Abstract
In a research environment dominated by reductionist approaches to brain disease mechanisms, gene network analysis provides a complementary framework in which to tackle the complex dysregulations that occur in neuropsychiatric and other neurological disorders. Gene-gene expression correlations are a common source of molecular networks because they can be extracted from high-dimensional disease data and encapsulate the activity of multiple regulatory systems. However, the analysis of gene coexpression patterns is often treated as a mechanistic black box, in which looming “hub genes” direct cellular networks, and where other features are obscured. By examining the biophysical bases of coexpression and gene regulatory changes that occur in disease, recent studies suggest it is possible to use coexpression networks as a multi-omic screening procedure to generate novel hypotheses for disease mechanisms. Because technical processing steps can affect the outcome and interpretation of coexpression networks, we examine the assumptions and alternatives to common patterns of coexpression analysis and discuss additional topics such as acceptable datasets for coexpression analysis, the robust identification of modules, disease-related prioritization of genes and molecular systems and network meta-analysis. To accelerate coexpression research beyond modules and hubs, we highlight some emerging directions for coexpression network research that are especially relevant to complex brain disease, including the centrality-lethality relationship, integration with machine learning approaches and network pharmacology.
Gene coexpression networks in complex disease research
Common brain diseases include dysfunction at the levels of genes, cells, brain regions and feedback between these networks at multiple biological scales. The overlapping activity and regulation of many systems can obscure the root pathogenic mechanisms when examining any single measurement. For example, major depressive disorder and other neuropsychiatric disorders involve changes in multiple genes, each conferring small and incremental risk that potentially converge in deregulated biological pathways, cellular functions, and local circuit changes, eventually scaling up to brain region pathophysiology (Belmaker & Agam, 2008, Sibille & French, 2013). In these conditions, when several hundred molecules in multiple biological pathways may be legitimately linked to pathogenesis, disease models face competing demands for conceptual clarity and biological accuracy.
What strategies are available to transform data from multi-scale brain diseases into testable hypotheses in cellular or animal disease models? Molecular pathway analysis of differentially expressed genes obtained from post-mortem tissue is constrained by the current state of molecular knowledge and does not provide a prioritization of molecules within the affected pathways. Network biology – an emerging discipline within systems biology - can catalog, integrate and quantify genome-scale molecular interactions, and by doing so can identify critical network features that are relevant to disease processes (Ma'ayan, 2009, Vidal et al., 2011). However, validating phenotype-level predictions from these brain-based models remains challenging. Neuronal simulations can accurately reproduce the dynamics of local and inter-regional brain networks (Izhikevich, 2007), but very rarely incorporate gene-regulation of ion-channels. At the level of genes, dynamic modeling approaches, such as probabilistic Boolean networks, can mimic processes involved in cellular decisions, such as stochastic switching of transcription factors that represent cellular decisions (Heinäniemi et al., 2013). In practice, dynamic simulations and modeling efforts are limited to small systems in which prediction can be easily verified (Choi et al., 2012). Notably, none of these techniques permit multi-system genome-scale dynamic simulations of disease processes, due to uncharacterized genetic and molecular dynamics-related parameters, computational limitations, and a paucity of biomarkers for intermediate phenotypes (Przytycka et al., 2010).
Gene coexpression networks offer genome-scale information and also have the potential to highlight specific molecular mechanisms in disease – particularly if the biophysical basis of coexpression is integrated into network analysis and if researchers examine network properties beyond modules and hubs. For instance, it is common to use coexpression links to identify highly connected genes (‘hub genes’) that are also disease-correlated, as putative mediators of pathology. While this approach has led to many valuable insights, it tends to focus attention on a few hub genes, and ignores the many other ways in which coexpression networks can be used to generate and translate systems biology insights into testable predictions. Coexpression networks have such tremendous potential because gene-gene correlations relate to core features of brain activity and structure, including spatial patterning, inter-tissue communication and epigenetic changes and other non-coding features of regulatory networks (Figure 1). The aggregation of multiple regulatory features into a single network provides a powerful tool to investigate cellular dysfunction, which can be traced back to deficits in specific molecular mechanisms, cell-types or inter-regional communication (Figures 2, 3).
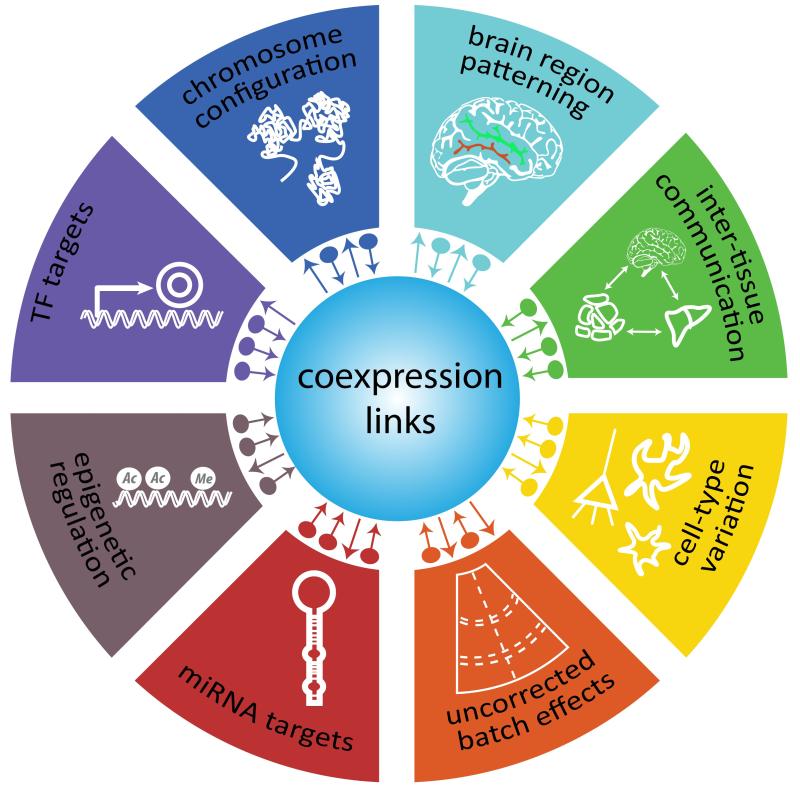
Figure 1. Summary of molecular, cellular, tissue and technical regulatory sources of observed gene-gene correlations/coexpression links.
Various biological activities (depicted in outer shapes) can influence the expression of two or more genes and yield correlated expression patterns, denoted as “coexpression links”. Hence coexpression links reflect the converging influences of these genetic, biochemical and environmental factors, and are thus informative of the biological state of an individual. The relative proportion of links from these various sources (depicted by small arrows) has not been surveyed in a consistent experimental system, and may vary for each gene. Furthermore, technical and cell-type variability can easily generate correlated expression patterns which are indistinguishable from “biological” sources of coexpression, such as epigenetic regulation. Therefore, when interpreting coexpression networks, it is helpful to separate gene-gene correlations with likely biological origins versus those which are related to overarching technical factors such as batch effects.
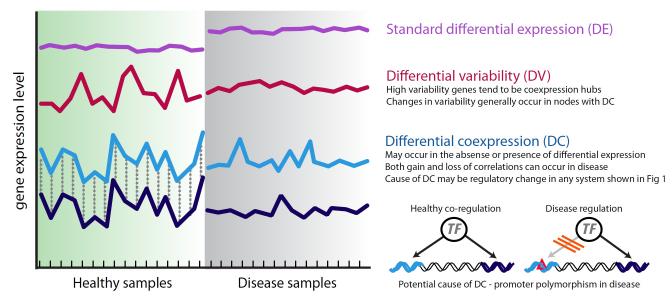
Figure 2. Gene expression patterns translate regulatory changes into networks links.
Gene expression patterns can change in several ways between control and disease samples, beyond standard differential expression (purple line). The variance of a gene’s expression may be altered in disease with or without differential expression (red gene expression profile) (Ho et al., 2008). Similarly altered gene-gene correlations in disease can occur with or without changes in expression (Hudson et al., 2009). A potential mechanism mediating the loss of gene-gene correlations in the disease state, through disrupted transcription factor (TF) binding, is shown on the right.
