynthesis. The genes hisA, encoding the 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino]imidazole-4 carboxamide (5′ProFAR) isomerase, and hisF, encoding one subunit of the imidazole glycerol phosphate synthase, were identified by complementation of corresponding histidine auxotrophic E. coli mutants (Jung et al., 1998). The gene hisH, coding for the second subunit of imidazole glycerol phosphate synthase (Kim and Lee, 2001), and the genes hisG and hisE, coding for the ATP phosphoribosyltransferase and phosphoribosyl-ATP pyrophosphatase, respectively, were identified by applying the same method (Kwon et al., 2000). The release of the complete genome sequence of the type strain C. glutamicum ATCC 13032 in 2003 (Ikeda and Nakagawa, 2003; Kalinowski et al., 2003) provided the opportunity for the reconstruction of various metabolic pathways, including histidine biosynthesis. The annotation of the genome led to the identification of genes coding for nine of the 10 enzymatic activities needed for histidine biosynthesis. In addition to the genes hisAEFGH, already known from C. glutamicum AS019, these were the genes hisI, encoding phosphoribosyl-AMP cyclohydrolase, hisB, coding for imidazoleglycerol-phosphate dehydratase, hisC, coding for histidinol-phosphate aminotransferase, and hisD, encoding histidinol dehydrogenase, which catalyses the final two steps of histidine biosynthesis in C. glutamicum. However, a gene encoding an enzyme with histidinol-phosphate phosphatase activity has neither been identified by automatic annotation of the genome sequence, nor by heterologous complementation of E. coli mutants. In 2006 a random mutagenesis approach using an IS6100-based transposon vector finally identified the gene encoding histidinol-phosphate phosphatase (Mormann et al., 2006). The gene was designated hisN, since the enzymatic activity is located on the N-terminal part of a bifunctional hisB gene product in S. typhimurium and E. coli (Houston, 1973a; Carlomagno et al., 1988). Additionally, the random transposon mutagenesis approach confirmed the involvement of the genes hisABDEFGI in histidine biosynthesis. Transposon insertion into either one of these genes resulted in histidine auxotrophy of the corresponding mutants (Mormann et al., 2006). Furthermore, participation of the genes hisBCD in histidine biosynthesis was again confirmed in complementation experiments with auxotrophic E. coli mutants (Jung et al., 2009). To sum up, C. glutamicum possesses ten histidine biosynthesis genes coding for nine enzymes which catalyse ten enzymatic reactions. This includes one bifunctional enzyme, the histidinol dehydrogenase (hisD), and one enzyme consisting of two subunits, the imidazoleglycerol-phosphate synthase (hisF and hisH). As a part of our own studies, each histidine gene has been deleted individually in C. glutamicum (Table 1). As for the transposon mutants, each single in frame deletion of one of the eight genes hisABCDEFGI resulted in histidine auxotrophy (R.K. Kulis-Horn, unpubl. obs.), confirming the essentiality of these genes. Interestingly, clear auxotrophies were not found for the deletions of hisH and hisN (discussed below).
Table 1.
Histidine biosynthesis genes in C. glutamicum ATCC 13032 and the effect of gene disruption on histidine-dependent growth in minimal medium
| Gene | Function | Effect of gene disruption | References |
|---|---|---|---|
| hisG | ATP phosphoribosyltransferase | auxotrophy | (1), (2), (3) |
| hisE | phosphoribosyl-ATP pyrophosphatase | auxotrophy | (1), (3) |
| hisI | phosphoribosyl-AMP cyclohydrolase | auxotrophy | (1), (3) |
| hisA | 5′ProFAR isomerase | auxotrophy | (1), (3) |
| hisF | synthase subunit of IGP synthase | auxotrophy | (1), (3) |
| hisH | glutaminase subunit of IGP synthase | no effect | (3) |
| hisB | imidazoleglycerol-phosphate dehydratase | auxotrophy | (1), (3) |
| hisC | histidinol-phosphate aminotransferase | auxotrophy | (3) |
| hisN | histidinol-phosphate phosphatase | auxotrophya | (1), (3)a |
| hisD | histidinol dehydrogenase | auxotrophy | (1), (3) |
| cg0911 | putative inositol monophosphatase | no effect | (3) |
| impA | putative inositol monophosphatase | no effect | (3) |
| cg2301 | putative antibiotic efflux permease, MFS-type | no effect | (3) |
| cg1305 | l-phenylalanine transporter | no effectb | (3), (4) |
Growth strongly reduced but still measurable (this study).
No growth of a Δcg1305 ΔhisG double mutant in histidine supplemented medium (this study).
(1) Mormann et al. (2006): transposon insertion.
(2) Zhang et al. (2012): substitution of hisG 3′ end with a chloramphenicol resistance gene.
(3) This study: in frame gene deletion.
(4) Zhao et al. (2011): in frame gene deletion.
ATP phosphoribosyltransferase (HisG)
ATP phosphoribosyltransferase (ATP-PRT) catalyses the first step of histidine biosynthesis, the condensation of ATP and PRPP to phosphoribosyl-ATP (PR-ATP) and pyrophosphate (PPi) (Alifano et al., 1996). ATP phosphoribosyltransferases can be divided into two subfamilies, the long and the short ATP-PRTs. Enzymes of the long subfamily are 280–310 amino acids in length and are present in lower eukaryotes and bacteria, like E. coli, S. typhimurium, or Mycobacterium tuberculosis (Zhang et al., 2012). The short forms of ATP-PRTs are lacking about 80 amino acids at their C-terminus. They are present in some bacteria, such as Bacillus subtilis, Lactococcus lactis, and Pseudomonas aeruginosa (Bond and Francklyn, 2000). These short ATP-PRTs require the presence of the hisZ gene product for their catalytic activity (Sissler et al., 1999). The HisZ protein has no sequence homology to the C-terminus of long ATP-PRTs, but is a paralogue of histidyl-tRNA synthetase (Sissler et al., 1999).
With a length of 281 amino acids, ATP-PRT from C. glutamicum (HisGCg) belongs to the long form of ATP-PRTs. Therefore, it is not surprising that the C. glutamicum genome lacks a paralogue of the hisZ gene. Kinetic parameters of HisGCg have been determined recently. The enzyme has a specific activity of 2.19 ± 0.09 μmol min−1 mg−1, a Km value for PRPP of 0.08 ± 0.01 mM, a Km value for ATP of 0.22 ± 0.02, and a kcat value of 1.91 ± 0.14 s−1 (Zhang et al., 2012). Comparison of crystal structures and structure-based multiple alignments of ATP-PRTs from bacteria, archaea, and baker's yeast revealed a common 3D structure of ATP-PRTs (Zhang et al., 2012). ATP-PRTs have no structural and sequence similarities to other phosphoribosyltransferases, besides the PRPP binding site. Therefore, ATP-PRT is considered a member of the new type IV class of phosphoribosyltransferases (Lohkamp et al., 2004; Zhang et al., 2012). The crystal structure of HisGCg is not available yet. However, a homology model based on the 3D structure of ATP-PRT from M. tuberculosis (HisGMt) (62% sequence identity and 89% sequence similarity) revealed an almost identical structure to HisGCg (Zhang et al., 2012). Knowledge about the 3D structure of HisGMt is therefore most likely also true for HisGCg. According to the predicted structure model, HisGCg is a L-shaped monomer composed of three distinct domains (Zhang et al., 2012). The first two domains form the catalytic core. The active site is located in a cleft between these two domains. The third domain is able to bind histidine and is therefore regarded as the regulatory domain (Cho et al., 2003; Zhang et al., 2012). The native HisG enzyme from E. coli and S. typhimurium is in equilibrium between a dimeric and hexameric form (Winkler, 1996). Gel filtration experiments with purified HisGCg confirmed this quaternary structure in C. glutamicum (Zhang et al., 2012). ATP-PRT is subject to feedback inhibition and its activity is also influenced by additional factors such as enzyme concentration or the energy status of the cell (Araki and Nakayama, 1974; Zhang et al., 2012). Since, the regulation of ATP-PRT is of great importance it will be discussed in more detail below.
Phosphoribosyl-ATP pyrophosphatase (HisE) and phosphoribosyl-AMP cyclohydrolase (HisI)
Phosphoribosyl-ATP pyrophosphatase catalyses the irreversible hydrolysis of PR-ATP to phosphoribosyl-AMP (PR-AMP) in the second step of histidine biosynthesis. Subsequently, in the third step PR-AMP cyclohydrolase opens the purine ring of PR-ATP releasing 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino) methylideneamino] imidazole-4 carboxamide (5′ProFAR) (Alifano et al., 1996). Both enzymatic activities are carried out by a single polypeptide chain in E. coli and S. typhimurium (Carlomagno et al., 1988). In C. glutamicum, the two activities are encoded by separate genes (Kalinowski et al., 2003). Bifunctional His(IE) enzymes exist in all eukaryotes and in several unrelated taxonomic bacterial lineages, but are absent in all Actinobacteria (Fani et al., 2007). Most likely, bifunctional His(IE) proteins in bacteria are the result of several independent fusion events and horizontal gene transfer (Fani et al., 2007). The native bifunctional His(IE) enzymes from E. coli and S. typhimurium act as dimers (Winkler, 1996). The crystal structure of phosphoribosyl-ATP pyrophosphatase from M. tuberculosis (HisEMt) was solved and revealed that it also forms a dimer (Javid-Majd et al., 2008). The amino acid sequences of HisECg and HisEMt share 62% identity and 90% similarity, assuming a very similar structure for both proteins. Based on this deduced 3D structure, native HisECg most likely acts as a dimer, too.
5′ProFAR isomerase (HisA)
The fourth step of histidine biosynthesis is performed by 5′ProFAR isomerase. This enzyme catalyses an internal redox reaction converting 5′ProFAR to 5-[(5-phospho-1-deoxyribulos-1-ylamino)methylideneamino]-1-(5-phosphoribosyl)imidazole-4-carboxamide (PRFAR) (Alifano et al., 1996). The native enzymes from E. coli and S. typhimurium act as monomers (Winkler, 1996). The crystal structure of 5′ProFAR isomerase from M. tuberculosis (PriAMt) encoded by the priA gene was solved recently (Due et al., 2011). Interestingly, PriAMt is also involved in tryptophan biosynthesis due to its phosphoribosylanthranilate isomerase activity. So far it cannot be excluded that 5′ProFAR isomerase from C. glutamicum (HisACg) is also involved in tryptophan biosynthesis. However, deletion of hisA resulted in histidine auxotrophy only (R.K. Kulis-Horn, unpubl. obs.), indicating that C. glutamicum must at least possess one additional gene coding for a phosphoribosylanthranilate isomerase. This enzyme activity is most likely exerted by the trp(CF) gene product, already annotated as a bifunctional phosphoribosylanthranilate isomerase/indoleglycerolphosphate synthase in C. glutamicum (Kalinowski et al., 2003). Nevertheless, the 3D structure of the bifunctional PriAMt enzyme, exhibiting 61% identity and 89% similarity on amino acid level, allows a deeper insight into the structure of 5′ProFAR isomerase from C. glutamicum (HisACg). Based on these data, native HisACg most likely acts as a monomer with an (α/β)8 barrel fold. [Corrections added on 09 October 2013, after first online publication: In the paragraph above, occurrences of the gene name “pirA” are now amended to “priA”.]
Imidazoleglycerol-phosphate synthase (HisFH)
The fifth step of histidine biosynthesis is the conversion of PRFAR to the next histidine intermediate imidazole-glycerol phosphate (IGP) and the byproduct 1-(5′-phosphoribosyl)-5-amino-4-imidazolecarboxamide (AICAR), an intermediate of de novo purine biosynthesis (Alifano et al., 1996). Glutamine is used as nitrogen donor in this amination step releasing glutamate (Smith and Ames, 1964). Mutations in either hisH or hisF result in histidine auxotrophy of S. typhimurium (Hartman et al., 1960). These genes were later linked to the fifth step of histidine biosynthesis, although both were initially assumed to code for independent enzymes catalysing different steps in the conversion of PRFAR to IGP and AICAR (Smith and Ames, 1964). The exact role of hisF and hisH gene products remained elusive for many years. It was finally demonstrated for hisF and hisH of E. coli that the two gene products act as a stable 1:1 dimeric complex which constitutes the IGP synthase holoenzyme (Klem and Davisson, 1993).
