Abstract
Proper protein folding is a prerequisite for protein stability and enzymatic activity. While directed evolution can be a powerful tool to investigate enzymatic function and to isolate novel activities, well-designed libraries of folded proteins are essential. In vitro selection methods are particularly capable of searching for enzymatic activities in libraries of trillions of protein variants, yet high-quality libraries of well-folded enzymes with such high diversity are lacking. We describe the construction and detailed characterization of a folding-enriched protein library based on the ubiquitous (β/α)8 barrel fold found in five of the six enzyme classes. We introduced seven randomized loops on the catalytic face of the monomeric, thermostable (β/α)8 barrel of glycerophosphodiester phosphodiesterase (GDPD) from Thermotoga maritima. We employed an in vitro folding selection based on protease digestion to enrich intermediate libraries containing three to four randomized loops for folded variants and then combined them to assemble the final library (1014 DNA sequences). The resulting library was analyzed using the in vitro protease assay and an in vivo GFP-folding assay and contains ~1012 soluble monomeric protein variants. We isolated six library members and demonstrated that these proteins are soluble, monomeric and show (β/α)8 barrel fold-like secondary and tertiary structure. The quality of the folding-enriched library improved up to 50-fold compared to a control library that was assembled without the folding selection. To the best of our knowledge, this work is the first example of combining the ultra-high throughput method mRNA display with a selection for folding. The resulting (β/α)8 barrel libraries provide a valuable starting point to study the unique catalytic capabilities of the (β/α)8 fold, and to isolate novel enzymes.
Keywords: (β/α)8 barrel, GFP-folding reporter, mRNA display, protein folding, protein engineering
Introduction
Directed evolution experiments have generated numerous commercially valuable enzymes and have helped gain insight into the origins and evolution of enzymatic function. The success of any directed evolution experiment fundamentally depends on the diversity and quality of the starting library of protein variants. A protein library is considered of high quality if a substantial fraction of the library consists of well-folded, soluble and stable proteins that contain a diverse set of mutations and potential active sites for a variety of desired activities. In vitro selection strategies generally outperform in vivo or screening approaches by several orders of magnitude with regard to library diversity and are preferred for the isolation of potentially very rare mutants, e.g. novel enzymes.[1] However, high quality enzymatic libraries that can harness the ultra-high throughput of in vitro methods are currently lacking.
The ubiquitous (β/α)8 or TIM barrel fold is a promising scaffold for a general-purpose protein library that could be used for the isolation of new enzymatic activities and the understanding of the origins of enzymatic function. This versatile fold is utilized in five of the six enzymatic classes and is highly favored by natural enzymes to catalyze a wide array of different reactions, in some cases at the diffusion rate limit.[2] In the (β/α)8 barrel fold, the main structural and catalytic elements are spatially separated. The barrel itself is formed by eight alternating alpha helices and beta strands and provides the structural foundation while the eight loops connecting helices and strands on one side of the barrel are responsible for substrate binding and catalysis and are known as the catalytic face of the barrel. These features are favorable for enzyme engineering since modification of functional elements is less likely to affect the structural stability of the overall scaffold.[3] In a few cases, the catalytic activities of (β/α)8 barrel enzymes were successfully swapped through protein engineering to understand how the (β/α)8 barrel fold could be recruited to perform new activities.[4] Although, in some other cases, desired activities could be obtained by altering the substrate specificity of existing enzymes via targeted mutagenesis,[5] the introduction of novel activities often necessitated more extensive protein remodeling.[1b, 1c, 6] In an effort to enable more divergent sequence exploration well beyond that obtainable by point mutations, the tolerance of (β/α)8 scaffolds to the insertion of different natural (β/α)8 loop fragments was investigated.[7] Furthermore, the enzymatic activity of existing (β/α)8 barrel proteins was improved or modified by a combination of rational design and directed evolution similar to proteins of other folds.[2c, 8] In addition, rational design approaches for de novo enzymes repeatedly favored the (β/α)8 barrel fold over others, likely due to its ability to appropriately position catalytic and substrate binding residues.[9] This is particularly significant, as despite recent success in the rational re-design of enzymes, the de novo design of enzymes is still considered a formidable task.[9b, 10] In summary, the combination of valuable (β/α)8 barrel protein features like catalytic versatility, efficiency, stability, structural modularity and plasticity make this fold an ideal scaffold for enzyme engineering.
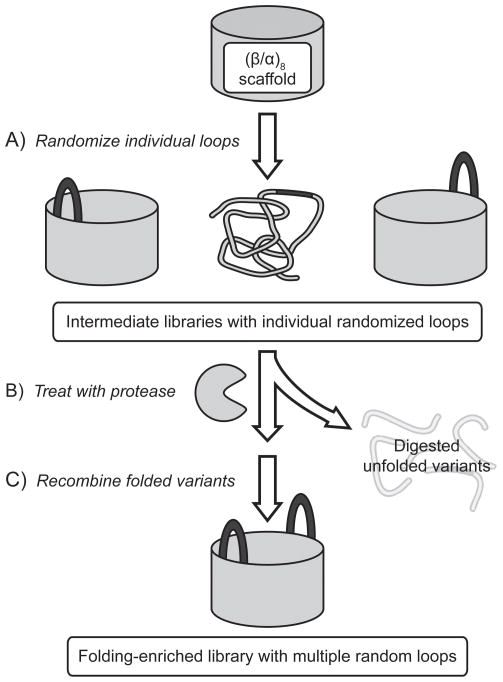
Herein we report the construction of a highly diverse (β/α)8 barrel library (~1014 unique DNA sequences) that contains seven randomized loops and is enriched for well-folded, soluble proteins. Unfortunately, the deleterious effect of mutations on stability is a major constraint in protein evolvability[11] and is implicated in limiting the speed of evolution in nature.[12] Previous studies predicted that the probability of a protein to retain its structure will decline exponentially with the number of mutations.[13] An additional concern during the creation of a highly diverse protein library is the unavoidable occurrence of frameshifts and unintended stop codons caused by imperfect chemical synthesis of the respective DNA library, which can greatly reduce the number of full-length library members.[14] To generate a high quality library, we employed two complementary strategies. The first strategy removed stop codons and frameshifts from shorter library cassettes via in vitro selection by mRNA display.[14] The second strategy selected for folded protein variants using protease digestion, which removed poorly folded proteins as they are more susceptible to proteolysis.[15] We combined these two strategies by assembling our final library in vitro and step-wise from intermediate libraries preselected for folded variants and the absence of frameshifts or premature stop-codons (Figure 1). While the selection procedures reduce the number of protein variants in the intermediate libraries, the diversity is regenerated in the final library by recombining these preselected intermediate libraries. Unlike the prior (β/α)8 library construction attempt where 49 amino acids were simultaneously inserted into all eight loops in the catalytic face of the (β/α)8 fold and likely caused unfolding of the substantial fraction of the final library,[14, 16] our conservative step-wise assembly approach aimed to significantly improve the overall library quality. In order to assess the impact of our folding selection, we additionally prepared a control library without the folding selection. The quality of the two libraries was assessed independently by orthogonal in vitro and in vivo folding assays. These libraries will be used for isolating de novo activities as well as for studying the origins of enzymatic function, the role of folding on the emergence of activity, and the adaptability of the omnipresent TIM barrel fold for different catalytic functions.
Figure 1.
General strategy for the stepwise construction of the folding-enriched library based on the (β/α)8 scaffold. A selection for folded proteins by protease digestion of unfolded variants is followed by recombination of folded variants to generate the final (β/α)8 library with seven randomized loops.
Results
Identification and characterization of a (β/α)8 scaffold protein and an unfolded control
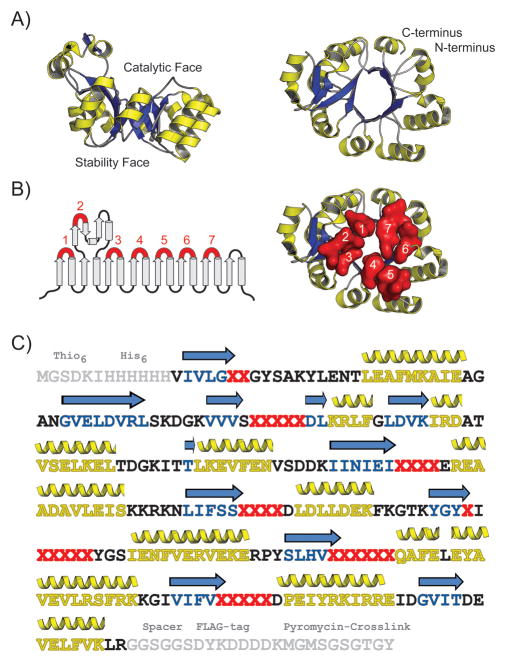
We first sought to identify a suitable (β/α)8 scaffold candidate as a starting point for the library design. We desired a highly stable, cysteine-free, monomeric protein with a known crystal structure and chose glycerophosphodiester phosphodiesterase (GDPD) from the hyperthermophile T. maritima as the starting scaffold that fits all those criteria (Figure 2). [17] We hypothesized that the overall structure of the GDPD protein would be sufficiently stable to tolerate the replacement of loops on the catalytic face of the barrel with random sequences and even the insertion of additional amino acids. The GDPD catalytic face consists mainly of short loops and could potentially accommodate larger active sites with minimal steric clashes, similar to recent experiments that changed TIM barrel activities.[7c] To optimize our protocols for folding assessment and selection that are essential to our library assembly strategy, we prepared a destabilized GDPD construct (GDPDmut) lacking the parental tertiary structure. In particular, two adjacent substitutions (G31R/V32E) were introduced to the (β/α)8 barrel to disrupt the parent GDPD structure (GDPDwt) via steric clashing and the insertion of unfavorable charge in the tightly packed core of the barrel.