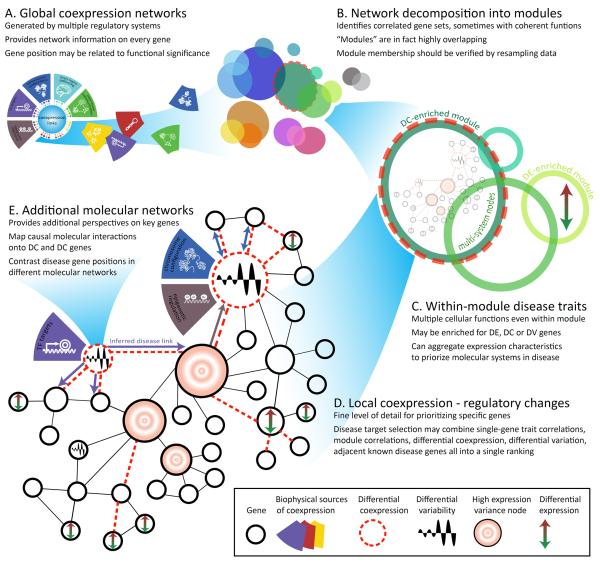
Figure 3. Multi-scale mapping of gene expression traits and coexpression networks.
Disease-related gene expression traits can be aggregated at various scales in the context of coexpression networks. A) Coexpression links stem from multiple sources, but aggregate into an approximately scale-free network. B) Global coexpression networks may be decomposed into groups of coexpressed gene through many different clustering methods. These clusters are overlapping and may be generated by multiple regulatory systems. C) Because coexpressed gene sets tend to have similar functions, they may be useful bins in which to assess the most disease-impacted systems. D) Final selection of disease or potential therapeutic targets can integrate information from all scales to identify genes at the center of complex regulatory changes. E) Changes in any of the regulatory systems that create coexpression may be reflected in differentially coexpressed links, genes or modules that are enriched in coexpressed links. Finding the source of differential coexpression requires additional data drawn from scientific literature or ideally assessed experimentally in the same model system (represented by color-coded arrows for disease-specific coexpression links).
Basics of gene-gene coexpression links
When the mRNA expression of two or more genes is correlated across multiple samples, these genes are said to be “coexpressed” (Figure 2). These coexpression links are generally inferred from large microarray or RNA sequencing studies with no reference to the mechanisms behind these correlations. Studies in multiples species, tissues and platforms have shown that coexpressed genes tend to be functionally related (Obayashi et al., 2008, Oldham et al., 2006). Analogously, gene sets that are densely interconnected by coexpression links within the global gene network are commonly known as clusters or “modules” (Fortunato, 2010, Langfelder & Horvath, 2008). If a significant fraction of genes in a module relate to a gene ontology category or canonical pathway, through guilt-by-association the remaining genes in the module are assumed to be related to that function (Gillis & Pavlidis, 2012, Wolfe et al., 2005). Thus a modular approach to gene function may circumvent knowledge limitations of biological databases that simply catalog items, although existing bias in ontology databases may still affect gene-node classification. Numerous studies have applied gene coexpression network analysis to associate coexpression modules with brain and psychiatric diseases (Chen et al., 2012, De Jong S & Janson E, 2012, Miller et al., 2008, Ponomarev et al., 2012, Torkamani et al., 2010, Voineagu et al., 2011, Zhang et al., 2013). Ironically, the practical utility of coexpression networks for identifying novel disease modules - for instance coexpression hubs within disease-associated modules – has pushed the molecular-mechanistic basis of coexpression into the background. By opening the “black box” that generates coexpression modules, it is possible to identify novel molecular mechanisms that are relevant to disease.
Interpreting coexpression networks that are composed of thousands of gene-gene correlations is challenging because these correlations can arise from several biological and non-biological sources that are mathematically indistinguishable (Figure 1). Any mechanism that synchronously regulates transcription of multiple genes may potentially generate coexpression relationships. For instance, transcription factors have unique DNA binding sites located in promoter regions of distinct sets of genes, and are hypothesized to be a major source of correlated gene expression (Allocco et al., 2004, Marco et al., 2009). The highly structured spatial configuration of chromosomes (Lieberman-Aiden et al., 2009) is an important determinant of gene expression patterns, through chromosome maps and transcriptional complexes (Homouz & Kudlicki, 2013). The linear sequence of DNA can also influence coexpression patterns, as polymerase binding may lead to synchronous transcription of several genes (Ebisuya et al., 2008). mRNA degradation may additionally play a role in observed coexpression networks, and pairs of miRNAs can themselves be coexpressed (Baskerville & Bartel, 2005, Dong et al., 2010) and co-vary with their targets (Bandyopadhyay & Bhattacharyya, 2009, Gennarino et al., 2012). Histone acetylation and methylation control gene expression on multiple segments of DNA and can contribute to coexpression of neighboring genes (Horvath et al., 2012, Numata et al., 2012).
In addition to these biophysical sources of expression variation, technical effects such as batch processing, RNA quality, etc. can produce non-biologically driven coexpression patterns and modules. Even when the exact source of systematic variation in microarrays is unknown, it is now common practice to identify and regress out the effects of such latent variables out of the gene expression dataset (Leek & Storey, 2007). These latent variables may be distinguished from biophysically coexpressed modules, in that they will account for a significant proportion of overall expression variance, but will not be associated with specific biological functions (as denoted by enrichment in annotations corresponding to functional categories). Removal of other covariates from the expression matrix depends on the biological goal of the analysis. For instance, up to 10% of all genes display age-correlated expression changes (Erraji-Benchekroun et al., 2005), so if the disease contrast is not age-related, it may improve results to remove the covariate of age, but if aging is a suspected component of the disease of interest, then it should not be removed.
Another source of coexpressed genes relates to the cellular admixture of the sampled tissue. Coexpression datasets which are not acquired from single cell populations – which is the case for the majority of brain datasets – must confront the influence of cellular heterogeneity on gene coexpression. Unmeasured cellular heterogeneity has both confounding and useful effects. If several cell-types are combined in a sample and the proportion of these cell-types varies randomly across samples, then it is possible to produce coexpression modules which are not driven by ongoing biophysical properties, but by variation in markers for various cell-types. This may create cell-type specific modules associated with oligodendrocytes, microglia and several classes of neurons (Hawrylycz et al., 2012, Oldham et al., 2008), as coexpression links within these modules are often driven by the covariance of cell-type markers. Accordingly, spatial patterns of gene expression across brain regions reflect the changes in cellular composition in addition to cell-based changes in transcriptional programs (Hawrylycz et al., 2012, Menashe et al., 2013). On the other hand, synchronous expression patterns across spatially separated brain regions (Gaiteri et al., 2010), or between a brain region and peripheral organs (Dobrin et al., 2009), may also indicate the presence of cross-tissue communication, likely mediated by circulating factors. Because systemic diseases such as diabetes and obesity can also affect the function of several brain regions and risk of Alzheimer’s disease, these cross-tissue coexpression links may provide novel observations on the spread of pathology. Unfortunately, multi-tissue datasets are rare due to high cost; however the Genome-Tissue Expression project (GTEx) data extracted from 30 tissues may expose novel multi-organ network (Lonsdale et al., 2013).
Differential coexpression represents altered regulatory network structure
Differential coexpression refers to changes in gene-gene correlations between two sets of phenotypically distinct samples (Figures 2, 3) (De La Fuente, 2010). Changes in gene-gene correlation may occur in the absence of differential expression, meaning that a gene may undergo changes in regulatory pattern that would be undetected by traditional differential expression analyses. This phenomenon has been shown in aging (Southworth et al., 2009), across corticolimbic regions in major depression (Gaiteri et al., 2010) and between miRNA’s in Alzheimer’s disease (Bhattacharyya & Bandyopadhyay, 2013). While tests for differential expression must be statistically corrected for the large number of genes measured by microarray, results from differential coexpression must endure a more extreme statistically correction, because identifying altered correlations involves a comparison between two matrices of pair-wise gene-gene correlations. Therefore it is sometimes useful to estimate aggregate differential expression on a gene-by-gene or module-by-module basis, to reduce the number of statistical tests, and to check for coherent correlation changes within a particular molecular system in the disease state (Amar et al., 2013, Kostka & Spang, 2004).