Corynebacterium glutamicum also possesses hisF and hisH genes. They exhibit 44% and 38% identity on amino acid level compared with enzymes from E. coli respectively. A genomic DNA fragment containing both genes from C. glutamicum AS019 was able to complement histidine auxotrophic hisF and hisH E. coli mutants, demonstrating that these two gene products have the same catalytic activities in both organisms (Jung et al., 1998; Kim and Lee, 2001). In accordance with these results, the deletion of hisF resulted in histidine auxotrophy in C. glutamicum. The deletion of hisH, however, did not have any effect on the growth behaviour of the mutant grown in minimal medium (R.K. Kulis-Horn, unpubl. result). This finding is also accordant with the results from the transposon mutagenesis approach where a transposon insertion in hisH was not observed in any of the histidine auxotrophic mutants (Mormann et al., 2006). There are different possible explanations for this surprising growth behaviour of the ΔhisH mutant on minimal medium. (1) The hisH gene in C. glutamicum might be wrongly annotated and another gene has the true hisH gene function. (2) There is a hisH paralogue which complements the gene function. (3) Unlike in E. coli and S. typhimurium, hisH is not essential for histidine biosynthesis in C. glutamicum.
Concerning hypotheses (1) and (2): There are no further genes within the genome of C. glutamicum encoding proteins with considerable sequence similarities to HisH (glutaminase subunit of IGP synthase). The two best blast hits are with pabAB (cg1134) and trpG (cg3360). The pabAB gene encodes a para-aminobenzoate synthase, an enzyme involved in folic acid biosynthesis (Stolz et al., 2007), and trpG, encoding the second subunit of anthranilate synthase, is involved in tryptophan biosynthesis (Heery and Dunican, 1993). It is known from studies with other organisms that these enzymes exhibit glutamine amidotransferase activity, which is also the reaction performed by HisH (Crawford and Eberly, 1986; Viswanathan et al., 1995). In theory, these two enzymes could take over the enzymatic activity of HisH. But this scenario seems rather unlikely, since it was demonstrated for IGP-synthase from E. coli that two perfectly matching HisF (synthase subunit of IGP synthase) and HisH monomers are needed for glutaminase acivity of HisH and channelling of ammonia to the catalytic centre of HisF (Klem et al., 2001; Amaro et al., 2005).
Concerning hypothesis (3): E. coli HisF is able to perform the fifth step of histidine biosynthesis without HisH activity in vitro in the presence of unphysiologically high ammonia concentrations and pH > 8 (Smith and Ames, 1964; Klem and Davisson, 1993). The HisH activity is only needed if glutamine is the only nitrogen donor in the in vitro reaction, since this subunit of the IGP synthase exhibits a glutamine amidotransferase activity (Klem and Davisson, 1993). However, glutamine seems to be the true nitrogen donor in vivo. Mutations in hisH result in histidine auxotrophy of S. typhimurium and E. coli despite the presence of ammonia in the minimal medium (Hartman et al., 1960). On the contrary, a C. glutamicum ΔhisH mutant still grows in ammonia containing minimal medium (R.K. Kulis-Horn, unpubl. obs.). The IGP synthase from C. glutamicum seems to have different properties than the enzymes from S. typhimurium, E. coli, and other species reported. The most probable explanation for this phenomenon is an ammonia-dependent substrate amination activity of HisFCg in vivo (Fig. 1). Our findings support this theory, since hisFCg is able to complement both, a hisF and a hisH deletion, in E. coli (R.K. Kulis-Horn and P. Humbert, unpubl. obs.). The other possibility, a glutamine amidotransferase activity already present in the HisF protein like observed in the monomeric IGP synthase HIS7 from Saccharomyces cerevisiae (Kuenzler et al., 1993), seems unlikely. HisFCg is only of the size of HisFEc and does not exhibit any sequence similarities to known amidotransferases. The overexpression of hisHCg is able to complement a hisH deletion in E. coli, demonstrating that the hisHCg gene product is functional though not needed in C. glutamicum (Jung et al., 1998). So far, no other IGP synthase has been reported being able to catalyse the fifth step of histidine biosynthesis without glutamine amidotransferase activity in vivo. These findings are very interesting especially in the view of the biotechnological application of C. glutamicum as histidine producer, since histidine production in this organism seems to be independent of glutamine biosynthesis.
Imidazoleglycerol-phosphate dehydratase (HisB)
The imidazoleglycerol-phosphate dehydratase catalyses the sixth step of histidine biosynthesis. The enzyme dehydrates IGP and the resulting enol is then ketonized non-enzymatically to imidazole-acetol phosphate (IAP) (Alifano et al., 1996). In S. typhimurium and E. coli this step is catalysed by a bifunctional enzyme comprising both, the imidazoleglycerol-phosphate dehydratase activity and the histidinol-phosphate phosphatase activity, catalysing the eighth step of biosynthesis (Loper, 1961; Houston, 1973a). In these two organisms the bifunctional enzyme is encoded by the his(NB) gene, comprising phosphatase activity at the N-terminus of the encoded protein and dehydratase activity at the C-terminus (Houston, 1973b; Rangarajan et al., 2006). There is evidence that this bifunctional his(NB) gene results from a rather recent gene fusion event in the γ-proteobacterial lineage (Brilli and Fani, 2004). In eukaryotes, archaea and most bacteria the two activities are encoded by separate genes (Fink, 1964; le Coq et al., 1999; Lee et al., 2008). This is also true for C. glutamicum, with IGP dehydratase being encoded by hisB and histidinol-phosphate phosphatase by hisN (Mormann et al., 2006; Jung et al., 2009).
Histidinol-phosphate aminotransferase (HisC)
The seventh step of histidine biosynthesis is the transamination of IAP to l-histidinol phosphate (Hol-P) using glutamate as amino group donor (Alifano et al., 1996). This step is catalysed by the pyridoxal 5′-phosphate (PLP) dependent histidinol-phosphate aminotransferase in C. glutamicum (Marienhagen et al., 2008). Like HisC from E. coli and S. typhimurium (Winkler, 1996), native HisCCg acts as a dimer (Marienhagen et al., 2008). Kinetic parameters of HisCCg were determined only for the back-reaction converting Hol-P and α-ketoglutarate into IAP and l-glutamate. The enzyme exhibits a Km value for Hol-P of 0.89 ± 0.1 mM, a kcat value of 1.18 ± 0.1 s−1 and a specific activity of 2.8 μmol min−1 mg−1 (Marienhagen et al., 2008). Interestingly, HisCCg shows also activity with the precursors of leucine and aromatic amino acids in in vitro assays, but the Km values are two orders of magnitude higher compared with those observed with the histidine precursor and HisCCg does not contribute to aromatic amino acid synthesis in vivo (Marienhagen et al., 2005; 2008). The crystal structure of HisCCg has been solved revealing a three-domain structure of the monomer, with a N-terminal arm, a large PLP binding domain, and a small C-terminal domain (Marienhagen et al., 2008). HisCCg dimerization occurs via extensive hydrophobic interactions and 24 intersubunit hydrogen bonds with the N-terminal arm contributing significantly to the intersubunit interface (Marienhagen et al., 2008). The active sites are made up almost exclusively of residues within one subunit, but the tight packing of the dimer shields the active sites from the solvent (Marienhagen et al., 2008). Site-directed mutagenesis experiments highlighted the importance of the conserved residue Tyr21 for Hol-P substrate specificity and Asn99 for the orientation of the cofactor PLP inside the active centre (Marienhagen et al., 2008). Recently, the structure of histidinol-phosphate aminotransferase from M. tuberculosis (HisC2Mt) has also been published (Nasir et al., 2012). Interestingly, in M. tuberculosis two genes (hisC1 and hisC2) are annotated encoding Hol-P aminotransferases (Camus et al., 2002). The first gene is clustered together with other histidine biosynthesis genes in the same order as in C. glutamicum. The second gene, however, is monocistronic and located in the genome distant from other his genes. The deduced amino acid sequence of hisC2 from M. tuberculosis is most similar to the aromatic amino acid aminotransferase encoded by aroT (cg0267) in C. glutamicum. HisCCg and AroTCg both exhibit high sequence similarity to Hol-P aminotransferases (McHardy et al., 2003). Whereas HisCCg is most similar to aminotransferases being exclusively involved in histidine biosynthesis, AroTCg is more similar to aminotransferases with a broader substrate spectrum being involved in histidine but also aromatic amino acid biosynthesis (McHardy et al., 2003). Enzyme assays with purified AroTCg demonstrated its involvement in synthesis of the aromatic amino acids tyrosine and phenylalanine (Marienhagen et al., 2005). Its function in histidine biosynthesis was not assayed. However, since the presence of the aroT gene is not able to prevent histidine auxotrophy in a hisC deletion mutant of C. glutamicum (R.K. Kulis-Horn, unpubl. obs.) it is very likely that aroTCg does not encode a Hol-P aminotransferase. The structure of the protein crystallized by Nasir and colleagues (2012) therefore does not give deeper insight into the 3D structure of Hol-P aminotransferase from C. glutamicum, but rather into the structure of AroTCg.
Histidinol-phosphate phosphatase (HisN)
During the eighth step of histidine biosynthesis Hol-P is dephosphorylated to l-histidinol. In E. coli and S. typhimurium this reaction is catalysed by a bifunctional enzyme comprising both, the Hol-P phosphatase activity and the IGP dehydratase activity catalysing the sixth step of the biosynthesis (see above). In C. glutamicum both activities are encoded by two genes, hisB encoding IGP phosphatase (Jung et al., 2009) and hisN encoding Hol-P phosphatase (Mormann et al., 2006). IGP phosphatases seem to be derived from a common ancestor in all organisms. But there is a difference in the origin of the Hol-P phosphatases being part of a bifunctional enzyme and those being encoded by a separate gene (Brilli and Fani, 2004). Bifunctional Hol-P phosphatases are members of the HAD family of the DDDD-superfamily of phosphatases. However, the monofunctional ones, present in, e.g. B. subtilis and L. lactis, belong to the PHP-superfamily (Brilli and Fani, 2004). The hisN gene product from C. glutamicum neither exhibits characteristics of the DDDD- nor the PHP-superfamily, thus representing a new class of Hol-P phosphatases. HisNCg is grouped into the family of bacterial-like inositol monophosphatases (IMPase), a member of the FIG-superfamily, based on search results within the Conserved Domain Database (Marchler-Bauer et al., 2010). Homologues of the monofunctional HisN from C. glutamicum can be found predominately in high GC Gram-positive bacteria (blastp). Almost all taxonomical orders of the class Actinobacteria contain genera with HisN homologues, including the Actinomycetales, Corynebacteriales, with the important families Corynebacteriaceae and Mycobacteriaceae, Frankiales, Micrococcales and Streptomycetales (data not shown). Due to the high sequence similarity to IMPase it is hard to decide on the basis of the sequence alone if a hisN homologue encodes a Hol-P phosphatase. Four genes exhibiting high sequence homology to hisNCg are already present in the genome of C. glutamicum. These genes are cg0911, cg2090 (suhB), cg2298 (impA), and cg0967 (cysQ), all encoding proteins with domains typical of inositol monophosphatases (Mormann et al., 2006).
Deletion of hisN was reported to result in histidine auxotrophy in C. glutamicum (Mormann et al., 2006). Contrary to this, Jung and colleagues (2009) reported the cloning and identification of all C. glutamicum his genes without mentioning the hisN gene and evidence for the need of such a gene by performing complementation studies with histidine auxotrophic E. coli mutants. This discrepancy can be explained by the E. coli mutants used in the study of Jung and colleagues (2009). The E. coli hisB463 mutant used had a deletion of the distal part of the hisB gene encoding the imidazoleglycerol-phosphate dehydratase activity, but the histidinol phosphate phosphatase activity is not affected in this strain (Struhl and Davis, 1977). We observed a strongly impaired growth of a C. glutamicum ΔhisN mutant on minimal medium, but no complete histidine auxotrophy, indicating the existence of at least one more gene encoding a protein with HisN activity (R.K. Kulis-Horn, unpubl. obs.). Most likely, one of the four hisNCg homologues present in C. glutamicum is able to partially complement the hisN deletion.