Figure 2.
Design of the (β/α)8 library based on the GDPD protein scaffold. A) Side view and top down view of crystal structure of the GDPD (β/α)8 scaffold that was used as a starting point for the library construction (PDB ID: 1O1Z). The α-helices and β-strands are shown in yellow and blue, respectively. B) Secondary structure representation and top down view of GDPD scaffold. The loops 1–7 that were randomized during library construction are numbered and shown in red. C) Sequence of the GDPD library. Positions randomized with the NNG/C codon are depicted as red “X”. Non-native residues added to the termini of the GDPD scaffold are shown in grey (purification tags, spacers and puromycin-crosslinking region needed for mRNA display). β-strands and α-helices are colored blue and yellow, respectively.
To ascertain that the mutant construct lacks the parent (β/α)8 structure, GDPDwt and GDPDmut were expressed, purified via His6-tag chromatography and characterized in solution. Unlike GDPDwt, GDPDmut did not express solubly but could subsequently be solubilized through purification under denaturing conditions followed by a refolding step. In contrast to the monomeric GDPDwt, GDPDmut exists almost exclusively as oligomeric species in solution, as shown by size exclusion chromatography (Figure S1A). Analysis of secondary structure by far-UV circular dichroism (CD) demonstrated that both constructs possess defined, yet differing, elements of secondary structure based on the similarities at 208 nm and differences at 222 nm, wavelengths associated with α-helical structure in the far-UV CD (Figure S1B). In order to gain greater insight into the overall folding of the two GDPD constructs, we probed the tertiary structure via 1-anilinonaphthalene-8-sulfonic acid (ANS) fluorescence and near-UV CD (Figure S1C and S1D). Both methods showed that GDPDmut has substantially less tertiary structure and more exposed hydrophobic surface area relative to GDPDwt. After establishing that GDPDmut lacks the tertiary and quaternary structure of the parent GDPD scaffold, the two constructs were used to establish and optimize the dynamic range of the protease digestion folding selection.
Optimization of the folding selection by in vitro protease digestion
In order to employ the protease digestion selection to reduce the fraction of poorly folded protein variants in our (β/α)8 library, we first optimized the selection conditions to successfully discriminate between GDPDwt and GDPDmut. Selections based on protease digestion using phage and ribosome display have successfully enriched protein libraries for folded members.[15a, 15c] Although primarily used to improve the stability of a single protein, in one case this approach was applied to improve qualities of de novo libraries based on specific secondary modules.[15b] Throughout our assembly protocol we utilized mRNA display, an in vitro selection and evolution method which employs the small molecule puromycin to covalently attach proteins to their own mRNA.[18] This method had been previously used to isolate an enzyme de novo from a non-catalytic scaffold with two randomized loops[1b, 1c, 19] and is excellently suited for the long term goal of isolating enzymatic activities from the large protein libraries described here.
In pilot experiments, mRNA-displayed proteins were first treated with several proteases (data not shown) known to have preferences for hydrophobic residues, and then His6-tag purified by immobilized metal affinity chromatography (Figure S2). We hypothesized that hydrophobic residues would serve as a good criterion for removing unfolded proteins from the library as such residues are preferentially buried in the protein core and less likely to be surface-exposed in well folded proteins.[20] Chymotrypsin, which cleaves adjacent to large hydrophobic residues, showed the largest discrimination between the two control constructs in the pilot experiments; the method was further optimized to yield ~140-fold enrichment of GDPDwt over GDPDmut (92 ± 1.2% vs. 0.67 ± 0.12% survival). Percent survival was defined as the ratio of His6-tag purification yields of protease-treated and untreated samples. Furthermore, mRNA-displayed fusions of GDPDmut and a GDPDmut control lacking the His6-tag were analyzed via the same protocol to determine the level of non-specific background binding. The optimized chymotrypsin protocol was utilized for the selection and analysis of the (β/α)8 based libraries.
Construction of intermediate libraries with randomized loops
Intermediate libraries with several randomized loops were used as building blocks during the step-wise assembly of the final folding-enriched library (Figure S3). To further increase the diversity of the libraries, we also inserted one to four additional amino acids into these loops, with the exception of loop 1 (Table 1). We generated seven libraries with a single randomized loop each, corresponding to loops 1 through 7 on the catalytic face of the scaffold. In the next step, these libraries were used to assemble intermediate libraries with multiple randomized loops (Figure S3A). Specifically, fragments of the GDPD gene were PCR amplified to introduce two to six NNS (S=G/C) randomized codons at the desired loop positions, the resulting fragments were digested with restriction enzyme and ligated together to generate libraries encoding full-length proteins that contain one or two random loops. Next, half-libraries with three or four random loops were generated by recombining PCR-amplified fragments of the libraries with one or two random loops. Loop 8 was omitted from the library assembly as its location is distant from the core of the (β/α)8 barrel and, therefore, loop 8 seemed unlikely to contribute to the formation of a potential active site with the rest of the randomized regions.
Table 1.
Comparison of loop length in the GDPDwt scaffold to the randomized loops used in assembling the (β/α)8 libraries
| Loop # | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| GDPDwt loop size | 2aa | 3aa | 1aa | 1aa | 5aa | 2aa | 4aa | 1aa |
| Library loop size | 2aa | 5aa | 4aa | 4aa | 6aa | 6aa | 5aa | Wild type |
The introduction of multiple loops into the GDPD protein was expected to substantially destabilize the starting scaffold and reduce the fraction of folded proteins in a given library. To guide the library assembly process and decide at which step to perform either the whole folding selection or the mRNA display alone, we first analyzed the protease digestion rates of several intermediate libraries as described in the next section. The mRNA display procedure removes unintended stop codons from the library, which are introduced by the use of NNS codons for randomization, and imperfections during DNA primer synthesis.[14] The mRNA display therefore increases the quality of a library, which is beneficial for a subsequent folding selection.
Folding selections of the intermediate libraries by in vitro protease digestion
To evaluate the tolerance of the GDPD scaffold to amino acid insertion and randomization, several libraries containing one or two randomized loops were treated with chymotrypsin to assess the fraction of surviving library members (Table 2). As expected, and likely due to steric clashes between random loops from different libraries, the survival rate for libraries with two randomized loops was lower than the product of the survival rates of the two parent libraries with a single randomized loop each. The survival rates observed for the libraries containing one or two randomized loops were significantly above the GDPDmut background (Table 2). Therefore, to preserve some spatial context of the randomized loops, we subjected those libraries only to mRNA display to remove stop codons, and then recombined them into the two half libraries, termed “L1-4” and “L5-7”, containing randomized loops 1–4 and 5–7, respectively. Our goal was to enrich these two libraries for folded proteins until the survival rate was well above that of GDPDmut in as few rounds of selection as possible to preserve library diversity. These libraries, possessing four and three randomized loops, respectively, were therefore subjected to the folding selection (Figure S3A). While L5-7 exhibited 52% survival rate, L1-4 showed a significantly lower 1.4% and the surviving variants were subjected to a second round of folding selection yielding a final survival rate of 9.2%. The increase in survival rates well above background implies that both half libraries were indeed enriched for folded sequences. Additional rounds of folding selection would decrease the diversity of enriched sequences without necessarily improving folding much further (Table 2).
Table 2.