In the same way that regulatory patterns within tissues may be altered across phenotypic states in manners that are reflected in altered coexpression networks, cross-tissues communication can be monitored via coexpression networks. For instance, a core feature of major depression is abnormal feedback between the amygdala and anterior cingulate cortex, mediating emotional reactivity (Kupfer et al., 2012). A study of gene coexpression across these two regions showed that more genes gained or lost coexpression links than expected at random, when comparing cross-tissue networks from post-mortem tissue of patients with major depression to healthy controls (Gaiteri et al., 2010), suggesting an orchestrated transcriptional reorganization affecting this neural network. In this particular example, biological pathways corresponding to the identified gene set suggested dysregulated functions for several hormone-type factors previously implicated in depression (insulin, interleukin-1, thyroid hormone, estradiol and glucocorticoids), indicating the presence of a distinct and integrated hormone-mediated corticolimbic homeostatic, although maladaptive and pathological, state in major depression. Hence, changes in coordinated gene expression across brain areas may represent a novel molecular probe for brain structure/function that is sensitive to disease condition.
Hubs and coexpression network topology
The structure of coexpression networks has particular properties that are relevant to the function of regulatory networks and disease resilience. Coexpression networks meet the definition of “small-world” networks (Watts & Strogatz, 1998) because they are highly clustered (connected nodes have common neighbors) yet maintain an overall short path length, meaning signals can traverse the entire network in only a few hops. The ability to efficiently transit within and between clusters is facilitated by ‘hubs”, which are connected to a large number of nodes. Hub genes have both theoretical and practical implications for coexpression networks. From a theoretical perspective, information flow through small-world, scale-free networks is unlikely to be affected by random node deletion, but is especially vulnerable to targeted hub attack (Albert et al., 2000). In a disease context, this is termed the “lethality-centrality” relationship (Jeong et al., 2001) and is supported by examples from multiple molecular and brain networks in which hub targeting leads to crucial functional impairment (Stam et al., 2007). Practically, hubs provide a specific focus for investigations into disease-correlated modules of genes (next section), (Miller et al., 2008, Ray et al., 2008, Torkamani et al., 2010, Voineagu et al., 2011). However, restricting experimental attention to coexpression hubs may discount other relevant molecules and is no guarantee of phenotypic effects, as coexpression links may represent a variety of causal or non-causal interactions (Figure 1).
Modules as functional markers of network activity
A “module” refers to a gene set whose expression patterns are mutually correlated (Langfelder & Horvath, 2008). Just as correlated genes tend to have similar biological functions, on a larger scale, modules tend to contain genes with similar biological functions (Lee et al., 2003) (Figure 3). Module membership can be compared between cases and controls, among different tissues, species, or other phenotypes or clinical traits (Cai et al., 2010, Kang & Kawasawa, 2011). Typical analysis of gene coexpression seeks to associate coexpression modules with disease or other phenotypic traits recorded in the same dataset. For instance if the average expression of a particular module is higher in patients with more severe pathology, then the activity of genes in that module is potentially linked to that pathological trait. While it would be desirable to identify causal molecular systems behind pathology, the trait-module association may be a downstream effect of the pathology. Module-trait correlation values tend to be relatively low (R<0.5), but statistically significant because they are sustained across hundreds of genes (Zhang et al., 2013). Moreover, the fraction of genes in a module relating to its main biological function is often under 20%, indicating modules contain diverse functions with a multidimensional relationship to measured traits. Thus a modular coexpression analysis can potentially highlight novel disease-relevant genes through guilt by association, but in reality modules are a complex mix of molecular functions (Gillis & Pavlidis, 2012) with limited, but hopefully robust, correlations to clinical traits (Langfelder et al., 2013).
A plethora of methods can identify putative coexpression modules (Fortunato, 2010, Jay et al., 2012, Langfelder & Horvath, 2008). Choosing the “best” clustering method is a balance between the mathematical ability to detect locally dense modules, the biological ability to find functionally enriched clusters and computational efficiency. Thus, depending on the data size and biological goals, the best method for a particular dataset may vary (Vega-Pons & Ruiz-Shulcloper, 2011). However, all clustering results can be evaluated through statistics on their reproducibility under data resampling and ability to find locally dense clusters (Fortunato, 2010). While clustering methods attempt to minimize links between modules, thousands of such links remain after clustering, which would be expected given the overlapping regulatory domains of systems that generate coexpression (Figures 1, 3) indicating that the concept of functionally and structurally independent modules is a convenient simplification of the structure of gene-gene correlations.
Practical coexpression network analysis, Part 1: Novel uses of coexpression networks for brain disease research
Coexpression networks provide a contextual biological framework for both discovery- and hypothesis-driven research with the goal of highlighting unifying features of suspected disease genes (Gulsuner et al., 2013). While many coexpression studies incorporate elements of gene modules and hubs, some studies have now advanced substantially beyond them to address the diverse biophysical sources of coexpression (Figure 1), additional coexpression-based changes in disease such as differential coexpression (Figure 2), or use experimental work to validate the predictions of coexpression networks, and incorporate systems biology perspectives to clarify the complex bases of brain diseases (Figure 3). To highlight this emerging potential and provide concrete examples of the complex questions and challenges that coexpression networks can address, we briefly discuss several key findings that have emerged from studies using coexpression networks in novel ways.
Changes in coexpression network structure identify candidate disease genes
Gene-gene correlations may be altered in disease and signal altered regulatory structure (Figure 2) without affecting differential expression (see section on differential connectivity). While differential coexpression is itself a novel tool, it is generally applied to find entire modules of genes with different connectivity (correlations) in the disease state (Zhang et al., 2013). Expanding on previous work in differential coexpression (Hudson et al., 2009), Rhinn et al. (2012), show how differential coexpression can be used to prioritize disease-related molecular targets. The alpha synuclein variant “aSynL”, containing a long 3′sUTR, was identified as the most differentially coexpressed gene in several Parkinson’s disease datasets; however, aSyl was not highly differentially expressed and thus would have likely been overlooked by traditional microarray analysis. Notably, all datasets used in that study to select and investigate aSynL are publically available, indicating that differential coexpression is an accessible and applicable technique for existing brain disease microarray data.
Coexpression networks track brain region differences and disease vulnerability
Integrating coexpression results with related datasets can increase the statistical confidence in the findings and show how these networks (which may include dozens of modules and hundreds of hub genes) fit within the broader context of research. Miller et al (2013) enhance their within-subject comparison of CA1 versus CA3 vulnerability during the progression of Alzheimer’s disease with statistical comparisons to related studies. These comparisons include module-module overlaps to other coexpression studies, rank-order comparisons to other differential expression studies and integration of cell-type signatures, all of which contribute to a high confidence set of disease genes and systems biology hypotheses of how region-specific expression relates to specific measures of Alzheimer’s disease progression and cell-type specific properties. This study illustrates that even when the primary dataset contains multiple brain regions, it is possible to substantially enhance the hypothesis generation from coexpression networks through integration of public data.
Coexpression networks unify heterogeneous molecular deficits in rare diseases
Gulsuner et al. (2013) provide a demonstration of how coexpression networks are useful in this context of highly heterogeneous pathology, by unifying de novo schizophrenia-associated mutations into more coherent mechanisms, in part by the coexpression relationships of the genes which harbor these mutations. They inferred coexpression relationships between genes using a pseudo time-series of 26 brains from a period of human development spanning 13 weeks of age to early adulthood in the Brainspan: Atlas of the developing human brain (www.brainspan.org). Then they counted the number coexpression and protein-protein interaction links between genes harboring these mutations and found a greater number of links than expected using sibling controls, with the most extreme difference found in the frontal cortex comparison. This indicates that mutated genes gain correlations in the disease state, and that disease state is not accompanied purely by loss of function at the coexpression level. By mapping a different data type (i.e., DNA sequences) directly to gene-gene correlations, this study shows how the apparently sporadic set of genes related to schizophrenia affect coherent molecular functions.