Histidinol dehydrogenase (HisD)
The last two steps of histidine biosynthesis are catalysed by a single enzyme. l-Histidinol is first oxidized by histidinol dehydrogenase to l-histidinal, which is further oxidized to l-histidine (Alifano et al., 1996). Both steps are catalysed by the same enzyme to prevent the decomposition of the unstable l-histidinal intermediate (Görisch and Hölke, 1985) and two molecules NAD+ (oxidized nicotinamide adenine dinucleotide) are reduced during the reaction (Adams, 1954). The native HisD enzyme from S. typhimurium (HisDSt) acts as a homodimer and both subunits are linked by disulfide bridges (Eccleston et al., 1979). HisDSt is Zn2+ dependent (Grubmeyer et al., 1989). Native histidinol dehydrogenase from M. tuberculosis (62% identity, 83% similarity to HisD from C. glutamicum) also acts as a homodimer and is metal dependent (Nunes et al., 2011). However, it remaines uncertain if Zn2+ or rather Mn2+ is the preferred metal ion. Nunes et al. also performed molecular homology modelling of HisDMt employing the crystal structure of histidinol dehydrogenase from E. coli (Barbosa et al., 2002) as template. Enzymes from both organisms have a very similar structure. Each homodimer comprises two identical active sites located at the interface of both subunits. Residues from both subunits form the binding sites for l-histidinol and the metal ion, whereas NAD+ binds only to residues from one subunit (Barbosa et al., 2002; Nunes et al., 2011). A Bi-Uni Uni-Bi ping-pong reaction mechanism was proposed for HisDMt. l-Histidinol binds first, followed by NAD+. NADH+H+ is released while l-histidinal stays enzyme-bound. Then the second NAD+ binds and is reduced, again releasing NADH+H+ and finally l-histidine (Nunes et al., 2011). This reaction mechanism most probably also reflects the HisDCg reaction mechanism.
Transcriptional organization of the histidine biosynthesis genes
The histidine gene cluster of S. typhimurium and E. coli was one of the model gene clusters leading to the development and approval of the operon theory (Alifano et al., 1996). In these two organisms all eight histidine biosynthesis genes are part of one operon and therefore trancribed and regulated as a single unit (Martin, 1963b; Fink and Martin, 1967; Carlomagno et al., 1988). This concentration of all histidine biosynthesis genes at one locus seems not to be the rule but rather an exception and restricted to the enterobacteria, since in other bacteria his genes are more scattered throughout the genome (Alifano et al., 1996).
Transcriptional organization of histidine genes in C. glutamicum
Jung and colleagues (2009) reported that the histidine genes in C. glutamicum AS019 are located and transcribed in two unlinked loci, hisEG and hisDCB-orf1-orf2-hisHA-impA-hisFI. As this study missed the hisN gene, the number of histidine loci increases to three (see above). The genes orf1 and orf2 correspond to genes cg2302 and cg2301 in C. glutamicum ATCC 13032 respectively. The release of the complete genome sequence of C. glutamicum (Kalinowski et al., 2003) revealed that the hisN, hisGE, and hisDCB-cg2302-cg2301-hisHA-impA-hisFI loci are each separated by several hundred kilobase pairs forming independent transcriptional units (Fig. 2). A closer look is needed to verify the operon structure of the hisDCB-cg2302-cg2301-hisHA-impA-hisFI locus. The conclusion that the genes hisDCB-orf1-orf2-hisHA-impA-hisFI form one transcriptional unit in C. glutamicum AS019 is based on results from RT-PCR analysis (Jung et al., 2009). In C. glutamicum ATCC 13032 the genes cg2301 and hisH are separated by a 1044 bp non-coding region. In contrast to the results from strain AS019, this large intergenic region implies two independent transcriptional units for hisDCB-cg2302-cg2301 and hisHA-impA-hisFI. Strand specific cDNA sequencing (RNA-Seq) was performed for the whole transcriptome of C. glutamicum ATCC 13032 in our group (K. Pfeifer-Sancar, A. Mentz, C. Rückert, and J. Kalinowski, manuscript in preparation). This study enabled for the first time the analysis of transcripts with a single nucleotide resolution for whole pathways in one experiment. The RNA-Seq data revealed transcription of all 10 histidine biosynthesis genes, but it did not give any evidence for transcripts spanning the non-coding region between cg2301 and hisH in C. glutamicum ATCC 13032 under all tested conditions (complex and minimal media, various stresses; R.K. Kulis-Horn, unpubl. data). Furthermore, the data showed transcription start sites in front of the hisD and the hisH gene. It appears that hisDCB-cg2032-cg2301 and hisHA-impA-hisFI represent two unlinked transcriptional units in C. glutamicum ATCC 13032 (Fig. 2). The genes cg2294, cg2301 and cg2302 are of unknown function. The deduced protein sequence of cg2301 shows characteristics of a permease of the major facilitator superfamily (blastp).
Fig. 2.
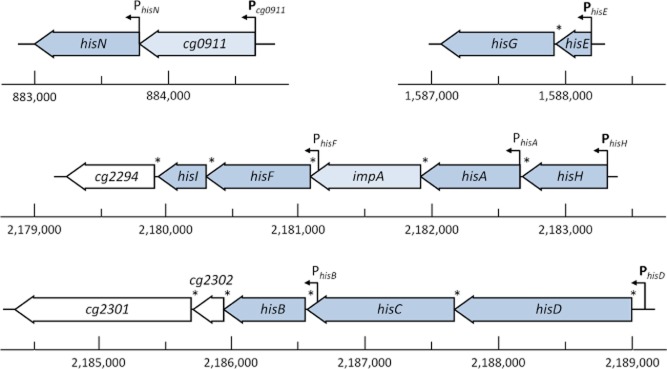
Structure of the four histidine operons in C. glutamicum. Canonical histidine biosynthesis genes are depicted in dark blue. Genes shown in light blue exhibit high sequence similarity to hisN. Genes shown in white have no apparent function in histidine biosynthesis. Arrows indicate the positions of putative primary and internal promoters. Presence of a SD sequence is marked with an asterisk. The ruler indicates the absolute position within the genome (based on the genome version by Kalinowski et al., 2003 RefSeq NC_006958.1).
Furthermore, the identification of transcription start sites by RNA-Seq revealed that the operons cg0911-hisN, hisEG, and hisHA-impA-hisFI-cg2294 are transcribed as leaderless mRNAs, meaning that the start of transcription is identical with the translational start site of the first gene (R.K. Kulis-Horn, unpubl. data). Leaderless transcripts are rarely found in Firmicutes and Proteobacteria, but are common in Actinobacteria where they represent on average 20% of all transcripts (Zheng et al., 2011). The hisDCB-cg2302-cg2301 operon on the other hand comprises a 5′ untranslated region (5′ UTR) with a classical Shine–Dalgarno (SD) sequence. The length of this 5′ UTR was determined by means of primer extension experiments to be 196 nucleotides in C. glutamicum AS019 (Jung et al., 2009). The RNA-Seq data for C. glutamicum ATCC 13032 revealed a shorter 5′ UTR comprising only 93 nucleotides (R.K. Kulis-Horn, unpubl. data). Although the DNA sequence of both C. glutamicum strains is identical in this particular region, there is no evidence for a transcription start site in C. glutamicum ATCC 13032 corresponding to the position mapped in C. glutamicum AS019 (data not shown).
Promoters
The putative promoter in front of hisD in C. glutamicum AS019 identified by primer extension experiments (Jung et al., 2009), so far was the only known his promoter determined in C. glutamicum. The RNA-Seq technique modified for the detection of transcription start sites in C. glutamicum ATCC 13032 enabled the search for further his promoter sequences (K. Pfeifer-Sancar, A. Mentz, C. Rückert, and J. Kalinowski, manuscript in preparation). Four primary promoters were identified in front of the four his operons (Pcg0911, PhisE, PhisH, PhisD). Additionally four internal promoters were observed (PhisN, PhisA, PhisF, PhisB). A −10 box hexamer fitting well to the consensus sequence TANaNT of sigma factor A (σA) dependent promoters in C. glutamicum (Pátek and Nešvera, 2011) was determined for all four primary promoters (Fig. 3). The spacing between the transcription start site and the −10 box is 7 ± 1 nucleotides. Only the −10 box of the hisE promoter exhibits the TGn extension which seems to enhance promoter strength (Vasicová et al., 1999). The −35 boxes located 17 ± 1 nucleotides upstream of the −10 box exhibit only low similarities to the −35 consensus sequence. However, −35 boxes are generally poorly conserved in C. glutamicum (Pátek and Nešvera, 2011).
Fig. 3.

Putative promoter sequences of histidine biosynthesis genes. Transcription start sites (+1) were determined by means of RNA-Seq (K. Pfeifer-Sancar, A. Mentz, C. Rückert, and J. Kalinowski, manuscript in preparation). Putative −10 and −35 boxes are shown in bold and underlined. Dashes indicate gaps of 1–2 nt introduced into the sequence to align the −10 and −35 boxes and the transcription start sites. The start codons are highlighted in italics. The promoter consensus sequences were calculated using either the sequence of all eight promoters, the four primary promoters (Pcg0911, PhisE, PhisH, PhisD), or the four internal promoters (PhisN, PhisA, PhisF, PhisB). The consensus sequence of sigma factor A (σA) dependent promoters from C. glutamicum (Pátek and Nešvera, 2011) is shown in addition. The consensus sequence represents nucleotides occurring in that particular position in more than 80% (uppercase letters) and 35% (lowercase letters) of analysed sequences.
RNA-Seq data indicated additional internal promoters within the four histidine operons (R.K. Kulis-Horn, unpubl. data). A start of transcription was observed in front of the hisN gene within the hisN-cg0911 operon. The data revealed a transcription start for a leaderless mRNA for hisN from the internal promoter PhisN. Furthermore, internal promoters were found in front the hisA (PhisA) and hisF (PhisF) genes within the hisHA-impA-hisFI-cg2294 operon. The hisA gene was shown to be transcribed leaderless and hisF with a 5′ UTR (61 nt). A fourth internal promoter (PhisB) was observed in front of hisB within the hisDCB-cg2302-cg2301 operon, resulting in a 5′ UTR (77 nt) in front of hisB. For all internal promoters a −10 box well-fitting the TANaNT consensus sequence is located 7 ± 1 nucleotides upstream of the transcription start site. The −35 boxes related to these promoters hardly fitted to any consensus sequence. Up to now, no data regarding the biological relevance of these internal promoters are available. However, they could play a role in regulation of histidine biosynthesis. In S. typhimurium and E. coli two internal promoters, hisp2 and hisp3, were identified within the histidine operon (Grisolia et al., 1983; Schmid and Roth, 1983; Carlomagno et al., 1988). Promoter hisp2 enables the additional transcription of the genes his(NB), hisH, hisA, hisF and his(IE), whereas hisp3 enables transcription only of the his(IE) gene. Alifano and colleagues (1996) speculated that such internal promoters might reinforce the expression of distal genes in large operons to counteract the effects of natural polarity. Another function might be to temporally allow expression of some of the genes organized in an operon under specific growth conditions (Schmid and Roth, 1983). Transcription starting from hisp2 is occluded by transcription from the primary promoter hisp1 under histidine limited conditions, but it increases if transcription from hisp1 does not occur at sufficient histidine supply (Alifano et al., 1992). In E. coli and S. typhimurium transcription from promoter hisp1 is known to be regulated by an attenuation mechanism in response to the availability of charged histidyl-tRNAs (Kasai, 1974; di Nocera et al., 1978; Johnston et al., 1980). As transcription from the internal promoters hisp2 and hisp3 is not affected by this attenuation mechanism, transcription of genes from these promoters may occur even in the presence of high levels of charged histidyl-tRNA. The biological function of such a transcriptional regulation, however, still remains unexplained.
Regulation of histidine gene expression
Regulation of biosynthetic pathways is of great importance for organisms to prevent wasting energy for the production of metabolites which are not needed under certain growth conditions. On the other hand, the regulation must also prevent the complete drainage of metabolites needed for survival and growth by temporally activating the biosynthesis. Such an accurate regulation is especially needed for the biosynthesis of amino acids as they are the building blocks of proteins and therefore needed for any enzymatic activity.