Results of the folding selection by in vitro protease digestion
| Digested species | % Survival[a] | |
|---|---|---|
| Control constructs | GDPDwt | 92 ± 1.2 |
| GDPDmut | 0.67 ± 0.12 | |
| GDPDmut (-His6)[b] | 0.4 | |
| L3 (-His6)[b] | 0.3 | |
| Analytical selections[c] | L3 | 28 |
| L4 | 78 | |
| L5 | 80 | |
| L3-4 | 10 | |
| Preparative selections[d] | L1-4 (1st round) | 1.4 |
| L1-4 (2nd round) | 9.2 | |
| L5-7 (1st round) | 52 | |
| Final libraries | Folding-enriched | 6.6 ± 1.1 |
| Control | 2.2 ± 0.3[e] | |
% survival is defined as fraction of mRNA-displayed species that are not digested during the chymotrypsin treatment and is calculated as the ratio (Ni-NTA purification yield of chymotrypsin treated species)/(Ni-NTA purification yield of undigested species).
Constructs lacking the His6-tag needed for Ni-NTA purification.
Small scale selections to assess tolerance of GDPDwt to the insertion of one or two loops to guide the library assembly process.
Preparative selections performed to generate intermediate libraries used for the assembly of the final folding-enriched library.
The % survival for the control library (loops 1–7 randomized) is higher than for library L1-4. This result is counter-intuitive and likely due to an artifact in the protease assay, potentially caused by unfolded proteins that escaped the protease digestion by aggregating (false-positives).
Assembly of the final folding-enriched library
The stop-codon free, folding-enriched variants from the libraries L1-4 and L5-7 that survived the protease digestion selection (~109 and 1010 sequences respectively) were used to assemble the final folding-enriched library with a total of 32 randomized amino acid positions. Although combining these intermediate libraries could theoretically produce ~1019 unique sequences, the physical amount is limited to sub-milligram quantities of DNA that can be synthesized in the lab. Our final library contains 1.6 × 1014 unique DNA sequences and is at the upper limit of library sizes compatible with in vitro selection methods such as mRNA and ribosome display.
Analysis of stability of folding-enriched library and comparison to control library using the protease assay (in vitro)
In order to assess the benefits of the folding selection, a control library was prepared from the same seven single loop libraries used during the construction of the folding-enriched library (Figure S3B). The resulting library shared the same randomized elements and a comparable 2.9 × 1014 complexity as the folding-enriched library, but had not been pre-selected to maintain the parent (β/α)8 fold. A single round of mRNA display was employed to remove the stop codons and frameshifts immediately prior to the final recombination step. Rather than using the full length GDPD gene, only half-gene fragments of the L1-4 and L5-7 libraries were subjected to the round of mRNA display. By using only these fragments instead of the whole parent scaffold, we aimed to avoid a bias of the randomized loops towards the folded parent structure and allow maximum diversification. To assess the impact of the folding selection by protease digestion, we directly compared a small fraction (~1010 sequences) of the control and folding-enriched libraries via our protease protocol. The folding-enriched library had a 6.6% survival rate, which is threefold higher than the control library assembled from the L1-4 and L5-7 fragments that had not been selected for folding (Table 2).
Assessment of folding of the final libraries via GFP-fused reporter assay (in vivo)
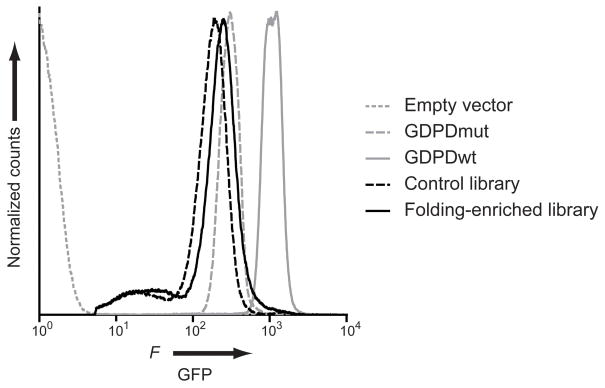
In order to confirm the efficacy of the protease digestion folding selection with an independent method, we analyzed a fraction of our libraries using a GFP-fused folding reporter system. In this system, the proteins are expressed as N-terminal fusions of GFP. The GFP fluorescence of the protein-GFP constructs is dependent on the soluble expression of the folded cargo protein and correlates with the stability to intracellular degradation.[21] This approach had been employed to enrich smaller protein libraries (up to 108) for folded variants in vivo and thus is an alternative to our in vitro folding selection.[21a] We first analyzed the several intermediate libraries that were used to construct the control library and compared them to the GDPDwt and GDPDmut controls (Figure S4). These intermediate libraries contained one to four randomized loops and had not been selected for folding. Since GDPDwt was shown to be solubly expressed and well behaved in solution, we were interested in the fraction of our libraries that exhibited fluorescence similar to GDPDwt-GFP fusions. In addition, we determined the mode of the GFP fluorescence as a qualitative metric for general library trends since GFP fluorescence correlates with intracellular stability. Flow cytometric analysis of E. coli BL21(DE3) cells expressing GDPD-GFP constructs showed a near base-line separation between GDPDwt and GDPDmut, both of which exhibit significantly higher fluorescence than cells transformed with an empty vector control plasmid. Analysis of the non-preselected libraries showed that libraries with randomized loops in the N-terminal half of the (β/α)8 barrel (libraries L1-2, L3-4 and L1-4) exhibit a lower GFP fluorescence and a lower GDPDwt-like fraction compared to the libraries with randomized loops in the C-terminal half of the (β/α)8 barrel (libraries L5, L6-7, L5-7) (Figure S4, Table S1). The folding-enriched and control libraries were analyzed in the E. coli BL21(DE3) Rosetta strain that provides enhanced eukaryotic protein expression since the assembly process potentially enriched for eukaryotic codons that are suboptimal for bacterial expression. We observed improvements in the folding-enriched library relative to the control library in both the mode of GFP fluorescence (the most frequently found fluorescence value) and the fraction of GDPDwt-like variants (Figure 3, Table 3).
Figure 3.
Assessment of folding by GFP-fusion assay. Fluorescence histograms of E. coli BL21(DE3) Rosetta cells containing library members or control proteins fused to GFP are shown. Empty vector (gray, dotted), control library (black, dashed), folding-enriched library (black, solid), GDPDmut (gray, dashed) and GDPDwt (gray, solid). The empty vector population was gated out on the histograms of cells transformed with the GDPD constructs.
Table 3.
GFP-fused in vivo folding assessment of the final (β/α)8 fold-based libraries.[a]
| Species | Mode of GFP fluorescence[b] | % cells with GDPDwt-GFP fluorescence[c] |
|---|---|---|
| GDPDmut | 24.6 | 0.01 |
| Control library | 15.4 | 1.4 |
| Folding-enriched library | 19.8 | 5.4 |
| GDPDwt | 100 | 98.5 |
Constructs were transformed into E. coli BL21(DE3) Rosetta cells. Prior to analysis, data were gated to exclude cell populations that matched fluorescence and scatter profiles of cells transformed with empty vector control plasmid.
Values normalized to the mode of GFP fluorescence of GDPDwt-GFP.
Wild type cells were gated on the forward scatter versus GFP contour plot to include ~98% of all wild type cells.
Isolation of well-folded members of the final libraries by cell sorting
To confirm that the soluble expression of GFP fusions is indeed closely correlated with the GDPDwt-like GFP fluorescence, control and folding-enriched libraries were sorted via fluorescence-activated cell sorting (FACS) (Figure S5). We subdivided the GDPDwt-like GFP fluorescence window into a low and a high GFP signal during sorting as the control library exhibited a discrete peak at high signal within this region (see gate H in Figure S5B). Cells with such high GFP profile could be false positives due to either insoluble aggregates or truncated proteins as noted in previous reports that used the GFP reporter system.[21a]
Analysis of soluble library-GFP fusions by Western blotting and SDS-PAGE
The four sorted populations (low and high GFP signal of each of the control and folding-enriched libraries in Figure S5) were re-grown in liquid culture under sorting conditions and the respective amount of soluble full-length library-GFP fusion proteins was compared by anti-GFP western blotting (Figure S6). A fraction of these cultures was also plated to isolate individual GFP-positive clones, express them, and analyze the soluble protein fraction of each clone by SDS-PAGE gel (data not shown). The SDS-PAGE and western blot results showed similar trends and were in good agreement with each other (Table S2). The folding-enriched library populations contained a higher fraction of soluble GFP fusions in both the low and high populations compared to the control library populations. Western blot analysis also showed that the high GFP populations for both libraries contained at least ~50% false positive clones that expressed GFP alone. Based on the fraction of full length, soluble library-GFP fusions in the FACS-sorted populations, we calculated that the soluble library members comprise between 1% and 1.2% of the folding enriched library and between 0.02% and 0.033% of the control library. This corresponds to an overall 35 to 50-fold improvement in the library quality based on the fraction of soluble, monomeric and folded sequences. Therefore, the final folding-enriched (β/α)8 fold library contains about 1012 soluble protein variants (Table S2).