Differentially expressed genes in some complex psychiatric diseases have low connectivity
In earlier work, we established that gene coexpression network topology, demonstrating both small-world and scale-free characteristics, is resilient to changes in diseased subjects across multiple brain regions (Gaiteri and Sibille 2011). As hub nodes are particularly vulnerable to perturbations in small-world networks, and standard pathological mechanisms for small-world networks involve attacks on central hubs, the finding that differentially expressed genes primarily reside on the periphery of coexpression networks for neuropsychiatric disorders such as depression, schizophrenia, and bipolar disorder was surprising, but consistent with the heterogeneous nature of these disorders. The low connectivity of differentially expressed genes suggests that modulating a single gene, or even category of genes, is likely to have a limited therapeutic effect, perhaps accounting for the low efficacy of current antidepressant treatments and providing a rationale for a treatment comprising a rational combination of mechanism-supported drugs.
Cross-tissue coexpression relationships affect brain gene expression
As shown in Dobrin et al. (2009), tissue-to-tissue coexpression networks can quantify inter-tissue interactions, even across the blood-brain barrier. Using microarrays from hypothalamus, liver and adipose tissue, they found that 40% of gene-gene correlations relate to cross-tissue interactions. Thus these cross-tissue interactions may account for a significant fraction of coexpression in other studies, but these contributions go undetected because peripheral tissues are not assessed simultaneously. This multi-tissue approach also has the potential of identifying more easily accessible peripheral regulators of brain processes. In this way, the structure of cross-tissue networks can be a hypothesis generator for diseases with suspected endocrine or inflammation involvement that would potentially synchronize gene expression across tissues and organs.
Practical coexpression network analysis, Part 2: Answers to common questions from experimental biologists
How many samples do I need for coexpression analysis?
The number of samples required for useful coexpression analysis depends on the genetic and environmental heterogeneity of the samples, their technical quality and the molecular severity of any disease contrasts. For instance, building coexpression networks from post-mortem brains of subjects with psychiatric disorders can be challenging because medication history and disease severity/onset are generally difficult to establish. Constructing networks from samples of pure cell populations or from mice of common genetic background allows coexpression networks to be constructed with fewer samples. Such pure cell populations will still have expression variation due to endogenous regulatory patterns (Basso et al., 2005, Clarke et al., 2011) or developmental regulation (Konopka et al., 2012). Networks inferred in culture systems avoid the confounding effects of opposite expression patterns that may occur in different cell types – which are intermingled in typical post-mortem brain samples.
For small sample sizes (~ n<20) it may be helpful to use robust correlations measures such as spearman’s correlation or the biweight correlation (Song et al., 2012) which limit the impact of a small set of outlying expression datapoints, that might drive high Pearson correlations, when the majority of datapoints are uncorrelated. However, as the number of samples increases, the strongest correlations identified by these robust methods become very similar to those identified by Pearson correlation. In order to identify differentially coexpressed links or genes, it is necessary to have sufficient samples for high-confidence network construction in each phenotype.
What about non-linear molecular interactions? Are those detected?
While early microarray analyses relied on Pearson correlation in part because it is very fast to compute, new efficient routines to compute all pair-wise mutual information and biweight correlations make it possible to test for non-linear relationships that are robust to outliers. Fortunately it appears that results from non-linear tests are dominated by linear relationships (Song et al., 2012, Steuer et al., 2002). Thus, if the strongest gene-gene interactions are prioritized to create coexpression networks, both linear and non-linear approaches tend to select similar set of interactions.
How do I know that gene modules are biologically real?
There are both statistical and biological approaches to the validity of gene expression modules. Statistical approaches focus on module reproducibility, while biological test of gene modules focus on the ability to consistently perturb entire gene modules. From a statistical perspective, if gene modules are not reproducible between similar cohorts or orthologous datasets, then the conclusions from coexpression analysis will not generalize. Therefore, it is important to quantify the reproducibility of gene-gene correlations and gene modules across multiple datasets. The gene-gene correlations that give rise to modules persistent across arrays, normalization procedures and species, making technical artifacts unlikely, as such effects are randomized across datasets (Obayashi et al., 2008). Within single datasets, spurious clusters can be generated if batch effects are not controlled (Leek & Storey, 2007), which might lead to spurious modules that will not be reproduced in other studies. While coexpression module might be expected to be more robust than specific gene-gene correlations, the process of generating modules may introduce noise because modules are highly overlapping and difficult to optimally define. For instance, if the clustering algorithm used to define gene modules is sensitive to various thresholds, this may also lead to “unstable” module definitions. It is possible to avoid this clustering instability by resampling the expression dataset and re-identifying modules many times to identify genes which robustly cluster together. This critical step to ensure module reproducibility is often skipped because some clustering algorithms take hours to generate a single set of gene modules.
Direct estimate of the reproducibility of modules across similar data sets, are rare, although a lone example from glioblastoma research supports ~50% overlap (Ivliev et al., 2010), which equates to extreme p-values for reproducibility. In the context of cross-species comparisons, this may be quite impressive (Langfelder et al., 2011, Oldham et al., 2006), but in the context of replicate cohorts which are expected to have similar results, it represents a high level of variability. The very concept of distinct modules is an intrinsic limitation to reproducibility, as there are many coexpression links between modules (Figure 3). To avoid the instability in clustering results due to overlapping modules it may be useful to employ clustering methods which produce overlapping clusters (Evans & Lambiotte, 2009) or methods that combine clusters from multiple techniques, harnessing diverse results to provide both robustness and unique insights. However the optimal way to define “consensus” modules based on the output of multiple clustering is an open mathematical question of equal complexity to the original clustering problem (Vega-Pons & Ruiz-Shulcloper, 2011).
While statistical techniques can prove that modules are co-regulated, “biologically real” also implies it is possible to operate on gene sets as units. To demonstrate biological coherence of a module, the effects of a perturbation should primarily be constrained to the genes within that module. Coexpression modules are not in fact completely modular – there are many correlations among the members of different modules. Therefore the effects of a perturbation may extend outside of a module, but should still be predicted by the network structure as in Zhang et al. (2013). The Connectivity Map (Lamb et al., 2006) and DrugMatrix (Natsoulis et al., 2008) databases offer libraries of perturbation microarrays, and indeed contain coexpressed modules that are generated by certain classes of drugs (Iskar et al., 2013), which indicates that perturbations tend to result in reproducible and bounded coexpression effects.
Do coexpression modules predict disease or disease severity?
If coexpression patterns are robustly related to cellular pathways activated in disease states, it would seem to follow that modules and hub gene expression should robustly predict disease status or severity. Many authors have noted the partial overlap between disease-associated coexpression hubs and known disease modulators or GWAS hits (Chen et al., 2012, De Jong S & Janson E, 2012, Miller et al., 2008, Ponomarev et al., 2012, Torkamani et al., 2010, Voineagu et al., 2011, Zhang et al., 2013). But the average correlation of coexpression modules with disease traits, is often less than R=0.5 (Zhang et al., 2013) although module-disease correlations are highly significant, as they are sustained across hundreds of genes. Another way to assess the predictive power of modules is by tracking the reproducibility of hub-disease correlations across replicate datasets, compared to standard meta-analysis for biomarker discovery. This comparison shows traditional meta-analysis techniques generally output more reproducible disease-correlated gene lists, except when hubs are carefully selected and the overall differential expression is weak (Langfelder et al., 2013). In these noisy cases, the redundancy inherent in coexpression networks helps to improve reproducibility compared to traditional measures. Therefore, while coexpression networks are a useful framework to drive experimental programs, their predictive performance on a module-by-module level makes it challenging to use them as disease classifiers.