The biosynthesis of histidine is associated with high energy costs for the cell. Brenner and Ames (1971) calculated a demand of 41 ATP equivalents for the synthesis of one histidine molecule in S. typhimurium. Unregulated histidine biosynthesis would waste about 2.5% of the bacterial cells metabolic energy (Brenner and Ames, 1971). Based on a genome-scale stoichiometric model of the C. glutamicum metabolism, the ATP demand for histidine biosynthesis was calculated to be 9.4 molATP molHis−1 (E. Zelle et al., pers. comm.). Since this ATP demand is the third highest for all proteinogenic amino acids exceeded only by arginine (12.0 molATP molArg−1) and tryptophan (13.0 molATP molTrp−1), the cellular demand for a strict regulation of histidine biosynthesis is obvious.
There are three general levels of regulation of a metabolic pathway: transcriptional or translational repression, and enzyme inhibition. All three possibilities will be discussed in the following chapters.
Transcriptional regulation
The transcriptional regulation is the first level in a regulatory cascade for metabolic pathways. Various studies concerning E. coli and S. typhimurium revealed changing mRNA levels of histidine genes with varying culture conditions (Winkler, 1996). This indicates regulation on transcriptional level, which has been also reported for C. glutamicum (Brockmann-Gretza and Kalinowski, 2006; Jung et al., 2009; 2010). The most common way of transcriptional regulation is the action of a regulatory protein binding to the operator region of a gene and thereby repressing or activating transcription (Huffman and Brennan, 2002). However, such regulatory proteins have not been identified in S. typhimurium or E. coli (Johnston et al., 1980). There is also no report of such a regulator in any other prokaryote, including C. glutamicum.
The transcription of histidine genes is under positive stringent control
Even though no regulatory protein is involved in transcription regulation of histidine biosynthesis genes, it is addressed by the stringent response in E. coli and S. typhimurium (Winkler, 1996). The stringent response is the answer to amino acid starvation in bacteria. The effector molecules of the stringent response, guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp), accumulate under starvation conditions (Chatterji and Ojha, 2001). On the one hand transcription of stable RNA species like tRNAs and rRNAs is repressed during Stringent Response, thereby downregulating protein synthesis. On the other hand transcription of amino acid biosynthesis genes is mostly upregulated (Chatterji and Ojha, 2001). The effector molecule (p)ppGpp is synthesized by the relA gene product, which catalyses phosphorylation of GDP or GTP using ATP as phosphate donor (Cashel, 1975). The spoT gene product was later identified to also participate in (p)ppGpp synthesis, most likely in the hydrolysis of (p)ppGpp (Laffler and Gallant, 1974; Jain et al., 2006). It was demonstrated for S. typhimurium that expression of his genes is stimulated 10-fold by addition of ppGpp in a relA deficient strain (Stephens et al., 1975). This stimulation is a result of enhanced transcription and not dependent on the regulatory elements needed for transcriptional attenuation (Stephens et al., 1975).
Corynebacterium glutamicum and other Actinobacteria possess a bifunctional Rel protein comprising both gene functions encoded by relA and spoT (Wehmeier et al., 1998; Avarbock et al., 1999). Analysis of a C. glutamicum Δrel mutant, not able to synthesize (p)ppGpp, revealed a growth requirement for histidine and serine. This result suggested that transcription of histidine and serine biosynthesis genes might by positively controlled by (p)ppGpp (Tauch et al., 2001). The stringent response can be induced artificially by addition of the serine analogue dl-serine hydroxamate (SHX) which inhibits the seryl-tRNA synthase (Tosa and Pizer, 1971). Real-time RT-PCR analysis revealed elevated transcript levels of all histidine genes in C. glutamicum organized in the three operons hisEG, hisHA-impA-HisFI-cg2294, and hisDCB-cg2302-cg2301 after treatment with SHX compared with untreated samples (Brockmann-Gretza and Kalinowski, 2006). The mRNA levels of his genes increased two to threefold 10 min after induction of the stringent response (Brockmann-Gretza and Kalinowski, 2006). These results clearly demonstrate that transcription of histidine biosynthesis genes is under positive stringent control in C. glutamicum. The cg0911-hisN operon was not identified to the time the study by Brockmann-Gretza and Kalinowski was conducted. It remains therefore unclear if this operon is also subject to positive stringent control in C. glutamicum.
Transcription of histidine biosynthesis genes in C. glutamicum is regulated by an attenuation mechanism
Next to the global transcriptional regulation of amino acid biosynthesis genes during stringent response, transcription of histidine genes in particular is regulated by an additional mechanism in S. typhimurium and E. coli. Research on the regulation of this pathway, in addition to tryptophan biosynthesis, led to the discovery of the transcriptional attenuation mechanism (Winkler, 1996). Escherichia coli and S. typhimurium possess a leader sequence between the hisp1 promoter and the first structural gene of the operon (Carlomagno et al., 1988). This leader sequence contains an open reading frame (ORF) coding for a short peptide (18 amino acids) with seven consecutive histidine residues. Transcription of the whole histidine operon is coupled to the translation of this leader peptide. During translation of the leader peptide the ribosome senses the availability of charged histidyl-tRNAs thereby influencing two possible alternative secondary structures of the nascent mRNA (Johnston et al., 1980). In brief, if enough charged histidyl-tRNAs are available to allow fast translation of the leader peptide, transcription of the operon is stopped due to the formation of a rho-independent terminator. On the other hand, a delay in translation due to lack of charged histidyl-tRNA promotes the formation of an anti-terminator allowing transcription of the whole operon (Johnston et al., 1980).
Jung and colleagues (2009) suggested a histidine-dependent transcription regulation of the hisDCB-orf1-orf2(-hisHA-impA-hisFI) operon in C. glutamicum AS019, since the corresponding mRNA was only detectable by RT-PCR if cells were grown in histidine free medium. Later, a 196 nt leader sequence in front of hisD was identified (Jung et al., 2010). Since no ORF coding for a short peptide containing several histidine residues is present in this leader sequence, a translation-coupled transcription attenuation mechanism like in E. coli and S. typhimurium can be excluded. Instead, a T-box mediated attenuation mechanism controlling the transcription of the hisDCB-orf1-orf2(-hisHA-impA-hisFI) operon has been proposed (Jung et al., 2010). Computational folding analysis of the 196 nt 5′ UTR from C. glutamicum AS019 revealed two possible stem-loop structures. In the first structure, the terminator structure, the SD sequence (−10 to −17 nt; numbering relative to hisD translation start site) is sequestered by formation of a hair pin structure. In the second structure, the anti-terminator structure, the SD sequence is available to ribosomes. Additionally, a histidine specifier CAU (−92 to −94 nt) and the binding site for uncharged tRNA 3′ ends UGGA (−58 to −61 nt) were identified. All these components are characteristics of T-box RNA regulatory elements. T-box RNAs are members of riboswitch RNAs commonly modulating the expression of genes involved in amino acid metabolism in Gram-positive bacteria (Gutierrez-Preciado et al., 2009). They were first discovered in B. subtilis regulating the expression of aminoacyl-tRNA synthases (Henkin, 1994). Uncharged tRNAs are able to concurrently bind to the specifier sequence and the UGGN-sequence of the T-box RNA via the tRNAs anti-codon loop and free CCA-3′ end, respectively, thereby influencing the secondary structure of the mRNA (Vitreschak et al., 2008). The T-box mechanism results in premature transcription termination due to the formation of a rho-independent transcription terminator hairpin structure in the absence of uncharged tRNAs (Henkin, 1994). Jung and colleagues (2010) showed that chloramphenicol acetyltransferase (CAT) activity decreases in response to histidine in the medium if the 196 nt 5′ UTR in front of hisD is transcriptionally fused to the chloramphenicol acetyltransferase (cat) gene, demonstrating its transcription termination ability. Additionally, the replacement of the UGGA sequence (−58 to −61 nt) reduced specific CAT activity even in the absence of histidine, strongly supporting the involvement of uncharged tRNAs in the regulatory mechanism (Jung et al., 2010).
To test the effect of histidine on the transcription of the remaining his operons we conducted real-time RT-PCR analysis of C. glutamicum ATCC 13032 grown on minimal medium without and with 1 mM histidine supplementation. Surprisingly, these experiments did not reveal any differences in transcription of his genes, neither organized in the hisDCB-cg2302-cg2301 operon nor in the three remaining operons (data not shown). The same holds true for a histidine dipeptide addition experiment analysed via subsequent real-time RT-PCR and micro-array analysis. No differences in transcription of his genes were observed after addition of the dipeptide, suggesting that, in contrast to C. glutamicum AS019, histidine does not affect transcription of the his genes in C. glutamicum ATCC 13032 (data not shown). The inconsistent results for the two C. glutamicum strains cannot be explained so far. Although the DNA-sequence upstream of the hisD seems to be identical in both C. glutamicum strains, it cannot the excluded completely that transcription of his genes is regulated differently in these two strains in respect to the effect of histidine. Strain-specific differences are already obvious in the different transcription start sites in front of the hisD genes in both organisms resulting in 5′ UTRs of different length. The hisD leader sequence in C. glutamicum ATCC 13032 is much shorter and consists of only 93 nt (R.K. Kulis-Horn, unpubl. obs.). Whereas the longer hisD 5′ UTR from C. glutamicum AS019 is clearly involved in transcriptional regulation, we suggest a translational control mechanism for the shorter hisD 5′ UTR from C. glutamicum ATCC 13032, which will be discussed in detail below.
Translational regulation
The translation process is part of the attenuation mechanism regulating transcription of his genes in E. coli and S. typhimurium (see above). Besides this, there is no report that translation of the his operon is regulated once the complete mRNA is transcribed in these two organisms. However, there is evidence for a translational regulation mechanism in C. glutamicum.
Jung and colleagues (2010) identified a 196 nt long 5′ UTR in front of the hisDCB-orf1-orf2 operon in C. glutamicum AS019 which can fold into two alternative secondary structures, one of them sequestering the SD sequence in a hair pin (see above). Although Jung and colleagues (2010) demonstrated that this T-box like RNA-element affects transcription of the hisDCB-orf1-orf2 operon, the possibility of an additional translational control by this process remains. T-box RNAs are usually known to regulate transcription only (Gutierrez-Preciado et al., 2009). However, a comparative analysis of RNA regulatory elements of amino acid metabolism genes from various bacteria revelled that some T-boxes from Actinobacteria might be involved in regulation of translation initiation instead (Seliverstov et al., 2005). The T-box RNA identified in front of the ileS gene of C. glutamicum for example sequesters the SD sequence in the alternative hairpin instead of forming a transcriptional terminator (Seliverstov et al., 2005). Yet, the bioinformatics analysis performed by Seliverstov et al. did not identify similar regulatory RNA elements in front of the his genes in C. glutamicum or other Actinobacteria. This might be attributed to the fact that translation-regulating T-boxes of Actinobacteria have a slightly different structure as compared with classical T-boxes. They are shorter and the specifier codon is located in the loop of the specifier hairpin and not in the bulge (Vitreschak et al., 2008). The RNA element in front of the hisDCB-orf1-orf2 proposed by Jung and colleagues (2010) is shorter as compared with classical T-boxes, since two of the conserved stems (stem II and III) are missing. For this reason, however, it is even much shorter than other known T-box RNAs from Actinobacteria. Moreover, the ‘T-box-sequence’, the 14 most highly conserved residues in T-box RNAs (Gutierrez-Preciado et al., 2009), is only moderately conserved and the histidine specifier is present in a bulge of the specifier hairpin and not in its loop. Consequently, the structure of this regulatory RNA element does not fit the characteristics of neither classical nor short T-box RNAs. It remains ambiguous if this RNA element really represents a T-box or another type of riboswitch.