Biophysical characterization of soluble library clones
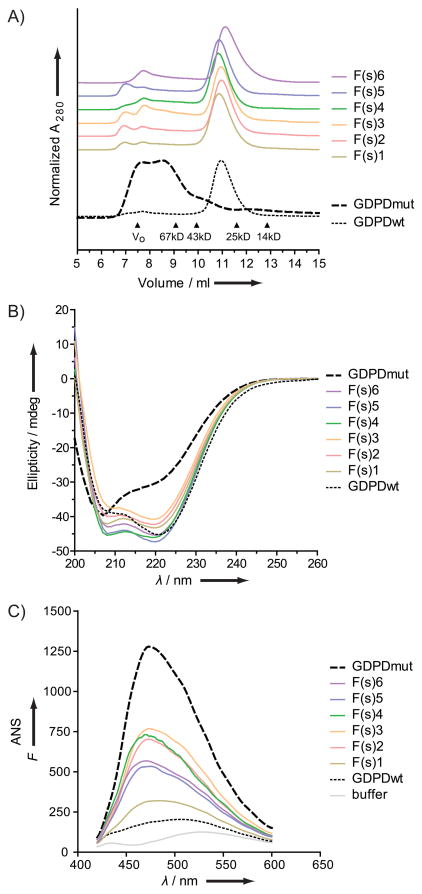
We sought to further investigate and compare the solubility of protein variants from the control and folding-enriched libraries selected at random, as well as the folding-enriched variants isolated by FACS (above). All constructs were cloned into a protein expression plasmid to express the FACS-sorted library-GFP constructs without the GFP. Only sequences from the FACS-sorted folding-enriched library produced soluble proteins (data not shown), six of which were purified for further characterization. Similar to the initial GDPDwt and GDPDmut characterization, we performed size exclusion chromatography and measured the near-UV CD and ANS fluorescence to investigate the quaternary, secondary and tertiary structure of these library variants (Figure 4). All of those proteins were monomeric in solution, maintained CD signatures similar to GDPDwt and ANS profiles intermediate between GDPDmut and GDPDwt.
Figure 4.
Biophysical characterization of six soluble folding-enriched library clones from the FACS-sorted high GFP population. GDPDwt and GDPDmut data included for reference as dotted and dashed lines, respectively. A) Size exclusion chromatography (quaternary structure). B) Far-UV circular dichroism spectroscopy (secondary structure). C) 1-Anilinonaphthalene-8-sulfonic acid (ANS) fluorescence measurements (tertiary structure).
Sequence analysis of library clones
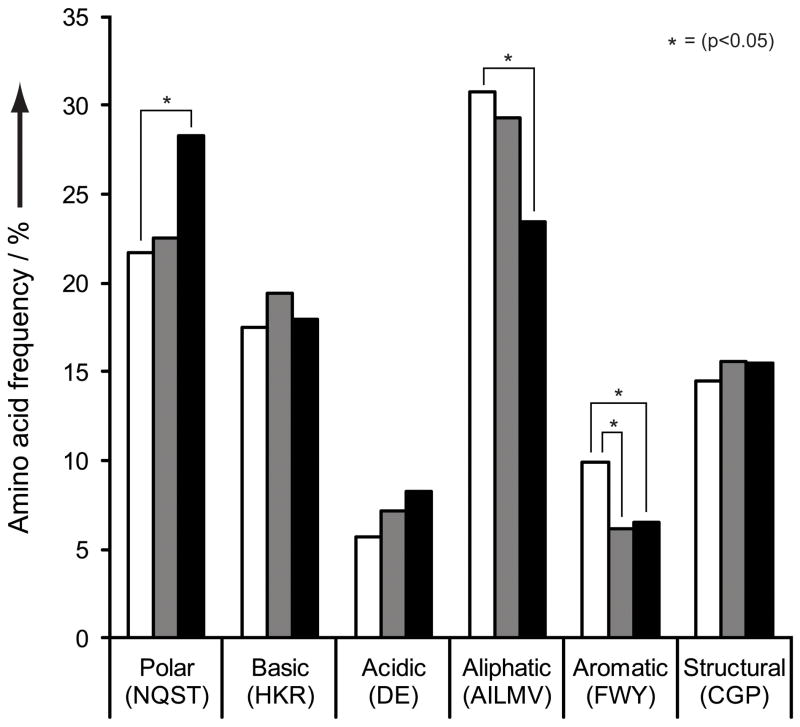
To better understand the underlying changes that occurred upon our selection for folding, we sequenced randomly chosen individual clones from the control and folding-enriched libraries, as well as the soluble folding-enriched library clones acquired by FACS sorting of the GFP-fused library. We analyzed the amino acid distribution of the 1,393 sequenced NNS codons and did not observe any stop codon, confirming that they were removed during the mRNA display step (Table 4). We further grouped the sequenced codons into classes of amino acids based on their properties and then compared the distributions of these classes for the control library (randomly chosen clones) and the folding-enriched library (randomly chosen clones, soluble clones) (Figure 5). To evaluate whether the detected distribution changes were statistically significant (p < 0.05), we performed pairwise t-test comparisons of the grouped codons from the folding-enriched library sequences (random and soluble clones) against the control library sequences (random clones). We observed a significant decrease in aromatic residues in the folding-enriched library relative to the control library. The soluble library clones from the folding-enriched library, isolated during FACS sorting experiment, exhibit the same decrease in aromatic residues, and, in addition, show an increase in polar residues at the expense of aliphatic residues.
Table 4.
Amino acid (aa) distribution for NNS codons, shown in %
| Amino acid | NNS (-stop)[a] | Control library[b] | Folding-enriched library[b] | ||
|---|---|---|---|---|---|
|
| |||||
| Random clones | Random clones | Soluble clones | |||
| Polar | Asn | 3.2 | 3.8 | 5.3 | 3.4 |
| Gln | 3.2 | 3.2 | 3.8 | 2.8 | |
| Ser | 9.7 | 7.0 | 8.4 | 15.5 | |
| Thr | 6.5 | 7.0 | 5.1 | 6.6 | |
| Basic | Arg | 9.7 | 10.2 | 10.1 | 11.0 |
| His | 3.2 | 3.2 | 4.2 | 3.1 | |
| Lys | 3.2 | 5.1 | 5.1 | 3.8 | |
| Acidic | Asp | 3.2 | 4.0 | 4.4 | 4.5 |
| Glu | 3.2 | 2.2 | 2.7 | 3.8 | |
| Aliphatic | Ala | 6.5 | 6.1 | 6.9 | 6.6 |
| Ile | 3.2 | 2.9 | 2.7 | 3.1 | |
| Leu | 9.7 | 9.7 | 11.2 | 8.3 | |
| Met | 3.2 | 4.3 | 2.9 | 2.4 | |
| Val | 6.5 | 6.7 | 5.5 | 3.1 | |
| Aromatic | Phe | 3.2 | 4.1 | 1.9 | 3.1 |
| Trp | 3.2 | 3.0 | 2.9 | 1.4 | |
| Tyr | 3.2 | 2.9 | 1.9 | 2.1 | |
| Structural | Cys | 3.2 | 1.3 | 1.3 | 1.4 |
| Gly | 6.5 | 5.4 | 6.3 | 8.3 | |
| Pro | 6.5 | 8.0 | 8.0 | 5.9 | |
| stop | 0 | Not observed | Not observed | Not observed | |
| Codons sequenced | 628 | 475[c] | 290[c] | ||
Theoretical aa distribution for NNS(-stop) was calculated from the expected NNS distribution lacking a stop codon.
Experimentally observed values from sequencing analysis of individual library clones.
Loops containing wild type sequences were omitted from analysis.
Figure 5.
Amino acid composition of randomized loop regions. Amino acids are grouped according to their chemical properties and the compositions were calculated from sequencing data. Control library, randomly picked clones (white); folding-enriched library, randomly picked clones (gray); folding-enriched library, soluble clones (black). Statistically significant differences, as determined by pairwise t-test, are indicated by a star.
Discussion
The objective of this study was to generate and characterize a high quality protein library based on the (β/α)8 fold by combining a stepwise assembly with an in vitro folding selection. We further sought to evaluate the efficacy of such an approach by comparing a representative fraction of members of the libraries using two orthogonal methods for the assessment of folding.