In light of these relatively low module-disease correlations, various robust regression and machine learning approaches likely offer better performance in classifying the disease status of microarray samples (Pirooznia et al., 2008). In contrast to coexpression approaches, which highlight the covariance structure of gene expression, common machine learning approaches tend to choose a single or small number of genes to exemplify correlated gene sets (Zou & Hastie, 2005). While machine learning techniques do not automatically place disease-associated molecules in a coherent biological framework, they can identify a limited set of predictive gene-features, which can be submitted for gene set enrichment analysis. The debate between coexpression versus disease biomarker detection goes beyond mathematical assessments, because these techniques generally take different scientific roles. The way in which coexpression networks reflect endogenous regulatory systems may set the stage for detailed set of molecular experiments that occur in a coherent molecular system, as in Rhinn (2012). However results from coexpression analysis are potentially less suited for identifying single-gene disease biomarkers (Langfelder et al., 2013). Thus while coexpression modules are significant predictors of disease, they are rarely used as pure predictors of disease and face a significant challenge from machine learning techniques in the search for biomarkers.
While machine learning and coexpression approaches to predicting gene expression have traditionally arisen from different conceptual approaches to biology, a rare example of the potential for hybrid approaches, the algorithm, Ontogenet, predicts major regulators associated with cell-types specific expression, using a combination of coexpression modules, machines learning inference and molecular interaction databases (Jojic et al., 2013). The results offer superior predictive performance versus elasticnet (Zou & Hastie, 2005) (a popular pure machine learning approach) while offering a more specific list of module regulators than is available by pure coexpression approaches. While such novel hybrid approaches are highly novel and have not been applied to brain disease datasets, they show an opportunity of maximize the predictive power and interpretability of coexpression modules.
How do you create network images?
Network visualization is an important step that allows researchers to intuitively explore the network topology and develop hypotheses. Cytoscape (Shannon et al., 2003) is a flexible and widely used software platform for visualizing networks and biological pathways and integrating these networks with annotations, gene expression profiles, and other data. It also contains various analysis tools as plugins that were contributed by other labs. Other visualization tools to illustrate biological networks include VisANT (Hu et al., 2005), Pajek (De Nooy W, 2005) and Gephi, and others reviewed elsewhere (Pavlopoulos et al., 2008). The choice of which visualization software to use depends on their ability to incorporate additional layers of biological information into network properties, such as node size and color (both Gephi and Cytoscape offer this extensively) as well as node layouts, which can radically alter the reader’s perception of networks. Unfortunately there is no generally optimal biological node layout; therefore it is helpful to rapidly try several layouts to determine an informative networks layout, as the base layer for additional node properties, node groupings, labels and experimental annotations.
It is challenging to visualize large and complex networks, because gene networks with thousands of dimensions are projected onto a 2D lane for publication, sometimes producing an uninformative “hair ball” effect. Traditionally multi-dimensional scaling (MDS), accessible in various R-packages, is used to visually maximize the distinctions between clusters (Langfelder & Horvath, 2008). More recent approaches to visualizing networks of thousands of nodes such as Biofabric give a new simple two dimensional line representation of the networks with additional clarity with row lines representing nodes and column lines representing links (Longabaugh, 2012), which allows a scalable and unambiguous presentation of the network edges. Another alternative network representation is the “hive plot”, which positions nodes on radically distributed axes based on network structural properties (Krzywinski et al., 2012).
Emerging topics and key issues for future coexpression network research
Comprehensive species, tissue and disease catalogue of coexpression modules
Comparisons of module membership across many datasets may increase confidence in the biological reality of modules and show novel cross-tissue communication, or similarities between modules found in different diseases. The basis for such a comparison would be a user-driven database of gene lists and their module assignments, annotated by tissue-type and disease status. These lists can be compiled even when the complete expression data is not publically available or restricted. Primary use cases would be researchers querying against the database for modules robustly associated with a given phenotype, or for overlap between their clustering results and all modules in the database. Such a database would include popular WGCNA-based results (Langfelder & Horvath, 2008), but also allow inputs from many clustering algorithms, which would further verify the existence of robustly correlated gene sets.
Identifying recurrent patterns across multiple networks could reveal important functional associations and increase the accuracy rather than focusing on single study analysis where random pattern could occur due to spurious correlations. A straightforward way to aggregating studies is simply to concatenate the gene expression matrices as in (Dunker et al., 2001, Mabbott et al., 2010) or to combine the evidence of gene interaction by vote counting or Fisher’s methods (Cancer Genome Atlas Research, 2011, Niida et al., 2010). However, combining in those ways could introduce false patterns from normalization and nature of heterogeneity between studies. Therefore, directly detecting frequent patterns in multiple networks is likely a better solution (Li et al., 2011).
Annotating coexpression links via causal molecular mechanisms
Differential coexpression is likely related to altered gene regulation (Figure 2), but in most cases the cause remains unknown. An ideal experiment to associate specific molecular mechanisms with differential coexpression would be to assess the regulatory structure of multiple systems in a disease model (Hudson et al., 2012). This would require multiple assays to be measured in pure cell populations, including chromosome interactions, ChIP-seq on at least several transcription factors, miRNA and methylation. Potential discoveries from this approach could determine if particular modules are generated predominantly by a single molecular mechanism, or if there are stereotypical inter-regulatory motifs (patterns of links between different regulatory systems, such as feed-forward inhibition) that have not been previously shown, but have been shown to occur in other networks (Gerstein et al., 2012, Jothi et al., 2009, Ma'ayan, 2009, Nazarov et al., 2013). Specific molecular mechanisms associated with disease states could be assessed by combining multiple aforementioned high-throughput methods with coexpression network structure (Figure 3E).
Beyond hubs: mapping the connectivity of differentially expressed and disease genes
If disease genes are characterized by a particular type of connectivity in molecular networks, it would be a powerful filtering mechanism to prioritize disease targets – simply examining the connectivity of various putative disease genes. Attempts to find such a disease-connectivity relationship suggest that if this relationship exists, it is sensitive to the definition of disease genes and molecular connectivity. For instance GWAS genes tend to be bottleneck nodes of high betweenness centrality in various networks (Lee et al., 2013) and genes with common cancer mutations tend to be protein hubs (Jonsson & Bates, 2006). This suggests that centrality and overall connectivity in a network are associated with disease activity. However, OMIM (Online Mendelian Inheritance in Man) genes do not occur with a characteristic connectivity in protein-protein interaction networks (Goh et al., 2007) and monogenic disease genes tend to have connectivity that is tightly constrained around average values (Feldman et al., 2008). Thus, different definitions of the set of disease genes can result in different conclusions about the expected connectivity of disease genes (hubs versus exact average connectivity). In addition to the definition of the set of disease genes, the type of network in which disease gene connectivity is measured can affect the disease-connectivity relationship. For instance, differentially expressed genes in Parkinson’s disease and schizophrenia tend to be hub nodes in protein networks (Mar et al., 2011), while differentially expressed genes from schizophrenia, bipolar disorder and major depression tend to be low-connected in coexpression networks (Gaiteri & Sibille, 2011). Thus, when evaluating the “meaning” of connectivity of a particular set of disease-related genes, it is useful to check the connectivity across multiple types of networks.
Disease severity may interact with molecular connectivity in such a way that places disease genes at different network locations. This interaction may explain some of the apparently conflicted results mentioned previously. Specifically, more severe diseases are associated with deficits in more central genes (Barrenas et al., 2009). Such a relationship could be tested in compiling differential gene expression signatures from diseases of varying severity/lethality and may be useful in understanding control mechanisms in complex diseases. The relationship of disease to network structure will likely be more complex than a linear relationship between the number of connections of disease-associated genes versus disease severity (Park & Kim, 2009). An example of a more complex network relationship that links connectivity to disease would be the way in which the severity of different cancers is related to the distribution of connectivity (known as degree heterogeneity) in KEGG pathways of all genes associated with a particular type of cancer (Breitkreutz et al., 2012). The sensitivity of the link between connectivity and disease activity is a cautionary note against exclusive focus on hub nodes in coexpression networks as disease-mediators.