We identified an even shorter 5′ UTR in front of the hisDCB-cg2302-cg2301 operon in C. glutamicum ATCC 13032 by means of RNA-Seq (see above). This 5′ UTR is only 93 nt in length. Like the 196 nt 5′ UTR identified in C. glutamicum AS019 (Jung et al., 2010), this short 5′ UTR is able to fold into two alternative secondary structures, one of them sequestering the SD sequence (Fig. 4). The binding site for uncharged tRNA 3′ ends UGGA (−58 to −61 nt; numbering relative to hisD translation start site) is present in this structure, too. However, the histidine specifier CAU described by Jung and colleagues (2010) is already located upstream of the transcribed region in C. glutamicum ATCC 13032 and is therefore not part of its 5′ UTR. But there is another histidine specifier CAC (−49 to −51, numbering relative to hisD translation start site) present in this short 5′ UTR. This CAC histidine specifier seems to be even better suited for interaction with histidyl-tRNAs as it is exactly complementary to the GUG anti-codon of the single histidyl-tRNA in C. glutamicum (Kalinowski et al., 2003). There is also a strong bias towards the presence of a C in the third position of the specifier sequence of most T-box RNAs (Gutierrez-Preciado et al., 2009). Nevertheless, a T-box regulatory mechanism seems to be even more unlikely than in the case of C. glutamicum AS019. The histidine specifier and the binding site for uncharged tRNA 3′ ends are separated by only seven nucleotides. The interaction of both binding sites with one histidyl-tRNA molecule at the same time seems to be improbable, if not impossible. Moreover, in all known T-box RNAs the specifier is always located upstream of the binding site for uncharged tRNA 3′ ends and not downstream as in this case (Vitreschak et al., 2008; Gutierrez-Preciado et al., 2009). Therefore, a T-box regulatory mechanism seems unlikely. However, it is still possible that histidyl-tRNAs function as effectors in another type of riboswitch mechanism, since elements for binding of histidyl-tRNAs are present and two alternative secondary structures are predicted. The sequestration of the SD sequence within a hairpin in one of these structures, together with the observation that histidine does not affect the transcription of his genes (see above), suggests a translational regulatory role of the 5′ UTR in front of hisDCB-cg2302-cg2301 operon in C. glutamicum ATCC 13032. Such a translational control would primary affect the expression of the hisD gene itself, since SD sequences are present in front of all genes in the operon. As long as these SD sequences are not sequestered by additional secondary mRNA structures, de novo initiation of translation of all downstream genes should still be possible even if hisD is not translated (Yoo and RajBhandary, 2008). Since HisD catalyses the final steps of histidine biosynthesis, the translational regulation of only this particular enzyme would enable a very rapid recovery of biosynthesis if histidine becomes limiting.
Fig. 4.
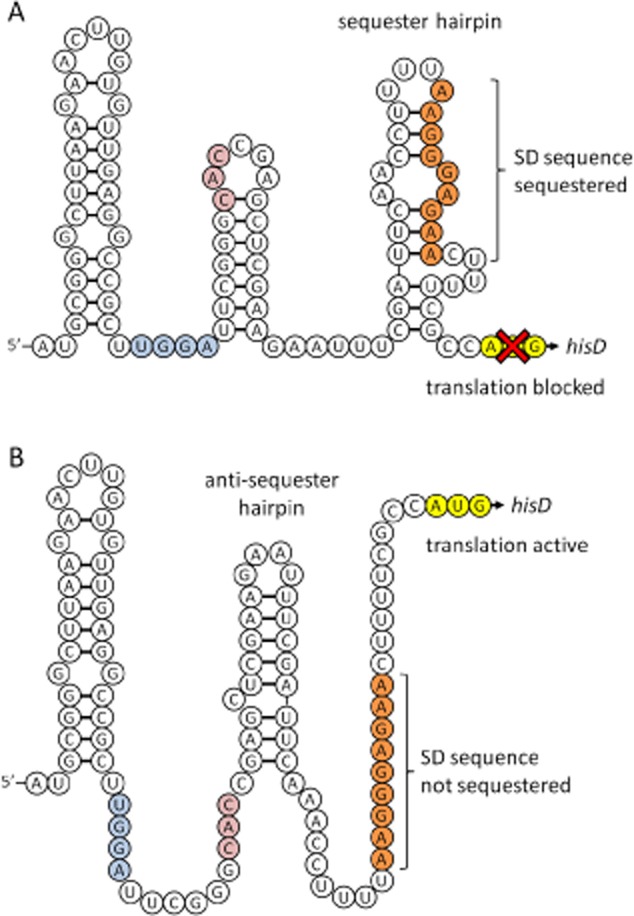
Secondary structure model of the 5′ UTR of the hisDCB-cg2302-cg2301 mRNA from C. glutamicum ATCC 13032. Nucleotides shown in orange and yellow represent the SD sequence and the hisD start codon respectively. The histidine specifier (CAC) is shown in red and the putative CCA binding site for uncharged tRNA 3′ ends (UGGA) is shown in blue. Both sequences might be involved in a histidyl-tRNA dependent riboswitch mechanism.A. SD sequester structure. The SD sequence is sequestered in a hairpin and not available to ribosomes. Translation of the hisD gene is blocked.B. SD anti-sequester structure. The formation of the anti-sequester hairpin prevents the formation of the sequester hairpin. The SD sequence is available to ribosomes and hisD is translated. Uncharged histidyl-tRNA interacting with the histidine specifier and the CCA binding site might be involved in the stabilization of the anti-sequester hairpin, resulting in a switch from the SD sequester to the SD anti-sequester structure.
Enzymatic regulation
The gene product of hisG, the ATP phosphoribosyltransferase (HisG), is the most important enzyme being regulated on enzymatic level in histidine biosynthesis. This enzyme catalyses the first step of histidine biosynthesis, the condensation of ATP and PRPP to PR-ATP. The regulation of this particular enzyme is of remarkable importance, as it prevents waste of ATP and also of PRPP. The latter is not only the substrate for the biosynthesis of histidine, but also used for the de novo synthesis of purines (Zhang et al., 2008) and pyrimidines (Garavaglia et al., 2012), the tryptophan biosynthesis (Sprenger, 2007), and for the synthesis of arabinogalactan, an important component of the corynebacterial cell wall (Alderwick et al., 2006).
HisG is affected by feedback inhibition in C. glutamicum
It has been demonstrated very early that HisG from S. typhimurium (HisGSt) is subject to histidine-mediated feedback inhibition in a non-competitive manner (Martin, 1963a) and the same holds true for HisG from E. coli (HisGEc) (Winkler, 1996). It has been suggested that ATP-PRT from C. glutamicum (HisGCg) is subject to histidine-mediated feedback inhibition, too, since the histidine analogues 2-thiazolyl-dl-alanine (2-TA) and 1,2,4-triazolyl-3-alanine (TRA) inhibit growth of C. glutamicum (Araki and Nakayama, 1971). These two analogues are known to be non-competitive inhibitors of ATP-PRT in S. typhimurium (Martin, 1963a). Analogue-resistant C. glutamicum mutants isolated by Araki and Nakayama (1971) accumulate histidine in the supernatant, indicating that these mutants are deregulated in histidine biosynthesis most likely due to loss of feedback inhibition. Later, by performing enzyme assays with cell-free extracts it was demonstrated that HisGCg is indeed inhibited by l-histidine (Araki and Nakayama, 1974), and recently, Zhang and colleagues (2012) confirmed the inhibition by histidine on the purified HisGCg enzyme. Histidine acts as non-competitive inhibitor of HisGCg having a Ki value of 0.11 ± 0.02 mM (Zhang et al., 2012). The enzyme is inhibited stronger by histidine than the corresponding ATP-PRTs from Thermotoga maritima, but less than those from S. typhimurium and L. lactis (Zhang et al., 2012). It was also demonstrated that, like in S. typhimurium (Martin, 1963a; Morton and Parsons, 1977a), AMP and ADP are competitive inhibitors with respect to ATP with Ki values of 1.29 ± 0.42 mM and 0.88 ± 0.35 mM respectively (Zhang et al., 2012). The inhibitory effect of these two substances with respect to PRPP was not tested. The inhibition of ATP-PRT by AMP and ADP enables to stop the highly energy-demanding histidine biosynthesis if the cells overall energy status is low.
d-Histidine and the histidine intermediates IGP, IAP, Hol-P, l-histidol, and l-histidinal show no inhibitory effect on HisGSt (Martin, 1963a), indicating that HisG inhibition is very specific. l-Histidine itself inhibits both, HisGSt and HisGCg, only as dipolar ion with a positively charged α-amino group, since the inhibitory effect is abolished under alkaline pH conditions (Martin, 1963a; Zhang et al., 2012). It is known from studies with S. typhimurium that ppGpp enhances the inhibitory effect of histidine, resulting in complete inhibition of enzyme activity already at moderate histidine concentrations (Morton and Parsons, 1977b). The alarmone ppGpp accumulates during general amino acid starvation and positively effects his operon transcription (see above). Therefore, the synergetic inhibition of HisGSt by ppGpp and histidine prevents unneeded histidine biosynthesis during stringent response induced by an amino acid different from histidine (Winkler, 1996). Since transcription of his genes in C. glutamicum is induced during stringent response, a synergetic inhibitory effect of ppGpp and l-histidine on HisGCg might exist, too, but has never been tested.
Gel filtration experiments with HisGCg demonstrated that it exists in a dimeric and a hexameric form (Zhang et al., 2012). It is already known for the highly similar HisGMt that it exists as homodimer in the absence of histidine and at low enzyme concentrations, but it forms hexamers or higher oligomers in the presence of histidine (Cho et al., 2003). This is in accordance with data obtained with HisGEc, whose dimer represents the active form of the enzyme whereas higher oligomers are inactive (Tébar et al., 1973). Due to the high structural similarity (Zhang et al., 2012) it is very likely that HisGCg acts in the same way, i.e. active in its dimeric form and inactive in a histidine-induced hexamer form. The histidine-induced change in quaternary structure from a dimeric to a hexameric form of HisGEc can be reversed by addition of the substrate PRPP (Tébar et al., 1973). This might also by true for HisGCg since the inhibitory effect of histidine is reduced by excess of PRPP (Araki and Nakayama, 1974). According to a predicted structure model, HisGCg monomers are L-shaped and composed of three distinct domains (Zhang et al., 2012). The first two domains are the catalytic domains and the third domain is able to bind histidine and therefore is regarded to be the regulatory domain (Cho et al., 2003; Zhang et al., 2012). It is known from the highly similar HisGMt that one histidine molecule interacts with residues of two different domains III at the same time. This led to the suggestion that direct interaction with histidine is responsible for aggregation of three active HisG dimers to one inactive hexamer (Cho et al., 2003). Binding of AMP additionally establishes intersubunit interactions that stabilize the histidine-bound HisGMt hexamer (Cho et al., 2003). This effect of AMP on HisG quaternary structure is also known for E. coli, where the same hexamer-stabilizing effect is observed additionally with the product PR-ATP and high enzyme concentrations (Klungsöyr and Kryvi, 1971). The crystal structure of HisGEc in complex with AMP demonstrated that AMP is not binding to the ATP binding site. Instead, the monophosphate and ribose moieties of AMP are binding to the PRPP binding site and only the adenine ring occupies the ATP binding site (Lohkamp et al., 2004), which explains the surprising observation that AMP acts as a competitive inhibitor with respect to both substrates ATP and PRPP (Morton and Parsons, 1977a).
The crystal structure of HisGMt in complex with histidine enabled the identification of amino acid residues interacting with histidine (Cho et al., 2003). These residues correspond to Gln215, Leu231, Thr235, Met250, and Ala270 in HisGCg. The role of the residues Gln215, Leu231, Thr235, and Ala270 in feedback inhibition of HisGCg has been confirmed by mutation studies, recently (Zhang et al., 2012). A mutated enzyme with three simultaneous substitutions (N215K/L231F/T235A) turned out to be least inhibited by histidine in this study. The mutated enzyme showed a Ki of 4.15 ± 0.21 mM without decreasing the specific enzyme activity (Zhang et al., 2012).