Our in vitro and in vivo folding assessment methods provided different metrics to measure folding stability, which are survival rates during protease digestion, the mode of fluorescence of GFP-fused library members, and the fraction of library members that behave like GDPDwt in the GFP assay. All three metrics displayed similar trends for the intermediate libraries, and showed a substantial improvement in the quality of the folding-enriched library compared to the control library, demonstrating the success of our folding selection. While those metrics were useful to characterize the libraries in bulk and assess the library construction process, they were only indirect measures for determining how much the library was enriched for soluble, well-folded protein variants that behaved like the starting (β/α)8 scaffold. To quantify directly the fraction of those desired library variants, we cloned and expressed 20 randomly chosen proteins from both libraries in E. coli. We did not obtain any soluble proteins from this small sample size, indicating that the fraction of soluble variants in each library was below 5%. We therefore sorted a fraction of the GFP-fused libraries via fluorescence-activated cell sorting (FACS) and were able to isolate library members that readily expressed in bacteria, are monomeric, and exhibit behavior similar to the GDPDwt scaffold in solution. Sequencing results suggested that the improved solubility correlates, as expected, with the increased presence of polar amino acids at the expense of aliphatic residues. Furthermore, the occurrence of aromatic amino acids was reduced in the folding-enriched library compared to the control library, which might in part be a result of the selection process (disfavoring those residues because of chymotrypsin’s preference to cleave next to aromatic amino acids). Based on the number of soluble, GDPDwt-like clones we obtained from the sorting experiment and the biochemical characterization of individual clones, we calculated that soluble, monomeric and folded sequences comprise about 1% of the folding-enriched library (~1012 variants), an increase over the control library of up to 50-fold.
The in vitro and in vivo folding methods employed in our work required the fusion of the (β/α)8 library proteins to either their own mRNA or a GFP reporter protein, which could, in principle, alter stability or solubility of the proteins. To minimize this potential issue during the in vitro protease digestion, the mRNA was reverse-transcribed to generate the linear mRNA-cDNA hybrid thereby preventing the mRNA from folding and affecting the digestion by obscuring protease sites. Furthermore, we assessed whether the fusion to GFP affected solubility of the library proteins by expressing soluble library-GFP constructs without the GFP fusion and analyzing them by SDS-PAGE – all proteins remained soluble in solution. Notably, during the FACS sorting experiment we encountered a substantial number of false-positive highly fluorescent cells resulting from clones that had lost their GDPD library cargo, leading to the expression of GFP alone. It has been proposed in earlier work that such false positives result from either truncated or highly aggregated and insoluble species.[21a] We were able to exclude these false-positives by analyzing the soluble fraction of the expressed proteins by gel electrophoresis. The folding selection by protease digestion likely also allowed for some false-positive protein variants to become selected. For example, we could envision certain unfolded proteins escaping the protease digest through aggregation as those proteins would be inaccessible to the protease enzyme. We counter-acted this possibility by including detergents and denaturants (Triton X-100 and SDS) in our buffers. Yet, as we cannot rule out a remaining selection bias of this kind, we deliberately chose not to further enrich the intermediate libraries L1-4 and L5-7 beyond the initial one or two rounds of selection. Finally, the biophysical characterization of individual soluble library members confirmed that our protease selection protocol successfully enriched for folded variants with a structure similar to the parental (β/α)8 scaffold.
The final folding-enriched library contains up to 32 randomized amino acid positions distributed over 7 loops. The soluble library variants isolated by FACS exhibited some variability in the location and number of loops that were randomized. Interestingly, randomization in loops 2 and 3 was disfavored in the folding-enriched library as we frequently recovered the parent GDPDwt sequence in these loops (~80% and ~40% parent sequence in randomly picked clones, respectively). All soluble clones isolated in the FACS experiments showed the parent sequence in loops 2 and 3, while containing other randomized loops. In addition, libraries that contained randomized loops 2 and/or 3 also exhibited lower protease survival rates and lower GFP-fluorescence, which is further evidence that their randomization is detrimental to the stability of the (β/α)8 barrel. We suspect that the wild type loops observed in the final library arose during the step-wise library assembly. While initially present only at very low levels in the intermediate libraries, these variants were enriched during the folding selections. However, the ~1012 soluble members of the folding-enriched library have at least 3 randomized loops and at least 13 randomized amino acids. For comparison, a recent study described the switch of one (β/α)8 scaffold enzyme to an unrelated (β/α)8 activity via a single loop insertion. [7c] If a new enzymatic activity can be found with the exchange of a single loop as those results suggest, our library of soluble proteins with three and more randomized loops likely has an even greater potential to contain different enzymatic activities. In addition, some of the less soluble library members may also be exploitable by in vitro selection methods as, for example, the mRNA display has been shown to help keep poorly soluble proteins in solution through the attachment of a large highly-soluble RNA molecule. However, the solubility of such proteins would subsequently need to be improved through directed evolution, in contrast to the ~1012 already soluble library members. In summary, we demonstrated that those soluble clones have retained most of the overall structural features of the parent (β/α)8 fold despite the introduction of multiple randomized stretches of amino acids. To the best of our knowledge, this is the first report of a high quality library based on the (β/α)8 enzyme fold with such a high complexity.
Our work also allowed us to make several observations regarding the behavior of the GDPDwt (β/α)8 fold, the role of randomized loop positions and the impact of combining individual loop libraries. We observed that single, entirely randomized loop insertions into the GDPDwt resulted in libraries with 30–80% survival in the protease digestion folding selection. Interestingly, prior in vivo work demonstrated that single known loops inserted into an unrelated (β/α)8 barrel resulted in similar tolerances with regards to folding. [7b] The authors suggested that it was the site of insertion and not the inserted sequence that had the greatest influence on the stability of the resulting protein chimera. The results we present here strongly support this notion and suggest that other (β/α)8 barrels may exhibit similar tolerances to single loop insertions, regardless of whether the loop sequence had been favored previously in nature or is entirely random. In fact, previous work suggests that random regions are beneficial in adapting known loops to the context of a new (β/α)8 barrel structure. [7a] When we combined two libraries with different folding stabilities, the resulting library displayed a lower folding stability than the less stable input library, as evidenced in both the protease digestion and the GFP-fusion assay for multiple libraries. We observed a general trend where the N-terminal half of the barrel appears more vital for folding stability than the C-terminal half. This finding was inferred from the low GFP fluorescence, the high protease digestion rates, and the sequencing results for libraries containing randomized N-terminal loops. Similar positional preferences were observed in previous experiments on another (β/α)8 scaffold.[7b] Although we were initially concerned that the introduction of several randomized loops into the GDPDwt scaffold would drastically unfold the (β/α)8 fold, by all our metrics, the data indicate that this scaffold is tolerant to multiple loop insertions, particularly in the C-terminal half of the barrel. In summary, our results support the hypothesis that the core of a hyperthermophile (β/α)8 barrel fold provides sufficient stability to offset the effects of destabilizing loops of the catalytic face, and render the (β/α)8 fold an attractive scaffold in enzyme engineering by loop insertion.
Conclusion
The high quality and complexity of the libraries reported here are expected to provide an invaluable starting point for the engineering of novel enzymes and the understanding of the origins of enzymatic function in the (β/α)8 fold. By introducing randomized elements onto a stable scaffold in step-wise fashion and enriching for folded variants, we have increased the probability of finding novel enzymes with diverse activities. These initial, potentially low enzymatic activities will subsequently be evolved further under appropriate selection conditions to give rise to more efficient specialist enzymes.[16, 22] Many (β/α)8 enzymes act on substrates with a phosphate group and some soluble variants of the folding-enriched library have retained the residues that compose the native phosphate binding site. This site can be used as a handle to improve substrate binding or to study the role of such handles in the evolution of enzymes. Furthermore, isolating novel activities from these libraries that are unrelated to the original GDPD function will help to elucidate whether the (β/α)8 barrel fold is predestined for certain activities, how it can be adapted to perform new functions, and what impact a library preselected for folding may have on isolation of enzymatic activity. Finally, an estimated 1% of our folding-enriched library contains sequences that are solubly expressed in E. coli while showing substantial diversity in the number and positioning of randomized loops. Our libraries are thus compatible with in vitro and in vivo evolution methods. Work is underway to interrogate the libraries for de novo enzymes using mRNA display and to study the (β/α)8 fold adaptability through bacterial selections.
Experimental Section
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise stated. All restriction enzymes, Calf Intestinal Alkaline Phosphatase, T7 RNA Polymerase, T4 DNA ligase and Phusion High Fidelity DNA polymerase were purchased from New England Biolabs (Ipswich, MA). All PCR reactions were performed with Phusion High Fidelity DNA polymerase. If available, high fidelity versions of the restrictions enzymes were employed. Gel extraction, PCR clean up and DNA mini-prep kits were purchased from Qiagen (Valencia, CA). Sequencing reactions were performed either by ACGT, Inc. (Wheeling, IL) or University of Minnesota BioMedical Genomics Center (St. Paul, MN).