Coexpression in network pharmacology
Neuroscience drug development is challenging because brain function-level phenotypes are difficult to simulate through in vitro systems, while animal models of common diseases including schizophrenia, major depression and Alzheimer’s disease do not generate the severe behavioral or molecular phenotypes of the human disease. Because coexpression networks encapsulate multiple molecular regulatory mechanisms in an unbiased manner, they may offer a framework to track connections between downstream disease effectors, to supply additional targets similar to existing targets that have been discarded for toxicology reasons or to indicate previously undetected aspects of pathology – for instance differential coexpression in the absence of differential expression (Csermely et al., 2013, Rhinn et al., 2012). Coexpression may even be useful in organizing compound libraries from a systems biology perspective. Using the Connectivity Map (Lamb et al., 2006) and DrugMatrix (Natsoulis et al., 2008) databases of drug-response microarrays from three human and one rat cell-line, Iskar et al. (2013) found reproducible coexpression modules that correspond to specific drug treatments. While finding coexpression patterns requires more samples than looking for differential expression among drug responses, it has the benefit of associating data-driven signaling pathways (coexpression modules) with each drug and identifying sets of drugs that activate related molecular systems.
However, the application of systems biology to drug discovery is impeded because most computational researchers do not have structures in place to perform validation experiments that “prove” their methods are correct. This conflict between pursuing “risky” experiments with no “guarantee” of positive results and the need to move beyond the single-gene, single-disease model, presents opportunities for coexpression analysis. There is no reason to limit network exploration purely to coexpression, but coexpression links should be compiled alongside protein-protein interactions, TF-binding, miRNA targets, chromosome contact maps into “meta-networks” which have been shown to collectively direct cellular activity (Gerstein et al., 2012, Ma'ayan, 2009). If the structure of networks in this hybrid database is compared to perturbation experiments, this would form the basis of new predictive methods to control target gene sets identified in human disease samples (Csermely et al., 2013, Hopkins, 2008) and to potentially identify critical regulatory elements hidden in gene coexpression networks as novel targets.
Acknowledgements
This work was supported by National Institute of Mental Health MH084060 (ES). The funding agency had no role in the study design, data collection and analysis, decision to publish and preparation of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIMH or the National Institutes of Health.
Glossary
- Betweenness centrality
is a measure of how central a node is in a network, i.e., the relative importance of the node to the network. It is equal to the fraction of the shortest paths that pass through a node, and counted over the shortest paths among all pairs of nodes.
- Biweight correlation, or “Bicor”
is the median-based correlation measure. It is more robust than the Pearson correlation and often more powerful than the Spearman correlation.
- Differential variability (DV)
measures the variance of a gene between two groups of samples; e.g., a gene which is highly variable in healthy samples may show less variation in disease samples.
- Differential coexpression (DC)
measures changes in coexpression between samples; e.g., genes which are coexpressed in healthy samples lose their correlation in diseased samples or vice versa.
- Differential expression
measures changes in expression levels of genes between two different groups of samples. For instance, gene expression measures for a particular gene in samples from healthy subjects may be lower compared to the expression of the same gene in samples from the disease group.
- Edge / link
is the connection between two nodes. An edge is usually based on correlation but sometimes utilizes physical binding or other forms of interactions.
- Guilt-by-association (GBA)
is a proposed biological principle stating that genes or proteins with the same or related cellular functions tend to share properties such as genetic or physical interactions. It is frequently used by computational biologists to assign function to genes.
- Hub genes
are genes with highest degree in a network.
- Nodes
are the fundamental units of the network which are linked by edges. For gene networks, nodes represent individual genes.
- OMIM (Online Mendelian Inheritance in Man)
as described by the OMIM website, “is a comprehensive, authoritative compendium of human genes and genetic phenotypes that is freely available and updated daily. OMIM is authored and edited at the McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, under the direction of Dr. Ada Hamosh. Its official home is omim.org.”
- Pearson correlation
is a measure of the extent of a linear relationship between two variables x and y. Pearson correlations are the most widely used correlation measures.
- Probabilistic Boolean networks
are models in which gene expression is quantized to either ‘on’ or ‘off’. Probabilistic Boolean networks create uncertainty in the functions that determine on/off behaviors.
- Scale-free networks
contain many nodes with very few connections and a small number of hubs with high connections. The logarithm of P(k) [the probability of a node to have degree k] is approximately inversely proportional to the logarithm of k [the degree of a node]. Modified from de la Fuente (2010).
- Small-world networks
exhibit high average clustering and small average distance between nodes. In small-world networks, nodes are typically strongly clustered into local communities that support biological sub-processes.
- Spearman’s correlations
are based on ranks, which measures the extent of a monotonic relationship between x and y. Spearman correlations are more robust measures of correlation than Pearson correlations.
References
- Albert R, Jeong H, Barabasi A-L. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- Allocco D, Kohane I, Butte A. Quantifying the relationship between co-expression, co-regulation and gene function. BMC Bioinformatics. 2004;5:18. doi: 10.1186/1471-2105-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amar D, Safer H, Shamir R. Dissection of regulatory networks that are altered in disease via differential co-expression. PLoS Comput Biol. 2013;9:e1002955. doi: 10.1371/journal.pcbi.1002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Bhattacharyya M. Analyzing miRNA co-expression networks to explore TF-miRNA regulation. BMC Bioinformatics. 2009;10:163. doi: 10.1186/1471-2105-10-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrenas F, Chavali S, Holme P, Mobini R, Benson M. Network properties of complex human disease genes identified through genome-wide association studies. PLoS One. 2009;4:e8090. doi: 10.1371/journal.pone.0008090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- Belmaker R, Agam G. Major depressive disorder. New England Journal of Medicine. 2008;358:55–68. doi: 10.1056/NEJMra073096. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya M, Bandyopadhyay S. Studying the differential co-expression of microRNAs reveals significant role of white matter in early Alzheimer's progression. Molecular bioSystems. 2013;9:457–466. doi: 10.1039/c2mb25434d. [DOI] [PubMed] [Google Scholar]
- Breitkreutz D, Hlatky L, Rietman E, Tuszynski JA. Molecular signaling network complexity is correlated with cancer patient survivability. Proceedings of the National Academy of Sciences. 2012;109:9209–9212. doi: 10.1073/pnas.1201416109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C, Langfelder P, Fuller T, Oldham M, Luo R, van den Berg L, Ophoff R, Horvath S. Is human blood a good surrogate for brain tissue in transcriptional studies? BMC Genomics. 2010;11:589. doi: 10.1186/1471-2164-11-589. -- [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Cheng L, Grennan K, Pibiri F, Zhang C, Badner JA, Members of the Bipolar Disorder Genome Study, C. Gershon ES, Liu C. Two gene co-expression modules differentiate psychotics and controls. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M, Shi J, Jung SH, Chen X, Cho K-H. Attractor landscape analysis reveals feedback loops in the p53 network that control the cellular response to DNA damage. Science signaling. 2012;5:ra83. doi: 10.1126/scisignal.2003363. [DOI] [PubMed] [Google Scholar]