To conclude, the HisG enzyme performing the first step in histidine biosynthesis is the only known enzyme in the biosynthesis that is controlled by a feedback inhibition mechanism involving the end-product of the biosynthesis, l-histidine. Therefore, it constitutes the most interesting target in the development of histidine-producing C. glutamicum strains.
l-Histidine uptake
Uptake systems for l-histidine are known from several microorganisms. Escherichia coli and S. typhimurium possess two main uptake systems, the high affinity HisJQMP ABC transport system specific for histidine and the low affinity AroP system transporting all aromatic amino acids (Winkler, 1996). The Km values for histidine transport in S. typhimurium are 8 × 10−8 M and 10−4 M for the HisJQMP permease and the AroP-system respectively (Ames and Roth, 2005). In B. subtilis the histidine-specific transporter is encoded by hutM as a part of the histidine utilization operon (Yoshida et al., 1995).
l-Histidine uptake system in C. glutamicum is encoded by cg1305
Corynebacterium glutamicum possesses a general uptake system for aromatic amino acids encoded by the gene aroP (Wehrmann et al., 1995). The transporter is able to import the three aromatic amino acids tyrosine, tryptophan, and phenylalanine. But, unlike the corresponding transporter from E. coli and S. typhimurium (Ames and Roth, 2005), it is unable to import histidine (Wehrmann et al., 1995). Therefore, a histidine uptake system in C. glutamicum still remains unidentified. Since the gene cg2301 exhibits characteristics of a transporter and is part of the hisDCB-cg2302-cg2301 operon, it can be regarded as a candidate to encode a l-histidine uptake system. However, the deletion of cg2301 did not affect growth of a histidine-auxotrophic ΔhisG mutant in minimal medium supplemented with histidine, demonstrating still functional histidine uptake (R.K. Kulis-Horn, unpubl. obs.). Further candidates for encoding the unknown l-histidine uptake system in C. glutamicum are the genes cg1305, cg0555, and aroP, since the amino acid sequence of the histidine transporter HutM of B. subtilis shows the highest similarity to their deduced amino acid sequences. The gene cg1305 has been recently reported to encode the l-phenylalanine-specific transporter (Zhao et al., 2011) and the gene product of cg0555 has been characterized as γ-aminobutyric acid uptake system (Zhao et al., 2012). Since deletion of aroP did not affect growth of a histidine auxotrophic ΔhisG mutant on minimal medium supplemented with histidine (R.K. Kulis-Horn, unpubl. obs.), the gene product of aroP, confirming the results of Wehrmann and colleagues (1995), does not encode the histidine uptake system in C. glutamicum. The same holds true for cg0555, since a deletion had no effect on growth of the ΔhisG mutant (R.K. Kulis-Horn, unpubl. obs.). The deletion of cg1305, however, resulted in a strongly reduced growth rate of the histidine auxotrophic mutant already on complex medium and growth of this mutant was almost completely inhibited on minimal medium supplemented with histidine (R.K. Kulis-Horn, unpubl. obs.). These results strongly suggest that cg1305 encodes a histidine uptake system, and probably that it is the only histidine importer in C. glutamicum. Recently, 14C-labelling experiments demonstrated that the transporter encoded by cg1305 is able to import l-phenylalanine (Zhao et al., 2011). Additionally, the uptake of labelled l-tyrosine, l-tryptophan, and l-proline was tested in this study, but does not occur via this transporter. The ability of importing labelled l-histidine was not tested, but strikingly unlabelled l-histidine does not compete with the uptake of labelled l-phenylalanine (Zhao et al., 2011). This surprising result is somehow inconsistent with our finding that cg1305 encodes the only histidine uptake system in C. glutamicum, since one would expect that unlabelled histidine slows down the uptake of labelled phenylalanine. A possible explanation is the existence of several uptake systems for l-phenylalanine in C. glutamicum (Cg1305, AroP, and at least one additional unknown) (Zhao et al., 2011). Although Zhao and colleagues (2011) used a ΔaroP strain in their study, the unknown third l-phenylalanine transporter might counteract the reduced phenylalanine uptake via Cg1305 in the presence of histidine, assuming that the unknown transporter does not additionally import histidine. Since our results with the C. glutamicum ΔhisG Δcg1305 did not indicate additional l-histidine uptake systems beside Cg1305, our observation and the results from Zhao et al. might still be consistent. However, the uptake of labelled l-histidine should be tested to undoubtedly confirm that cg1305 encodes the l-histidine uptake system in C. glutamicum.
l-Histidine export
To our knowledge no histidine export system has been described in any organism. Exporters for other amino acids, however, are well known in E. coli and C. glutamicum, including efflux systems for l-lysine, l-arginine, l-threonine, l-cysteine, l-leucine, l-isoleucine, and l-valine (Eggeling and Sahm, 2003). Hashimoto et al. recently showed that l-glutamate, l-aspartate and l-phenylalanine are secreted via a mechano-sensitive channel by passive diffusion in C. glutamicum (Hashimoto et al., 2012). In the past, the export of amino acids by bacteria was believed to be an artificial result of industrial overproduction and to have no biological relevance. But, next to regulation of the biosynthesis of an amino acid and degradation, the corresponding export might be an important possibility to maintain amino acid homoeostasis, especially in peptide-rich environments (Eggeling and Sahm, 2003). Genes for histidine utilization, which are present in several pathogenic Corynebacterium species, are missing in C. glutamicum (Schröder et al., 2012). However, Bellmann and colleagues (2001) demonstrated the ability of C. glutamicum to export histidine, which may allow to maintain histidine homoeostasis in an environment rich in histidine-containing peptides. Addition of 2 mM His-Ala dipeptide to a C. glutamicum culture resulted in a steady increase of external histidine concentration (Bellmann et al., 2001). The export, however, seems to be rather inefficient as internal histidine concentration rises from zero to 200 mM after addition of the dipeptide (Bellmann et al., 2001). Since C. glutamicum does not secrete any peptidases (Erdmann et al., 1993), the only explanation for the rising external histidine concentration is export of histidine that was cleaved of from the dipeptide itracellularly. However, no candidate gene encoding the exporter has been proposed so far. Interestingly, histidine acts as a co-inducers of lysE transcription, a gene encoding the l-lysine and l-arginine efflux system in C. glutamicum, although histidine is not exported by LysE (Bellmann et al., 2001). There is no explanation, why histidine acts as co-inducer of the exporter, which is unable to export l-histidine. In fact, this might cause a disadvantageous situation for the cell as high histidine concentrations might trigger efflux of l-lysine and l-arginine although their concentrations are low. This negative effect, however, might somehow be counteracted by the high Km value of 20 mM for l-lysine export (Bröer and Krämer, 1991).
Acknowledgments
R. K. Kulis-Horn is supported by a CLIB-GC (Graduate Cluster Industrial Biotechnology) Phd grant co-funded by the Ministry of Innovation, Science and Research of the federal state of North Rhine-Westphalia (MIWF). This work was part of the SysEnCor research project (Grant 0315598E) funded by the German Federal Ministry of Education and Research (BMBF). We thank Katharina Pfeifer-Sancar and Dr. Christian Rückert for providing unpublished RNA-Seq data for C. glutamicum. Additional thanks goes to Elisabeth Zelle (Research Centre Jülich) for help with metabolic modelling of C. glutamicum.
Conflict of interest
None declared.
References
- Adams E. The enzymatic synthesis of histidine from histidinol. J Biol Chem. 1954;209:829–846. [PubMed] [Google Scholar]
- Alderwick LJ, Dover LG, Seidel M, Gande R, Sahm H, Eggeling L, Besra GS. Arabinan-deficient mutants of Corynebacterium glutamicum and the consequent flux in decaprenylmonophosphoryl-d-arabinose metabolism. Glycobiology. 2006;16:1073–1081. doi: 10.1093/glycob/cwl030. [DOI] [PubMed] [Google Scholar]
- Alifano P, Piscitelli C, Blasi V, Rivellini F, Nappo AG, Bruni CB, Carlomagno MS. Processing of a polycistronic mRNA requires a 5′ cis element and active translation. Mol Microbiol. 1992;6:787–798. doi: 10.1111/j.1365-2958.1992.tb01529.x. [DOI] [PubMed] [Google Scholar]
- Alifano P, Fani R, Lió P, Lazcano A, Bazzicalupo M, Carlomagno MS, Bruni CB. Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol Rev. 1996;60:44–69. doi: 10.1128/mr.60.1.44-69.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro RE, Myers RS, Davisson VJ, Luthey-Schulten ZA. Structural elements in IGP synthase exclude water to optimize ammonia transfer. Biophys J. 2005;89:475–487. doi: 10.1529/biophysj.104.058651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames GF, Roth JR. Histidine and aromatic permeases of Salmonella typhimurim. J Bacteriol. 1968;96:1742–1749. doi: 10.1128/jb.96.5.1742-1749.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Nakayama K. Studies on histidine fermentation: Part I. l-histidine production by histidine analog-resistant mutants from several bacteria. Agric Biol Chem. 1971;35:2081–2088. [Google Scholar]
- Araki K, Nakayama K. Feedback-resistant phosphoribosyl-ATP pyrophosphorylase in l-histidine-producing mutants of Corynebacterium glutamicum. Agric Biol Chem. 1974;38:2209–2218. [Google Scholar]
- Avarbock D, Salem J, Li L-S, Wang ZM, Rubin H. Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene. 1999;233:261–269. doi: 10.1016/s0378-1119(99)00114-6. [DOI] [PubMed] [Google Scholar]
- Barbosa JARG, Sivaraman J, Li Y, Larocque R, Matte A, Schrag JD, Cygler M. Mechanism of action and NAD+-binding mode revealed by the crystal structure of l-histidinol dehydrogenase. Proc Natl Acad Sci USA. 2002;99:1859–1864. doi: 10.1073/pnas.022476199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Wittmann C. Systems and synthetic metabolic engineering for amino acid production – the heartbeat of industrial strain development. Curr Opin Biotechnol. 2011;23:1–9. doi: 10.1016/j.copbio.2011.12.025. [DOI] [PubMed] [Google Scholar]
- Becker J, Wittmann C. Bio-based production of chemicals, materials and fuels – Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol. 2012;23:631–640. doi: 10.1016/j.copbio.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Becker J, Zelder O, Hafner S, Schroder H, Wittmann C. From zero to hero – design-based systems metabolic engineering of Corynebacterium glutamicum for l-lysine production. Metab Eng. 2011;13:159–168. doi: 10.1016/j.ymben.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Bellmann A, Vrljic M, Pátek M, Sahm H, Krämer R, Eggeling L. Expression control and specificity of the basic amino acid exporter LysE of Corynebacterium glutamicum. Microbiology. 2001;147:1765–1774. doi: 10.1099/00221287-147-7-1765. [DOI] [PubMed] [Google Scholar]
- Blombach B, Seibold GM. Carbohydrate metabolism in Corynebacterium glutamicum and applications for the metabolic engineering of l-lysine production strains. Appl Microbiol Biotechnol. 2010;86:1313–1322. doi: 10.1007/s00253-010-2537-z. [DOI] [PubMed] [Google Scholar]
- Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, Wendisch VF, Eikmanns BJ. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol. 2011;77:3300–3310. doi: 10.1128/AEM.02972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond JP, Francklyn C. Proteobacterial histidine-biosynthetic pathways are paraphyletic. J Mol Evol. 2000;50:339–347. doi: 10.1007/s002399910037. [DOI] [PubMed] [Google Scholar]