Cloning and expression of GDPDwt and GDPDmut constructs
The synthetic gene encoding GDPD flanked by purification tags, optimized for dual expression in rabbit reticulocyte and E. coli, was purchased from GenScript (Piscataway, NJ). Specifically, the gene coded for Thio6His6 tag–GDPDwt–(GGS)2 spacer–FLAG epitope–pyromycin crosslinking region. This construct was PCR amplified and cloned into pET28a vector (Novagen). GDPDmut was generated using standard mutagenesis protocols using pET28/GDPDwt as template. For protein expression, plasmids were transformed into BL21(DE3) Rosetta E. coli strains (Novagen) and grown on LB media in presence of kanamycin (34 mg/l) and chloramphenicol (34 mg/l). Overnight cultures were diluted 1:1,000 into fresh LB media and grown to OD600 = 1 prior to induction with IPTG (1 mM). Cells were grown an additional 4 hours at 37 °C prior to harvesting and storage at −20 °C. Frozen cell pellets were resuspended in lysis buffer (50 mM Tris-HCl pH 8.0, 50 mM NaCl) and lysed using an S-450D Digital Sonifier (Branson). Cell debris was removed by centrifugation and the His-tagged proteins were purified by affinity chromatography using Ni-NTA Superflow resin (Qiagen) under native conditions for GDPDwt and buffers containing denaturant (guanidinium chloride 6 M)s for GDPDmut according to the manufacturer recommendation. Elution fractions containing GDPDmut were dialyzed to remove denaturants by first diluting 1:4 in dialysis buffer (50 mM Tris-HCl, 100 mM NaCl, pH 7.5) then dialyzing overnight in 7 kDa MWCO Snake Skin Dialysis Tubing (Pierce) in dialysis buffer. The protein purification was evaluated by SDS-PAGE on precast 4–12% gradient gels (Invitrogen).
Circular Dichroism (CD) spectroscopy
All CD experiments were performed on a Jasco J-815 spectropolarimeter. For far-UV experiments, ellipticity of protein samples (20 μM protein in 10 mM Tris-HCl, 20 mM NaCl, pH 7.5) was measured from 190 to 260 nm at 50 nm/min using a quartz cuvette with a 1 mm path length. Each spectrum represents the average of 10 accumulations. For near-UV experiments, ellipticity was measured the same as far-UV except a quartz cuvette with a 10 mm path length was used and wavelengths ranged from 260 to 350 nm.
1-Anilinonaphthalene-8-sulfonic acid (ANS) fluorescence measurements
ANS is an environmentally sensitive dye which exhibits increased fluorescence upon interaction with hydrophobic protein surfaces and has been previously used to indirectly report on protein tertiary structure.[23] Measurements were performed on either the SpectraMax M2 or M5 plate readers (Molecular Devices) in black flat-bottom 96-well NUNC Maxisorp® plates. Samples containing protein (5 μM) and ANS (1 mM) in dialysis buffer (50 mM Tris-HCl, 100 mM NaCl, pH 7.5), were excited at 403 nm, monitoring emission at 430–600 nm in 1 nm intervals. Data was smoothed with Kaleidograph software.
Size exclusion chromatography
Ni-NTA purified protein samples were loaded onto a 10 mm × 300 mm column (Tricorn) packed with Superdex 75 resin (GE Healthcare) and analyzed on the AKTA FPLC system (GE Healthcare) in dialysis buffer (50 mM Tris-HCl, 100 mM NaCl, pH 7.5). Column was calibrated using Amersham low molecular weight calibration kit (GE Healthcare).
Library assembly
All loop libraries were assembled via a three step process of PCR amplification, restriction digest and ligation (Figure S3). All PCR reactions employed a constant primer at the 5′ and 3′ termini and internal primers containing a restriction site (Tables S3 and S4). Loop randomization and insertion was carried out at the single or double loop library level by amplifying two fragments of GDPDwt from the pET28/GDPDwt template and introducing the randomized NNS codons via one of the primers. Assembly of half libraries and final libraries was performed using internal primers that did not introduce any randomized nucleotides. Following PCR amplification, DNA was phenol-chloroform extracted and ethanol-precipitated following standard molecular biology protocols.[24] DNA was digested with appropriate restriction enzyme (Table S4) and purified on 2% agarose gel. Purified digested fragments were ligated with T4 DNA ligase at 16 °C overnight. The ligation product was purified on 2% agarose gel and PCR amplified with external primers to generate ~10 copies of the full length template to be used for the next set of library construction. During the construction of folding-enriched library, the L1-2, L3-4, L5 and L6-7 libraries were subjected to a single round of mRNA display (below) to remove stop codons and frameshifts and then recombined to generate L1-4 and L5-7 libraries. These libraries were subjected to protease based selection (below) and then recombined to assemble the final folding-enriched library. During the control library assembly, half gene fragments of the L1-4 and L5-7 libraries were mRNA-displayed to minimize artifacts related to folding. In the final assembly step for both the control and folding-enriched libraries, ~109–1010 L1-4 and L5-7 DNA sequences were amplified on 20 ml scale to generate ~5×1014 starting sequences. Due to increased scale of the BsaI-HF digests and final ligation reaction, DNA purification was performed via 4.5% native PAGE gel, extracted under UV-shadowing and electroeluted on S&S Elutrap (Schleicher & Schuell).
mRNA display
Creation of mRNA displayed fusions was performed similarly to previously published[18b] but with the following alteration. RNA was produced from the DNA library with T7 RNA polymerase (5 nM DNA template, 200 mM HEPES, 35 mM MgCl2, 2 mM spermidine, 5 mM dNTP (each), 0.1 mg/ml BSA, 40 mM DTT, 1 U/ml inorganic pyrophosphatase, 150 U/ml RNaseOUT (Invitrogen), 50 U/ml T7 RNA Polymerase, pH 7.5) and incubated at 37 °C for 3 hours. RNA was precipitated by LiCl (1/3 equivalent of 8M LiCl) at −20 °C for at least 30 min. The RNA pellet was washed with ice cold 70% ethanol and dissolved in water. RNA was photo crosslinked (3 μM RNA, 20 mM HEPES, 100 mM KCl, 1 mM Spermidine, 1 mM EDTA, 7.5 μM oligo, pH 7.5) with a Psoralen-puromycin oligo (5′-X(tagccggtg)AAAAAAAAAAAAAAAZZACCP-3′ X = psoralen C6, lower case letters = 2′-OMe, Z = spacer 9, P = puromycin, stretch of A’s and ACC = DNA) under 365 nm light on ice for 20 min with an efficiency of approximately 50%. Crosslinked RNA was ethanol precipitated and dissolved in water. A 200 μl or 1 ml translation (200 nM crosslinked RNA, 40% nuclease treated rabbit reticulocyte lysate (Promega), 25 μM amino acid mix), 25 nM, >1000 Ci/mmol, 35S-methionine (PerkinElmer) with additional KCl and Mg (OAc)2 to a final concentration of 120 mM and 0.6 mM respectively) was incubated at 30 °C for 10 min followed by high-salt incubation (550 mM KCl, 50 mM MgCl2) for 5 min at RT. The translation mixture was diluted ten-fold into oligo (dT) binding buffer (20 mM Tris-HCl, 10 mM EDTA, 1 M NaCl, 0.2% Triton X-100, pH 8) and incubated with oligo (dT) cellulose (GE Healthcare, 40 mg) with rotation for 15 minutes at 4 °C. The oligo (dT) cellulose was washed on a chromatography column (Bio-Rad) with more oligo (dT) binding buffer, oligo (dT) wash buffer (20 mM Tris-HCl, 0.3 M NaCl, pH 8) and eluted with elution buffer (2 mM Tris-HCl, pH 8). The eluent was spin filtered through a 0.45 μm filter (Millipore) to remove any additional oligo (dT) cellulose and mixed with 10X phosphate buffered saline (PBS) with 0.1% Triton X-100 to a final concentration of 1x. The mixture was added to Anti-Flag M2-Agarose Affinity Gel (25 μl, equilibrated according to the manufacturer’s instructions) and incubated with rotation for at least 1 h at 4 °C. Flag resin was washed on a chromatography column (Bio-Rad) with PBS w/0.01% Triton X-100 followed by Flag wash buffer (50 mM HEPES, 150 mM NaCl, 0.01% Triton X-100, pH 7.4) where the final wash was performed in batch in a microcentrifuge tube. Elution was performed by incubating Flag resin with Flag peptide (56 μM in Flag wash buffer) for 10 min at 4 °C with rotation and filtered through a 0.45 μm filter (Millipore) to remove any additional Flag resin. Eluent was diluted with Flag elution buffer until mRNA displayed fusions reached 3×108 fusions/μl as measured by scintillation counting (LS6500 multipurpose scintillation counter; Beckman). This was followed by reverse transcription with Superscript II (1.5×108 fusions/μl, 50 nM RT-primer (5′-TTTTTTTTTTTTTTTNCCAGATCCAGACATTCCCAT-3′), 50 mM Tris-HCl, 3 mM MgCl2, 10 mM 2-mercaptoethanol, 0.5 mM dCTP, dGTP, dTTP, 5 μM dATP, 100 U/ml RNaseOUT (Invitrogen), 500 U/ml Superscript II (Invitrogen), pH 8.3). A 10 μl sample was removed to serve as a non-radiolabeled control prior to the addition of α-32P-dATP (Perkin Elmer, 3000 Ci/mmol; 16 μM final concentration) to the reverse transcription. Both tubes were incubated at 42 °C for 30 min and the control was stored at −20 °C. The reverse transcription was treated with calf intestinal alkaline phosphatase (30 U/mL) at 37 °C for 10 min. Reverse transcribed fusions were then dialyzed in a 20K MWCO Slide-A-Lyzer (Pierce) 3–4 times against dialysis buffer (50 mM Tris-HCl, 100 mM NaCl, pH 7.5) until all unincorporated 32P had been removed.