- Clarke C, Doolan P, Barron N, Meleady P, O'Sullivan F, Gammell P, Melville M, Leonard M, Clynes M. Large scale microarray profiling and coexpression network analysis of CHO cells identifies transcriptional modules associated with growth and productivity. Journal of biotechnology. 2011;155:350–359. doi: 10.1016/j.jbiotec.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Csermely P, Korcsmáros T, Kiss HJ, London G, Nussinov R. Structure and dynamics of molecular networks: A novel paradigm of drug discovery: A comprehensive review. Pharmacology & therapeutics. 2013 doi: 10.1016/j.pharmthera.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong S BM, Fuller TF, Strengman E, Janson E. A Gene Co-Expression Network in Whole Blood of Schizophrenia Patients Is Independent of Antipsychotic-Use and Enriched for Brain-Expressed Genes. PLoS ONE. 2012;7(6):e39498. doi: 10.1371/journal.pone.0039498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente A. From 'differential expression' to 'differential networking' - identification of dysfunctional regulatory networks in diseases. Trends Genet. 2010;26:326–333. doi: 10.1016/j.tig.2010.05.001. [DOI] [PubMed] [Google Scholar]
- de Nooy W MA, Batagelj V. Exploratory social network analysis with pajek (structural analysis in the social sciences) Cambridge University Press; Cambridge: 2005. [Google Scholar]
- Dobrin R, Zhu J, Molony C, Argman C, Parrish ML, Carlson S, Allan MF, Pomp D, Schadt EE. Multi-tissue coexpression networks reveal unexpected subnetworks associated with disease. Genome Biol. 2009;10:R55. doi: 10.1186/gb-2009-10-5-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Luo L, Hong S, Siu H, Xiao Y, Jin L, Chen R, Xiong M. Integrated analysis of mutations, miRNA and mRNA expression in glioblastoma. BMC Systems Biology. 2010;4:163. doi: 10.1186/1752-0509-4-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Ebisuya M, Yamamoto T, Nakajima M, Nishida E. Ripples from neighbouring transcription. Nat Cell Biol. 2008;10:1106–1113. doi: 10.1038/ncb1771. [DOI] [PubMed] [Google Scholar]
- Erraji-Benchekroun L, Underwood MD, Arango V, Galfalvy H, Pavlidis P, Smyrniotopoulos P, Mann JJ, Sibille E. Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biol Psychiatry. 2005;57:549–558. doi: 10.1016/j.biopsych.2004.10.034. [DOI] [PubMed] [Google Scholar]
- Evans T, Lambiotte R. Line graphs, link partitions, and overlapping communities. Physical Review E. 2009;80:016105. doi: 10.1103/PhysRevE.80.016105. [DOI] [PubMed] [Google Scholar]
- Feldman I, Rzhetsky A, Vitkup D. Network properties of genes harboring inherited disease mutations. Proceedings of the National Academy of Sciences. 2008;105:4323–4328. doi: 10.1073/pnas.0701722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato S. Community detection in graphs. Physics Reports. 2010;486:75–174. [Google Scholar]
- Gaiteri C, Guilloux JP, Lewis DA, Sibille E. Altered gene synchrony suggests a combined hormone-mediated dysregulated state in major depression. PLoS One. 2010;5:e9970. doi: 10.1371/journal.pone.0009970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiteri C, Sibille E. Differentially expressed genes in major depression reside on the periphery of resilient gene coexpression networks. Frontiers in Neuroscience. 2011;5 doi: 10.3389/fnins.2011.00095. -- [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino VA, D'Angelo G, Dharmalingam G, Fernandez S, Russolillo G, Sanges R, Mutarelli M, Belcastro V, Ballabio A, Verde P, Sardiello M, Banfi S. Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Res. 2012;22:1163–1172. doi: 10.1101/gr.130435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, Min R, Alves P, Abyzov A, Addleman N, Bhardwaj N, Boyle AP, Cayting P, Charos A, Chen DZ, Cheng Y, Clarke D, Eastman C, Euskirchen G, Frietze S, Fu Y, Gertz J, Grubert F, Harmanci A, Jain P, Kasowski M, Lacroute P, Leng J, Lian J, Monahan H, O'Geen H, Ouyang Z, Partridge EC, Patacsil D, Pauli F, Raha D, Ramirez L, Reddy TE, Reed B, Shi M, Slifer T, Wang J, Wu L, Yang X, Yip KY, Zilberman-Schapira G, Batzoglou S, Sidow A, Farnham PJ, Myers RM, Weissman SM, Snyder M. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis J, Pavlidis P. “Guilt by association” is the exception rather than the rule in gene networks. PLoS computational biology. 2012;8:e1002444. doi: 10.1371/journal.pcbi.1002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh K-I, Cusick ME, Valle D, Childs B, Vidal M, Barabasi A-L. The human disease network. Proceedings of the National Academy of Sciences. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, Rippey C, Shahin H, Nimgaonkar VL, Go RC, Savage RM, Swerdlow NR, Gur RE, Braff DL, King MC, McClellan JM. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein S, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinäniemi M, Nykter M, Kramer R, Wienecke-Baldacchino A, Sinkkonen L, Zhou JX, Kreisberg R, Kauffman SA, Huang S, Shmulevich I. Gene-pair expression signatures reveal lineage control. Nature methods. 2013;10:577–583. doi: 10.1038/nmeth.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JW, Stefani M, dos Remedios CG, Charleston MA. Differential variability analysis of gene expression and its application to human diseases. Bioinformatics. 2008;24:i390–i398. doi: 10.1093/bioinformatics/btn142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homouz D, Kudlicki AS. The 3D organization of the yeast genome correlates with co-expression and reflects functional relations between genes. PLoS One. 2013;8:e54699. doi: 10.1371/journal.pone.0054699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nature chemical biology. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- Horvath S, Zhang Y, Langfelder P, Kahn RS, Boks MP, van Eijk K, van den Berg LH, Ophoff RA. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012;13:R97. doi: 10.1186/gb-2012-13-10-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Mellor J, Wu J, Yamada T, Holloway D, DeLisi C. VisANT: data-integrating visual framework for biological networks and modules. Nucleic Acids Research. 2005;33:W352–W357. doi: 10.1093/nar/gki431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson NJ, Dalrymple BP, Reverter A. Beyond differential expression: the quest for causal mutations and effector molecules. BMC Genomics. 2012;13:356. doi: 10.1186/1471-2164-13-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson NJ, Reverter A, Dalrymple BP. A differential wiring analysis of expression data correctly identifies the gene containing the causal mutation. PLoS computational biology. 2009;5:e1000382. doi: 10.1371/journal.pcbi.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskar M, Zeller G, Blattmann P, Campillos M, Kuhn M, Kaminska KH, Runz H, Gavin AC, Pepperkok R, van Noort V, Bork P. Characterization of drug-induced transcriptional modules: towards drug repositioning and functional understanding. Mol Syst Biol. 2013;9:662. doi: 10.1038/msb.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivliev AE, AC't Hoen P, Sergeeva MG. Coexpression network analysis identifies transcriptional modules related to proastrocytic differentiation and sprouty signaling in glioma. Cancer Research. 2010;70:10060–10070. doi: 10.1158/0008-5472.CAN-10-2465. [DOI] [PubMed] [Google Scholar]
- Izhikevich EM. Dynamical systems in neuroscience. The MIT press; 2007. [Google Scholar]
- Jay JJ, Eblen JD, Zhang Y, Benson M, Perkins AD, Saxton AM, Voy BH, Chesler EJ, Langston MA. A systematic comparison of genome-scale clustering algorithms. BMC Bioinformatics. 2012;13:S7. doi: 10.1186/1471-2105-13-S10-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabási A-L, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Jojic V, Shay T, Sylvia K, Zuk O, Sun X, Kang J, Regev A, Koller D, Best AJ, Knell J, Goldrath A, Cohen N, Brennan P, Brenner M, Kim F, Rao TN, Wagers A, Heng T, Ericson J, Rothamel K, Ortiz-Lopez A, Mathis D, Benoist C, Bezman NA, Sun JC, Min-Oo G, Kim CC, Lanier LL, Miller J, Brown B, Merad M, Gautier EL, Jakubzick C, Randolph GJ, Monach P, Blair DA, Dustin ML, Shinton SA, Hardy RR, Laidlaw D, Collins J, Gazit R, Rossi DJ, Malhotra N, Kreslavsky T, Fletcher A, Elpek K, Bellemare-Pelletier A, Malhotra D, Turley S. Identification of transcriptional regulators in the mouse immune system. Nat Immunol. 2013;14:633–643. doi: 10.1038/ni.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson PF, Bates PA. Global topological features of cancer proteins in the human interactome. Bioinformatics. 2006;22:2291–2297. doi: 10.1093/bioinformatics/btl390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothi R, Balaji S, Wuster A, Grochow JA, Gsponer J, Przytycka TM, Aravind L, Babu MM. Genomic analysis reveals a tight link between transcription factor dynamics and regulatory network architecture. Mol Syst Biol. 2009;5:294. doi: 10.1038/msb.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka G, Wexler E, Rosen E, Mukamel Z, Osborn GE, Chen L, Lu D, Gao F, Gao K, Lowe JK, Geschwind DH. Modeling the functional genomics of autism using human neurons. Mol Psychiatry. 2012;17:202–214. doi: 10.1038/mp.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostka D, Spang R. Finding disease specific alterations in the co-expression of genes. Bioinformatics. 2004;20(Suppl 1):i194–199. doi: 10.1093/bioinformatics/bth909. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Birol I, Jones SJ, Marra MA. Hive plots--rational approach to visualizing networks. Brief Bioinform. 2012;13:627–644. doi: 10.1093/bib/bbr069. [DOI] [PubMed] [Google Scholar]