- Brenner M, Ames BN. The histidine operon and its regulation. In: Greenberg DM, editor. Metabolic Pathways. New York, USA: Academic Press; 1971. pp. 349–387. [Google Scholar]
- Brilli M, Fani R. Molecular evolution of hisB genes. J Mol Evol. 2004;58:225–237. doi: 10.1007/s00239-003-2547-x. [DOI] [PubMed] [Google Scholar]
- Brinkrolf K, Schröder J, Pühler A, Tauch A. The transcriptional regulatory repertoire of Corynebacterium glutamicum: reconstruction of the network controlling pathways involved in lysine and glutamate production. J Biotechnol. 2010;149:173–182. doi: 10.1016/j.jbiotec.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Brockmann-Gretza O, Kalinowski J. Global gene expression during stringent response in Corynebacterium glutamicum in presence and absence of the rel gene encoding (p)ppGpp synthase. BMC Genomics. 2006;7:230. doi: 10.1186/1471-2164-7-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröer S, Krämer R. Lysine excretion by Corynebacterium glutamicum: 1. Identification of a specific secretion carrier system. Eur J Biochem. 1991;202:131–135. doi: 10.1111/j.1432-1033.1991.tb16353.x. [DOI] [PubMed] [Google Scholar]
- Camus JC, Pryor MJ, Medigue C, Cole ST. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology. 2002;148:2967–2973. doi: 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
- Carlomagno MS, Chiariotti L, Alifano P, Nappo AG, Bruni CB. Structure and function of the Salmonella typhimurium and Escherichia coli K-12 histidine operons. J Mol Biol. 1988;203:585–606. doi: 10.1016/0022-2836(88)90194-5. [DOI] [PubMed] [Google Scholar]
- Cashel M. Regulation of bacterial ppGpp and pppGpp. Annu Rev Microbiol. 1975;29:301–318. doi: 10.1146/annurev.mi.29.100175.001505. [DOI] [PubMed] [Google Scholar]
- Chapman LF, Nester EW. Gene-enzyme relationships in histidine biosynthesis in Bacillus subtilis. J Bacteriol. 1969;97:1444–1448. doi: 10.1128/jb.97.3.1444-1448.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji D, Ojha AK. Revisiting the stringent response, ppGpp and starvation signaling. Curr Opin Microbiol. 2001;4:160–165. doi: 10.1016/s1369-5274(00)00182-x. [DOI] [PubMed] [Google Scholar]
- Cho Y, Sharma V, Sacchettini JC. Crystal structure of ATP phosphoribosyltransferase from Mycobacterium tuberculosis. J Biol Chem. 2003;278:8333–8339. doi: 10.1074/jbc.M212124200. [DOI] [PubMed] [Google Scholar]
- le Coq D, Fillinger S, Aymerich S. Histidinol phosphate phosphatase, catalyzing the penultimate step of the histidine biosynthesis pathway, is encoded by ytvPhisJ) in Bacillus subtilis. J Bacteriol. 1999;181:3277–3280. doi: 10.1128/jb.181.10.3277-3280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford IP, Eberly L. Structure and regulation of the anthranilate synthase genes in Pseudomonas aeruginosa: I. Sequence of trpG encoding the glutamine amidotransferase subunit. Mol Biol Evol. 1986;3:436–448. doi: 10.1093/oxfordjournals.molbev.a040408. [DOI] [PubMed] [Google Scholar]
- Due AV, Kuper J, Geerlof A, Kries JP, Wilmanns M. Bisubstrate specificity in histidine/tryptophan biosynthesis isomerase from Mycobacterium tuberculosis by active site metamorphosis. Proc Natl Acad Sci USA. 2011;108:3554–3559. doi: 10.1073/pnas.1015996108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccleston ED, Thayer ML, Kirkwood S. Mechanisms of action of histidinol dehydrogenase and UDP-Glc dehydrogenase: evidence that the half-reactions proceed on separate subunits. J Biol Chem. 1979;254:11399–11404. [PubMed] [Google Scholar]
- Eggeling L, Bott M, editors. Handbook of Corynebacterium glutamicum. Boca Raton, FL, USA: Taylor & Francis; 2005. [Google Scholar]
- Eggeling L, Sahm H. New ubiquitous translocators: amino acid export by Corynebacterium glutamicum and Escherichia coli. Arch Microbiol. 2003;180:155–160. doi: 10.1007/s00203-003-0581-0. [DOI] [PubMed] [Google Scholar]
- Erdmann A, Weil B, Krämer R. Lysine secretion by wild-type Corynebacterium glutamicum triggered by dipeptide uptake. J Gen Microbiol. 1993;139:3115–3122. [Google Scholar]
- Fani R, Brilli M, Fondi M, Lió P. The role of gene fusions in the evolution of metabolic pathways: the histidine biosynthesis case. BMC Evol Biol. 2007;7:S4. doi: 10.1186/1471-2148-7-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink GR. Gene-enzyme relations in histidine biosynthesis in yeast. Science. 1964;146:525–527. doi: 10.1126/science.146.3643.525. [DOI] [PubMed] [Google Scholar]
- Fink GR, Martin RG. Translation and polarity in the histidine operon II: polarity in the histidine operon. J Mol Biol. 1967;30:97–107. doi: 10.1016/0022-2836(67)90246-x. [DOI] [PubMed] [Google Scholar]
- Gao B, Gupta RS. Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol Mol Biol Rev. 2012;76:66–112. doi: 10.1128/MMBR.05011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavaglia M, Rossi E, Landini P, Marinus MG. The pyrimidine nucleotide biosynthetic pathway modulates production of biofilm determinants in Escherichia coli. PLoS ONE. 2012;7:e31252. doi: 10.1371/journal.pone.0031252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB. Bergey's Manual® of Systematic Bacteriology: Volume Five The Actinobacteria, Part A. New York, USA: Springer; 2012. [Google Scholar]
- Görisch H, Hölke W. Binding of histidinal to histidinol dehydrogenase. Eur J Biochem. 1985;150:305–308. doi: 10.1111/j.1432-1033.1985.tb09021.x. [DOI] [PubMed] [Google Scholar]
- Grisolia V, Riccio A, Bruni CB. Structure and function of the internal promoter (hisBp) of the Escherichia coli K-12 histidine operon. J Bacteriol. 1983;155:1288–1296. doi: 10.1128/jb.155.3.1288-1296.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubmeyer C, Skiadopoulos M, Senior AE. l-Histidinol dehydrogenase, a Zn2+-metalloenzyme. Arch Biochem Biophys. 1989;272:311–317. doi: 10.1016/0003-9861(89)90224-5. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol Mol Biol Rev. 2009;73:36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman PE, Loper JC, Serman D. Fine structure mapping by complete transduction between histidine-requiring Salmonella mutants. J Gen Microbiol. 1960;22:323–353. doi: 10.1099/00221287-22-2-323. [DOI] [PubMed] [Google Scholar]
- Hashimoto KI, Murata J, Konishi T, Yabe I, Nakamatsu T, Kawasaki H. Glutamate is excreted across the cytoplasmic membrane through the NCgl1221 channel of Corynebacterium glutamicum by passive diffusion. Biosci Biotechnol Biochem. 2012;76:1422–1424. doi: 10.1271/bbb.120366. [DOI] [PubMed] [Google Scholar]
- Heery DM, Dunican LK. Cloning of the trp gene cluster from a tryptophan-hyperproducing strain of Corynebacterium glutamicum: identification of a mutation in the trp leader sequence. Appl Environ Microbiol. 1993;59:791–799. doi: 10.1128/aem.59.3.791-799.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin TM. tRNA-directed transcription antitermination. Mol Microbiol. 1994;13:381–387. doi: 10.1111/j.1365-2958.1994.tb00432.x. [DOI] [PubMed] [Google Scholar]
- Houston LL. Purification and properties of a mutant bifunctional enzyme from the hisB gene of Salmonella typhimurium. J Biol Chem. 1973a;248:4144–4149. [PubMed] [Google Scholar]
- Houston LL. Specialized subregions of the bifunctional hisB gene of Salmonella typhimurium. J Bacteriol. 1973b;113:82–87. doi: 10.1128/jb.113.1.82-87.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman JL, Brennan RG. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr Opin Struct Biol. 2002;12:98–106. doi: 10.1016/s0959-440x(02)00295-6. [DOI] [PubMed] [Google Scholar]
- Ikeda M. Amino acid production processes. Adv Biochem Eng Biotechnol. 2003;79:1–35. doi: 10.1007/3-540-45989-8_1. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Nakagawa S. The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl Microbiol Biotechnol. 2003;62:99–109. doi: 10.1007/s00253-003-1328-1. [DOI] [PubMed] [Google Scholar]
- Jain V, Kumar M, Chatterji D. ppGpp: stringent response and survival. J Microbiol. 2006;44:1–10. [PubMed] [Google Scholar]
- Javid-Majd F, Yang D, Ioerger TR, Sacchettini JC. The 1.25 Å resolution structure of phosphoribosyl-ATP pyrophosphohydrolase from Mycobacterium tuberculosis. Acta Crystallogr D Biol Crystallogr. 2008;64:627–635. doi: 10.1107/S0907444908007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston HM, Barnes WM, Chumley FG, Bossi L, Roth JR. Model for regulation of the histidine operon of Salmonella. Proc Natl Acad Sci USA. 1980;77:508–512. doi: 10.1073/pnas.77.1.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Chun JY, Yim S-H, Lee S-S, Cheon C-I, Song E, Lee M-S. Transcriptional regulation of histidine biosynthesis genes in Corynebacterium glutamicum. Can J Microbiol. 2010;56:178–187. doi: 10.1139/w09-115. [DOI] [PubMed] [Google Scholar]
- Jung S-I, Han MS, Kwon JH, Cheon C-I, Min KH, Lee M-S. Cloning of the histidine biosynthetic genes of Corynebacterium glutamicum: organization and sequencing analysis of the hisAimpA, and hisF gene cluster. Biochem Biophys Res Commun. 1998;247:741–745. doi: 10.1006/bbrc.1998.8850. [DOI] [PubMed] [Google Scholar]
- Jung S-I, Chun JY, Yim S-H, Cheon C-I, Song E, Lee S-S, Lee M-S. Organization and analysis of the histidine biosynthetic genes from Corynebacterium glutamicum. Genes Genom. 2009;31:315–323. [Google Scholar]
- Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, et al. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J Biotechnol. 2003;104:5–25. doi: 10.1016/s0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- Kasai T. Regulation of the expression of the histidine operon in Salmonella typhimurium. Nature. 1974;249:523–527. doi: 10.1038/249523a0. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee M-S. Molecular cloning and analysis of the hisH gene encoding glutamine amidotransferase from Corynebacterium glutamicum. Korean J Genet. 2001;23:121–127. [Google Scholar]
- Kinoshita S, Nakayama S, Akita S. Taxonomic study of glutamic acid accumulating bacteria, Micrococcus glutamicusnov. sp. Bull Agric Chem Soc Jpn. 1958;22:176–185. [Google Scholar]
- Klem TJ, Davisson VJ. Imidazole glycerol phosphate synthase: the glutamine amidotransferase in histidine biosynthesis. Biochemistry. 1993;32:5177–5186. doi: 10.1021/bi00070a029. [DOI] [PubMed] [Google Scholar]
- Klem TJ, Chen Y, Davisson VJ. Subunit interactions and glutamine utilization by Escherichia coli imidazole glycerol phosphate synthase. J Bacteriol. 2001;183:989–996. doi: 10.1128/JB.182.3.989-996.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klungsöyr L, Kryvi H. Sedimentation behaviour of phosphoribosyladenosine triphosphate synthetase: effects of substrates and modifiers. Biochim Biophys Acta. 1971;227:327–336. doi: 10.1016/0005-2744(71)90064-7. [DOI] [PubMed] [Google Scholar]
- Kuenzler M, Balmelli T, Egli CM, Paravicini G, Braus GH. Cloning, primary structure, and regulation of the HIS7 gene encoding a bifunctional glutamine amidotransferase:cyclase from Saccharomyces cerevisiae. J Bacteriol. 1993;175:5548–5558. doi: 10.1128/jb.175.17.5548-5558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon JH, Chun JY, Lee HS, Cheon C-I, Song E-S, Min KH, Lee M-S. Cloning of the histidine biosynthetic genes from Corynebacterium glutamicum: organization and analysis of the hisG and hisE genes. Can J Microbiol. 2000;46:848–855. [PubMed] [Google Scholar]
- Laffler T, Gallant J. spoT, a new genetic locus involved in the stringent response in E. coli. Cell. 1974;1:27–30. [Google Scholar]