In vitro folding selection by protease digestion
The dialyzed fusions were subjected to our folding selection. Triton X-100 and sodium dodecyl sulfate were added to 0.1% and 0.05% (w/v) respectively. Fusions were incubated with Chymotrypsin (Princeton Separations, 6 μg/ml) at 30 °C for 5 min, the digest was stopped by the sequential addition of phenylmethylsulfonyl fluoride (2 mM) and KCl (final concentration of 5 mM) and incubated on ice for 10 minutes. The potassium dodecyl sulfate precipitate was removed via Ultrafree-MC 0.45 μm Spinfilter (Millipore) at 4 °C followed by addition of 3 volumes of Ni-NTA binding buffer (100 mM Phosphate, 10 mM Tris-HCl, 250 mM NaCl, 6 M guanidinium hydrochloride (Amresco), 0.1% Triton X-100, pH 8). The mixture was added to 1 volume Ni-NTA agarose (Qiagen) pre-equilibrated to Ni-NTA binding buffer and incubated with rotation for at least 1 hour at 4 °C. The Ni-NTA agarose was washed on a chromatography column (Bio-Rad) with more Ni-NTA binding buffer followed by a gradient of increasing amounts of Ni-NTA native wash buffer (10 mM Tris-HCl, 250 mM NaCl, 0.01% Triton X-100, pH 8) followed by elution by Ni-NTA elution buffer (50 mM Tris-HCl, 50 mM NaCl, 500 mM imidazole, 0.01% Triton X-100, pH 8). The eluent was concentrated to a third its original volume using a SpeedVac concentrator, ethanol precipitated and dissolved in 10 mM Tris-HCl pH 8 by heating to 80 °C. cDNA was amplified by PCR with Phusion polymerase and primers to add a 5′-UTR (untranslated region) for the next round of mRNA display. Yields from each purification step were determined via scintillation or Cerenkov counting on the Beckman LS6500 multipurpose scintillation counter.
GFP-based folding assay
The GFP-based folding assay is based on the pER13a reporter plasmid previously employed to isolate protein variants with improved folding and contains an out of frame GFP. [21a] A fraction of the library of interest (~108–109 sequences) was PCR amplified with Phusion polymerase and cloned into pER13a plasmid using NdeI and NotI restriction sites to generate N-terminal fusions to GFP. Libraries were ligated into the digested pER13a plasmid using T4 DNA ligase. Ligation reactions were purified via spin columns (PCR Purification Kit, Qiagen) prior to electroporation into electrocompetent NEB 5-alpha cells (New England Biolabs). Following 1 hour incubation at 37 °C, cells were plated and grown overnight on kanamycin containing agar plates. Approximately 104–105 independent colonies were washed off the plates and their plasmids were isolated (QIAprep Spin Miniprep kit, Qiagen). BL21(DE3) and BL21(DE3) Rosetta cells (Novagen) were used for GFP-fused expression of intermediate (Table S1) and final libraries (Table 3), respectively. Electrocompetent cells prepared using standard molecular biology protocols, [24] were transformed with ~108 DNA sequences and grown overnight at 37 °C in LB media (50 ml) supplemented with kanamycin (75 mg/l ) and chloramphenicol (34 mg/l). Overnight culture was used to inoculate the same medium (10 ml) and cells were grown approximately to OD600 = 0.6, transferred to 30 °C for 30 min prior to addition of IPTG (0.5 mM). Growth was continued for 6 h at 30 °C. An aliquot of the cells (1.5 ml) was pelleted by centrifugation (Eppendorf 5415R, 3 min, 4,500 rpm, room temperature), washed with phosphate-buffered saline (PBS, 1 ml) and resuspended in PBS (500 μl). Flow cytometry experiments were performed at the University Flow Cytometry Resource (University of Minnesota, Twin-Cities). Samples were analyzed on FACSCalibur (BD Biosciences) using 488 nm excitation and monitoring emission by a 530/30 nm bandpass filter. FlowJo software package (TreeStar Inc) was used for data analysis. The population of cells transformed with the empty vector was gated out from all experiments before determining the GFP mode (most frequently found fluorescence value) for the remaining cells. Cell sorting experiments were performed on FACSAria (BD Biosciences). Sorting gates were defined on the side scatter vs. GFP fluorescence dot plots. The GDPDwt-like population gate was set based on the cells transformed with GDPDwt-GFP construct while gates for low GFP and high GFP populations were set based on the cells transformed with the GFP-fused control library. Sorted cells were used to inoculate LB medium containing kanamycin and chloramphenicol and re-grown again under sorting conditions as above for Western blot analysis. An aliquot of the re-grown cells was removed prior to IPTG induction and plated on LB agar plates containing kanamycin and chloramphenicol. Individual clones picked at random from these plates were grown in liquid culture and analyzed by SDS-PAGE for soluble expression of library-GFP variants. Six clones from the high GFP population of the GFP-fused folding-enriched library (out of 20 soluble clones identified by SDS-PAGE) were subcloned into pET28a and expressed for further characterization, as above for the GDPD control constructs.
Western blot analysis
Cell pellets were lysed using BugBuster protein extraction reagent (Novagen) according to manufacturer’s recommendations. Insoluble fraction was pelleted and resuspended in the original volume of the BugBuster reagent. Samples were mixed with equal volume of 2X Laemmli sample buffer (BioRad), heated for 5 min at 95 °C and spun down. A fraction of all samples was removed, diluted 10-fold and run on 4–12% gradient gel (Invitrogen). Western blotting was performed according to standard protocols [24] using affinity-purified polyclonal rabbit anti-GFP primary antibody (Abcam, cat. no 290) at 1:5,000 dilution. Anti-rabbit secondary antibody labeled with the DyLight 800 infrared dye (Cell Signaling, cat. no 5151) was used at 1:20,000 dilution and visualized using the Li-Cor Odyssey infrared imaging system. Images were analyzed using Image J software package (NIH) to quantify intensities of anti-GFP stained bands.
Supplementary Material
Acknowledgments
We thank the R. Sterner lab for providing the pER13a plasmid, P. Champoux for assistance with the FACS experiments, members of J. M. Ervasti lab for access to the Li-Cor imaging system, and B. J. Hackel, R. J. Kazlauskas, M. D. Lane, D. Morrone, K. N. Golynskiy and M. B. Quin for comments on the manuscript. This work was supported in part by the US National Institutes of Health (NIH) (T32 GM08347 to J.C.H.), the Biocatalysis Initiative of the BioTechnology Institute at the University of Minnesota (to M.V.G. and B.S.) and the Minnesota Medical Foundation (to B.S.).
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org or from the author.
Trillions of barrels: The catalytically versatile (β/α)8 fold is a highly favored scaffold for natural enzymes. As a resource for enzyme engineering, we generated a library of 1014 proteins based on this fold. We show that despite the introduction of multiple randomized loops, our step-wise assembly and a folding selection by protease digestion enriched for soluble, monomeric and folded proteins.