- Kupfer DJ, Frank E, Phillips ML. Major depressive disorder: new clinical, neurobiological, and treatment perspectives. Lancet. 2012;379:1045–1055. doi: 10.1016/S0140-6736(11)60602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7:e1001057. doi: 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Mischel PS, Horvath S. When is hub gene selection better than standard meta-analysis? PLoS One. 2013;8:e61505. doi: 10.1371/journal.pone.0061505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Research. 2003;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Li H, Li J, Rebman E, Achour I, Regan KE, Gamazon ER, Chen JL, Yang XH, Cox NJ. Network models of genome-wide association studies uncover the topological centrality of protein interactions in complex diseases. Journal of the American Medical Informatics Association. 2013;20:619–629. doi: 10.1136/amiajnl-2012-001519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. Plos Genet. 2007;3:e161. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Liu C-C, Zhang T, Li H, Waterman MS, Zhou XJ. Integrative analysis of many weighted co-expression networks using tensor computation. PLoS computational biology. 2011;7:e1001106. doi: 10.1371/journal.pcbi.1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longabaugh WJ. Combing the hairball with BioFabric: a new approach for visualization of large networks. BMC Bioinformatics. 2012;13:275. doi: 10.1186/1471-2105-13-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N. The Genotype-Tissue Expression (GTEx) project. Nature genetics. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma'ayan A. Insights into the organization of biochemical regulatory networks using graph theory analyses. Journal of Biological Chemistry. 2009;284:5451–5455. doi: 10.1074/jbc.R800056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabbott NA, Kenneth Baillie J, Hume DA, Freeman TC. Meta-analysis of lineage-specific gene expression signatures in mouse leukocyte populations. Immunobiology. 2010;215:724–736. doi: 10.1016/j.imbio.2010.05.012. [DOI] [PubMed] [Google Scholar]
- Mar JC, Matigian NA, Mackay-Sim A, Mellick GD, Sue CM, Silburn PA, McGrath JJ, Quackenbush J, Wells CA. Variance of gene expression identifies altered network constraints in neurological disease. PLoS Genet. 2011;7:e1002207. doi: 10.1371/journal.pgen.1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco A, Konikoff C, Karr TL, Kumar S. Relationship between gene co-expression and sharing of transcription factor binding sites in Drosophila melanogaster. Bioinformatics. 2009;25:2473–2477. doi: 10.1093/bioinformatics/btp462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menashe I, Grange P, Larsen EC, Banerjee-Basu S, Mitra P. Co-expression Profiling of Autism Genes in the Mouse Brain. PLoS Comput Biol. 2013;9:e1003128. doi: 10.1371/journal.pcbi.1003128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J, Oldham M, Geschwind D. A systems level analysis of transcriptional changes in Alzheimer's disease and normal aging. J Neurosci. 2008;28:1410–1420. doi: 10.1523/JNEUROSCI.4098-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Woltjer RL, Goodenbour JM, Horvath S, Geschwind DH. Genes and pathways underlying regional and cell type changes in Alzheimer's disease. Genome medicine. 2013;5:48. doi: 10.1186/gm452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsoulis G, Pearson CI, Gollub J, B PE, Ferng J, Nair R, Idury R, Lee MD, Fielden MR, Brennan RJ, Roter AH, Jarnagin K. The liver pharmacological and xenobiotic gene response repertoire. Mol Syst Biol. 2008;4:175. doi: 10.1038/msb.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarov PV, Reinsbach SE, Muller A, Nicot N, Philippidou D, Vallar L, Kreis S. Interplay of microRNAs, transcription factors and target genes: linking dynamic expression changes to function. Nucleic Acids Research. 2013;41:2817–2831. doi: 10.1093/nar/gks1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida A, Imoto S, Nagasaki M, Yamaguchi R, Miyano S. A novel meta-analysis approach of cancer transcriptomes reveals prevailing transcriptional networks in cancer cells. Genome informatics. International Conference on Genome Informatics. 2010;22:121–131. [PubMed] [Google Scholar]
- Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, Wininger M, Colantuoni C, Weinberger DR, Kleinman JE, Lipska BK. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet. 2012;90:260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Hayashi S, Shibaoka M, Saeki M, Ohta H, Kinoshita K. COXPRESdb: a database of coexpressed gene networks in mammals. Nucleic Acids Research. 2008;36:D77–D82. doi: 10.1093/nar/gkm840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham M, Horvath S, Geschwind D. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci USA. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K, Kim D. Localized network centrality and essentiality in the yeast–protein interaction network. Proteomics. 2009;9:5143–5154. doi: 10.1002/pmic.200900357. [DOI] [PubMed] [Google Scholar]
- Pavlopoulos GA, Wegener AL, Schneider R. A survey of visualization tools for biological network analysis. BioData mining. 2008;1:12. doi: 10.1186/1756-0381-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirooznia M, Yang JY, Yang MQ, Deng Y. A comparative study of different machine learning methods on microarray gene expression data. BMC Genomics. 2008;1(9 Suppl):S13. doi: 10.1186/1471-2164-9-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene Coexpression Networks in Human Brain Identify Epigenetic Modifications in Alcohol Dependence. The Journal of Neuroscience. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przytycka TM, Singh M, Slonim DK. Toward the dynamic interactome: it's about time. Briefings in bioinformatics. 2010;11:15–29. doi: 10.1093/bib/bbp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray M, Ruan J, Zhang W. Variations in the transcriptome of Alzheimer's disease reveal molecular networks involved in cardiovascular diseases. Genome Biology. 2008;9:R148. doi: 10.1186/gb-2008-9-10-r148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn H, Qiang L, Yamashita T, Rhee D, Zolin A, Vanti W, Abeliovich A. Alternative alpha-synuclein transcript usage as a convergent mechanism in Parkinson's disease pathology. Nat Commun. 2012;3:1084. doi: 10.1038/ncomms2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibille E, French B. Biological substrates underpinning diagnosis of major depression. Int J Neuropsychopharmacol. 2013;16:1893–1909. doi: 10.1017/S1461145713000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Langfelder P, Horvath S. Comparison of co-expression measures: mutual information, correlation, and model based indices. BMC Bioinformatics. 2012;13:328. doi: 10.1186/1471-2105-13-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southworth LK, Owen AB, Kim SK. Aging Mice Show a Decreasing Correlation of Gene Expression within Genetic Modules. Plos Genet. 2009;5 doi: 10.1371/journal.pgen.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam C, Jones B, Nolte G, Breakspear M, Scheltens P. Small-World Networks and Functional Connectivity in Alzheimer's Disease. Cerebral Cortex. 2007;17:92–99. doi: 10.1093/cercor/bhj127. [DOI] [PubMed] [Google Scholar]
- Steuer R, Kurths J, Daub CO, Weise J, Selbig J. The mutual information: detecting and evaluating dependencies between variables. Bioinformatics. 2002;18:S231–S240. doi: 10.1093/bioinformatics/18.suppl_2.s231. [DOI] [PubMed] [Google Scholar]
- Torkamani A, Dean B, Schork N, Thomas E. Coexpression network analysis of neural tissue reveals perturbations in developmental processes in schizophrenia. Genome Research. 2010;20:403–412. doi: 10.1101/gr.101956.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Pons S, Ruiz-Shulcloper J. A survey of clustering ensemble algorithms. International Journal of Pattern Recognition and Artificial Intelligence. 2011;25:337–372. [Google Scholar]
- Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of `small-world' networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Wolfe CJ, Kohane IS, Butte AJ. Systematic survey reveals general applicability of. BMC Bioinformatics. 2005;6:227. doi: 10.1186/1471-2105-6-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, Fluder E, Clurman B, Melquist S, Narayanan M, Suver C, Shah H, Mahajan M, Gillis T, Mysore J, Macdonald ME, Lamb JR, Bennett DA, Molony C, Stone DJ, Gudnason V, Myers AJ, Schadt EE, Neumann H, Zhu J, Emilsson V. Integrated Systems Approach Identifies Genetic Nodes and Networks in Late-Onset Alzheimer's Disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2005;67:301–320. [Google Scholar]