- Lee HS, Cho Y, Lee J-H, Kang SG. Novel monofunctional histidinol-phosphate phosphatase of the DDDD superfamily of phosphohydrolases. J Bacteriol. 2008;190:2629–2632. doi: 10.1128/JB.01722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsanov B, Brocker M, Bott M. Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl Environ Microbiol. 2012;78:3325–3337. doi: 10.1128/AEM.07790-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohkamp B, McDermott G, Campbell SA, Coggins JR, Lapthorn AJ. The structure of Escherichia coli ATP-phosphoribosyltransferase: identification of substrate binding sites and mode of AMP inhibition. J Mol Biol. 2004;336:131–144. doi: 10.1016/j.jmb.2003.12.020. [DOI] [PubMed] [Google Scholar]
- Loper JC. Enzyme complementation in mixed extracts of mutants from the Salmonella histidine B locus. Proc Natl Acad Sci USA. 1961;47:1440–1450. doi: 10.1073/pnas.47.9.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHardy AC, Tauch A, Rückert C, Pühler A, Kalinowski J. Genome-based analysis of biosynthetic aminotransferase genes of Corynebacterium glutamicum. J Biotechnol. 2003;104:229–240. doi: 10.1016/s0168-1656(03)00161-5. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2010;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marienhagen J, Kennerknecht N, Sahm H, Eggeling L. Functional analysis of all aminotransferase proteins inferred from the genome sequence of Corynebacterium glutamicum. J Bacteriol. 2005;187:7639–7646. doi: 10.1128/JB.187.22.7639-7646.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marienhagen J, Sandalova T, Sahm H, Eggeling L, Schneider G. Insights into the structural basis of substrate recognition by histidinol-phosphate aminotransferase from Corynebacterium glutamicum. Acta Crystallogr D Biol Crystallogr. 2008;64:675–685. doi: 10.1107/S0907444908009438. [DOI] [PubMed] [Google Scholar]
- Martin RG. The first enzyme in histidine biosynthesis: the nature of feedback inhibition by histidine. J Biol Chem. 1963a;238:257–268. [Google Scholar]
- Martin RG. The one operon-one messenger theory of transcription. Cold Spring Harb Symp Quant Biol. 1963b;28:357–361. [Google Scholar]
- Martin RG, Berberich MA, Ames BN, Davis WW, Goldberger RF, Yourno JD. [147] Enzymes and intermediates of histidine biosynthesis in Salmonella typhimurium. In: Tabor H, Tabor CW, editors. Methods in Enzymology: Metabolism of Amino Acids and Amines Part B. New York, USA: Academic Press; 1971. pp. 3–44. [Google Scholar]
- Mizukami T, Hamu A, Ikeda M, Oka T, Katsumata R. Cloning of the ATP phosphoribosyl transferase gene of Corynebacterium glutamicum and application of the gene to l-histidine production. Biosci Biotechnol Biochem. 1994;58:635–638. doi: 10.1271/bbb.58.635. [DOI] [PubMed] [Google Scholar]
- Mormann S, Lömker A, Rückert C, Gaigalat L, Tauch A, Pühler A, Kalinowski J. Random mutagenesis in Corynebacterium glutamicum ATCC 13032 using an IS6100-based transposon vector identified the last unknown gene in the histidine biosynthesis pathway. BMC Genomics. 2006;7:205. doi: 10.1186/1471-2164-7-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton DP, Parsons SM. Inhibition of ATP phosphoribosyltransferase by AMP and ADP in the absence and presence of histidine. Arch Biochem Biophys. 1977a;181:643–648. doi: 10.1016/0003-9861(77)90270-3. [DOI] [PubMed] [Google Scholar]
- Morton DP, Parsons SM. Synergistic inhibition of ATP phosphoribosyltransferase by guanosine tetraphosphate and histidine. Biochem Biophys Res Commun. 1977b;74:172–177. doi: 10.1016/0006-291x(77)91390-0. [DOI] [PubMed] [Google Scholar]
- Nasir N, Vyas R, Chugh C, Ahangar MS, Biswal BK. Molecular cloning, overexpression, purification, crystallization and preliminary X-ray diffraction studies of histidinol phosphate aminotransferase (HisC2) from Mycobacterium tuberculosis. Acta Crystallogr F. 2012;68:32–36. doi: 10.1107/S1744309111045386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DL, Cox MM. Lehninger Biochemie. Berlin, Germany: Springer; 2009. [Google Scholar]
- di Nocera PP, Blasi F, Lauro R, Frunzio R, Bruni CB. Nucleotide sequence of the attenuator region of the histidine operon of Escherichia coli K-12. Proc Natl Acad Sci USA. 1978;75:4276–4280. doi: 10.1073/pnas.75.9.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes JES, Ducati RG, Breda A, Rosado LA, de Souza BM, Palma MS, et al. Molecular, kinetic, thermodynamic, and structural analyses of Mycobacterium tuberculosis hisD-encoded metal-dependent dimeric histidinol dehydrogenase (EC 1.1.1.23) Arch Biochem Biophys. 2011;512:143–153. doi: 10.1016/j.abb.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Pátek M, Nešvera J. Sigma factors and promoters in Corynebacterium glutamicum. J Biotechnol. 2011;154:101–113. doi: 10.1016/j.jbiotec.2011.01.017. [DOI] [PubMed] [Google Scholar]
- Polgár L. The catalytic triad of serine peptidases. Cell Mol Life Sci. 2005;62:2161–2172. doi: 10.1007/s00018-005-5160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan ES, Proteau A, Wagner J, Hung M-N, Matte A, Cygler M. Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway. J Biol Chem. 2006;281:37930–37941. doi: 10.1074/jbc.M604916200. [DOI] [PubMed] [Google Scholar]
- Rebek J. On the structure of histidine and its role in enzyme active sites. Struct Chem. 1990;1:129–131. [Google Scholar]
- Sawada K, Zen-in S, Wada M, Yokota A. Metabolic changes in a pyruvate kinase gene deletion mutant of Corynebacterium glutamicum ATCC 13032. Metab Eng. 2010;12:401–407. doi: 10.1016/j.ymben.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Schmid MB, Roth JR. Internal promoters of the his operon in Salmonella typhimurium. J Bacteriol. 1983;153:1114–1119. doi: 10.1128/jb.153.2.1114-1119.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder J, Maus I, Meyer K, Wördemann S, Blom J, Jaenicke S, et al. Complete genome sequence, lifestyle, and multi-drug resistance of the human pathogen Corynebacterium resistens DSM 45100 isolated from blood samples of a leukemia patient. BMC Genomics. 2012;13:141. doi: 10.1186/1471-2164-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seliverstov AV, Putzer H, Gelfand MS, Lyubetsky VA. Comparative analysis of RNA regulatory elements of amino acid metabolism genes in Actinobacteria. BMC Microbiol. 2005;5:54. doi: 10.1186/1471-2180-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sissler M, Delorme C, Bond J, Ehrlich SD, Renault P, Francklyn C. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc Natl Acad Sci USA. 1999;96:8985–8990. doi: 10.1073/pnas.96.16.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DW, Ames BN. Intermediates in the early steps of histidine biosynthesis. J Biol Chem. 1964;239:1848–1855. [PubMed] [Google Scholar]
- Sprenger GA. Aromatic amino acids. In: Wendisch VF, editor. Amino Acid Biosynthesis – Pathways, Regulation and Metabolic Engineering. Berlin, Germany: Springer; 2007. pp. 93–127. [Google Scholar]
- Stepansky A, Leustek T. Histidine biosynthesis in plants. Amino Acids. 2006;30:127–142. doi: 10.1007/s00726-005-0247-0. [DOI] [PubMed] [Google Scholar]
- Stephens JC, Artz SW, Ames BN. Guanosine 5′-diphosphate 3′-diphosphate (ppGpp): positive effector for histidine operon transcription and general signal for amino-acid deficiency. Proc Natl Acad Sci USA. 1975;72:4389–4393. doi: 10.1073/pnas.72.11.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz M, Peters-Wendisch P, Etterich H, Gerharz T, Faurie R, Sahm H, et al. Reduced folate supply as a key to enhanced l-serine production by Corynebacterium glutamicum. Appl Environ Microbiol. 2007;73:750–755. doi: 10.1128/AEM.02208-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K, Davis RW. Production of a functional eukaryotic enzyme in Escherichia coli: cloning and expression of the yeast structural gene for imidazole-glycerolphosphate dehydratase (his3. Proc Natl Acad Sci USA. 1977;74:5255–5259. doi: 10.1073/pnas.74.12.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauch A, Wehmeier L, Götker S, Pühler A, Kalinowski J. Relaxed rrn expression and amino acid requirement of a Corynebacterium glutamicum rel mutant defective in (p)ppGpp metabolism. FEMS Microbiol Lett. 2001;201:53–58. doi: 10.1111/j.1574-6968.2001.tb10732.x. [DOI] [PubMed] [Google Scholar]
- Tébar AR, Fernández VM, MartinDelRío R, Ballesteros AO. Studies on the quaternary structure of the first enzyme for histidine biosynthesis. Experientia. 1973;29:1477–1479. doi: 10.1007/BF01943865. [DOI] [PubMed] [Google Scholar]
- Tosa T, Pizer LI. Biochemical bases for the antimetabolite action of l-serine hydroxamate. J Bacteriol. 1971;106:972–982. doi: 10.1128/jb.106.3.972-982.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasicová P, Pátek M, Nešvera J, Sahm H, Eikmanns B. Analysis of the Corynebacterium glutamicum dapA promoter. J Bacteriol. 1999;181:6188–6191. doi: 10.1128/jb.181.19.6188-6191.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan VK, Green JM, Nichols BP. Kinetic characterization of 4-amino 4-deoxychorismate synthase from Escherichia coli. J Bacteriol. 1995;177:5918–5923. doi: 10.1128/jb.177.20.5918-5923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitreschak AG, Mironov AA, Lyubetsky VA, Gelfand MS. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA. 2008;14:717–735. doi: 10.1261/rna.819308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehmeier L, Schäfer A, Burkovski A, Krämer R, Mechold U, Malke H, et al. The role of the Corynebacterium glutamicum rel gene in (p)ppGpp metabolism. Microbiology. 1998;144:1853–1862. doi: 10.1099/00221287-144-7-1853. [DOI] [PubMed] [Google Scholar]
- Wehrmann A, Morakkabati S, Krämer R, Sahm H, Eggeling L. Functional analysis of sequences adjacent to dapE of Corynebacterium glutamicum reveals the presence of aroP, which encodes the aromatic amino acid transporter. J Bacteriol. 1995;177:5991–5993. doi: 10.1128/jb.177.20.5991-5993.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendisch VF, editor. Amino Acid Biosynthesis – Pathways, Regulation and Metabolic Engineering. Berlin, Germany: Springer; 2007. [Google Scholar]
- Winkler M. Biosynthesis of histidine. In: Neidhardt FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC, USA: ASM Press; 1996. pp. 485–505. [Google Scholar]
- Yoo J-H, RajBhandary UL. Requirements for translation re-initiation in Escherichia coli: roles of initiator tRNA and initiation factors IF2 and IF3. Mol Microbiol. 2008;67:1012–1026. doi: 10.1111/j.1365-2958.2008.06104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K-I, Sano H, Seki S, Oda M, Fujimura M, Fujita Y. Cloning and sequencing of a 29 kb region of the Bacillus subtilis genome containing the hut and wapA loci. Microbiology. 1995;141:337–343. doi: 10.1099/13500872-141-2-337. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Morar M, Ealick SE. Structural biology of the purine biosynthetic pathway. Cell Mol Life Sci. 2008;65:3699–3724. doi: 10.1007/s00018-008-8295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shang X, Deng A, Chai X, Lai S, Zhang G, Wen T. Genetic and biochemical characterization of Corynebacterium glutamicum ATP phosphoribosyltransferase and its three mutants resistant to feedback inhibition by histidine. Biochimie. 2012;94:829–838. doi: 10.1016/j.biochi.2011.11.015. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Ding J-Y, Li T, Zhou N-Y, Liu S-J. The ncgl1108phePCg) gene encodes a new l-phe transporter in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2011;90:2005–2013. doi: 10.1007/s00253-011-3245-z. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Ding J-Y, Ma W-H, Zhou N-Y, Liu S-J. Identification and characterization of γ-aminobutyric acid uptake system GabPCg (NCgl0464) in Corynebacterium glutamicum. Appl Environ Microbiol. 2012;78:2596–2601. doi: 10.1128/AEM.07406-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Hu G-Q, She Z-S, Zhu H. Leaderless genes in bacteria: clue to the evolution of translation initiation mechanisms in prokaryotes. BMC Genomics. 2011;12:361. doi: 10.1186/1471-2164-12-361. [DOI] [PMC free article] [PubMed] [Google Scholar]