References
- 1.a) Leemhuis H, Stein A, Griffiths D, Hollfelder F. Curr Opin Struct Biol. 2005;15:472–478. doi: 10.1016/j.sbi.2005.07.006. [DOI] [PubMed] [Google Scholar]; b) Golynskiy MV, Haugner JC, III, Morelli A, Morrone D, Seelig B. Meth Mol Biol. 2013;978:73–92. doi: 10.1007/978-1-62703-293-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Seelig B, Szostak JW. Nature. 2007;448:828–831. doi: 10.1038/nature06032. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Chao F-A, Morelli A, Haugner JC, III, Churchfield L, Hagmann LN, Shi L, Masterson LR, Sarangi R, Veglia G, Seelig B. Nat Chem Biol. 2013;9:81–83. doi: 10.1038/nchembio.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Nagano N, Orengo CA, Thornton JM. J Mol Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]; b) Wierenga RK. FEBS Lett. 2001;492:193–198. doi: 10.1016/s0014-5793(01)02236-0. [DOI] [PubMed] [Google Scholar]; c) Sterner R, Höcker B. Chemical Reviews. 2005;105:4038–4055. doi: 10.1021/cr030191z. [DOI] [PubMed] [Google Scholar]; d) Blacklow SC, Raines RT, Lim WA, Zamore PD, Knowles JR. Biochemistry. 1988;27:1158–1167. doi: 10.1021/bi00404a013. [DOI] [PubMed] [Google Scholar]
- 3.Gerlt JA, Raushel FM. Curr Opin Chem Biol. 2003;7:252–264. doi: 10.1016/s1367-5931(03)00019-x. [DOI] [PubMed] [Google Scholar]
- 4.a) Höcker B, Claren J, Sterner R. Proc Nat Acad Sci. 2004;101:16448–15453. doi: 10.1073/pnas.0405832101. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Leopoldseder S, Claren J, Jürgens C, Sterner R. J Mol Biol. 2004;337:871–879. doi: 10.1016/j.jmb.2004.01.062. [DOI] [PubMed] [Google Scholar]; c) Claren J, Malisi C, Höcker B, Sterner R. Proc Nat Acad Sci. 2009;106:3704–3709. doi: 10.1073/pnas.0810342106. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Evran S, Telefoncu A, Sterner R. Prot Eng Des Sel. 2012;25:285–293. doi: 10.1093/protein/gzs015. [DOI] [PubMed] [Google Scholar]
- 5.Bornscheuer UT, Huisman GW, Kazlauskas RJ, Lutz S, Moore JC, Robins K. Nature. 2012;485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 6.a) Park HS, Nam SH, Lee JK, Yoon CN, Mannervik B, Benkovic SJ, Kim HS. Science. 2006;311:535–538. doi: 10.1126/science.1118953. [DOI] [PubMed] [Google Scholar]; b) Tawfik DS. Science. 2006;311:475–476. doi: 10.1126/science.1123883. [DOI] [PubMed] [Google Scholar]; c) Heinis C, Johnsson K. Meth Mol Biol. 2010;634:217–232. doi: 10.1007/978-1-60761-652-8_16. [DOI] [PubMed] [Google Scholar]
- 7.a) Ochoa-Leyva A, Barona-Gómez F, Saab-Rincón G, Verdel-Aranda K, Sánchez F, Soberón X. J Mol Biol. 2011;411:143–157. doi: 10.1016/j.jmb.2011.05.027. [DOI] [PubMed] [Google Scholar]; b) Ochoa-Leyva A, Soberón X, Sánchez F, Argüello M, Montero-Morán G, Saab-Rincón G. J Mol Biol. 2009;387:949–964. doi: 10.1016/j.jmb.2009.02.022. [DOI] [PubMed] [Google Scholar]; c) Saab-Rincón G, Olvera L, Olvera M, Rudiño-Piñera E, Benites E, Soberón X, Morett E. J Mol Biol. 2012;416:255–270. doi: 10.1016/j.jmb.2011.12.042. [DOI] [PubMed] [Google Scholar]; d) Ma H, Penning TM. Proc Nat Acad Sci. 1999;96:11161–11166. doi: 10.1073/pnas.96.20.11161. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Campbell E, Chuang S, Banta S. Prot Eng Des Sel. 2013;26:181–186. doi: 10.1093/protein/gzs095. [DOI] [PubMed] [Google Scholar]
- 8.a) Griffiths AD, Tawfik DS. EMBO J. 2003;22:24–35. doi: 10.1093/emboj/cdg014. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Vick JE, Schmidt DMZ, Gerlt JA. Biochemistry. 2005;44:11722–11729. doi: 10.1021/bi050963g. [DOI] [PubMed] [Google Scholar]; c) Tsai PC, Fox N, Bigley AN, Harvey SP, Barondeau DP, Raushel FM. Biochemistry. 2012;51:6463–6475. doi: 10.1021/bi300811t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Jiang L, Althoff EA, Clemente FR, Doyle L, Röthlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, Hilvert D, Houk KN, Stoddard BL, Baker D. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Röthlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]; c) Privett HK, Kiss G, Lee TM, Blomberg R, Chica RA, Thomas LM, Hilvert D, Houk KN, Mayo SL. Proc Nat Acad Sci. 2012;109:3790–3795. doi: 10.1073/pnas.1118082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Siegel JB, Zanghellini A, Lovick HM, Kiss G, Lambert AR, StClair JL, Gallaher JL, Hilvert D, Gelb MH, Stoddard BL, Houk KN, Michael FE, Baker D. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Baker D. Protein Sci. 2010;19:1817–1819. doi: 10.1002/pro.481. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Golynskiy MV, Seelig B. Trends Biotechnol. 2010;28:340–345. doi: 10.1016/j.tibtech.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 11.a) Tokuriki N, Tawfik DS. Curr Opin Struct Biol. 2009;19:596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]; b) Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Proc Nat Acad Sci. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeldovich KB, Chen P, Shakhnovich EI. Proc Nat Acad Sci. 2007;104:16152–16157. doi: 10.1073/pnas.0705366104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bloom JD, Silberg JJ, Wilke CO, Drummond DA, Adami C, Arnold FH. Proc Nat Acad Sci. 2005;102:606–611. doi: 10.1073/pnas.0406744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho G, Keefe AD, Liu R, Wilson DS, Szostak JW. J Mol Biol. 2000;297:309–319. doi: 10.1006/jmbi.2000.3571. [DOI] [PubMed] [Google Scholar]
- 15.a) Sieber V, Pluckthun A, Schmid FX. Nat Biotech. 1998;16:955–960. doi: 10.1038/nbt1098-955. [DOI] [PubMed] [Google Scholar]; b) Matsuura T, Plückthun A. Origins Life Evol B. 2004;34:151–157. doi: 10.1023/b:orig.0000009836.86519.eb. [DOI] [PubMed] [Google Scholar]; c) Matsuura T, Plückthun A. FEBS Lett. 2003;539:24–28. doi: 10.1016/s0014-5793(03)00178-9. [DOI] [PubMed] [Google Scholar]; d) Schmid FX. ChemBioChem. 2012;12:1501–1507. doi: 10.1002/cbic.201100018. [DOI] [PubMed] [Google Scholar]
- 16.Khersonsky O, Tawfik DS. Annu Rev Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 17.Santelli E, Schwarzenbacher R, McMullan D, Biorac T, Brinen LS, Canaves JM, Cambell J, Dai X, Deacon AM, Elsliger MA, Eshagi S, Floyd R, Godzik A, Grittini C, Grzechnik SK, Jaroszewski L, Karlak C, Klock HE, Koesema E, Kovarik JS, et al. Proteins. 2004;56:167–170. doi: 10.1002/prot.20120. [DOI] [PubMed] [Google Scholar]
- 18.a) Roberts RW, Szostak JW. Proc Nat Acad Sci. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Seelig B. Nat Protocols. 2011;6:540–552. doi: 10.1038/nprot.2011.312. [DOI] [PubMed] [Google Scholar]
- 19.Cho GS, Szostak JW. Chem Biol. 2006;13:139–147. doi: 10.1016/j.chembiol.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 20.Moelbert S, Emberly E, Tang C. Protein Sci. 2004;13:752–762. doi: 10.1110/ps.03431704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.a) Seitz T, Thoma R, Schoch GA, Stihle M, Benz J, D’Arcy B, Wiget A, Ruf A, Hennig M, Sterner R. J Mol Biol. 2010;403:562–577. doi: 10.1016/j.jmb.2010.08.048. [DOI] [PubMed] [Google Scholar]; b) Graziano JJ, Liu W, Perera R, Geierstanger BH, Lesley SA, Schultz PG. J Am Chem Soc. 2008;130:176–185. doi: 10.1021/ja074405w. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Pédelacq JD, Piltch E, Liong EC, Berendzen J, Kim CY, Rho BS, Park MS, Terwilliger TC, Waldo GS. Nat Biotech. 2002;20:927–932. doi: 10.1038/nbt732. [DOI] [PubMed] [Google Scholar]
- 22.a) Amar D, Berger I, Amara N, Tafa G, Meijler MM, Aharoni A. J Mol Biol. 2012;416:21–32. doi: 10.1016/j.jmb.2011.12.013. [DOI] [PubMed] [Google Scholar]; b) Copley SD. J Biol Chem. 2012;287:3–10. doi: 10.1074/jbc.R111.254714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stryer L. J Mol Biol. 1965;13:482–495. doi: 10.1016/s0022-2836(65)80111-5. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.